ABSTRACT
Mobile colistin resistance gene mcr-1 and extended-spectrum β-lactamase gene blaCTX-M are highly prevalent in human – and pet-derived bacteria. Isolation of identical strains of mcr-1-positive Escherichia coli (MCRPEC) or blaCTX-M-positive E. coli (CTX-MPEC) from pets and humans highlighted the potential for co-colonization of antibiotic-resistant bacteria which can be a risk for dissemination of resistance genes. In this study, the prevalence of mcr-1 and blaCTX-M carriage from rectal swabs in 299 families (dogs and their owners) were 2.7 and 5.3%, respectively. We identified a significant association of mcr-1 carriage between dogs and their owners. Whilst antibiotic use in the previous three months was associated with blaCTX-M carriage in dogs. Only one instance of dog and owner carrying identical CTX-MPEC was observed. Although the prevalence of identical strains in one family is rare, the huge number of dog ownership worldwide suggest that this threat should not be underestimated.
KEYWORDS: Prevalence, risk factors, gene transfer, MCR-1, CTX-M
Introduction
Animals as pets are always kept for company, entertainment or as act of compassion for humans. Currently, more than 223 million pets are estimated to be owned worldwide [1]. In China, 55.03 million dogs and 44.12 million cats were kept in cities in 2019 [2]. In Beijing, nearly four million people raised pet dogs in 2019 [2]. Most pet owners in China (59.1%) treat pets as their “children”, and 27.8% of owners consider their pets family members [2]. However, close contact between humans and pets enhances the risk of pathogens transmission such as bacteria even antimicrobial-resistant bacteria (AR bacteria) [3]. Previous studies showed that antimicrobial-resistant bacteria (AR bacteria) are frequently isolated from companion animals, including dogs, cats, and birds [4] and some AR bacteria (i.e. mcr-1-positive Escherichia coli; MCRPEC, extended-spectrum β-lactamase (ESBL)-producing and carbapenem-resistant Enterobacteriaceae) may also be exchanged between companion animals and humans, are an urgent threat to public health and becoming a major concern [5,6].
Both mcr-1 and ESBL-encoding gene blaCTX-M have high prevalence in bacteria from humans and pets. The prevalence of mcr-1-positive bacteria in humans ranges from 0.02 to 28.3% worldwide [7], with an average value 15% in China in 2016 [8]. The prevalence of MCRPEC in Chinese pets ranged from 6.1 to 14.3% [9,10]. The average global prevalence of ESBL class A (mainly blaCTX-M)-positive bacterial colonization of humans was 14% [11], but varied from 18 to 74% in China [12,13]. The prevalence of blaCTX-M among bacteria derived from pets in different countries (including China) ranged from 10 to 21% [14–16]. The high prevalence of mcr-1 and blaCTX-M raise concerns of AR bacteria transmission between pets to humans via close contact [3,17].
Previous studies support the potential for transmission of mcr-1 – and blaCTX-M-carrying bacteria between humans and pets. For instance, the MCRPEC showing identical pulsed-field gel electrophoresis (PFGE) patterns were recovered from a 50-year-old pet store employee and four dogs and two cats from the same store [17]. Similarly, PFGE revealed that the same CTX-M group-9-producing E. coli strain was present in two owners and two dogs from the same household [3]. However, there is a lack of population-based studies assessing the risk of AR bacterial transfer between humans and their pets. Therefore, we conducted a population-based cross-sectional study at the largest veterinary teaching hospital in Beijing, China, to investigate this public health-related risk and the factors associated with MCRPEC and blaCTX-M-positive E. coli (CTX-MPEC) carriage by pet dogs and their owners.
Materials and method
Epidemiological investigation
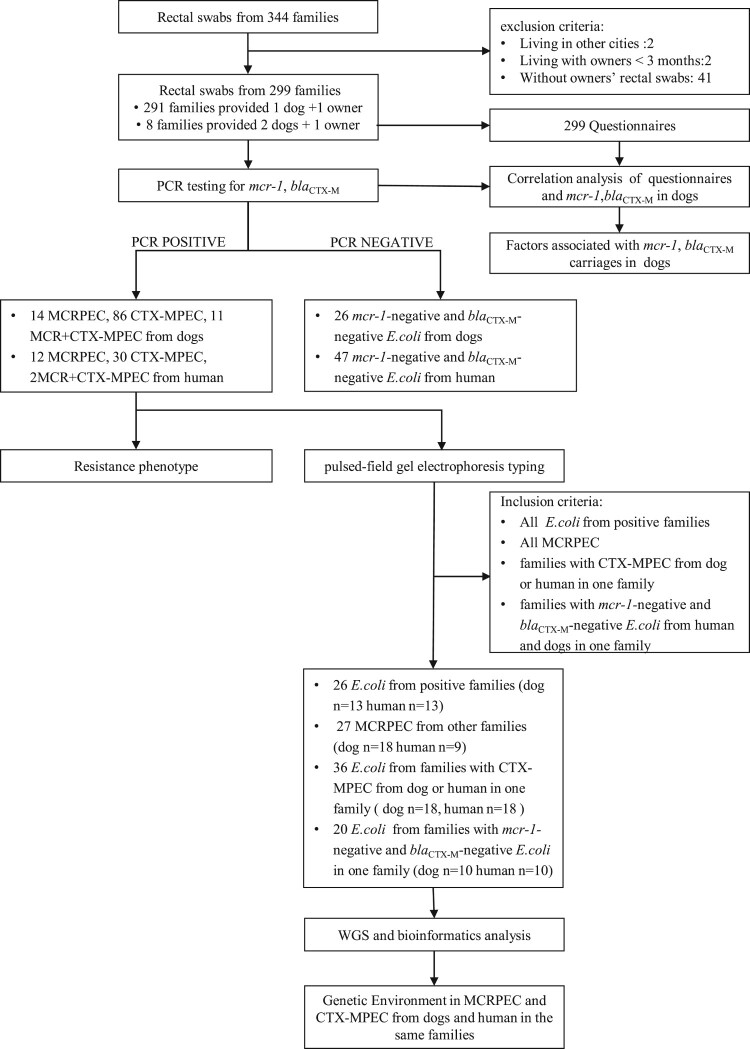
We recruited participants together with their dogs amongst visitors to the Veterinary Teaching Hospital of China Agricultural University, Beijing, between March and November 2017. After obtaining consent, rectal swabs were collected from dogs and their owners. Owners also completed a questionnaire (Supplementary Table S1) covering the sex, age, habits, diet, address, antibiotic use, and interaction with veterinary hospitals or clinics within the previous 3 months for both the dog(s) and their owner(s). We targeted at Beijing residents who lived with their dog(s) for over three months. We excluded dogs that had intestinal diseases to reduce data bias (Figure 1). In this study, a “family” was defined as a unit consisting of a pet owner and his/her dog(s).
Figure 1.
Workflow of this study.
Laboratory tests
All samples were screened for the presence of antibiotic resistance (AR) genes mcr-1 and blaCTX-M by PCR (Supplementary Table S2) [18]. E. coli was selected as a typical species for the characterization of AR bacteria in humans and their pet dogs, and one E. coli isolate was selected from each sample. Bacterial isolation procedures was presented in Appendices. The presence of resistance genes in each of the colonies was confirmed by PCR. Species were identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and 16S rRNA gene sequencing.
All E. coli isolates were subjected to antimicrobial susceptibility testing. Isolates were screened using seven clinical antimicrobial agents (colistin, ceftiofur, cefotaxime, amoxicillin-clavulanic acid, gentamicin, ciprofloxacin, and meropenem). Results were interpreted according to the European Committee on Antimicrobial Susceptibility Testing breakpoints [19] and Clinical and Laboratory Standards Institute documents M100-ED29 and VET08 [20,21].
All E. coli isolates were analysed using PFGE. The PFGE plugs were made according to the standard PlusNet operating procedure and were treated with XbaI before being run on a CHEF Mapper apparatus. Using the criteria listed in Figure 1, 109 E. coli strains were selected from dogs and their owners from different families for whole-genome sequencing. Genomic DNA was extracted using a HiPure Bacterial DNA Kit. Indexed DNA libraries were prepared using a KAPA Hyper Prep Kit and sequenced using the Illumina HiSeq X platform with a 150-bp paired-end strategy.
Draft genomes were assembled using SPAdes [22]. Sequence types (STs), AR genes, and virulence genes were identified using SRST2 toolkit [23]. Minimum spanning trees of MCRPEC and CTX-MPEC were generated in BioNumerics. A phylogenetic tree was produced by RedDog (https://github.com/katholt/RedDog) and IQ-TREE [24] using core-genome alignments and was visualized using iTOL [25]. mcr-1-containing contigs and blaCTX-M-containing contigs were examined to determine Inc types using PlasmidFinder [26]. Three CTX-MPEC and three MCRPEC, in which the assembled contigs are too short to analyse, were subjected to Nanopore sequencing to determine the genetic environments of the blaCTX-M and mcr-1 genes. All raw data in this study have been deposited in the GenBank and under Bioproject accession no. PRJNA650157.
Statistical analysis
Data was consolidated in EXCEL 2016. Univariable analysis was conducted using χ2 tests inSPSS® software (version 23.0). Variables with a P-value < 0.2 were kept for multivariable analysis. Multivariable logistic regression model adopted the backward stepwise process. Variables with a P-value< 0.05 were considered as a risk factor.
Results
From March to November 2017, we collected rectal swabs from 344 families but excluded 45 families because they reside out of Beijing, their dog(s) had lived with family for less than three months, or their owners’ rectal swabs were missing. A total of 299 families were included, providing 299 rectal swabs from humans and 307 rectal swabs from their dogs (Figure 1). PCR testing of the stool samples revealed that 49 dogs (16.0%, 95% CI: 12.0–20.5%) and 21 humans (7.0%, 95% CI: 4.4–10.5%) were positive for mcr-1. blaCTX-M was identified in 126 samples from dogs (41.0%, 95% CI: 35.5–46.8%) and 45 samples from humans (15.1%, 95% CI: 11.2–19.6%). The prevalence of positive families (i.e. those in which both the human and their dog(s) showed positive results) was low: Eight families were positive for mcr-1 (2.7%, 95% CI: 1.2–5.2%) and 16 were for blaCTX-M (5.3%, 95% CI: 3.1–8.5%). Univariable analysis revealed seven variables in the risk analysis of mcr-1 positivity in dogs (Table 1), but the multivariable logistic analysis kept two (P < 0.05), dog with ovariectomy/castration (OR = 2.29, 95% CI: 1.00–5.20) and an owner with mcr-1-positivity (OR = 4.19, 95% CI: 1.59–11.05) (Table 2). Univariable analysis revealed that six variables in the risk analysis of blaCTX-M-positivity in dogs (Table 3), but multivariable analysis identified only one risk factor (P < 0.05), antibiotic treatment within the previous three months (OR = 2.02, 95% CI: 1.27–3.22) (Table 4).
Table 1.
Univariable analysis of mcr-1-positivity in dogs of interest.
| Variable | Categories | Percentage | Prevalence % (95% CI) | OR (95% CI) | P |
|---|---|---|---|---|---|
| Ovariectomy/castration | Yes | 28.0% | 9.3 (4.1–17.5) | 0.45 (0.20–1.01) | 0.047* |
| No | 72.0% | 18.6 (13.7–24.3) | 1.0 | ||
| Rummage the trash bin | Yes | 28.0% | 17.4 (10.1–27.1) | 1.16 (0.60–2.26) | 0.659 |
| No | 72.0% | 15.4 (10.9–20.8) | 1.0 | ||
| Lick/Kiss owner | Yes | 80.5% | 14.2 (10.1–19.2) | 0.54 (0.27–1.09) | 0.082* |
| No | 19.5% | 23.3 (13.4–36.0) | 1.0 | ||
| Play in the bed | Yes | 45.0% | 15.2 (9.7–22.3) | 0.90 (0.49–1.67) | 0.748 |
| No | 55.0% | 16.6 (11.3–23.0) | 1.0 | ||
| Commercial dog food | Yes | 84.0% | 14.3 (10.3–19.2) | 0.52 (0.25–1.08) | 0.075* |
| No | 16.0% | 24.5 (13.3–38.9) | 1.0 | ||
| Immunization in past 3 months | Yes | 21.5% | 21.2 (12.1–33.0) | 1.59 (0.79–3.16) | 0.189* |
| No | 78.5% | 14.5 (10.3–19.6) | 1.0 | ||
| Have medicine/Infusion in past 3 months | Yes | 38.1% | 18.8 (12.2–27.1) | 1.40 (0.75–2.59) | 0.286 |
| No | 61.9% | 14.2 (9.6–20.0) | 1.0 | ||
| Surgery/Hospitalization in past 3 months | Yes | 11.7% | 8.3 (1.8–22.5) | 0.45 (0.13–1.51) | 0.184* |
| No | 88.3% | 17.0 (12.7–22.0) | 1.0 | ||
| Have medicine for human use | Yes | 27.0% | 18.1 (10.5–28.0) | 1.23 (0.63–2.40) | 0.539 |
| No | 73.0% | 15.2 (10.7–20.6) | 1.0 | ||
| Use of antibiotic in past three months | Yes | 45.6% | 15.7 (10.1–22.8) | 0.97 (0.52–1.79) | 0.914 |
| No | 54.4% | 16.2 (10.9–22.6) | 1.0 | ||
| Owner with mcr-1 positivity | Yes | 6.8% | 38.1 (18.1–61.6) | 3.68 (1.44–9.42) | 0.010* |
| No | 93.2% | 14.3 (10.5–18.9) | 1.0 |
*Variables with P < 0.20 were entered into the multivariable model.
Table 2.
Multivariable logistic regression analysis of factors associated with mcr-1-positivity in dogs.
| Variable | OR (95% CI) | P |
|---|---|---|
| Without ovariectomy/castration | 2.29 (1.00–5.20) | 0.049* |
| Owner with mcr-1 positivity | 4.19 (1.59–11.05) | 0.004* |
| Without Lick/Kiss owner | 1.87 (0.92–3.83) | 0.085 |
*Variables with P < 0.05 were considered significantly associated with positivity.
Table 3.
Univariable analysis of blaCTX-M-positivity in dogs.
| Variable | Categories | Percentage | Prevalence % (95% CI) | OR (95% CI) | P |
|---|---|---|---|---|---|
| Ovariectomy/castration | Yes | 28.0% | 41.9 (31.3–53.0) | 1.05 (0.63–1.74) | 0.856 |
| No | 72.0% | 40.7 (34.2–47.5) | 1 | ||
| Rummage the trash bin | Yes | 28.0% | 44.2 (33.5–55.3) | 1.20 (0.72–1.98) | 0.485 |
| No | 72.0% | 39.8 (33.3–46.6) | 1 | ||
| Lick/Kiss owner | Yes | 80.5% | 41.3 (35.1–47.7) | 1.06 (0.59–1.88) | 0.855 |
| No | 19.5% | 40.0 (27.6–53.5) | 1 | ||
| Play in the bed | Yes | 45.0% | 40.6 (32.3–49.3) | 0.97 (0.61–1.53) | 0.882 |
| No | 55.0% | 41.4 (33.9–49.2) | 1 | ||
| Commercial dog food | Yes | 84.0% | 38.4 (32.4–44.6) | 0.51 (0.28–0.94) | 0.029* |
| No | 16.0% | 55.1 (40.2–69.3) | 1 | ||
| Immunization in past 3 months | Yes | 21.5% | 31.8 (20.9–44.4) | 0.60 (0.34–1.10) | 0.086* |
| No | 78.5% | 43.6 (37.2–50.1) | 1 | ||
| Have medicine/Infusion in past 3 months | Yes | 38.1% | 47.9 (38.5–57.3) | 1.57 (0.99–2.51) | 0.057* |
| No | 61.9% | 36.8 (30.0–44.1) | 1 | ||
| Surgery/Hospitalization in past 3 months | Yes | 11.7% | 52.8 (35.5–69.6) | 1.71 (0.85–3.44) | 0.128* |
| No | 88.3% | 39.5 (33.6–45.6) | 1 | ||
| Have medicine for human use | Yes | 27.0% | 48.2 (37.1–59.4) | 1.49 (0.90–2.48) | 0.121* |
| No | 73.0% | 38.4 (32.0–45.1) | 1 | ||
| Use of antibiotic in past 3 months | Yes | 45.6% | 50.7 (42.1–59.3) | 2.10 (1.32–3.33) | 0.002* |
| No | 54.4% | 32.9 (25.9–40.6) | 1 | ||
| Owner with blaCTX-M positivity | Yes | 14.7% | 35.6 (21.9–51.2) | 0.76 (0.40–1.47) | 0.418 |
| No | 85.3% | 42.0 (35.9–48.2) | 1 |
*Variables with P < 0.20 were entered into the multivariable model.
Table 4.
Multivariable logistic regression analysis of factors associated with blaCTX-M-positivity in dogs.
| Variable | OR (95% CI) | P |
|---|---|---|
| No Commercial dog food | 1.82 (0.97–3.41) | 0.060 |
| Use of antibiotic in past 3 months | 2.02 (1.27–3.22) | 0.003* |
*Variables with P < 0.05 were considered significantly associated with positivity.
We isolated 228 E. coli from dogs (n = 137) and their owners (n = 91). These included 26 MCRPEC, 116 CTX-MPEC, 13 MCR + CTX-MPEC, and 73 E. coli negative for both mcr-1 and blaCTX-M (Figure 1). Three families had both the dog and human carrying MCRPEC and nine carrying CTX-MPEC, one family carried MCR + CTX-MPEC. blaCTX-M genes identified in the 126 isolates predominantly belonged to the CTX-M-1 (41 isolates from dogs and eight from humans) or CTX-M-9 (53 isolates from dogs and 24 from humans) groups. Three isolates from separate dogs showed two CTX-M genotypes (CTX-M-1 and CTX-M-9). The most prevalent blaCTX-M subtype was blaCTX-M-14 (35 isolates from dogs and 19 from humans) (Supplementary Figure S1), which was found in five blaCTX-M-positive families. The strains isolated from dogs and their owners in the remaining five blaCTX-M-positive families harboured different blaCTX-M genotypes.
Susceptibility testing showed that the resistance profiles of all E. coli isolates. Isolates from dogs exhibited higher resistance to all antimicrobials except amoxicillin-clavulanate compared with those from humans (Supplementary Table S3). Isolates harbouring mcr-1 (n = 25) from dogs were more often resistant to ceftiofur, cefotaxime and ciprofloxacin than isolates harbouring mcr-1 (n = 14) from humans (P < 0.05), while blaCTX-M-positive isolates (n = 97) from dogs were more frequently resistant to gentamicin compared with blaCTX-M-positive isolates (n = 32) from humans (P < 0.05) (Supplementary Table S4). E. coli negative for both mcr-1 and blaCTX-M from dogs showed no significance with those from humans. Isolates from the dog and human in either mcr-1-positive or blaCTX-M-positive families usually showed different resistance phenotypes; however, the two blaCTX-M-positive E. coli isolates from one family (one from the human and one from the dog) exhibited identical resistance profiles.
We used PFGE analysis to examine the similarity among the 228 E. coli isolates from humans and their pet dogs. Except for 15 non-typeable isolates, 213 E. coli isolates (85 from humans and 128 from dogs) showed diverse PFGE profiles. Isolates from the same host origin showed greater similarity than those from the other host origin. For example, 23 dog isolates from 23 families showed eight PFGE patterns with a similarity value ≥85%. Moreover, five human isolates from five families showed two PFGE patterns with a similarity value ≥85%. The paired isolates from humans (n = 54) and their dogs (n = 54) in each of 54 families showed different PFGE profiles, except for the two blaCTX-M-positive E. coli from one family which showed a similarity value ≥85% (Supplementary Figure S2). Five patterns displayed by isolates (six from humans and five from dogs) from different families had a similarity value ≥85%. Although these isolates from dogs and humans showed a PFGE similarity value ≥85%, the isolates were recovered from families that lived >15 km from each other based on information provided by the owners.
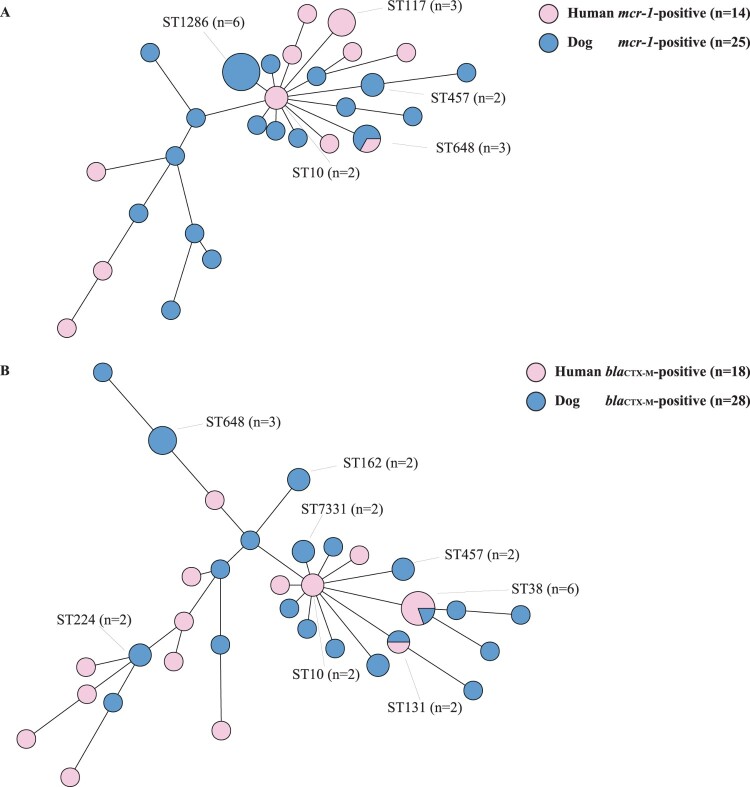
The 109 E. coli isolates selected for whole-genome sequencing shown as Figure 1. The minimum spanning tree showed that MCRPEC from humans and their dogs had diverse STs (Figure 2(A)). MCRPEC from dogs (n = 25) showed 18 STs, and those from humans (n = 14) showed 11 STs. ST648 was the only ST demonstrated by isolates from both humans (one case) and dogs (two cases) from three different families. CTX-MPEC isolates from dogs (n = 28) showed 21 STs, and those from humans (n = 18) showed 14 different STs (Figure 2(B)). Only ST38 and ST131 were identified in both dogs and humans: ST131 was shared by one dog-derived isolate and one human-derived isolate from different families, but ST38 isolates were recovered from one dog and its owner, as well as from three humans from other families.
Figure 2.
Mining spanning trees based on multi-locus sequence types and seven housekeeping gene alleles of (A) mcr-1-positive Escherichia coli (MCRPEC) and (B) blaCTX-M-positive E. coli (CTX-MPEC) from humans and their dogs.
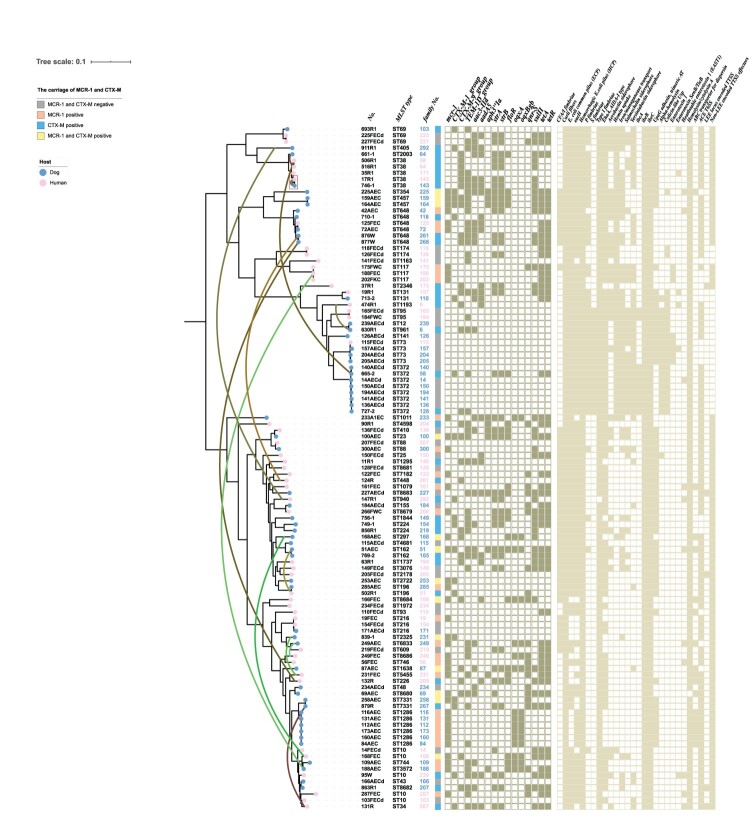
Core-genome-based phylogenetic analysis of the 109 whole-genome-sequenced E. coli isolates showed that, in most cases, the isolates from humans and dogs clustered separately (Figure 3). For instance, isolates from dogs (ST162, ST457, ST372, ST1286, or ST7331) or humans (ST174, ST117, ST95, or ST69) showed high similarity to each other. Although isolates with STs 38, 648, 131, 88, 196, 216, and 73 were found in both humans and dogs and displayed high similarity, the dogs and owners carrying these isolates belonged to different families. Meanwhile, strains from dogs and owners from the same family, including MCRPEC in three families (curved green lines link MCRPEC from humans and dogs in the same family), CTX-MPEC in eight families (the curved brown lines link CTX-MPEC from humans and dogs in the same family), and MCR + CTX-MPEC from dog and human in one family, displayed low similarity. Among the 299 families included in this study, only one woman and her dog from one blaCTX-M-positive family carried identical E. coli isolates which SNPs less than 50 (Figure 3).
Figure 3.
Phylogenetic tree of Escherichia coli isolated from humans and their pet dogs.
Considering resistance genes, sulphonamide resistance gene sul2 was highly prevalent in both humans and dogs, while quinolone resistance genes oqxAB was more prevalent in dogs than humans (P < 0.05). Virulence genes associated with S fimbriae, iron/manganese transport, and α-hemolysin were more commonly present in strains of dog origin than in those of human origin (P < 0.05), while other virulence genes, such as toxin genes associated with hemolysin/cytolysin A, secretion system genes associated with the ace T6 secretion system, adherence genes associated with type I fimbriae, and eaeH, were prevalent in isolates from both dogs and humans (Supplementary Table S6). Almost all isolates from humans and dogs within a family showed different virulence gene profiles, except for the CTX-MPEC isolate shared by one woman and her dog (see above).
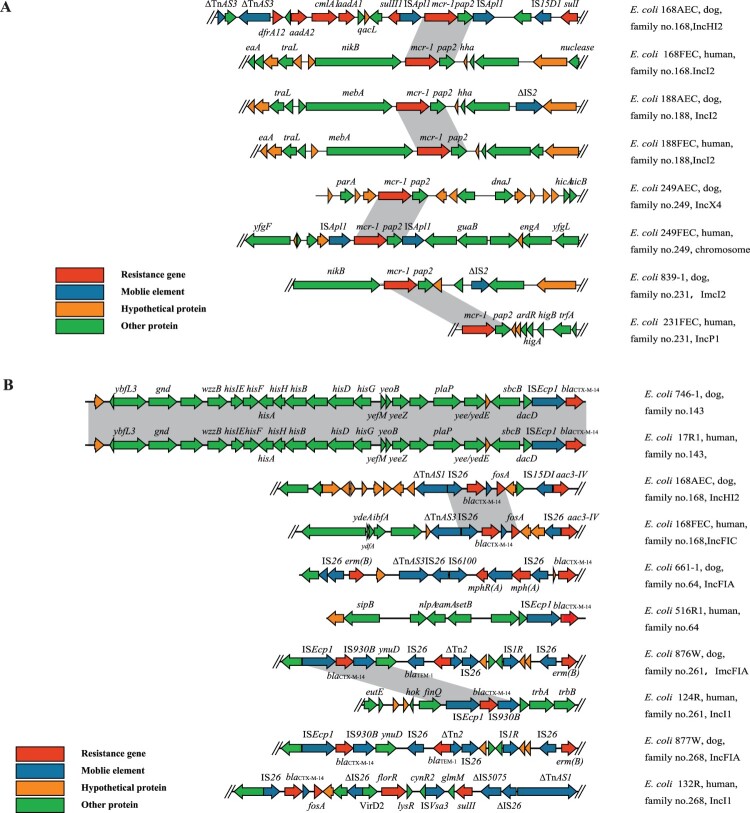
Different genetic environments were observed for resistance genes in isolates from dogs (n = 4) and humans (n = 4) from three mcr-1-positive families and one mcr-1 and blaCTX-M-positive family (Figure 4(A)). In the four strains from dogs, mcr-1 was located on plasmids with different Inc types (two IncI2, one IncX4, and one IncHI2). In the four strains from humans, mcr-1 was located on the chromosome in one case and on plasmids (one IncP1 and two IncI2) in the other three cases. Although mcr-1-positive IncI2 plasmids were identified in isolates from both humans and dogs, they were derived from different families or showed different genetic environments in isolates from the same family. The genetic environments of mcr-1 in isolates from the four dogs and four humans could be divided into two types: one (n = 2) contained mcr-1 together with pap2 and flanked by two ISApl1 elements, while the other (n = 6) contained mcr-1 flanked by genes encoding a hypothetical protein and nikB/mebA.
Figure 4.
(A) Genetic environment of MCRPEC from different origins. (B) Genetic environment of CTX-MPEC from different origins.
Different from strains of mcr-1-positive families, the blaCTX-M-positive isolates from humans and dogs within a family in four of the five families had different Inc types and showed different genetic structures (Figure 4(B)). In the four isolates from dogs, blaCTX-M-14 with different Inc types (one IncHI2, three IncFIA). The Inc types of the four isolates from the corresponding humans were IncI1 (n = 2), IncFIC (n = 1), and unknown (n = 1). Isolates from two families had different IS elements flanking blaCTX-M-14, another two families had the same IS elements flanking blaCTX-M-14 but contained different genes along with the IS elements. blaCTX-M-14 was associated with IS elements in the strains from dogs (n = 4): two ISEcp1 and IS930B, and two IS26. However, in the four strains from the corresponding humans, blaCTX-M-14 was found in association with ISEcp1 (n = 1), IS26 (n = 2), and ISEcp1 and IS930B (n = 1). Only one family had two blaCTX-M-14-carrying E. coli isolates with blaCTX-M-14 in an identical genetic environment with ISEcp1 nearby blaCTX-M-14.
Discussion
We analysed possible risk factors associated with AR gene carriage in dogs to help to design the applicable control measures. A significant association of mcr-1 carriage between dogs and their owners (OR = 4.19, 95% CI: 1.59–11.05), suggesting the mcr-1 bacteria in humans would increase the risk of mcr-1 carriage in their dogs or vice versa. Sharing common living areas and having similar diet would be the explanation of such risk. Others found similar or indistinguishable Methicillin-resistant Staphylococcus pseudintermedius in human, companion animals and environmental samples isolated from the same household [27]. Food consumed by humans and pets has a direct impact on their gut microbiome [28,29]. Previous studies showed that animal products in China posed direct risk of mcr-1 positivity to the health people in China due to the drug resistance gene transmission along food chain [8,30]. Our study indicated that the prevalence of mcr-1 in dogs sharing their owner’s food (24.5%) was higher than in those ate commercial dog food (14.3%). Therefore, mcr-1 in dogs may also sources from their owner’s meat meals. Since this study was conducted in the pet hospital, no environmental samples were collected from the pets living areas. Further studies focusing on the source of mcr-1 in dogs and the genetic similarity of mcr-1 isolated from pets, owners and their living areas were strongly recommended for a deep understanding of the transmission of mcr-1 in one health approach.
Ovariectomy/castration of dogs may not directly associate with the mcr-1 positivity in dogs but function as a confounding factor. The colonization of AR genes could be influenced by hormonal changes or behaviour changes caused by ovariectomy/castration, as these changes had impacts on the composition and diversity of gut microbiota, such as ovariectomy/castration resulted in decreasing in Bacteroidetes or increasing in Firmicutes [31,32] which further enhanced the colonization resistance of bacteria [33]. Normal dogs are more active and more likely to contact other dogs than ovariectomized/castrated dogs in estrus, which would encourage the dogs without ovariectomy/castration acquired mcr-1-positive bacteria from dogs and environment [34].
Not surprisingly, the antibiotic usage is the risk factor for blaCTX-M positivity in both dogs and humans [35,36]. The questionnaires in our study showed that usage of cephalosporins and fluoroquinolones more than other antimicrobial agents which may influence blaCTX-M-positivity in dogs. Li et al. [37] found that AR bacteria can be transmitted between humans and backyard animals, while another study suggested that pets can act as potential reservoirs of AR bacteria [38]. Thus, pets may play an important role in the transmission of AR bacteria to humans because they live in close confines with their owners and inevitably have an impact on humans. It is highly recommended that measures should be taken to control the potential risk from dogs such as reducing and standardizing antibiotic usage in clinics, and protecting living areas from being contacted by AR bacteria. Another highly practicable measure is to control the diet of dogs to prevent AR bacterial colonization such as feeding commercial dog food and reducing raw meat and owners food.
Our study revealed diverse MCRPEC and CTX-MPEC profiles in human and dogs. Family level prevalence of mcr-1 and blaCTX-M was 2.7 and 5.3%, respectively. Apart from the one case (0.3%) in which a dog and its owner carried high similarity strain of blaCTX-M-14-positive E. coli, the isolates from humans and their dogs showed low similarity in both PFGE and whole-genome sequencing analysis. Previous studies have reported sporadic interspecies dissemination of AR bacteria among pets and humans [3, 17]. We hypothesized that technical difficulties in the detection of AR bacterial colonization and selection bias during sample collection and bacterial isolation may contribute to the lack of correlation between isolates from humans and dogs in this study, despite we found correlation of mcr-1 in human and dogs in statistical analysis. We previously demonstrated a strong association between meat production, consumption, and daily intake and human carriage of MCRPEC [8]. The similar dietary structure of the different families enrolled in this study may explain the high level of similarity among isolates recovered from different families. The prevalence of MCRPEC and CTX-MPEC positive families was fairly low in our survey. However, considering the large population of pet families worldwide, the number of MCRPEC and CTX-MPEC positive families cannot be ignored. There is likely to be a significant number of cases of AR bacterial transmission and colonization in humans and dogs belong to one family. The risk of AR bacteria dissemination among pets, pet owners and their living areas should be highlighted seriously since this public health issue is threating one health approach. We acknowledge that our study has several limitations. Except for the limitations we already mentioned, we only collected and studied samples from one hospital in Beijing. However, this institute, the Veterinary Teaching Hospital of China Agricultural University, is the largest small animal hospital in China and serves 100–150 patients per day from Beijing city and even surrounding provinces. Second, we only collected one human sample per family, whilst we confirmed that the selected individual had the greatest interaction with the dog(s) but we ignored the impact of other household members and community members on dogs. Third, we haven’t collected environmental samples of tested families, the role of environment in AR bacteria transmission is unknown.
Supplementary Material
Funding Statement
This work was supported in part by National Natural Science Foundation of China [Grant numbers 81861138051 and 81991535], and UK MRC DETER-XDR-China-HUB [Grant number MR/S013768/1].
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Companion animals responsible ownership Statiscs on Dogs[Internet] . (2012). [cited 2019 November 3]. Available from: https://www.carodog.eu/statistics-on-cats-and-dogs/.
- 2.Tian LL. Chinese pets industry white paper-consumer report (consumer report). Beijing: Dog Network; 2019. [Google Scholar]
- 3.Gronthal T, Osterblad M, Eklund M, et al. Sharing more than friendship - transmission of NDM-5 ST167 and CTX-M-9 ST69 Escherichia coli between dogs and humans in a family, Finland, 2015. Euro Surveill. 2018 Jul;23(27):pii=1700497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Köck R, Daniels-Haardt I, Becker K, et al. Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: a systematic review. Clin Microbiol Infec. 2018 Dec;24(12):1241–1250. [DOI] [PubMed] [Google Scholar]
- 5.Bassetti M, Poulakou G, Ruppe E, et al. Antimicrobial resistance in the next 30 years, humankind, bugs and drugs: a visionary approach. Intens Care Med. 2017 Oct;43(10):1464–1475. [DOI] [PubMed] [Google Scholar]
- 6.Andersson DI, Hughes D.. Selection and transmission of antibiotic-resistant bacteria. Microbiol Spectr. 2017 Jul;5(4). MTBP-0013-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elbediwi M, Li Y, Paudyal N, et al. Global burden of colistin-resistant bacteria: mobilized colistin resistance genes study (1980-2018). Microorganisms. 2019 Oct;7(10):461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen Y, Zhou H, Xu J, et al. Anthropogenic and environmental factors associated with high incidence of mcr-1 carriage in humans across China. Nat Microbiol. 2018 Sep;3(9):1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lei L, Wang Y, Schwarz S, et al. mcr-1 in Enterobacteriaceae from companion animals, Beijing, China, 2012–2016. Emerg Infect Dis. 2017 Apr;23(4):710–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Huang XY, Xia YB, et al. Clonal spread of Escherichia coli ST93 carrying mcr-1-harboring IncN1-IncHI2/ST3 plasmid among companion animals, China. Front Microbiol. 2018;9:2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karanika S, Karantanos T, Arvanitis M, et al. Fecal colonization With extended-spectrum Beta-lactamase–producing Enterobacteriaceae and risk factors among healthy individuals: a systematic review and metaanalysis. Clin Infect Dis. 2016 Aug;63(3):310–318. [DOI] [PubMed] [Google Scholar]
- 12.Hu YY, Cai JC, Zhou HW, et al. Molecular typing of CTX-M-producing Escherichia coli isolates from environmental water, swine feces, specimens from healthy humans, and human patients. Appl Environ Microb. 2013 Oct;79(19):5988–5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, Zhou Y, Guo S, et al. High prevalence and risk factors of fecal carriage of CTX-M type extended-spectrum beta-lactamase-producing Enterobacteriaceae from healthy rural residents of Taian, China. Front Microbiol. 2015;6:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abbas G, Khan I, Mohsin M, et al. High rates of CTX-M group-1 extended-spectrum beta-lactamases producing Escherichia coli from pets and their owners in Faisalabad, Pakistan. Infect Drug Resist. 2019;12:571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Liu H, Li Y, et al. High prevalence of beta-lactamase and plasmid-mediated quinolone resistance genes in extended-spectrum cephalosporin-resistant Escherichia coli from dogs in Shaanxi, China. Front Microbiol. 2016;7:1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zogg AL, Simmen S, Zurfluh K, et al. High prevalence of extended-spectrum β-lactamase producing enterobacteriaceae among clinical isolates from cats and dogs admitted to a veterinary hospital in Switzerland. Front Vet Sci. 2018;5:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang XF, Doi Y, Huang X, et al. Possible transmission of mcr-1 –harboring Escherichia coli between companion animals and human. Emerg Infect Dis. 2016 Sep;22(9):1679–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu YY, Wang YL, Sun QL, et al. Colistin resistance gene mcr-1 in gut flora of children. Int J Antimicrob Agents. 2017 Oct;50(4):593–597. [DOI] [PubMed] [Google Scholar]
- 19.The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 9.0. 2019.
- 20.CLSI . Performance standards for antimicrobial susceptibility testing. 29th ed. CLSI supplement M100S. Wayne (PA: ): Clinical and Laboratory Standards Institute; 2019. [Google Scholar]
- 21.CLSI . Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. 4th ed. CLSI suplement VET08. Wayne (PA: ): Clinical and Laboratory Standards Institute; 2018. [Google Scholar]
- 22.Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012 May;19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inouye M, Dashnow H, Raven LA, et al. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6(11):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen LT, Schmidt HA, von Haeseler A, et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015 Jan;32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Letunic I, Bork P.. Interactive tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007 Jan;23(1):127–128. [DOI] [PubMed] [Google Scholar]
- 26.Carattoli A, Zankari E, Garcia-Fernandez A, et al. In Silico detection and typing of plasmids using plasmid finder and plasmid multilocus sequence Typing. Antimicrob Agents Chemother. 2014 Jul;58(7):3895–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laarhoven LM, de Heus P, van Luijn J, et al. Longitudinal study on methicillin-resistant Staphylococcus pseudintermedius in households. PLoS one. 2011;6(11):e27788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schauf S, de la Fuente G, Newbold CJ, et al. Effect of dietary fat to starch content on fecal microbiota composition and activity in dogs1. J Anim Sci. 2018 Sep;96(9):3684–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh RK, Chang HW, Yan D, et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017 Apr;15(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Zhang R, Li J, et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbol. 2017 Feb;2:16260. [DOI] [PubMed] [Google Scholar]
- 31.Shin JH, Park YH, Sim M, et al. Serum level of sex steroid hormone is associated with diversity and profiles of human gut microbiome. Res Microbiol. 2019;170(4-5):192–201. [DOI] [PubMed] [Google Scholar]
- 32.Org E, Mehrabian M, Parks BW, et al. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes. 2016 Jul;7(4):313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buffie CG, Pamer EG.. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013 Nov;13(11):790–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGuire B. Effects of gonadectomy on scent-marking behavior of shelter dogs. J Vet Behav. 2019;30:16–24. [Google Scholar]
- 35.Leangapichart T, Tissot-Dupont H, Raoult D, et al. Risk factors for acquisition of CTX-M genes in pilgrims during Hajj 2013 and 2014. J Antimicrob Chemother. 2017 Sep;72(9):2627–2635. [DOI] [PubMed] [Google Scholar]
- 36.Luvsansharav UO, Hirai I, Nakata A, et al. Prevalence of and risk factors associated with faecal carriage of CTX-M -lactamase-producing Enterobacteriaceae in rural Thai communities. J Antimicrob Chemother. 2012 Jul;67(7):1769–1774. [DOI] [PubMed] [Google Scholar]
- 37.Li J, Bi Z, Ma S, et al. Inter-host transmission of carbapenemase-producing Escherichia coli among humans and backyard animals. Environ Health Persp. 2019 Oct;127(10):107009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lloyd DH. Reservoirs of antimicrobial resistance in pet animals. Clin Infect Dis. 2007 Sep;45(Suppl 2):S148–S152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.