Abstract
The integral membrane M2 protein is a 97-residue membrane protein that assembles as a tetramer to conduct protons at a slow rate (102−103/s) when activated by low pH. The proton conductance mechanism has been extensively debated in the literature, but it is accepted that the proton conductance is facilitated by hydrogen bonds involving the His37 residues. However, the hydrogen bonding partnership remains unresolved. Here, we report on the measurement of 15N−15N J-couplings of 15N His37-labeled full length M2 (M2FL) protein from Influenza A virus embedded in synthetic liquid crystalline lipid bilayers using two-dimensional J-resolved NMR spectroscopy. We experimentally observed the hydrogen-bond mediated J-couplings between Nδ1 and Nε2 of adjacent His37 imidazole rings, providing direct evidence for the existence of various imidazolium-imidazole hydrogenbonding geometries in the histidine tetrad at low pH, thus validating the proton conduction mechanism in the M2FL protein by which the proton is transferred through the breaking and reforming of the hydrogen bonds between pairs of His37 residues.
The M2 protein from the Influenza A virus is a 97-residue membrane protein with a 24-residue N-terminal and a 51-residue C-terminal segment connected by a single trans-membrane (TM) helix of 22 residues. It assembles as a tetrameric bundle to form a channel that is activated at low pH to conduct protons at a slow rate (102 − 103/s), essential for the viral life cycle.1,2 The α-helical TM domain (residues 25–46) is responsible for proton conductance triggering the release of viral RNA into the host cells. This tetrameric TM domain is an important drug target.3–6 The four His37 residues reside near the center of the TM helix and are known to be the heart of the proton conducting channel, the key to the proton transport mechanism.7 Several different constructs reconstituted in various lipid environments have been the subject of intensive high-resolution structural studies in the past decade using both magic-angle-spinning (MAS)8–15 and oriented sample3,16–19 solid-state NMR, as well as solution NMR20–23 and X-ray crystallography.24,25 It has been clear so far that the proton conductance in the M2 channel is facilitated through hydrogen bonds with the His37 residues. But two possible models for His37 hydrogen bonding partners have been presented and debated in the literature (Figure 1). The imidazolium-imidazole bonding of His-His+ pairs (c.f. Figure 1a) that form low-barrier hydrogen bonds (LBHB) as previously suggested with 1H frequencies as high as 18.7 ppm and protonated 15N frequencies as high as 195 ppm; i.e., the proton is transferred through protonating the τ state of one His-His+ pair to form a +3 state for the His tetrad prior to deprotonation and reforming the +2 state of the His tetrad.7,17,26 If the His37 residue does not form His-His hydrogen bonded pairs, but only hydrogen bonds with water (cf. Figure 1b), a proton shuttling mechanism is proposed; i.e., the His37 shuttles protons through imidazole ring reorientations and exchanging protons with water without forming an intermonomer hydrogen bond between the His37 residues.27–29
Figure 1.
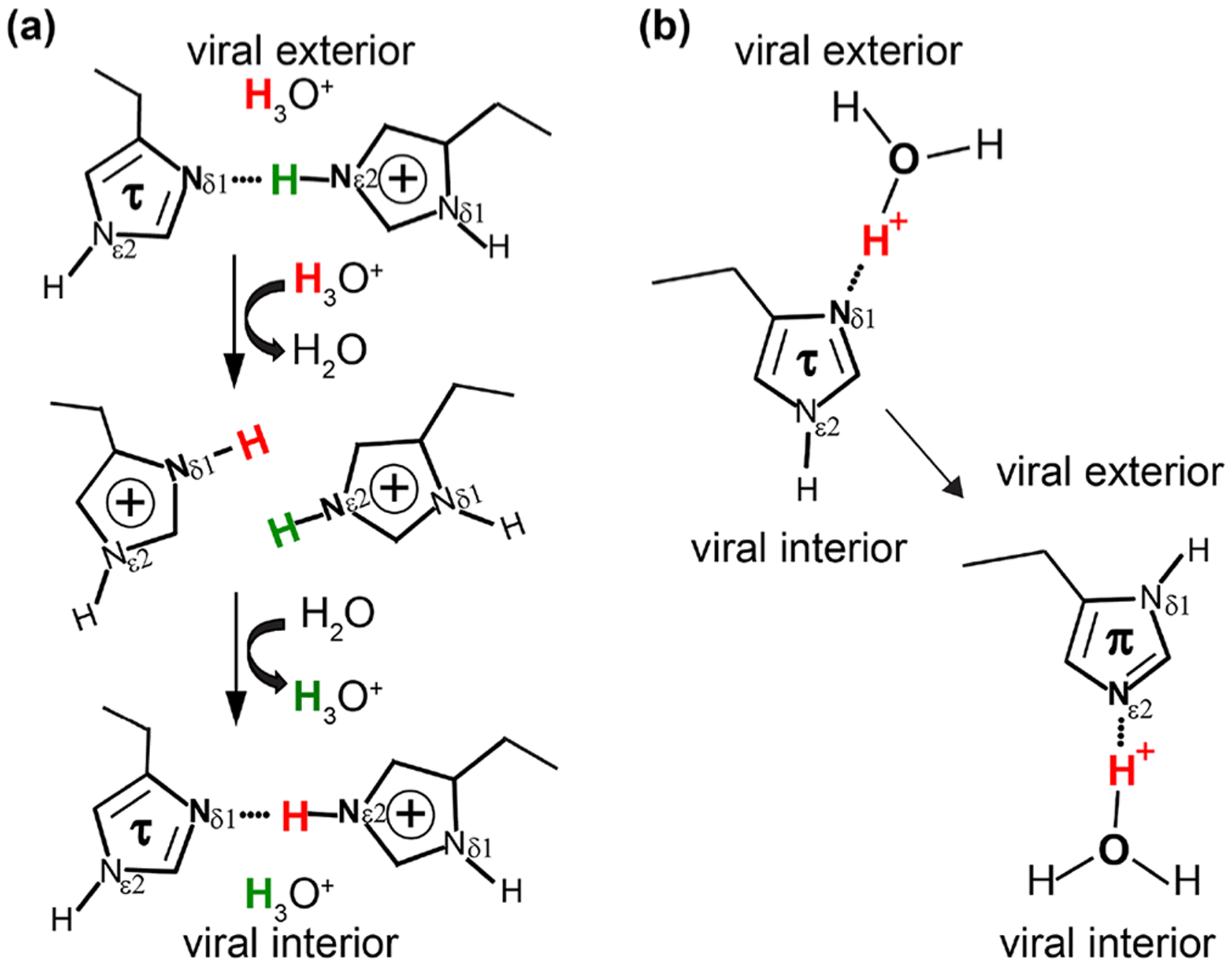
Two possible hydrogen bonding partnerships in the His37 tetrad of the M2 proton channel. (a) One of two pairs of His37 residues showing the low-barrier hydrogen bond model, in which the proton is transferred through the breaking and reforming of the intermonomer imidazole-imidazolium hydrogen bonds between His37 residues; (b) the water-His37 hydrogen bonding model, in which the His37 shuttles protons through imidazole ring reorientations and exchanging protons with water.
To support the shuttling mechanism, two-dimensional (2D) 1H−15N heteronuclear correlation (HETCOR) experiments were performed to see if there were any correlations between the water protons and the nitrogens in the His37 side chains. It has been shown in the 1H−15N HETCOR spectra28 of an M2 transmembrane construct, M2(22–46), in a virus-envelopemimetic lipid membrane that the 15N peaks were in the range 160–180 ppm, correlating to 1H chemical shifts of 8–12 ppm, similar to a typical backbone amide 1H chemical shift range, and correlating to the 1H peak of water at ~5 ppm. No imidazole-imidazolium cross peaks were observed in the 13C−13C correlation spectra of the +2 charged state of M2(22–46).27 However, the correlated peaks between 15N and water protons can arise either from direct water-His proton transfer (cf. Figure 1b) or from additional protonation to form a transient +3 state for the His tetrad prior to deprotonation, potentially from a water in the viral interior (i.e., indirect transfer through the chemical exchange between hydronium ions and the protons of the histidine side chain NH bonds),26 as illustrated in Figure 1a. Recently, more experimental evidence has been obtained as alluded to above from the full length M2 (M2FL) protein in lipid bilayers that support the LBHB model: (i) both high 1H and 15N frequencies extend up to 19 and 190 ppm, respectively, in the 2D 1H−15N HETCOR spectra, and the nonprotonated 15N resonances extending from 250 ppm down to 235 ppm—such extreme frequencies clearly indicate the formation of short imidazole-imidazolium hydrogen bonds in the His37 tetrad;26,30 (ii) the observation that the hydronium ions are in chemical exchange on a submsec time scale with the protons of the imidazole-imidazolium hydrogen bonds;31 (iii) the observations of 13C−13C and 15N−15N chemically exchange cross peaks between His37 residues in the neutral and charged states;32 and (iv) no evidence for the π state under even mild acidic pH conditions when characterizing the full length protein.26 Nevertheless, these results may still be considered as indirect evidence to affirm the LBHB model. Here, we seek direct and definitive evidence for the imidazole-imidazolium hydrogen bonding partner by measuring through-bond scalar coupling, i.e. J-coupling, between the two nitrogen sites.
A hydrogen bond is primarily an electrostatic attraction between two polar groups containing highly electronegative atoms such as nitrogen, oxygen, and fluorine that are bridged by a hydrogen atom. However, charge transfer (covalency) and dispersive energies are also important, especially when considering strong hydrogen bonds.33 It has been well-known that the electron sharing within covalent bonds results in J-couplings. Similarly, the redistribution of electron densities upon hydrogen bond formation acts as electron sharing between the two nuclei involved in the hydrogen bond, although they are not covalently bonded.34 As a result, there exist hydrogen-bond mediated J-couplings between the two electronegative atoms involved.35 Such J-couplings across the hydrogen bonds have been observed in both solution36–40 and solid-state NMR41–43 spectra and are considered as a direct detection of hydrogen bonds. Here, the LBHB model (cf. Figure 1a) involves two electronegative nitrogen atoms, one of which is protonated; thus, there should exist homonuclear hydrogen-bond mediated 15N−15N J-couplings 2hJ(Nδ1⋯H⋯Nε2) between the two involved nitrogen atoms. In the water-His37 hydrogen bonding model (cf. Figure 1b), the two involved electronegative atoms are nitrogen and oxygen, meaning that there should be heteronuclear hydrogen-bond mediated 15N−17O J-couplings between the nitrogen and oxygen, but not homonuclear 15N−15N J-couplings. Therefore, we use 15N 2D J-resolved spectroscopy in high-resolution solid-state NMR to differentiate the hydrogen bonding partners at the heart of the His37 tetrad of the M2FL proton channel.
As demonstrated in the literature,41 the rotor-synchronized spin–echo (CP − t1/2 − π − t1/2 − t2) sequence can be used to measure homonuclear J-couplings, where CP stands for cross-polarization from 1H to 15N, t1 and t2 represent the time evolution in the indirect and observed dimension, respectively, where t1/2 has to be set to a multiple of spinning periods. High power 1H decoupling is applied during the entire t1 and t2 dimensions. Because there are two nitrogen sites in the imidazole ring of histidine separated by two covalent bonds, we need to first examine whether or not there exists a two-bond J-coupling (i.e., 2J(Nδ1⋯Nε2)) between these two nitrogen atoms. Figure 2 shows the 15N 2D J-resolved spectrum for the histidine sample lyophilized from a pH 6.3 solution. At this pH, there are two tautomeric states, the neutral τ state and the charged state; the π state is not present at this pH.44 From the slices in the right panel, it is clear that the protonated 15N sites (especially Nε2) have a slightly broader line width (~9 Hz) than the nonprotonated 15N site (~7 Hz), owing to some residual 1H−15N interactions for the protonated 15N sites that are not fully refocused by the spin echo sequence. Nevertheless, no J-splitting is observed for the histidine sample, implying that there is no detectable 2J(Nδ1⋯Nε2) between the Nδ1 and Nε2 sites that are separated by two chemical bonds in the imidazole rings for both charged and neutral τ states.
Figure 2.
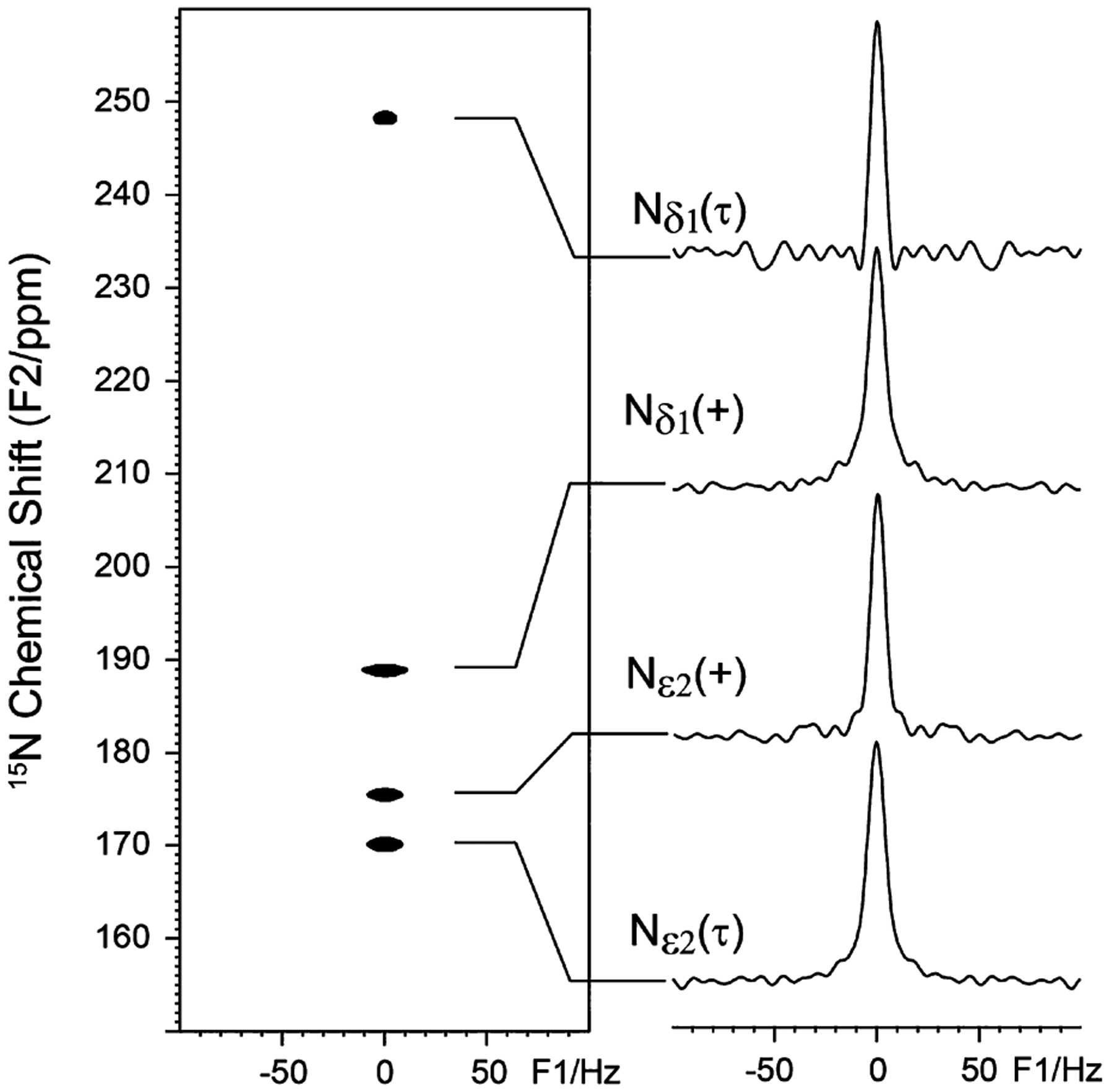
15N 2D J-resolved spectrum (left) of the histidine powder sample (at pH 6.3) and their corresponding slices (right).
Figure S1 shows the 15N CPMAS NMR spectrum of the His37-labeled M2FL (H57,90Y, pH 6.2) in DOPC/DOPE liposomes. It is noteworthy that, in the uniform 13C and 15N labeling media for this protein expression, unlabeled Phe, Tyr, and Trp amino acids were added to the bacterial cultures to suppress these aromatic amino acid resonances. Therefore, the observed signals from 150 to 260 ppm, as shown in the expanded spectrum in Figure S1 and in refs 26, 31, and 32 are solely from the His37 tetrad. Furthermore, it is important to emphasize that the M2 spectra display a broad distribution of 15N chemical shifts for both the protonated and nonprotonated 15N His37 side chain resonances.26 These resonances have been shown to be heterogeneously and not homogeneously broadened.26,32 In addition, the heterogeneity largely results from transmembrane helix–helix packing. This proton channel is formed by a tetramer of a single transmembrane helix, and unlike many transmembrane proteins that have small residues at the helix–helix interface, M2 has large side chains, such as Leu38, that can take on different χ2 rotameric states resulting in slightly different 15N−15N distances for the imidazole-imidazolium hydrogen bonded state leading to the broad distribution of the 15N, 13C, and 1H chemical shifts observed for His37.26,32 Such heterogeneity may be the key to the instability of the imidazole-imidazolium hydrogen bonds that permit proton conductance, albeit on the very slow millisecond time scale. Such a slow time scale is not explained by the mechanism illustrated in Figure 1b.
Figure 3 shows the 15N 2D J-resolved spectrum of the His37-labeled M2FL protein at pH 6.2 in DOPC/DOPE liposomes together with extracted rows at specific 15N chemical shifts. The natural line width for all 15N chemical shift positions from 165 to 190 ppm appears to be ~60 Hz. This implies, as described above, that the broad 15N resonance bands observed from 165 to 190 ppm are the result of inhomogeneous broadening, a distribution of chemically distinguishable His37 conformations, rather than exchange broadening at a rate comparable to the chemical shift difference between the protonated and nonprotonated Nδ1 and Nε2 sites. The slices through the 2D data set (Figure 3) do not have enough sensitivity owing to having just four 15Nδ1 and four 15Nε2 sites whose signals are each spread over more than 25 ppm between 160 and 200 ppm in this full length protein in liquid crystalline lipid bilayers. But the transverse dephasing times (T′2)45 from fittings (Figure 3) decrease steadily at a higher ppm position, all shorter than that of the amide nitrogen (Figure S2). Such a decrease in T′2 could indicate the presence of J-coupling (eq S1). It is known that the homonuclear J-coupling can be refocused by applying a π/2 pulse in the middle of double spin-echoes (Figure S3).46,47 Figure 4 shows the doubly spin-echoed 15N spectra of the M2FL sample with and without J-refocusing. At t1 = 20.48 ms, the J-refocused signals (in red) from 170 to 185 ppm are consistently higher than those (in black) without applying the π/2 pulse, as clearly indicated by the integration curve (blue). These results document the existence of 2hJ(Nδ1⋯H⋯Nε2) as suggested previously through the 15N−1H correlation spectra with 1H frequencies up to 18.7 ppm and 15N frequencies up to 195 ppm.26 Using t1 = 32.0 ms, the J-refocused signal at 174 ppm had a much higher intensity (above the noise level) than without the J-refocusing, whose ratio yielded a 2hJ(Nδ1⋯H⋯Nε2) value of 8.1 Hz from eqs S1 and S2, which represents typical 2hJ(N⋯H⋯N) values observed in other compounds.41–43 This confirms the existence of imidazole-imidazolium hydrogen bonds in the His37 tetrad of the M2 protein, since the control spectrum (Figure 2) indicates no two-bond J-coupling between the Nδ1 and Nε2 sites within the imidazole ring. The further decrease in the T′2 values at the higher 15N chemical shifts might indicate a larger 2hJ(Nδ1⋯H⋯Nε2) and thus could imply stronger hydrogen bonds as the protonated 15N chemical shift moves to lower field.26,32 Thus, the large 2hJ(Nδ1⋯H⋯Nε2) values hypothesized here are consistent with the observation of high 1H and 15N chemical shifts observed in the 1H−15N HETCOR spectra for the same sites, all reflecting on the broad range of the imidazole-imidazolium hydrogen-bond strengths in the His37 tetrad.26,30,32 These relatively infrequent strong hydrogen bonds may be responsible for the low M2 proton conductance rate. Furthermore, this range of J-couplings and chemical shifts is consistent with the explanation that packing of various large hydrophobic side chains in the helix–helix interface results in a subtle range of distances between the imidazole and imidazolium side chains due to slight shifts in the Cα−Cα separation of the backbone.26
Figure 3.
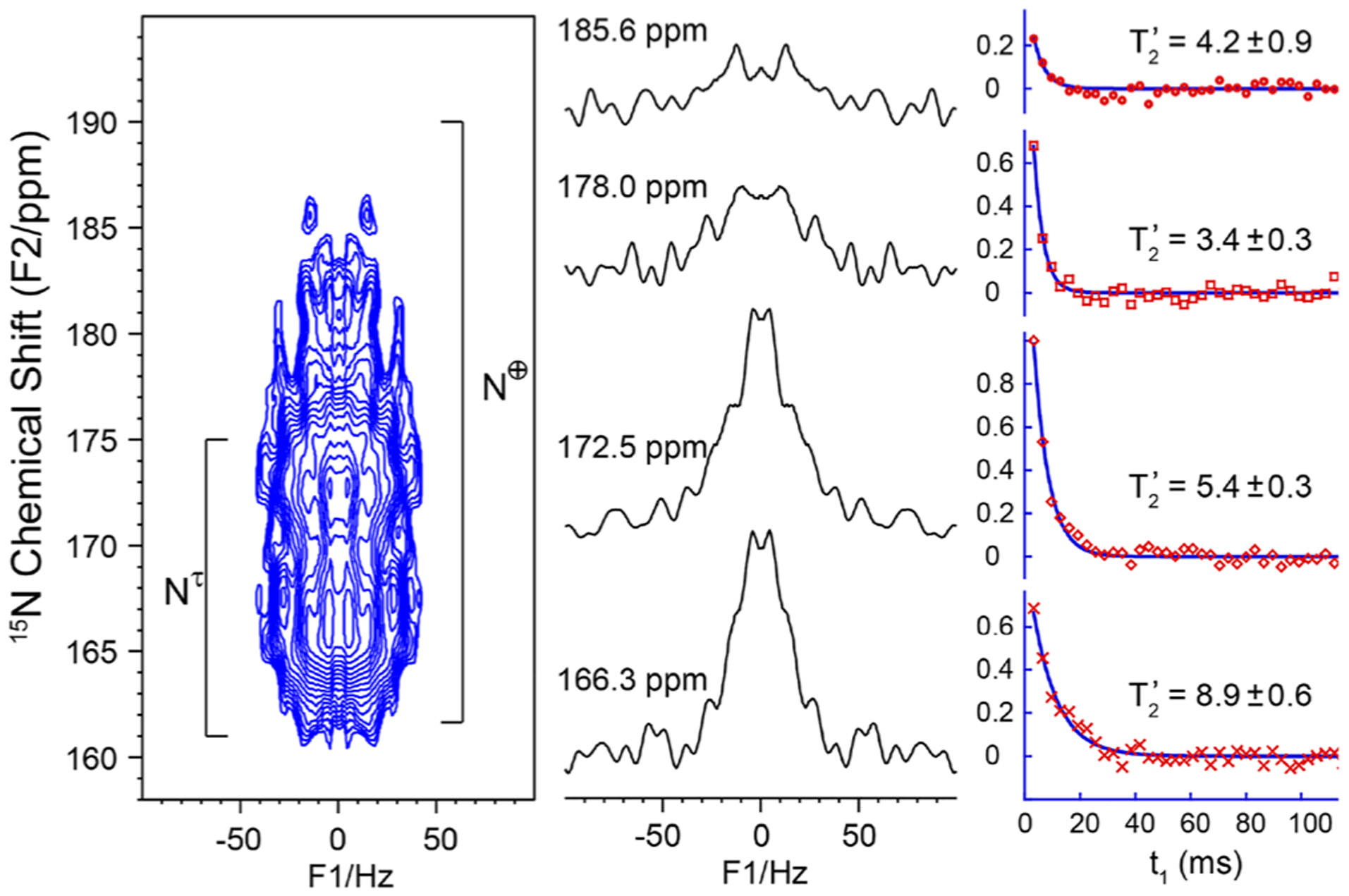
Expanded 15N 2D J-resolved spectrum (left) of the His37-labeled M2FL (pH 6.2) in DOPC/DOPE liposomes and the slices (middle panel) and the normalized echo signal intensities as a function of spin-echo time t1 (right panel) at different chemical shift positions. This 2D spectrum was recorded in 109 h on a 600 MHz NMR spectrometer where the Larmor frequencies are 600.1 and 60.8 MHz for 1H and 15N, respectively. The sample was spun at 12.5 kHz. The expected protonated 15N chemical shift range for the τ and charge states are marked in the 2D spectrum.
Figure 4.
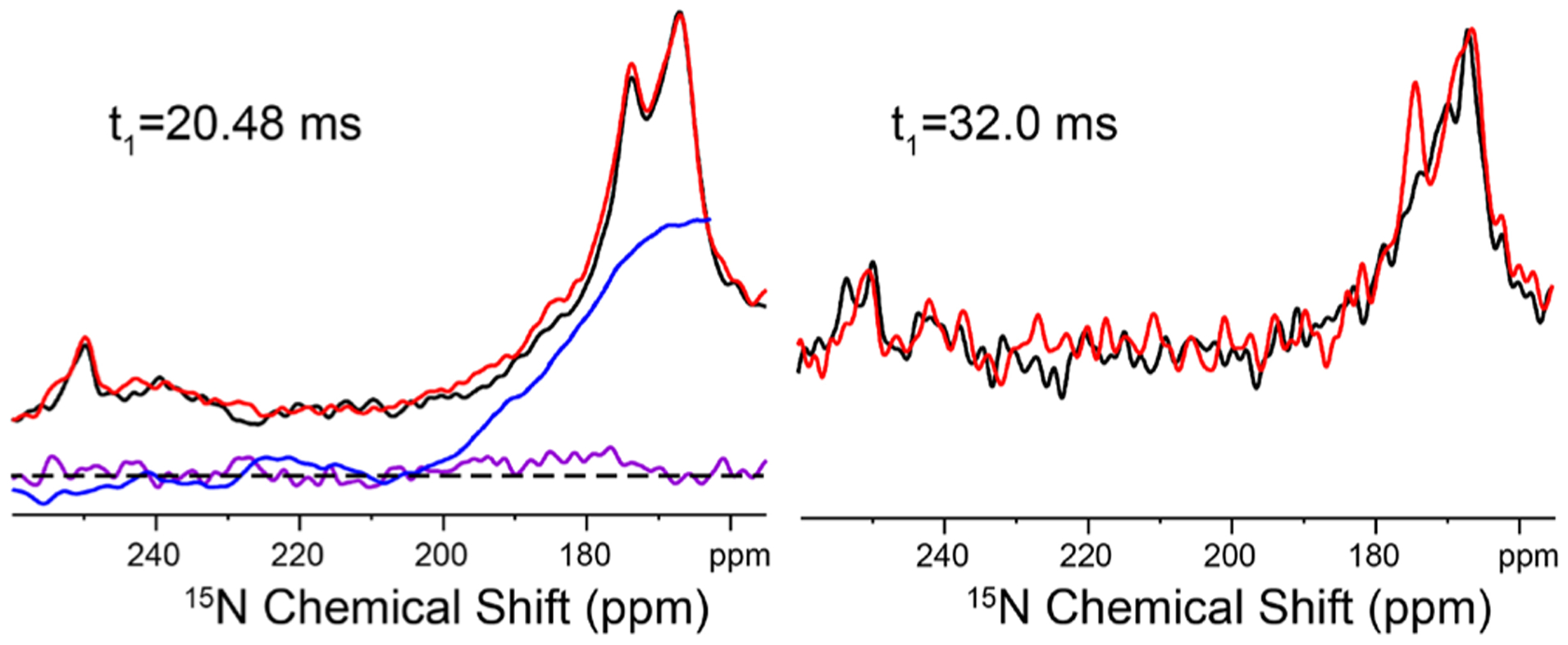
Expanded 15N doubly spin–echoed spectra of the His37-labeled M2FL (pH 6.2) in DOPC/DOPE liposomes with (red) and without (black) the J-refocusing π/2 pulse at different spin–echo time t1. The subtraction of the black from red spectra is shown in purple with dashed lines indicating the zero reference. The blue line shows the integration of the difference spectrum from 260 to 164 ppm.
To conclude, we used the 2D J-resolved NMR spectrum to measure the hydrogen-bond mediated 15N−15N J-couplings at the heart of the His37 tetrad of the M2 protein. The observed 2hJ(Nδ1⋯H⋯Nε2) provides direct observation of the heterogeneous imidazole-imidazolium hydrogen bonds in the His37 tetrad of the M2 protein, thus validating the LBHB proton conductance mechanism at the heart of the M2FL proton channel. Furthermore, these J-couplings are highly correlated with distributions of chemical shifts, as in disordered solids,48,49 and could be used to refine detailed local structural models in such heterogeneous environments through theoretical calculations.35,50
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH Grants AI023007 and AI119178. All NMR experiments were carried out at the National High Magnetic Field Lab (NHMFL) supported by the NSF Cooperative Agreement DMR-1644779 and the State of Florida.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.9b09985.
Materials and experimental details; 1D 15N CPMAS spectrum of the His37-labeled M2FL sample; 1D spectra taken along the F1 dimension of the 2D J-resolved spectrum in Figure 3; demonstration for the double spin–echo sequence used for J-refocusing and the full doubly spin–echoed spectra of the M2FL sample at different echo times (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/jacs.9b09985
The authors declare no competing financial interest.
Contributor Information
Riqiang Fu, National High Magnet Field Lab, Tallahassee, Florida 32310, United States.
Timothy A. Cross, National High Magnet Field Lab, Tallahassee, Florida 32310, United States; Department of Chemistry and Biochemistry, Florida State University, Tallahassee, Florida 32306, United States
REFERENCES
- (1).Sugrue RJ; Hay AJ Structural characteristics of the M2 protein of influenza-A viruses - evidence that it forms a tetrameric channel. Virology 1991, 180, 617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Sakaguchi T; Tu QA; Pinto LH; Lamb RA The active oligomeric state of the minimalistic influenza virus M-2 ion channel is a tetramer. Proc. Natl. Acad. Sci. U. S. A 1997, 94, 5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Hu J; Asbury T; Achuthan S; Li C; Bertram R; Quine JR; Fu R; Cross TA Backbone structure of the amantadine-blocked trans-membrane domain M2 proton channel from influenza A virus. Biophys. J 2007, 92, 4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Nishimura K; Kim SG; Zhang L; Cross TA The closed state of a H+ channel helical bundle combining precise orientational and distance restraints from solid state NMR. Biochemistry 2002, 41, 13170. [DOI] [PubMed] [Google Scholar]
- (5).Lamb RA; Zebedee SL; Richardson CD Influenza virus-M2 protein is an integral membrane-protein expressed on the infected-cell surface. Cell 1985, 40, 627. [DOI] [PubMed] [Google Scholar]
- (6).Grambas S; Bennett MS; Hay AJ Influence of amantadine resistance mutations on the pH regulatory function of the M2-protein of influenza-A viruses. Virology 1992, 191, 541. [DOI] [PubMed] [Google Scholar]
- (7).Hu J; Fu R; Nishimura K; Zhang L; Zhou HX; Busath DD; Vijayvergiya V; Cross TA Histidines, heart of the hydrogen ion channel from influenza A virus: Toward an understanding of conductance and proton selectivity. Proc. Natl. Acad. Sci. U. S. A 2006, 103, 6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Miao Y; Qin H; Fu R; Sharma M; Can TV; Hung I; Luca S; Gor’kov PL; Brey WW; Cross TA M2 Proton Channel Structural Validation from Full-Length Protein Samples in Synthetic Bilayers and E. coli Membranes. Angew. Chem 2012, 124, 8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Can TV; Sharma M; Hung I; Gor’kov PL; Brey WW; Cross TA Magic Angle Spinning and Oriented Sample Solid-State NMR Structural Restraints Combine for Influenza A M2 Protein Functional Insights. J. Am. Chem. Soc 2012, 134, 9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Cady SD; Schmidt-Rohr K; Wang J; Soto CS; DeGrado WF; Hong M Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature 2010, 463, 689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Cady SD; Mishanina TV; Hong M Structure of amantadine-bound M2 transmembrane peptide of influenza A in lipid bilayers from magic-angle-spinning solid-state NMR: the role of Ser31 in amantadine binding. J. Mol. Biol 2009, 385, 1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Andreas LB; Eddy MT; Pielak RM; Chou JJ; Griffin RG Magic angle spinning NMR investigation of Influenza A M218–60: Support for an allosteric mechanism of inhibition. J. Am. Chem. Soc 2010, 132, 10958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Andreas LB; Eddy MT; Chou JJ; Griffin RG Magic-angle-spinning NMR of the drug resistant S31N M2 proton transporter from Influenza A. J. Am. Chem. Soc 2012, 134, 7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Colvin MT; Andreas LB; Chou JJ; Griffin RG Proton association constants of His 37 in the Influenza-A M218–60 dimerof-dimers. Biochemistry 2014, 53, 5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Andreas LB; Reese M; Eddy MT; Gelev V; Ni QZ; Miller EA; Emsley L; Pintacuda G; Chou JJ; Griffin RG Structure and mechanism of the Influenza A M218–60 Dimer of Dimers. J. Am. Chem. Soc 2015, 137, 14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Du J; Cross TA; Zhou HX Recent progress in structure-based anti-influenza drug design. Drug Discovery Today 2012, 17, 1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Sharma M; Yi M; Dong H; Qin H; Peterson E; Busath DD; Zhou HX; Cross TA Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science 2010, 330, 509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Wang J; Wu Y; Ma C; Fiorin G; Wang J; Pinto LH; Lamb RA; Klein ML; Degrado WF Structure and inhibition of the drug-resistant S31N mutant of the M2 ion channel of influenza A virus. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wang JF; Kim SG; Kovacs F; Cross TA Structure of the transmembrane region of the M2 protein H+ channel. Protein Sci. 2001, 10, 2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Schnell JR; Chou JJ Structure and mechanism of the M2 proton channel of influenza A virus. Nature 2008, 451, 591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Pielak RM; Schnell JR; Chou JJ Mechanism of drug inhibition and drug resistance of influenza A M2 channel. Proc. Natl. Acad. Sci. U. S. A 2009, 106, 7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Pielak RM; Oxenoid K; Chou JJ Structural investigation of rimantadine inhibition of the AM2-BM2 chimera channel of influenza viruses. Structure 2011, 19, 1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Pielak RM; Chou JJ Solution NMR structure of the V27A drug resistant mutant of influenza A M2 channel. Biochem. Biophys. Res. Commun 2010, 401, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Stouffer AL; Acharya R; Salom D; Levine AS; Di Costanzo L; Soto CS; Tereshko V; Nanda V; Stayrook S; DeGrado WF Structural basis for the function and inhibition of an influenza virus proton channel. Nature 2008, 451, 596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Acharya R; Carnevale V; Fiorin G; Levine BG; Polishchuk AL; Balannik V; Samish I; Lamb RA; Pinto LH; DeGrado WF; Klein ML Structure and mechanism of proton transport through the transmembrane tetrameric M2 protein bundle of the influenza A virus. Proc. Natl. Acad. Sci. U. S. A 2010, 107, 15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Miao Y; Fu R; Zhou HX; Cross TA Dynamic short hydrogen bonds in histidine tetrad of full length M2 proton channel reveal tetrameric structural heterogeneity and functional mechanism. Structure 2015, 23, 2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Hu F; Schmidt-Rohr K; Hong M NMR Detection of pH-Dependent Histidine-Water Proton Exchange Reveals the Conduction Mechanism of a Transmembrane Proton Channel. J. Am. Chem. Soc 2012, 134, 3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Hong M; Fritzsching KJ; Williams JK Hydrogen-Bonding Partner of the Proton-Conducting Histidine in the Influenza M2 Proton Channel Revealed From 1H Chemical Shifts. J. Am. Chem. Soc 2012, 134, 14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Liang R; Li H; Swanson JMJ; Voth GA Multiscale simulation reveals a multifaceted mechanism of proton permeation through the influenza A M2 proton channel. Proc. Natl. Acad. Sci. U. S. A 2014, 111, 9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Eckert H; Yesinowski JP; Silver LA; Stolper EM Water in silicate glasses: Quantitation and structural stidies by proton solid echo and magic angle spinning NMR methods. J. Phys. Chem 1988, 92, 2055. [Google Scholar]
- (31).Fu R; Miao Y; Qin H; Cross TA Probing Hydronium Ion Histidine NH Exchange Rate Constants in the M2 Channel via Indirect Observation of Dipolar-Dephased 15N Signals in Solid-State Magic-Angle-Spinning NMR. J. Am. Chem. Soc 2016, 138, 15801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Qin H; Miao Y; Cross TA; Fu R Beyond structural biology to functional biology: Solid-state NMR experiments and strategies for understanding the M2 proton channel conductance. J. Phys. Chem. B 2017, 121, 4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Grabowski SJ What Is the Covalency of Hydrogen Bonding? Chem. Rev 2011, 111, 2597. [DOI] [PubMed] [Google Scholar]
- (34).Dingley AJ; Cordier F; Grzesiek S An Introduction to Hydrogen Bond Scalar Couplings. Concepts Magn. Reson 2001, 13, 103. [Google Scholar]
- (35).Joyce SA; Yates JR; Pickard CJ; Brown SP Density Functional Theory Calculations of Hydrogen-Bond-Mediated NMR J Coupling in the Solid State. J. Am. Chem. Soc 2008, 130, 12663. [DOI] [PubMed] [Google Scholar]
- (36).Blake JB; Adams MWW; Summers MF Novel Observation of NH-S(Cys) Hydrogen-Bond-Mediated Scalar Coupling in 113Cd-Substituted Rubredoxin from Pyrococcus furiosus. J. Am. Chem. Soc 1992, 114, 4931. [Google Scholar]
- (37).Dingley AJ; Grzesiek S Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide 2JNN couplings. J. Am. Chem. Soc 1998, 120, 8293. [Google Scholar]
- (38).Pervushin K; Ono A; Fernandez C; Szyperski T; Kainosho M; Wuthrich K NMR scalar couplings across Watson!Crick base pair hydrogen bonds in DNA observed by transverse relaxation-optimized spectroscopy. Proc. Natl. Acad. Sci. U. S. A 1998, 95, 14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Cornilescu G; Hu JS; Bax A Identification of the hydrogen bonding network in a protein by scalar couplings. J. Am. Chem. Soc 1999, 121, 2949. [Google Scholar]
- (40).Hennig M; Geierstanger BH Direct detection of a histidinehistidine side chain hydrogen bond important for folding of apomyoglobin. J. Am. Chem. Soc 1999, 121, 5123. [Google Scholar]
- (41).Brown SP; Perez-Torralba M; Sanz D; Claramunt RM; Emsley L Determining hydrogen-bond strengths in the solid state by NMR: the quantitative measurement of homonuclear J couplings. Chem. Commun 2002, 1852. [DOI] [PubMed] [Google Scholar]
- (42).Brown SP; Perez-Torralba M; Sanz D; Claramunt RM; Emsley L The Direct Detection of a Hydrogen Bond in the Solid State by NMR through the Observation of a Hydrogen-Bond Mediated 15N-15N J Coupling. J. Am. Chem. Soc 2002, 124, 1152. [DOI] [PubMed] [Google Scholar]
- (43).Pham TN; Griffin JM; Masiero S; Lena S; Gottarelli G; Hodgkinson P; Filip C; Brown SP Quantifying hydrogen-bonding strength: the measurement of 2hJNN couplings in self-assembled guanosines by solid-state 15N spin-echo MAS NMR. Phys. Chem. Chem. Phys 2007, 9, 3416. [DOI] [PubMed] [Google Scholar]
- (44).Miao Y; Cross TA; Fu R Differentiation of histidine tautomeric states using 15N selectively filtered 13C solid-state NMR spectroscopy. J. Magn. Reson 2014, 245, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Lesage A; Emsley L; Penin F; Bockmann A Investigation of Dipolar-Mediated Water-Protein Interactions in Microcrystalline Crh by Solid-State NMR Spectroscopy. J. Am. Chem. Soc 2006, 128, 8246. [DOI] [PubMed] [Google Scholar]
- (46).Takegoshi K; Ogura K; Hikichi K A Perfect Spin Echo in a Weakly Homonuclear J-Coupled Two Spin-1/2 System. J. Magn. Reson 1989, 84, 611. [Google Scholar]
- (47).van Zijl PCM; Moonen CTW; von Kienlin M Homonuclear J refocusing in Echo Spectroscopy. J. Magn. Reson 1990, 89, 28. [Google Scholar]
- (48).Cadars S; Lesage A; Trierweiler M; Heux L; Emsley L NMR measurements of scalar-coupling distributions in disordered solids. Phys. Chem. Chem. Phys 2007, 9, 92. [DOI] [PubMed] [Google Scholar]
- (49).Guerry P; Smith ME; Brown SP 31P MAS Refocused INADEQUATE Spin-Echo (REINE) NMR Spectroscopy: Revealing J Coupling and Chemical Shift Two-Dimensional Correlations in Disordered Solids. J. Am. Chem. Soc 2009, 131, 11861. [DOI] [PubMed] [Google Scholar]
- (50).Hughes CE; Reddy GNM; Masiero S; Brown SP; Williams PA; Harris KD M. Determination of a complex crystal structure in the absence of single crystals: analysis of powder X-ray diffraction data, guided by solid-state NMR and periodic DFT calculations, reveals a new 2’-deoxyguanosine structural motif. Chem. Sci 2017, 8, 3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


