ABSTRACT
Background: We investigated the association between prenatal GDM exposure and offspring DNA methylation (DNAm) age at 3–10 years of age in the Tianjin GDM Observational Study.
Methods: This study included 578 GDM and 578 non-GDM mother-child pairs. Children underwent an exam with anthropometric measurements and blood draw for DNAm analysis (Illumina 850 K array) at a median age of 5.9 years (range 3.1–10.2). DNAm age was calculated using two epigenetic clock algorithms (Horvath and Hannum). The residual resulting from regressing DNAm age on chronological age was used as a metric for age acceleration.
Results: Chronological age was positively correlated with Horvath DNAm age (r = 0.53, p < 0.0001) and Hannum DNAm age (r = 0.38, p < 0.0001). Offspring age acceleration was higher in the GDM group than non-GDM group after adjustment for potential confounders (Horvath: 4.96 months higher, adjusted for sex, pre-pregnancy BMI, cell-type proportions, and technical bias, p = 0.0002; Hannum: 11.2 months higher, adjusted for cell-type proportions and technical bias, p < 0.0001). Increased offspring DNAm age acceleration was associated with increased offspring weight-for-age Z-score, BMI-for-age-Z-score, waist circumference, body fat percentage, subscapular skinfold, suprailiac skinfold, upper-arm circumference, and blood pressure; findings were stronger in the GDM group.
Conclusions: We found that offspring of women with GDM exhibit accelerated epigenetic age compared to control participants, independent of other maternal factors. Epigenetic age in offspring was associated with cardiometabolic risk factors, suggesting that GDM and GDM-associated factors may have long-term effects on offspring epigenetic age and contribute to health outcomes.
KEYWORDS: Gestational diabetes mellitus, DNA methylation, epigenetic age
Background
The prevalence of gestational diabetes mellitus (GDM), a condition in which a woman without diabetes presents with elevated blood glucose levels during pregnancy, has increased on a global scale [1,2]. Women with GDM are at an increased risk of adverse complications during pregnancy and delivery [3,4]. Growing evidence also indicates that intrauterine exposure to GDM places offspring at an increased risk for metabolic disorders later in life [5–7], in line with the Developmental Origins of Health and Disease (DOHaD) hypothesis [8–10]. While DOHaD was originally developed to conceptualize the relationship between foetal undernutrition and heart disease in early life, evidence of long-term GDM effects is consistent with a broader DOHaD hypothesis that factors acting early in life can have profound effects on predisposition to disease later in adult life.
Prior reports have linked GDM exposure to shortened telomere length, a clinical biomarker of ageing, in offspring in childhood [11]. Epigenetic modifications, including genome-wide DNA methylation (DNAm), have been studied as a potential mechanism linking maternal GDM with offspring metabolic risk in later life [12–16]. DNAm may also have value as a biomarker of biological ageing. Several ‘epigenetic clocks’ have been proposed using DNAm levels at ageing-relevant CpG sites from multiple tissues to predict chronological age with high accuracy [17–19]. ‘Accelerated aging’ can be assessed by evaluating if DNAm age is greater than chronological age, and recent studies have linked accelerated ageing to cardiovascular risk and all-cause mortality [20–24]. Based on this evidence, we hypothesized that offspring exposed to GDM with increased risk of metabolic disease in later life may exhibit evidence of accelerated ageing by DNAm age in early childhood.
To our knowledge, no studies have explored the association between GDM exposure and accelerated epigenetic age in offspring. This is an important area to study, as a recent study found that accelerated epigenetic age in adolescents was associated with inflammation, BMI measured 5 years later, and probability of middle-age cardiovascular disease [25]. Thus, this study aimed to evaluate the association between prenatal GDM exposure and offspring DNAm age at 3–10 years of age in the Tianjin GDM Observational Study and to examine if DNAm age is associated with childhood cardiometabolic health.
Methods
Study design and population
This study uses data from a large observational study of 1156 women with and without GDM and their offspring conducted in Tianjin, China [26,27]. In brief, women who underwent a two-hour oral glucose tolerance test (OGTT) with a 75 g glucose load and had a result confirming either diabetes (fasting glucose >7 nmol/l or two-hour OGTT >11.1 nmol/l) or impaired glucose tolerance (two-hour glucose >7.8 and <11.1 nmol/l) based on the 1999 WHO criteria were defined as having GDM [28]. Women diagnosed with GDM were invited to participate in the Tianjin GDM Prevention Program, completing a baseline survey from 2009–2011 and followed up for surveys at 1 and 2 years. Of women who finished the follow-up surveys and also had blood samples, 578 GDM mother-child pairs were randomly selected as GDM cases. Additional 578 women without GDM and their paired children were randomly recruited, frequency matched to the cases on the child’s birth date and sex. Written informed consent was collected from all participants and this study was approved by the Human Subjects Committee of Tianjin Women’s and Children’s Health Centre and the Institutional Review Boards of Columbia University Medical Centre and Pennington Biomedical Research Centre.
Measurements
At the childhood baseline study visit, a questionnaire on socio-demographic, pregnancy, and child characteristics in early infancy and at the visit was completed by mothers, and most above information was also available from healthcare records for both women and children available electronically [29]. Socio-demographic characteristics included maternal age at delivery, marital status, maternal education, and family income. Pregnancy characteristics included weight gain during pregnancy, pre-pregnancy BMI, family history of diabetes, and lifestyle in the past year, such as smoking status, passive smoking, and drinking status. Pre-pregnancy BMI calculations were based on self-reported pre-pregnancy weight and measured height at baseline survey and categorized as <24 (not overweight or obese), 24–27.9 (overweight) and ≥28 kg/m2 (obese) according to the Chinese BMI cut-offs [30]. Child characteristics in early infancy included sex, mode of delivery, birth weight, birth length, gestational age, and infant feeding within the first 6 months. Characteristics at the childhood visit included age and routine activities (indoor and outdoor activities, TV watching time and sleep duration). In addition, children underwent a standardized physical examination at the baseline study visit that included anthropometric measurements. Body weight was measured with a beam balance scale to the nearest 0.1 kg; height was measured by a stadiometer to the nearest 0.1 cm. Waist circumference was measured midway between the 10th rib and the top of the iliac crest to the nearest 0.1 cm. Body fat was measured by a body composition analyser (InBody J20) to the nearest 0.1%. Triceps skinfold, subscapular skinfold, and suprailiac skinfold were measured by skinfold calliper to the nearest 0.5 cm. BMI was obtained by dividing weight in kilograms by the square of height in metres. Weight-for-age, height-for-age, and BMI-for age Z-scores were calculated based on growth references from the World Health Organization. Overweight and obesity were defined as Z-score ≥1.035 and Z-score ≥1.645, respectively [31].
DNA methylation data and data pre-processing
DNA was extracted from whole blood samples from the childhood visit using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA). DNA methylation was measured using the Infinium MethylationEPIC BeadChip Kit (Illumina, San Diego, CA) at the University of Chicago Genomics Core Facility according to the manufacturer’s protocol. Quality control and data pre-processing were conducted using the R packages minfi [32]. First, for quality control, low-quality methylation measurements were identified by detection p-value <10−6 or number of beads <3. We excluded 10,861 CpGs with a detection rate <95% and 11 samples with a percentage of low-quality methylation measurements >5% or extremely low intensity of bisulphite conversion probes. No sample outliers were identified based on total intensity across CpGs. The remaining samples were pre-processed using preprocessNoob function in minfi package, as recommended in the online epigenetic clock calculator (http://dnamage.genetics.ucla.edu/) [32]. To account for cell composition variability we estimated the proportions of CD4 + T lymphocytes, CD8 + T lymphocytes, B lymphocytes, natural killer cells, monocytes, and granulocytes using the Houseman et al. method [33]. To account for experimental batch effects and other technical biases, we derived surrogate variables from intensity data for non-negative internal control probes using principal components (PCs) analysis. The top 16 PCs explain 95.03% of the variation across the non-negative internal control probes. The final dataset contained 856,146 CpG probes and 1145 samples (572 in the GDM group and 573 in the non-GDM group).
Statistical analysis
Means and standard deviations (SD) or median and interquartile ranges were calculated for all maternal child characteristics to describe the study population overall and stratified by maternal GDM status. We used t-tests and chi-square tests or Fisher’s exact tests to compare the general characteristics (continuous and categorical variables) of both mothers and children by maternal GDM status.
Using the online epigenetic clock calculator we obtained predicted DNAm age using both the Horvath and the Hannum methods [17,18]. The Horvath method is based on 353 CpG sites while the Hannum method is based on 71 CpG sites. The correlation between predicted DNAm age and chronological age was evaluated by Spearman correlation coefficients. The residual resulting from regressing DNAm age on chronological age was used as a metric for DNAm age acceleration and the primary outcome variable.
Linear regression models were used to investigate bivariate associations between GDM status, as well as covariates associated with GDM status, and DNAm age acceleration. Multivariable linear regression models were used to investigate the association between GDM status and DNAm age acceleration, controlling for covariates that were associated with GDM status and with age acceleration, as well as estimated cell-type proportions and the 16 PCs to control for technical bias. Finally, generalized linear regression models were used to evaluate the association between cardiometabolic risk factors in childhood and DNAm age acceleration in all children, as well as stratified by GDM status, adjusted for age, sex, and pre-pregnancy BMI. Data analyses were performed by SAS version 9.4 (SAS Institute Inc., Cary, NC, USA), and all statistical tests were two-sided with p-values of <0.05.
Results
Descriptive statistics of the study participants are shown overall and stratified by GDM status in Table 1. Overall, compared to non-GDM mothers, GDM mothers were older at delivery and had less advanced education and less income. More GDM mothers were overweight and obese before pregnancy, with less gestational weight gain during pregnancy compared to non-GDM mothers. Fewer GDM mothers were current drinkers. Children born to GDM mothers had higher birth weight and more were born by C-section.
Table 1.
Characteristics of participants according to maternal diabetes mellitus (GDM) status
| All (N = 1145) |
Non-GDM (N = 573) | GDM (N = 572) |
P | |
|---|---|---|---|---|
| Socio-demographic characteristics | ||||
| Maternal age at delivery (years), mean ± SD | 30.1 ± 3.29 | 29.7 ± 2.90 | 30.5 ± 3.59 | <0.001 |
| Maternal age at delivery (years), n (%) 20–30 30-40 ≥40 |
653 (57.0) 479 (41.8) 13 (1.1) |
357 (62.3) 214 (37.4) 2 (0.4) |
296 (51.8) 265 (46.3) 11 (1.9) |
<0.001 |
| Marital status, n (%) Married Divorced/widowed |
1133 (99.0) 12 (1.0) |
566 (98.8) 7 (1.2) |
567 (99.1) 5 (0.9) |
0.56 |
| Maternal education level, n (%) Up to secondary school Up to senior high school (<13 years) Up to bachelor (13–16 years) Up to master (≥16 years) |
16 (1.4) 161 (41.1) 849 (74.2) 119 (10.4) |
5 (0.9) 57 (10.0) 433 (75.6) 78 (13.6) |
11 (1.9) 104 (18.2) 416 (72.7) 41 (7.2) |
<0.001 |
| Family income class (yuan/RMB), n (%) <5000 5000–8000 ≥8000 |
173 (15.4) 283 (25.1) 671 (59.5) |
13 (2.3) 90 (16.2) 452 (81.4) |
160 (28.0) 193 (33.7) 219 (38.3) |
<0.001 |
| Pregnancy characteristics | ||||
| Weight gain during pregnancy (kg), mean ± SD | 17.4 ± 6.3 | 18.3 ± 6.7 | 16.6 ± 5.8 | <0.001 |
| Parity, n (%) 1 2 |
1129 (98.6) 16 (1.4) |
567 (99.0) 6 (1.0) |
562 (98.3) 10 (1.7) |
0.31 |
| Pre-pregnancy BMI (kg/m2), mean ± SD | 22.1 ± 3.1 | 22.4 ± 2.9 | 22.9 ± 3.1 | <0.001 |
| Pre-pregnancy BMI (kg/m2), n (%) <24 24–27 ≥28 |
881 (76.9) 207 (18.1) 57 (5.0) |
491 (85.7) 61 (10.7) 21 (3.7) |
390 (68.2) 146 (25.5) 36 (6.3) |
<0.001 |
| Smoking status during pregnancy (%), n (%) Never Past/Current |
1091 (95.3) 54 (4.7) |
543 (94.8) 30 (5.2) |
548 (95.8) 24 (4.2) |
0.41 |
| Passive smoking exposure in past year (%), n (%) No Yes |
527 (46.0) 618 (54.0) |
259 (45.2) 314 (54.8) |
268 (46.9) 304 (53.1) |
0.57 |
| Drinking status during pregnancy, n (%) No Yes |
837 (73.1) 308 (26.9) |
391 (68.2) 182 (31.8) |
446 (78.0) 126 (22.0) |
<0.001 |
| Child characteristics in early infancy | ||||
| Sex, n (%) Male Female |
598 (52.2) 547 (47.8) |
300 (52.4) 273 (47.6) |
298 (52.1) 274 (47.9) |
0.93 |
| Mode of delivery, n (%) Vaginal C-Section |
310 (27.1) 834 (72.9) |
183 (32.0) 389 (68.0) |
127 (22.2) 445 (77.8) |
<0.001 |
| Birth weight (g), mean ± SD | 3479 ± 488 | 3400 ± 451 | 3543 ± 507 | <0.001 |
| Birth length (cm), mean ± SD | 50.7 ± 2.0 | 50.6 ± 2.1 | 50.7 ± 1.9 | 0.62 |
| Gestational age (weeks), median (IQR) | 39.0 (38.0–40.0) |
39.0 (38.0–40.0) |
39.0 (38.0–40.0) |
0.91 |
| Preterm, n (%) Yes No |
41 (3.6) 1104 (96.4) |
22 (3.8) 551 (962) |
19 (3.3) 553 (96.7) |
0.64 |
| Feeding patterns in first 6 months, n (%) Breast feeding Mixed feeding Formula feeding |
488 (42.7) 500 (43.7) 156 (13.6) |
233 (40.7) 255 (44.6) 84 (14.7) |
255 (44.6) 245 (42.8) 72 (12.6) |
0.35 |
| Child characteristics at childhood visit | ||||
| Age (years), mean ± SD | 5.88 ± 1.25 | 5.87 ± 1.24 | 5.87 ± 1.25 | 0.96 |
| Family history of diabetes, n (%) | 364 (31.8) | 160 (27.9) | 204 (35.7) | 0.005 |
| Total outdoor time (hours), mean ± SD | 3.19 ± 1.4 | 3.05 ± 1.4 | 3.33 ± 1.5 | <0.001 |
| Total indoor time (hours), mean ± SD | 7.45 ± 0.95 | 7.40 ± 0.93 | 7.50 ± 0.97 | 0.08 |
| Sleep duration (hours/day), n (%) ≤8 9–10 ≥11 |
150 (13.1) 792 (69.2) 202 (17.7) |
65 (11.4) 383 (67.0) 124 (21.7) |
85 (14.9) 409 (71.5) 78 (13.6) |
<0.001 |
| Weight-for-age Z-score, mean ± SD | 0.58 ± 1.24 | 0.45 ± 1.21 | 0.70 ± 1.26 | 0.008 |
| Height-for-age Z-score, mean ± SD | 0.72 ± 1.01 | 0.69 ± 1.01 | 0.76 ± 1.02 | 0.26 |
| Body mass index (kg/m2), mean ± SD | 15.9 ± 2.47 | 15.7 ± 2.31 | 16.2 ± 2.61 | <0.001 |
| BMI for age Z score, mean ± SD | 0.18 ± 1.33 | 0.02 ± 1.28 | 0.34 ± 1.36 | <0.001 |
| Overweight, n (%) | 262 (22.9) | 107 (18.7) | 155 (27.1) | <0.001 |
| Obese, n (%) | 158 (13.8) | 61 (10.7) | 97 (17.0) | 0.002 |
| Waist circumference (cm), mean ± SD | 55.5 ± 6.52 | 54.7 ± 6.11 | 56.3 ± 6.81 | <0.001 |
| Body fat (%), mean ± SD | 20.0 ± 7.81 | 19.1 ± 7.45 | 20.9 ± 8.05 | <0.001 |
| Triceps skinfold (mm), mean ± SD | 12.8 ± 5.62 | 12.8 ± 5.37 | 12.7 ± 5.86 | 0.73 |
| Subscapular skinfold (mm), mean ± SD | 7.68 ± 4.17 | 7.24 ± 3.83 | 8.12 ± 4.44 | <0.001 |
| Suprailiac skinfold (mm), mean ± SD | 10.7 ± 6.59 | 9.70 ± 5.98 | 11.6 ± 7.03 | <0.001 |
| Upper arm circumference (cm), mean ± SD | 18.3 ± 2.7 | 18.0 ± 2.5 | 18.7 ± 2.8 | <0.001 |
| Haemoglobin (g/L), mean ± SD | 131.7 ± 8.41 | 132.4 ± 7.95 | 131.1 ± 8.79 | 0.007 |
| Diastolic BP (mmHg), mean ± SD | 60.1 ± 7.44 | 59.9 ± 6.59 | 60.3 ± 8.20 | 0.37 |
| Systolic BP (mmHg), mean ± SD | 95.8 ± 8.74 | 94.4 ± 8.32 | 97.2 ± 8.92 | <0.001 |
| Total cholesterol (mmol/L), mean ± SD | 4.42 ± 0.74 | 4.43 ± 0.73 | 4.41 ± 0.74 | 0.76 |
| HDL cholesterol (mmol/L), mean ± SD | 1.45 ± 0.28 | 1.40 ± 0.26 | 1.50 ± 0.30 | <0.001 |
| LDL cholesterol (mmol/L), mean ± SD | 2.26 ± 0.60 | 2.27 ± 0.59 | 2.25 ± 0.61 | 0.57 |
| Triglycerides (mmol/L), mean ± SD | 0.78 ± 0.33 | 0.76 ± 0.31 | 0.81 ± 0.34 | 0.009 |
The childhood visit, including blood draw for DNAm analysis, took place at a mean age of 5.9 years (range 3.1–10.2 years). At this visit, children born to GDM mothers had higher weight-for-age Z-score (0.70 vs. 0.45, p = 0.008), BMI-for-age Z-score (0.34 vs. 0.02, p < 0.001), waist circumference (56.3 vs. 54.7 cm, p < 0.001), body fat (20.9 vs. 19.1%, p < 0.001), subscapular skinfold (8.12 vs. 7.24 mm, p < 0.001), suprailiac skinfold (11.6 vs. 9.70 mm, p < 0.001), and upper arm circumference (18.7 vs 18.0 cm, p <.001) compared to non-GDM-exposed children. Diastolic blood pressure was similar between groups but systolic blood pressure was higher in the GDM group than the non-GDM group (97.2 vs 94.4 mmHg, p < 0.01). In addition, triglycerides were higher in the GDM group than the non-GDM group (0.81 vs 0.76 mmol/L, p = 0.009).
Chronological age was positively correlated with Horvath DNAm age (r = 0.53, p < 0.0001) and Hannum DNAm age (r = 0.38, p < 0.0001). When stratified by GDM group, positive correlations were observed for non-GDM (Horvath: r = 0.62, p < 0.0001; Hannum: r = 0.39, p < 0.0001) and GDM groups (Horvath: r = 0.46, p < 0.0001; Hannum: r = 0.39, p < 0.0001) (Figure 1). There was a significant interaction between GDM status and chronological age on Horvath DNAm age (interaction term: p = 0.0002) but not on Hannum DNAm age (interaction term: p = 0.3425).
Figure 1.
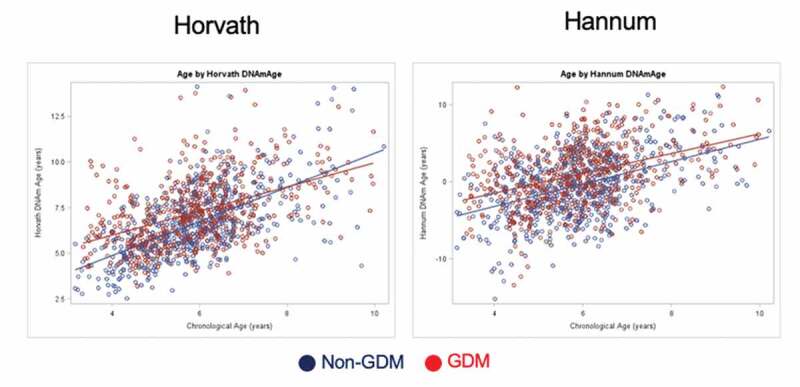
Scatterplots of predicted DNA methylation age and chronological age, by GDM group
Average DNAm age was higher in the GDM group compared to the non-GDM group by the Horvath method (7.23 ± 1.82 vs. 6.63 ± 1.87, p < 0.0001) as well as by the Hannum method (0.91 ± 4.04 vs. −0.49 ± 4.17, p < 0.0001). As shown in Figure 2, offspring DNAm age acceleration was greater in the GDM exposed group than the non-GDM-exposed group (Horvath: 7.2 months greater, p < 0.0001; Hannum: 16.8 months greater, p < 0.0001).
Figure 2.
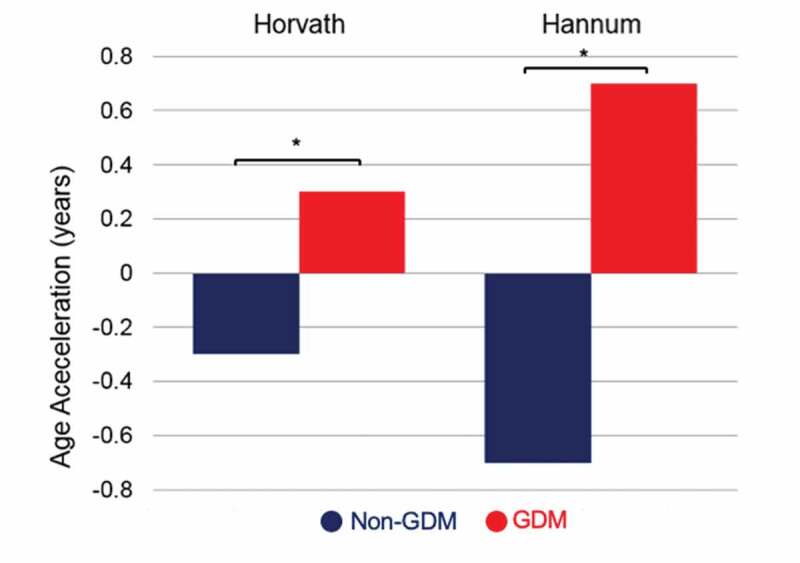
Average DNA methylation age acceleration, by GDM group
Of the covariates associated with GDM status, only pre-pregnancy BMI and sex were associated with DNAm age acceleration by the Horvath method in bivariate regression models (Table 2). No covariates were associated with DNAm age acceleration by the Hannum method in bivariate regression models. Offspring age acceleration remained higher in the GDM group than the non-GDM group after adjustment for potential confounders (Horvath: 4.96 months higher, adjusted for sex, pre-pregnancy BMI, estimated cell-type proportions, and technical bias PC, p = 0.0002; Hannum: 11.2 months higher, adjusted for estimated cell-type proportions and technical bias PC, p < 0.0001). Pre-pregnancy BMI and sex were no longer significantly associated with DNAm age acceleration by the Horvath method in the multivariable regression model.
Table 2.
Associations of GDM status, socio-demographic characteristics, pregnancy characteristics, and child characteristics during infancy with DNA methylation age acceleration
| DNA methylation age acceleration |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Horvath |
Hannum |
||||||||
| Unadjusted |
Adjusteda | Unadjusted |
Adjusteda |
||||||
| Characteristic | Categories | Coef (SE) | p-value | Coef (SE) | p-value | Coef (SE) | p-value | Coef (SE) | p-value |
| GDM status | GDM Non-GDM |
0.61 (0.09) Ref. |
<0.0001- | 0.41 (0.11) Ref. |
0.0002- | 1.40 (0.22) Ref. |
<0.0001- | 0.76 (0.20) Ref. |
0.0001- |
| Maternal age at delivery (years) | - | −0.02 (0.01) | 0.15 | −0.01 (0.03) | 0.80 | ||||
| Maternal education level | Secondary school Senior HS (<13) Bachelor (13–16) Master (≥16) |
−0.13 (0.40) 0.24 (0.14) 0.13 (0.15) Ref |
0.74 0.08 0.39- |
0.20 (0.96) 0.66 (0.33) -0.30 (0.37) Ref. |
0.84 0.04 0.42- |
||||
| Family income class (yuan/RMB) | <5000 5000–8000 ≥8000 |
0.15 (0.13) 0.14 (0.11) Ref. |
0.28 0.20- |
0.59 (0.32) 0.44 (0.29) Ref. |
0.07 0.10- |
||||
| Weight gain during pregnancy (kg) | - | −0.002 (0.007) | 0.84 | 0.001 (0.02) | 0.94 | ||||
| Pre-pregnancy BMI (kg/m2) | - | 0.05 (0.01) | 0.002 | 0.01 (0.01) | 0.50 | 0.04 (0.04) | 0.32 | ||
| Drinking status during pregnancy | Yes No |
0.03 (0.11) Ref. |
0.78- | −0.21 (0.25) Ref. |
0.41- | ||||
| Sex | Male Female |
0.41 (0.09) Ref. |
<0.001- | 0.20 (0.26) Ref. |
0.46- | 0.23 (0.22) Ref. |
0.30- | ||
| Mode of delivery | Vaginal C-Section |
|
0.90- | −0.34 (0.25) Ref. |
0.18- | ||||
| Birth weight (g) | - | 0.00 (0.00) | 0.76 | 0.00 (0.00) | 0.09 | ||||
aAdjusted for covariates with p < 0.05 in unadjusted analysis, estimated cell-type proportions, and 16 PCs to control for technical bias
The associations between cardiometabolic risk factors in childhood and DNAm age acceleration, adjusted for age, sex, and pre-pregnancy BMI, in all children and stratified by GDM group, are presented in Table 3. In all children, increased offspring DNAm age acceleration by both the Horvath and Hannum methods was associated with increased offspring weight-for-age Z-score, BMI-for-age Z-score, waist circumference, body fat percentage, subscapular skinfold, suprailiac skinfold, upper arm circumference, and systolic and diastolic blood pressure. When stratified by GDM group, findings were primarily seen in the GDM group but not the non-GDM group for weight-for-age Z-score, BMI-for-age Z-score, body fat percentage, suprailiac skinfold, upper arm circumference, and blood pressure.
Table 3.
Association between cardiometabolic risk factors in childhood with DNA methylation age acceleration in all children and by GDM group, adjusted for age, sex, and pre-pregnancy BMI
| DNA methylation age acceleration | |||||||
|---|---|---|---|---|---|---|---|
| All |
Non-GDM |
GDM |
|||||
| Cardiometabolic risk factor | Coef (SE) | p-value | Coef (SE) | p-value | Coef (SE) | p-value | |
| Weight-for-age Z-score | Horvath | 0.12 (0.04) | 0.002 | 0.09 (0.05) | 0.09 | 0.15 (0.06) | 0.006 |
| Hannum | 0.19 (0.09) | 0.0495 | 0.07 (0.14) | 0.63 | 0.25 (0.13) | 0.054 | |
| Height-for-age Z-score | Horvath | 0.13 (0.05) | 0.006 | 0.13 (0.06) | 0.03 | 0.13 (0.07) | 0.046 |
| Hannum | −0.00 (0.11) | 0.95 | −0.04 (0.16) | 0.81 | −0.01 (0.15) | 0.95 | |
| BMI-for-age Z-score | Horvath | 0.09 (0.04) | 0.01 | 0.04 (0.05) | 0.41 | 0.13 (0.05) | 0.01 |
| Hannum | 0.25 (0.09) | 0.005 | 0.11 (0.13) | 0.37 | 0.32 (0.12) | 0.008 | |
| Waist circumference (cm) | Horvath | 0.02 (0.01) | 0.04 | 0.02 (0.01) | 0.14 | 0.01 (0.01) | 0.29 |
| Hannum | 0.05 (0.02) | 0.01 | 0.03 (0.03) | 0.34 | 0.05 (0.03) | 0.07 | |
| Body fat (%) | Horvath | 0.02 (0.01) | 0.007 | 0.01 (0.01) | 0.13 | 0.02 (0.01) | 0.046 |
| Hannum | 0.05 (0.01) | 0.002 | 0.02 (0.02) | 0.30 | 0.06 (0.02) | 0.003 | |
| Triceps skinfold (mm) | Horvath | −0.00 (0.01) | 0.73 | 0.01 (0.01) | 0.78 | 0.01 (0.01) | 0.64 |
| Hannum | 0.23 (0.02) | 0.26 | 0.03 (0.03) | 0.35 | 0.04 (0.03) | 0.16 | |
| Subscapular skinfold (mm) | Horvath | 0.03 (0.01) | 0.01 | 0.03 (0.02) | 0.03 | 0.02 (0.02) | 0.15 |
| Hannum | 0.08 (0.03) | 0.008 | 0.04 (0.04) | 0.36 | 0.09 (0.04) | 0.02 | |
| Suprailiac skinfold (mm) | Horvath | 0.02 (0.01) | 0.006 | 0.01 (0.01) | 0.56 | 0.03 (0.01) | 0.006 |
| Hannum | 0.07 (0.02) | <0.001 | 0.03 (0.03) | 0.23 | 0.09 (0.02) | 0.001 | |
| Upper arm circumference (cm) | Horvath | 0.05 (0.02) | 0.01 | 0.04 (0.03) | 0.20 | 0.05 (0.03) | 0.07 |
| Hannum | 0.14 (0.05) | 0.006 | 0.06 (0.07) | 0.43 | 0.15 (0.06) | 0.017 | |
| Haemoglobin (g/L) | Horvath | −0.00 (0.01) | 0.68 | 0.00 (0.01) | 0.87 | 0.00 (0.01) | 0.92 |
| Hannum | −0.00 (0.01) | 0.90 | −0.00 (0.02) | 0.94 | 0.01 (0.02) | 0.56 | |
| Systolic BP (mmHg) | Horvath | 0.02 (0.01) | 0.004 | 0.01 (0.01) | 0.20 | 0.01 (0.01) | 0.15 |
| Hannum | 0.04 (0.01) | 0.009 | −0.00 (0.02) | 0.94 | 0.04 (0.02) | 0.02 | |
| Diastolic BP (mmHg) | Horvath | 0.01 (0.01) | 0.04 | 0.00 (0.01) | 0.68 | 0.02 (0.01) | 0.04 |
| Hannum | 0.03 (0.02) | 0.03 | 0.01 (0.02) | 0.71 | 0.04 (0.02) | 0.02 | |
| Total cholesterol (mmol/L) | Horvath | 0.05 (0.06) | 0.44 | −0.03 (0.08) | 0.74 | 0.13 (0.09) | 0.17 |
| Hannum | −0.27 (0.15) | 0.08 | −0.44 (0.22) | 0.046 | −0.08 (0.21) | 0.72 | |
| HDL cholesterol (mmol/L) | Horvath | −0.19 (0.17) | 0.26 | 0.04 (0.21) | 0.86 | −0.08 (0.27) | 0.75 |
| Hannum | −1.40 (0.40) | 0.001 | −1.45 (0.54) | 0.01 | −0.48 (0.61) | 0.44 | |
| LDL cholesterol (mmol/L) | Horvath | 0.03 (0.08) | 0.68 | −0.06 (0.11) | 0.60 | 0.19 (0.11) | 0.09 |
| Hannum | −0.11 (0.19) | 0.54 | −0.36 (0.28) | 0.20 | 0.22 (0.26) | 0.39 | |
| Triglycerides (mmol/L) | Horvath | 0.23 (0.14) | 0.11 | 0.14 (0.20) | 0.49 | 0.19 (0.20) | 0.34 |
| Hannum | 0.36 (0.35) | 0.31 | 0.56 (0.53) | 0.29 | 0.00 (0.46) | 0.99 | |
Discussion
To our knowledge, this is the first study to examine the effects of GDM exposure on blood measures of epigenetic age in children 3–10 years of age. Based on previous studies of GDM exposure and changes in offspring DNA methylation profiles as well as GDM exposure and shortened offspring telomere length, we hypothesized that offspring of mothers with GDM would have greater DNAm age acceleration compared to offspring of mothers without GDM. We found evidence to support our hypothesis by two markers of epigenetic age, after accounting for measured confounders. Our findings are consistent with other studies examining the association between GDM exposure and telomere length, another molecular biomarker linked to ageing, which found shorter telomeres in cord blood as well as peripheral blood cells at 9–16 years of age in offspring of mothers with GDM compared to offspring of mothers without GDM [34].
We also examined whether epigenetic age was associated with cardio-metabolic outcomes, finding associations between accelerated DNAm age and offspring anthropometrics in children (3–10 years of age) both with and without GDM exposure. Findings were stronger in the GDM group. Of note, a recent study found that epigenetic age acceleration in adolescence (17 years of age) was associated with inflammation, BMI measured 5 years later, as well as the probability of cardiovascular disease in middle age by Framingham algorithms [25,35]. Although it is unknown how these associations may shift as the children age into adolescence and young adulthood, there may be long-term consequences associated with accelerated ageing in early life, especially if the trajectory of DNAm ageing is set before adulthood [36]. Therefore, an early signal of accelerated epigenetic age using DNAm markers may allow for early identification of those at greater risk of future cardiovascular and/or metabolic disease. This information could contribute to tools for accurate risk stratification and potential early interventions [16].
Accumulating evidence from animal and human studies suggests intrauterine hyperglycaemia may lead to persistent epigenetic changes in developmentally important genes, including those that affect energy metabolism and metabolic signalling and regulation [37–40]. Our findings indicate GDM exposure may also influence ageing as predicted by the epigenetic clock. Prior work has suggested that the prenatal environment may affect ageing. A study by Simpkin and colleagues found that a number of infants and maternal characteristics, including sex, birth weight, caesarean section birth, maternal exposure to selenium, maternal smoking in pregnancy, maternal weight, maternal BMI, and maternal selenium and cholesterol levels, were associated with accelerated epigenetic age in offspring during childhood (age 7) and adolescence (ages 15–17) [41]. We adjusted for potential measured confounders of the association between GDM and offspring epigenetic age. However, we were unable to account for unmeasured variables in our study, such as environmental factors or genetic factors that could potentially be associated with GDM and accelerated epigenetic age [16].
Our study had a number of limitations. First, this study was observational in nature and DNAm age was only measured at one time point. Second, DNA was extracted from whole blood and future studies should consider other cell types or tissues, such as adipose tissue. Third, we adjusted for estimated cell-type proportions using estimates made from the DNAm data, rather than flow cytometry approaches. Therefore, it is unknown if we were adequately able to account for differences in cell-type proportions. We were able to consider results using both the Horvath and Hannum epigenetic age estimation methods. Others have reported that the Hannum method may be more influenced by differences in cell types than the Horvath method [42]. Our finding of an interaction between GDM status and chronological age on Horvath DNAm age but not on Hannum DNAm age is difficult to interpret given that we only have a single cross-sectional time point, rather than repeated measures to indicate actual change over time. Future longitudinal studies with repeated measures of predicted DNAm age from both methods are needed to investigate the rate of change of epigenetic age more closely. Finally, in our cross-sectional analysis of the relationship between DNAm age and offspring cardiometabolic health, there is a risk of false-positive findings given the large number of cardiometabolic measures, and additional studies in larger cohorts will be necessary to add validity to our findings.
Conclusion
In this unique cohort study of mother-child pairs in Tianjin, China, we found that offspring of women with GDM exhibit accelerated epigenetic ageing compared to control participants, independent of other maternal factors. Epigenetic ageing in offspring was also associated with cardiometabolic risk factors, suggesting that GDM and GDM-associated factors may have long-term effects on offspring epigenetic ageing and contribute to health outcomes. These findings warrant further investigation across the life course using longitudinal samples to study the association with later onset of adult metabolic diseases, including type 2 diabetes and cardiovascular disease.
Funding Statement
This study was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (R01DK100790).
Availability of data and materials
The datasets generated and/or analysed during the current study are not publicly available due to protection of individual privacy but are available from the corresponding author on reasonable request.
Authors’ contributions
GH designed the study. LW, HL, WL, JL, RG conducted the field research. GH, LH, and AAB designed and obtained funding for the DNA methylation study. SS, BJ, AD, YZ, YS conducted data organization and analysis. SS drafted the manuscript. All authors provided critical revisions to the manuscript and approved the final manuscript.
Consent for publication
Not applicable for this study.
Disclosure statement
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Written informed consent was collected from all participants and this study was approved by the Human Subjects Committee of Tianjin Women’s and Children’s Health Center and the Institutional Review Boards of Columbia University Medical Center and Pennington Biomedical Research Center.
References
- [1].Zhu Y, Zhang C.. Prevalence of gestational diabetes and risk of progression to type 2 diabetes: a global perspective. Curr Diab Rep. 2016;16:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nguyen CL, Pham NM, Binns CW, et al. Prevalence of gestational diabetes mellitus in Eastern and Southeastern Asia: a systematic review and meta-analysis. J Diabetes Res. 2018;2018:6536974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Crowther CA, Hiller JE, Moss JR, et al. Effect of treatment of gestational diabetes mellitus on pregnancy outcomes. N Engl J Med. 2005;352:2477–2486. [DOI] [PubMed] [Google Scholar]
- [4].Farrar D, Simmonds M, Bryant M, et al. Hyperglycaemia and risk of adverse perinatal outcomes: systematic review and meta-analysis. BMJ. 2016;354:i4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dabelea D. The predisposition to obesity and diabetes in offspring of diabetic mothers. Diabetes Care. 2007;30(Suppl 2):S169–174. [DOI] [PubMed] [Google Scholar]
- [6].Reece EA, Leguizamón G, Wiznitzer A. Gestational diabetes: the need for a common ground. Lancet. 2009;373:1789–1797. [DOI] [PubMed] [Google Scholar]
- [7].Kaseva N, Vääräsmäki M, Sundvall J, et al. Gestational diabetes but not prepregnancy overweight predicts for cardiometabolic markers in offspring twenty years later. J Clin Endocrinol Metab. 2019;104:2785–2795. [DOI] [PubMed] [Google Scholar]
- [8].Barker DJP. The origins of the developmental origins theory. J Intern Med. 2007;261:412–417. [DOI] [PubMed] [Google Scholar]
- [9].Hales CN, Barker DJ, Clark PM, et al. Fetal and infant growth and impaired glucose tolerance at age 64. BMJ. 1991;303:1019–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yan J, Yang H. Gestational diabetes mellitus, programing and epigenetics. J Matern Fetal Neonatal Med. 2014;27:1266–1269. [DOI] [PubMed] [Google Scholar]
- [11].Xu J, Ye J, Wu Y, et al. Reduced fetal telomere length in gestational diabetes. PLoS ONE. 2014;9:e86161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hjort L, Martino D, Grunnet LG, et al. Gestational diabetes and maternal obesity are associated with epigenome-wide methylation changes in children. JCI Insight. 2018. 3 (17): e122572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kim E, Kwak SH, Chung HR, et al. DNA methylation profiles in sibling pairs discordant for intrauterine exposure to maternal gestational diabetes. Epigenetics. 2017;12:825–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Weng X, Liu F, Zhang H, et al. Genome-wide DNA methylation profiling in infants born to gestational diabetes mellitus. Diabetes Res Clin Pract. 2018;142:10–18. [DOI] [PubMed] [Google Scholar]
- [15].Moen G-H, Sommer C, Prasad RB, et al. MECHANISMS IN ENDOCRINOLOGY: epigenetic modifications and gestational diabetes: a systematic review of published literature. Eur J Endocrinol. 2017;176:R247–67. [DOI] [PubMed] [Google Scholar]
- [16].Elliott HR, Sharp GC, Relton CL, et al. Epigenetics and gestational diabetes: a review of epigenetic epidemiology studies and their use to explore epigenetic mediation and improve prediction. Diabetologia. 2019;62:2171–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Levine ME, Lu AT, Quach A, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY). 2018;10:573–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Horvath S, Gurven M, Levine ME, et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 2016;17:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Perna L, Zhang Y, Mons U, et al. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin Epigenetics. 2016;8:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen BH, Marioni RE, Colicino E, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY). 2016;8:1844–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Roetker NS, Pankow JS, Bressler J, et al. Prospective study of epigenetic age acceleration and incidence of cardiovascular disease outcomes in the ARIC study (atherosclerosis risk in communities). Circ Genom Precis Med. 2018;11:e001937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Irvin MR, Aslibekyan S, Do A, et al. Metabolic and inflammatory biomarkers are associated with epigenetic aging acceleration estimates in the GOLDN study. Clin Epigenetics. 2018;10:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Huang R-C, Lillycrop KA, Beilin LJ, et al. Epigenetic age acceleration in adolescence associates with BMI, inflammation, and risk score for middle age cardiovascular disease. J Clin Endocrinol Metab. 2019;104:3012–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang J, Wang L, Liu H, et al. Maternal gestational diabetes and different indicators of childhood obesity - a large study. Endocr Connect. 2018. DOI: 10.1530/EC-18-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Liang Z, Liu H, Wang L, et al. Maternal MTNR1B genotype, maternal gestational weight gain, and childhood obesity. Am J Clin Nutr. 2020;111:360–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].World Health Organization (WHO) . Part 1: diagnosis and classification of diabetes mellitus. WHO/NCD/NCS/99.2 ed. Definition, Diagnosis and Classification of Diabetes Mellitus and its Complications. Geneva: World Health Organization; 1999. [Google Scholar]
- [29].Wang J, Pan L, Liu E, et al. Gestational diabetes and offspring’s growth from birth to 6 years old. Int J Obes (Lond). 2019;43:663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Li N, Liu E, Guo J, et al. Maternal prepregnancy body mass index and gestational weight gain on offspring overweight in early infancy. PLoS ONE. 2013;8:e77809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ogden CL, Carroll MD, Kit BK, et al. Prevalence of obesity and trends in body mass index among US children and adolescents, 1999-2010. JAMA. 2012;307:483–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive bioconductor package for the analysis of infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hjort L, Vryer R, Grunnet LG, et al. Telomere length is reduced in 9- to 16-year-old girls exposed to gestational diabetes in utero. Diabetologia. 2018;61:870–880. [DOI] [PubMed] [Google Scholar]
- [35].Pencina MJ, D’Agostino RB, Larson MG, et al. Predicting the 30-year risk of cardiovascular disease: the framingham heart study. Circulation. 2009;119:3078–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kananen L, Marttila S, Nevalainen T, et al. The trajectory of the blood DNA methylome ageing rate is largely set before adulthood: evidence from two longitudinal studies. Age (Omaha). 2016;38:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jiang Y, Yu Y-C, Ding G-L, et al. Intrauterine hyperglycemia induces intergenerational Dlk1-Gtl2 methylation changes in mouse placenta. Oncotarget. 2018;9:22398–22405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Reichetzeder C, Dwi Putra SE, Pfab T, et al. Increased global placental DNA methylation levels are associated with gestational diabetes. Clin Epigenetics. [Internet]. 2016. [cited 2019 July16];8. Available from. [];. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4960714/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bouchard L, Thibault S, Guay S-P, et al. Leptin gene epigenetic adaptation to impaired glucose metabolism during pregnancy. Diabetes Care. 2010;33:2436–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bouchard L, Hivert M-F, Guay S-P, et al. Placental adiponectin gene DNA methylation levels are associated with mothers’ blood glucose concentration. Diabetes. 2012;61:1272–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Simpkin AJ, Hemani G, Suderman M, et al. Prenatal and early life influences on epigenetic age in children: a study of mother-offspring pairs from two cohort studies. Hum Mol Genet. 2016;25:191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Marioni RE, Shah S, McRae AF, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analysed during the current study are not publicly available due to protection of individual privacy but are available from the corresponding author on reasonable request.


