Abstract
Techniques used to prepare clinical samples have been perfected for use in diagnostic testing in a variety of clinical situations, e.g., to extract, concentrate, and purify respiratory virus particles. These techniques offer a high level of purity and concentration of target samples but require significant equipment and highly trained personnel to conduct, which is difficult to achieve in resource-limited environments where rapid testing and diagnostics are crucial for proper handling of respiratory viruses. Microfluidics has popularly been utilized toward rapid virus detection in resource-limited environments, where most devices focused on detection rather than sample preparation. Initial microfluidic prototypes have been hindered by their reliance on several off-chip preprocessing steps and external laboratory equipment. Recently, sample preparation methods have also been incorporated into microfluidics to conduct the virus detection in an all-in-one, automated manner. Extraction, concentration, and purification of viruses have been demonstrated in smaller volumes of samples and reagents, with no need for specialized training or complex machinery. Recent devices show the ability to function independently and efficiently to provide rapid, automated sample preparation as well as the detection of viral samples with high efficiency. In this review, methods of microfluidic sample preparation for the isolation and purification of viral samples are discussed, limitations of current systems are summarized, and potential advances are identified.
INTRODUCTION
Respiratory viruses are found globally and are capable of infecting both the upper and lower respiratory tracts of humans, leading to respiratory distress, which holds significant potential for morbidity, particularly in children and elderly individuals.1,2 The prevalence of these viruses can remain persistent or vary seasonally. In general, these respiratory viruses continually hold potential for larger dissemination globally, as seen with previous outbreaks [2009 H1N1, severe acute respiratory syndrome (SARS), and Middle East respiratory syndrome (MERS)] and currently with the coronavirus disease 2019 (COVID-19) pandemic.2,3–5 Significant mitigation, management, and treatment of these viruses relies heavily on the ability to quickly identify those at risk and test biological samples of interest for virus presence. Often, the rapid and sensitive sensing of these samples occurs in resource-limited environments without laboratory machinery or highly trained personnel. These biological and clinical samples must be processed to result in highly pure, concentrated virus outputs. The collection methods of biological samples as well as the sample storage and processing steps required vary based on the virus to be detected and on the sample matrix that is most appropriate for detection of the virus.2,6 Discussion of these matrices and locations is crucially important in determining what samples must be collected for optimal early detection of respiratory viruses. Furthermore, the sample types inform the sample preparation microfluidic device requirements.
Respiratory viruses
There are a significant number of respiratory viruses. Given their mutagenicity and prevalence globally, hundreds of serotypes must be tested for.1 These viruses generally create symptoms of respiratory distress and fever and result in morbidities, particularly in children and elderly individuals.2,7 As a result of their pathology and burden, testing has been developed for many of these viruses that are commonly encountered.1 The viruses that are the most common in humans include respiratory syncytial virus (RSV), human parainfluenza virus (HPIV), human metapneumovirus (HMPV), human rhinovirus (HRV), coronaviruses, adenoviruses, bocavirus, enterovirus, and influenza A and B.1,3,8 These viruses are not the only respiratory viruses that exist, but they cover the largest range of clinically relevant respiratory viruses.1,3,8 Particularly, 2009 H1N1 (novel influenza A/H1N1; also known as swine flu), SARS (severe acute respiratory syndrome; caused by SARS-coronavirus or SARS-CoV), MERS (Middle East respiratory syndrome; caused by MERS-coronavirus or MERS-CoV), and most recently COVID-19 (coronavirus disease 2019; caused by SARS-CoV-2) have resulted in a worldwide pandemic initially with neither adequate treatment options nor vaccines, where the rapid and low-cost detection are the keys to successful containment.
Clinical samples
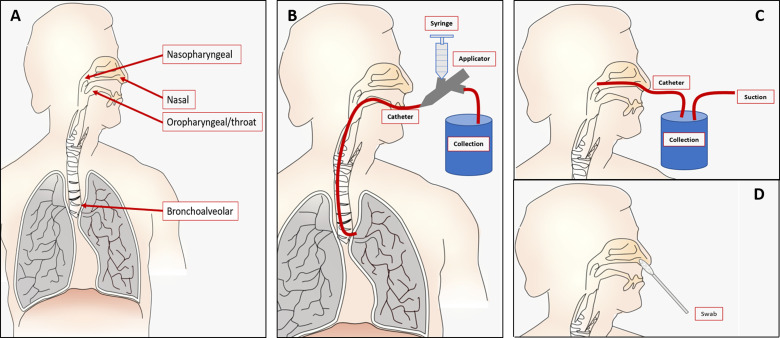
The biological origin of clinical samples and time since recovery from patients greatly impact the accurate and early detection of viruses.3 For example, not all viruses initially result in the production of sputum (mucus from coughs), so swabs may be necessary, although they are not able to reach as deep as other sampling methods, such as aspirate collection (in which the sample is collected via a vacuum pump catheter system).9 Additionally, viruses accumulate in different regions at different times throughout incubation in the body. For these reasons, appropriate testing requires various samples that are recovered through a variety of means, either aided by suction, swab, or natural means of recovery. The clinical samples recovered for these viruses are from (1) nasal, (2) nasopharyngeal, (3) throat (oropharyngeal), (4) bronchoalveolar, (5) sputum, (6) stool, (7) cerebral spinal fluid, and (8) blood components such as whole blood and serum. Samples can be collected via (1) lavages (washes), (2) swabs, (3) aspirates, and (4) phlebotomy (needle withdrawal, especially for blood). These can be seen graphically in Fig. 1.
FIG. 1.
(a) Various sample locations displayed in the body, including nasal, nasopharyngeal, oropharyngeal/throat, and bronchoalveolar. (b) Lavage: example of bronchoalveolar lavage (BAL) in which a catheter is placed in the desired location, saline is rinsed via an applicator, and an effluent is collected. (c) Aspiration is collection of samples via a suction powered catheter. An example is shown in nasopharyngeal aspiration. (d) Swabs are inserted into the location to collect the sample, as shown in the nasal swab example.
The sample matrices and tests must be carefully selected for the virus that is being targeted. Previous studies have shown for RSV that nasopharyngeal aspirates provide laboratory samples that allow for higher specificity for certain diagnostic procedures (i.e., indirect immunofluorescent antibody techniques) than nasopharyngeal swabs.5 In early detection, the sample collection method also determines the number of viruses in the given sample matrix, which is dictated in part by where the virus accumulates in the body.10 Therefore, the sample collection method should be determined by the sample matrix optimal for the virus to be detected and the number of viruses in the sample. We should note that detection methods also vary based on the sample available for early detection. As a result, there may be more than one collection method used for each virus. A review of the literature shows that similar methods are used for the same virus (i.e., throat lavages and throat swabs are used for adenovirus detection).5,10–12 Additionally, clinical samples that require processing at locations other than where they are collected must also be placed in either viral or universal transport medium or stored cold (2/8 °C for short-term or −70/−80 °C for long-term storage).11,13–18 This handling step is important in that it preserves the viral samples for later processing and applies to all sample types facing long lead times from sample collection to use. Table I summarizes the sample collection methods for each respiratory virus as well as common detection methods used.
TABLE I.
Sample collection and detection methods for respiratory viruses. RT-PCR, reverse transcription polymerase chain reaction; LFIA, lateral flow immunochromatographic assay; ELISA, enzyme-linked immunosorbent assay.
| Respiratory virus | Reference | Clinical sample | Detection methods |
|---|---|---|---|
| Respiratory syncytial virus (RSV) | 5, 6, and 19 | Nasopharyngeal aspirate | Viral culture |
| Nasopharyngeal swab | Fluorescence immunoassay | ||
| Rapid antigen test (LFIA) | |||
| RT-PCR | |||
| Parainfluenza virus (HPIV) | 5, 13, and 20 | Throat swab | RT-PCR |
| Bronchoalveolar lavage | |||
| Nasopharyngeal aspirate | |||
| Human metapneumovirus (HMPV) | 15–17 | Nasopharyngeal aspirate | RT-PCR |
| Throat swab | Virus culture | ||
| Sputum | |||
| Bronchoalveolar lavage | |||
| Rhinovirus (HRV) | 18, 21, and 22 | Nasopharyngeal swab | RT-PCR |
| Virus culture | |||
| DNA sequencing | |||
| Coronaviruses | 9, 11, 23, and 24 | Nasal aspirate | Virus culture |
| Nasal wash | RT-PCR | ||
| Nasopharyngeal swab | Fluorescence immunoassay | ||
| Bronchoalveolar lavage | |||
| Sputum | |||
| Adenovirus | 25 and 26 | Throat swab | Virus culture |
| Throat wash | ELISA | ||
| Blood | RT-PCR | ||
| Blood serum | |||
| Stool | |||
| Bocavirus | 27–29 | Nasopharyngeal swab | RT-PCR |
| Nasopharyngeal aspirates | Virus culture | ||
| Nasopharyngeal wash | |||
| Stool | |||
| Sputum | |||
| Enterovirus | 12, 30, and 31 | Stool | RT-PCR |
| Nasopharyngeal swab | |||
| Mouth/nose vesicles | |||
| Cerebral spinal fluid | |||
| Influenza A and B | 32–36 | Throat lavage | RT-PCR |
| Nasal swab | DNA microarray | ||
| Throat swab | |||
| Nasopharyngeal swab and suction |
Standard purification and extraction methods
Once the biological sample is acquired using the appropriate method, samples are preprocessed to perform various concentration and purification procedures. Samples such as blood, stool, and other larger or more viscous samples are pretreated in ways such that they can be processed. These include the use of standard anticoagulants in the case of blood draws (often found in the collection tubes), or dilution in buffers as is the case with stool, so that they can freely be pipetted. (As mentioned earlier, in the case of detection methods using genetic materials, samples collected away from processing areas must be added to viral transfer medium to maintain viral RNA stability.) These include, but not limited to, a lysing step either through chemical or physical lysing involving the use of various buffers,37,38 concentration and purification steps utilizing filter membranes or centrifugation,39 and lysing/isolation through temperature changes.40 These are vital steps in the process in that it releases and isolates viral samples found in cells and works to inactivate nucleases that would otherwise degrade the sample. Beyond detection methods utilizing genetic materials, there may also be preprocessing for protein-based detection methods (such as rapid antigen tests and lateral flow immunochromatographic assays) involving the addition of various buffers (e.g., virus transport media or phosphate buffered saline) and enzymes (such as proteinase K40), and/or the addition of a heating step. In immunoassays such as ELISA, pretreatment steps are contained mostly to the antibodies used on microwell plates, where the sample is simply diluted to useable range.
Once appropriate lysis steps are completed, viral RNA can be extracted particularly for PCR-based detection methods using one of three general methods: aqueous phase-based, silica-based, or magnetic bead-based separation. These methods are all heavily used in the field today, often through various commercial kits, and have a long track record of use. For example, the single step method of RNA isolation by guanidinium thiocyanate-phenol-chloroform described by Chomczynski and Sacchi has amassed over 66 000 citations over its 30-year history.41
Aqueous phase-based extraction relates to the use of various chemicals that create a phase separation between RNA and proteins, such as guanidinium thiocyanate-phenol-chloroform extraction. This method, for example, has been used for over 30 years to extract RNA and requires the addition of relevant chemicals and a centrifugation step, whereby total RNA remains soluble in the upper acidic phase.41 While the protocol is simple, there is still the need for a fume hood, an autoclave, a pH meter, a vortexer, cold centrifuge, and an automatic pipet41 as well other laboratory basics. Commercial kits have emerged using similar principles, such as those by Peqlab38 and others who have simplified the handling of buffers and other toxic chemicals. Despite this advance, there is still a certain level of complexity to extract RNA that is of high quality and free of any leftover elution buffers, such as isopropanol alcohol, which may act as a PCR inhibitor and interfere with downstream amplifications and analysis of samples.
Silica-based methods rely on the selective binding of nucleic acids (RNA) to a silica membrane, as other cell debris and proteins are discarded via centrifugation.39 These kits involve the addition of sample and various buffers to the upper compartment of a spin tube, a centrifugation step to flow through, and discarding the waste.37,42 The nucleic acids are then eluted from the membrane and stored for later analysis. This process is efficient and limited mainly by the careful treatment of the silica membrane and the assay time.
Magnetic bead-based assays, such as the popularly used commercial kits developed by Roche,40 use selective binding of RNA to surface activated magnetic beads to separate nucleic acids. Magnetic beads can be functionalized in a variety of ways, for example, surface coated with oligos or coated with silica glass.43 RNA is attached to the beads and then separated by the addition of an external magnet. Limitations with this procedure include bead binding efficiency and bead wash-out.
MICROFLUIDIC APPROACHES
Advances in microfluidics and lab-on-a-chip (LOC) devices have allowed for higher throughput virus sample purification that is often automated and requires smaller volumes of starting clinical samples.44,45 These portable devices are made at low unit cost and have final purification efficiencies as good or better than those of standard laboratory protocols. Importantly, these devices must extract, purify, and concentrate viral samples from the clinical sample in such a way to avoid damage to the viral nucleic acids and avoid potentially inhibitory effects in downstream detection and quantification. These microfluidic devices use various principles to extract, purify, and concentrate viral samples from biological matrices. These techniques can be broadly classified into (1) bead-based, (2) droplet-based, (3) structure-based, and (4) fluid property-based methods. The classification of these methods is based on the major component resulting in the capture of the virus sample.
Bead-based methods
Bead-based methods involve the addition of antibody-coated, biotinylated, or otherwise functionalized micrometer or submicrometer particles to clinical samples to capture target viruses from various clinical samples. These beads can be centrifuged to remove them from the solution because they have a larger size and density than the unbound viruses or separated by filtering using a membrane, leaving the rest of the waste to flow out. Centrifugation can be difficult to achieve on microfluidic platforms, as this requires a more complex microfluidic design, in several cases performed by external devices (i.e., placement into a computer's CD drive). Although this is the case with several devices,46–48 and the devices described have high yields of viral RNA, the supplementary devices necessary render them less practical than other approaches.
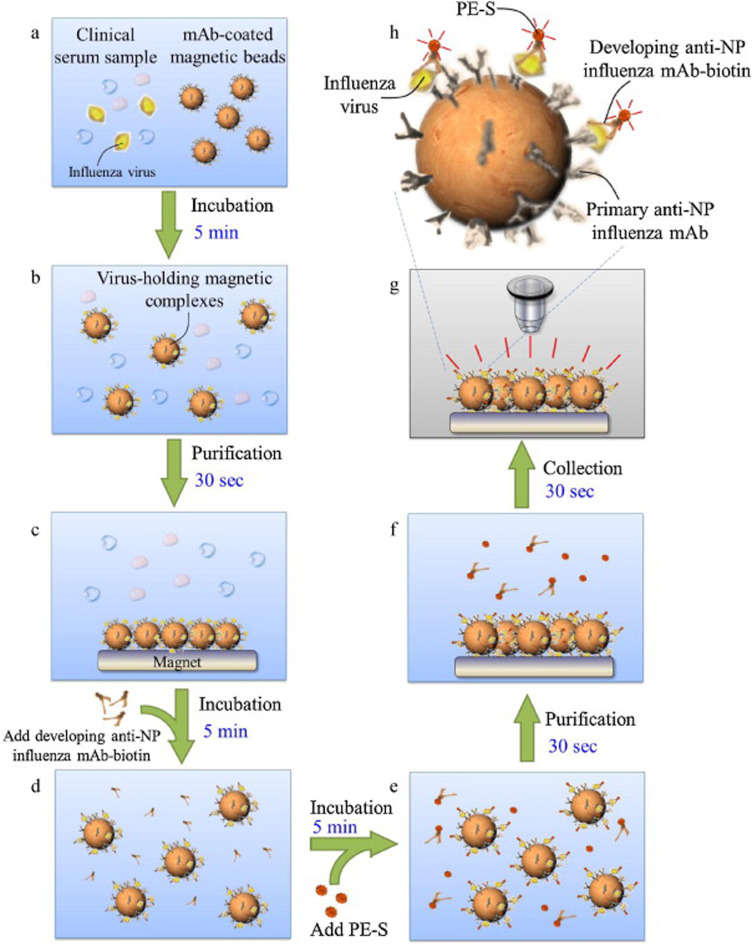
Other bead-based methods depend on magnetic functionalized beads to filter viral samples from clinical samples. These magnetic beads do not require centrifugation as an external magnetic field in the system acts on them to remove them from solution, as seen in Fig. 2. The external fields are induced by magnets placed over the reaction chamber containing the beads. This method can easily be implemented on microfluidic devices. The microfluidic chamber size, the solution flow rate, and the reaction volume are determined by the strength of the applied magnetic field. For ease of use, these magnets generating fields are often permanent, meaning fixed in location and intensity. There are many examples of these, such as those described in Refs. 44 and 49–52, yet permanently fixed magnets can result in lower sample flow rates. When a stronger field must be used to collect magnetic beads, either an inducible magnet driven by an electrical current or other field concentrating components can be introduced. These have been incorporated into devices such as those described in Refs. 53 and 54 yet are less attractive than permanent magnets due to their higher complexity.
FIG. 2.
Example of bead-based purification using a magnetic concentrating field. Reproduced with permission from Lien et al., Biosens. Bioelectron. 26, 3900 (2011). Copyright 2011 Elsevier B.V.50
Droplet-based methods
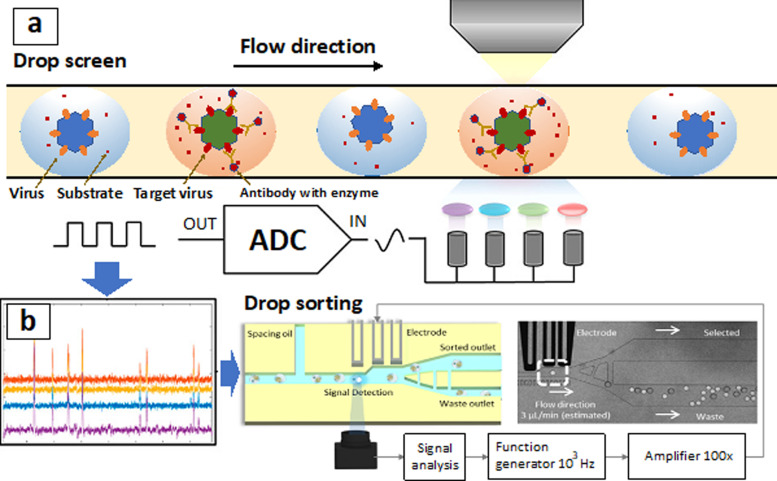
To sort the target pathogens in a sample for assaying, the droplet microfluidic technology was developed.55 By emulsifying the aqueous phase containing the pathogens in an oil phase with surfactants, single cell/bacteria/virus compartmentalization could be achieved. A large number of monodispersed water-in-oil droplets were produced with a frequency ∼1000 droplets per second. As the pathogens and reagents were encapsulated in the micro-droplets (the diameter ∼30 μm and the volume ∼5 pl), the concentration was increased to enhance the detection sensitivity. Additionally, by using droplets to isolate the pathogens, most random molecules and contaminations could be encapsulated in separate droplets to minimize background noises. For example, a single-virus droplet-based microfluidics device was fabricated before to sort millions of HIV-1 particles with high efficiency.55 Based on the expression of epitopes recognized by broadly neutralizing antibodies, enrichment of functional HIV-1 virus was approached. Notably, unlike the conventional flow cytometry, the enzyme reactions triggered by surface bindings could be processed within the droplets to amplify the fluorescence signals for sorting. By labeling rare surface epitopes via antibodies with an enzyme alkaline phosphatase (AP), the target virus could be sorted to distinguish the wild type HIV-1 Env protein and HIV-1 mutant Env protein (T303A). Approximately 1900-fold enrichment of viral particles displaying neutralizing epitopes was approached via droplet-based microfluidics.55 Similar droplet-based methods were conducted to sort the cancer cells (Fig. 3),56–58 immune cells,59 and functional mutants60 by the enzymatic reactions within the droplets.
FIG. 3.
(a) The pathogen extraction can be approached by using a drop screen technology.55,56 By encapsulating the single pathogens within the droplets, enzymatic reactions were processed. The fluorescence intensities were automatically evaluated for sorting. (b) The fluorescence signals could be converted to be electrical signals.56 The threshold signals could be automatically counted for droplet sorting. Reproduced with permission from Ng et al., Anal. Chem. 91, 1277 (2019). Copyright 2019 American Chemical Society.56
The fluorescence signals of pathogens in the droplets were characterized for end timepoint measurements.61 An optical system capable of high-speed fluorescence analysis of the single pathogen in the droplets and a high rate of data transmission between modules and software were developed for continuous flow-cell analysis. For example, the analog voltage signals from the photomultiplier tubes (PMTs) could be converted into digital form by a data acquisition (DAQ) system at a sampling rate of 12 500 samples/s.56 The droplets with desirable pathogens such as viral particles could be incubated for a certain period so that the fluorescence intensity reached an optimized regime for the identification (Fig. 3).56–58 These droplets could then be uploaded into a screening microfluidic channel for continuous screening for another hour. Spacer oil could be introduced from the side of the straight channel to space the droplets and maintain the cell order during screening.60 The droplets would be kept in the order in which they were generated, and the last measurement timepoint could be fixed after encapsulation to enable calibration of the increasing fluorescence slopes. Droplets with fluorescence signals reflecting to pathogen's activities would flow past a charge-coupled device (CCD) camera connected to a computer for end point fluorescence measurements (Fig. 3).56–58 A throughput of screening ∼100–1000 droplets per second was achieved before, allowing for the collection of ∼18 × 105 data points within 30 min (Fig. 3).56 To determine the fluorescence levels of individual pathogens, real-time analysis would be processed through image processing. Above the advantages, after droplet-based sorting, it is necessary to remove the oils by intensive washing, which might affect the pathogen's survivability and activity.61,62
Structure-based methods
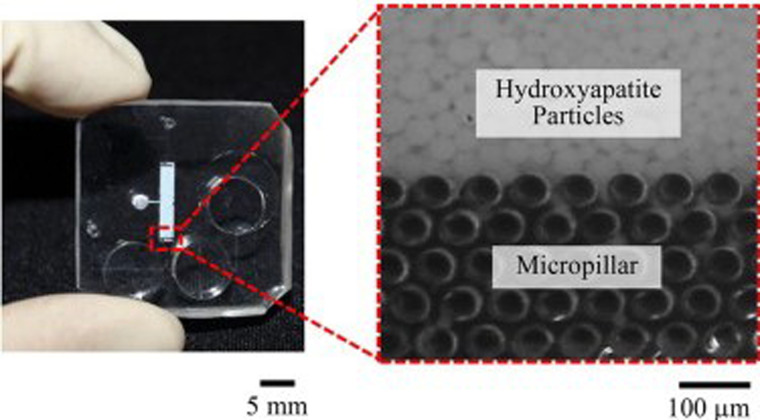
These techniques broadly include solid phase extraction-based and size-based filtering methods. Generally, the physical structures used in these microfluidic devices allow for size or affinity separation of viruses from bulk samples. The most common form of viral extraction in this category uses silicon microbeads for solid phase extraction of viruses, such as those found in Refs. 63 and 64 that replicate in large part the gold standard solid phase extraction method but use smaller volumes and little-to-no external processing, as demonstrated in microfluidic devices as a whole. These solid phase extraction methods, as discussed previously, work by the reversible binding of viruses to silica particles. In these cases, the silica particles are trapped in the microfluidic reaction chamber by the chip dimension63 or by secondary structures, such as microcolumns (Fig. 4),65 to simplify washing/elution steps and remove the need for centrifugation. The reversible binding of viruses to substrates can be incorporated in other ways as well, most notably using membranes.66,67 While DNA is present in higher concentrations in blood and can be captured directly using binders, viral RNA is less present and requires the use of filters, as demonstrated in Ref. 66. Ultimately, these methods differ only by the addition of extra filters to sort whole blood components from the desired viral material but maintain the use of reversible binding regions of the microfluidic chip to do so.
FIG. 4.
Visualization of micrometer and submicrometer structures used in microfluidic sample purification. Reproduced with permission from Niimi et al., Sens. Actuators B Chem. 201, 185 (2014). Copyright 2014 Elsevier B.V.65
Other researchers have used direct size-based sorting techniques to separate viruses from other clinical sample components. These nanoscale structures can be manufactured in a variety of ways but generally serve to capture submicrometer structures in a label-free manner. Although this size-based separation is complicated given the additional sample matrix components, capture efficiencies ranging from 50%68 to 99%69 have been demonstrated. Additionally, certain nanostructures can be dissolved directly to release viral material for further downstream applications.
Fluid property-based methods
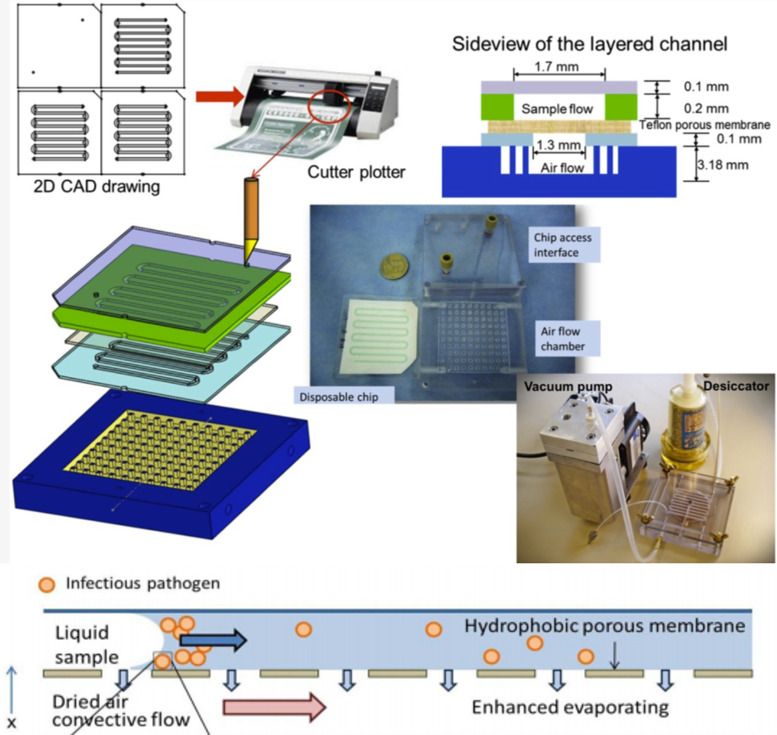
The methods described here rely on practices that change the solubility, viscosity, or other fluid properties of the sample compared to that of background components. Examples of methods that fall into this category include using aqueous two-phase extraction and electrophoresis. These methods are less common than the other two categories, as they rely on arguably less “standard” ways to extract virions, shown in the more limited number of publications available. These methods do not require the addition of other micrometer-sized particles on submicrometer-sized reaction chambers. Zhang et al.70 used a permeable membrane exposed to air with constant fluid flow on top to evaporate liquid and slowly concentrate the virus particles into one leading edge of the fluid with the highest interfacial tension (Fig. 5). Jacinto et al.71 utilized aqueous two-phase extraction, which is “formed when two immiscible compounds, such as two polymers or a polymer and a salt, are mixed in an aqueous solution above a certain critical concentration and spontaneously separate into two immiscible phases.” This method allows for biocompatible and selective separation of virus particles from samples, although it is not yet optimized for the small volumes found in microfluidic devices. Additionally, the mechanism of separation of biomolecules is poorly understood and although mathematical models have been created to quantify this separation, Negrete et al.72 acknowledge that “none of these models has been applicable yet due to the limited knowledge of the complex partition behavior of biomolecules.” Overall, these fluid property-based innovative approaches show good promise yet vary somewhat in the properties modulated for virus separation.
FIG. 5.
One example of fluid properties utilized to purify and extract virus from sample. Top portion of image shows the microfluidic chip design and fabrication process while bottom portion demonstrates working principle of evaporation and interfacial tension. Zhang et al., Diagnostics 3, 155 (2013).70 Copyright 2013 Author(s), licensed under a Creative Commons Attribution (CC BY license).
SAMPLE PURIFICATION METHODS DEMONSTRATED TOGETHER WITH DETECTION ON MICROFLUIDICS
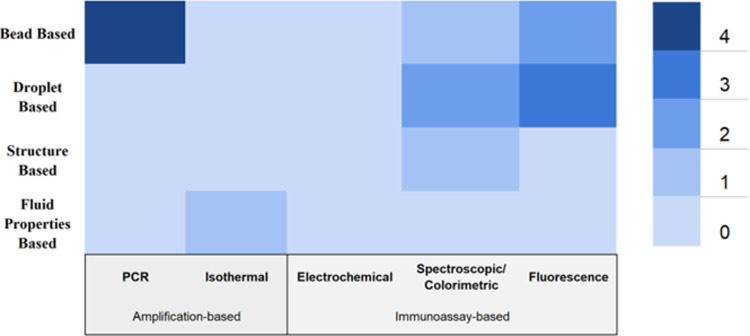
Although the focus of this review is to discuss the microfluidic approaches to the preparation of necessary biological samples, microfluidic sample preparation components are often demonstrated alongside detection methods on a single microfluidic platform toward highly efficient, fully automated, small-volume microfluidic diagnostic systems for resource-poor environments. The sample preparation method is highly dependent on the choice of detection method. Therefore, there must be some discussion regarding the microfluidic detection methods in the context of how these detection methods affect and are informed by the microfluidic sample preparation methods. There are many challenges with this approach as concurrent demonstration of both purification and detection in a single microfluidic chip will certainly increase the fabrication complexity and potentially compromise the assay performance when compared to the laboratory-based purification methods commonly used for microfluidic virus detection. Therefore, the number of publications demonstrating both in a single microfluidic chip is still low in number. However, there is a strong need to accomplish this toward true point-of-care diagnostic devices that can really be used in resource-poor environments. There are many microfluidic platforms that are applicable for the detection of respiratory viruses, but within the scope of this review, it is important to note that various detection methods are also incorporated into microfluidic chips that deal mainly with sample purification and extraction. These methods are primarily amplification based either through PCR44 or isothermal amplification,73 yet there are certain designs that generally utilize fluorescence signals from antibody-based immunoassays44,49,50,68 transduced in a variety of ways to aid in this detection. Table II summarizes the microfluidic methods demonstrating both sample preparation and detection together, sorted by the year of publication, including information on the assay time and minimum sample volume. It is important to note that not all publications list sample volumes or assay times for sample preparation modules and detection modules separately. Immunoassay-based methods include electrochemical, fluorescence, and spectroscopic/colorimetric detection. Amplification-based methods include PCR and isothermal detection. In addition to this table, Fig. 6 shows a matrix of the prevalence of certain sample preparation methods for a specific detection method, or vice versa.
TABLE II.
Sample purification and detection coupled on microfluidic chips.
| Virus isolation method | Detection method | Year of publication | Reference | Minimum sample volume | Assay time |
|---|---|---|---|---|---|
| Bead based | Immunoassay (fluorescence) | 2008 | 44 | 100 μl | 40 min |
| Bead based | Immunoassay (fluorescence) | 2011 | 50 | 25 μl | 15 min |
| Bead based | Amplification (PCR) | 2011 | 74 | 20 μl | 39 min (detection only) |
| Bead based | Amplification (PCR) | 2011 | 75 | 35 μl | ∼3.5 h |
| Bead based | Amplification (PCR) | 2014 | 53 | ∼0.1 μl | 1 min (extraction only) |
| Bead based | Amplification (PCR) | 2019 | 48 | 500 μl | 48 min |
| Bead based | Immunoassay (spectroscopic/colorimetric) | 2017 | 76 | 45 μl | ∼1 h |
| Droplet based | Immunoassay (spectroscopic/colorimetric) | 2018 | 58 | … | 1 h |
| Droplet based | Immunoassay (spectroscopic/colorimetric) | 2019 | 62 | … | … |
| Droplet based | Immunoassay (fluorescence) | 2010 | 60 | <150 μl | <10 h |
| Droplet based | Immunoassay (fluorescence) | 2019 | 56 | … | 2 h |
| Droplet based | Immunoassay (fluorescence) | 2020 | 61 | … | 1.5 h (detection only) |
| Structure based | Immunoassay (spectroscopic/colorimetric) | 2017 | 68 | 100 μl | 30 min |
| Fluid property based | Amplification (isothermal) | 2018 | 73 | 100 μl | <1 h |
FIG. 6.
A matrix showing the prevalence of different sample purification methods with detection methods coupled on chip through number of publications.
With regard to sample preparation microfluidic modules, there are several areas of emerging interest that can be discussed further, given the context of currently used detection methods. Although bead-based methods have been more popular in the past due to more standard/cross platform availability of reagents and protocols, droplet-based methods are gaining popularity due to their ability to perform multiplex experiments at very low volumes with increased sensitivity. Structure-based methods have seen limitations with regard to chip design and ease of use, although this method has shown great utility and must be explored further to overcome these limits. Amplification-based detection of samples is largely popular, as RT-PCR can be used for very early detection of virus particles. This requires methods that do not involve harmful or degrading chemicals of enzymes found in some bead-based extractions, for example, and as droplet microfluidic approaches evolve, there will be continued opportunity for amplification-based detection methods to be coupled on chip—including isothermal amplification methods. Application of novel isothermal amplification techniques to these sample preparation and detection devices is limited, and a worthy area of future research given its potential for lower limit of detection and higher sensitivity. Additionally, spectroscopic/colorimetric detection-based methods are deeply impacted by wastes left by imperfect sample preparation methods. In the future, these methods hold great potential for their ease of use as well as their sensitivity and specificity respectively, but more work needs to be done in perfecting sample preparation modules. Notably, there are extremely few examples of electrochemical detection coupled on-chip with microfluidic sample preparation modules. This may be, in part, due to accuracy issues attributed to sample wastes and other factors but should be investigated further. Overall, it can be seen that there are several significant areas of advancement possible in the coupling of on-chip virus detection methods with sample preparation modules.
CONCLUSION
Recent advances in microfluidic devices coupled with well-established gold standard purification and/or extraction protocols have opened the door for the development of highly efficient extraction systems that require smaller, clinically relevant sample volumes. Appropriate sample purification, concentration, and complete sample preparation provide applications to resource-poor environments to tackle respiratory virus emergence and management. Currently, challenges still exist with these devices in the form of variably efficient capture mechanisms and complex device fabrication, yet the discovery of novel methods and further advances coupling these sample preparation methods with rapid and novel detection methods will allow for a more complete sample to answer devices currently lacking with regard to point of care devices for respiratory viruses.
DATA AVAILABILITY
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Note: This paper is part of the special issue on Microfluidic Detection of Viruses for Human Health.
Contributor Information
Chia-Hung Chen, Email: .
Jeong-Yeol Yoon, Email: .
REFERENCES
- 1.Weston S. and Frieman M. B., Reference Module in Biomedical Sciences (Elsevier, 2018), pp. 85–101. [Google Scholar]
- 2.Pitkäranta A. and Hayden F. G., Ann. Med. 30, 390 (1998). 10.3109/07853899709002600 [DOI] [PubMed] [Google Scholar]
- 3.Lessler J., Reich N. G., Brookmeyer R., Perl T. M., Nelson K. E., and Cummings D. A., Lancet Infect. Dis. 9, 291 (2009). 10.1016/S1473-3099(09)70069-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van den Hoogen B. G., Osterhaus D. M. E., and Fouchier R. A. M., Pediatr. Infect. Dis. J. 23, 1893 (2004). 10.1097/01.inf.0000108190.09824.e8 [DOI] [PubMed] [Google Scholar]
- 5.SARS, MERS and Other Viral Lung Infections, edited by Hui D. S., Rossi G. A., and Johnston S. L. (European Respiratory Society, 2016), p. 84. [Google Scholar]
- 6.Allen K. E., Beekmann S. E., Polgreen P., Poser S., St. Pierre J., Santibañez S., Gerber S. I., and Kim L., Diagn. Microbiol. Infect. Dis. 92, 206 (2018). 10.1016/j.diagmicrobio.2017.12.011 [DOI] [PubMed] [Google Scholar]
- 7.Gill P. J., Richardson S. E., Ostrow O., and Friedman J. N., JAMA Pediatr. 171, 798 (2017). 10.1001/jamapediatrics.2017.0786 [DOI] [PubMed] [Google Scholar]
- 8.O’Callaghan K. and Jones K., Infect. Dis. Health 24, 147 (2019). 10.1016/j.idh.2019.05.001 [DOI] [PubMed] [Google Scholar]
- 9.Loeffelholz M. J. and Tang Y.-W., Emerging Microbes Infect. 9, 747 (2020). 10.1080/22221751.2020.1745095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahluwalia G., Embree J., McNicol P., Law B., and Hammond G. W., J. Clin. Microbiol. 25, 763 (1987). 10.1128/JCM.25.5.763-767.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Elden L. J. R., van Loon A. M., van Alphen F., Hendriksen K. A. W., Hoepelman A. I. M., van Kraaij M. G. J., Oosterheert J., Schipper P., Schuurman R., and Nijhuis M., J. Infect. Dis. 189, 652 (2004). 10.1086/381207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harvala H., Calvert J., van Nguyen D., Clasper L., Gadsby N., Molyneaux P., Templeton K., McWilliams Leitch C., and Simmonds P., Eurosurveillance 19, 20772 (2014). 10.2807/1560-7917.ES2014.19.15.20772 [DOI] [PubMed] [Google Scholar]
- 13.Terlizzi M. E., Massimiliano B., Francesca S., Sinesi F., Rosangela V., Stefano G., Costa C., and Rossana C., J. Virol. Methods 160, 172 (2009). 10.1016/j.jviromet.2009.04.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao N., Duan Z., Xie Z., Zhong L., Zeng S., Huang H., Gao H., and Zhang B., J. Med. Virol. 88, 2085 (2016). 10.1002/jmv.24580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yahia S., Kandeel A. Y., Hammad E., and El-Gilany A.-H., Indian J. Pediatr. 79, 1323 (2012). 10.1007/s12098-011-0677-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Y.-W., Schmitz J. E., Persing D. H., and Stratton C. W., J. Clin. Microbiol. 58, e00512–20 (2020). 10.1128/JCM.00512-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Hoogen B. G., van Doornum G. J. J., Fockens J. C., Cornelissen J. J., Beyer W. E. P., de Groot R., Osterhaus A. D. M. E., and Fouchier R. A. M., J. Infect. Dis. 188, 1571 (2003). 10.1086/379200 [DOI] [PubMed] [Google Scholar]
- 18.Chen W.-J., Arnold J. C., Fairchok M. P., Danaher P. J., McDonough E. A., Blair P. J., Garcia J., Halsey E. S., Schofield C., Ottolini M., Mor D., Ridoré M., Burgess T. H., and Millar E. V., J. Clin. Virol. 64, 74 (2015). 10.1016/j.jcv.2015.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffiths C., Drews S. J., and Marchant D. J., Clin. Microbiol. Rev. 30, 277 (2017). 10.1128/CMR.00010-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu W.-K., Liu Q., Chen D.-H., Liang H.-X., Chen X.-K., Huang W.-B., Qin S., Yang Z.-F., and Zhou R., BMC Infect. Dis. 13, 28 (2013). 10.1186/1471-2334-13-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bochkov Y. A. and Gern J. E., Microbes Infect. 14, 485 (2012). 10.1016/j.micinf.2011.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savolainen C., Blomqvist S., and Hovi T., Paediatr. Respir. Rev. 4, 91 (2003). 10.1016/S1526-0542(03)00030-7 [DOI] [PubMed] [Google Scholar]
- 23.Myint S., Johnston S., Sanderson G., and Simpson H., Mol. Cell. Probes 8, 357 (1994). 10.1006/mcpr.1994.1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corman V. M., Eckerle I., Bleicker T., Zaki A., Landt O., Eschbach-Bludau M., van Boheemen S., Gopal R., Ballhause M., Bestebroer T. M., Muth D., Müller M. A., Drexler J. F., Zambon M., Osterhaus A. D., Fouchier R. M., and Drosten C., Eurosurveillance 17, 20285 (2012). 10.2807/ese.17.39.20285-en [DOI] [PubMed] [Google Scholar]
- 25.Wong S., Pabbaraju K., Pang X. L., Lee B. E., and Fox J. D., J. Med. Virol. 80, 856 (2008). 10.1002/jmv.21136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heim A., Ebnet C., Harste G., and Pring-Åkerblom P., J. Med. Virol. 70, 228 (2003). 10.1002/jmv.10382 [DOI] [PubMed] [Google Scholar]
- 27.Chow B. D. W. and Esper F. P., Clin. Lab. Med. 29, 695 (2009). 10.1016/j.cll.2009.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arthur J. L., Higgins G. D., Davidson G. P., Givney R. C., and Ratcliff R. M., PLoS Pathog. 5, e1000391 (2009). 10.1371/journal.ppat.1000391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koskenvuo M., Möttönen M., Waris M., Allander T., Salmi T. T., and Ruuskanen O., Eur. J. Pediatr. 167, 1011 (2008). 10.1007/s00431-007-0631-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blomqvist S., Paananen A., Savolainen-Kopra C., Hovi T., and Roivainen M., J. Clin. Microbiol. 46, 2410 (2008). 10.1128/JCM.00313-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanden S. M. G., Koopmans M. P. G., and Avoort H. G. A. M., Eur. J. Clin. Microbiol. Infect. Dis. 32, 1525 (2013). 10.1007/s10096-013-1906-9 [DOI] [PubMed] [Google Scholar]
- 32.Wright K. E., Wilson G. A., Novosad D., Dimock C., Tan D., and Weber J. M., J. Clin. Microbiol. 33, 1180 (1995). 10.1128/JCM.33.5.1180-1184.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu C., Cheng X., He J., Lv X., Wang J., Deng R., Long Q., and Wang X., J. Virol. Methods 148, 81 (2008). 10.1016/j.jviromet.2007.10.023 [DOI] [PubMed] [Google Scholar]
- 34.Quan P.-L., Palacios G., Jabado O. J., Conlan S., Hirschberg D. L., Pozo F., Jack P. J. M., Cisterna D., Renwick N., Hui J., Drysdale A., Amos-Ritchie R., Baumeister E., Savy V., Lager K. M., Richt J. A., Boyle D. B., Garcia-Sastre A., Casas I., Perez-Brena P., Briese T., and Lipkin W. I., J. Clin. Microbiol. 45, 2359 (2007). 10.1128/JCM.00737-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi Y. K., Goyal S. M., Kang S. W., Farnham M. W., and Joo H. S., J. Virol. Methods 102, 53 (2002). 10.1016/S0166-0934(01)00442-6 [DOI] [PubMed] [Google Scholar]
- 36.Katz J. M., Wang M., and Webster R. G., J. Virol. 64, 1808 (1990). 10.1128/JVI.64.4.1808-1811.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quiagen, see https://www.qiagen.com/us/resources/download.aspx?id=c80685c0-4103-49ea-aa72-8989420e3018&lang=en for “QIAamp® Viral RNA Mini Handbook” (last accessed May 2020).
- 38.Peqlab, see https://uk.vwr.com/assetsvc/asset/en_GB/id/17035116/contents for “TriFast: Isolation of RNA, DNA and Protein Simultaneously” (last accessed May 2020).
- 39.Roche, see https://lifescience.roche.com/en_us/articles/high-pure-technology-and-silica-adsorption-kits.html for “High Pure Technology and Silica Adsorption Kits” (last accessed May 2020).
- 40.Roche, see https://lifescience.roche.com/documents/MagNA-Pure-Compact-System-Versatile-Nucleic-Acid-Purification.pdf for “MagnaPure: Versatile Nucleic Acid Purification” (last accessed May 2020).
- 41.Chomczynski P. and Sacchi N., Anal. Biochem. 162, 156 (1987). 10.1016/0003-2697(87)90021-2 [DOI] [PubMed] [Google Scholar]
- 42.Qiagen, see https://www.qiagen.com/us/resources/download.aspx?id=62a200d6-faf4-469b-b50f-2b59cf738962&lang=en for “QIAamp DNA Mini Blood Mini Handbook—EN” (last accessed May 2020).
- 43.Loens K., Bergs K., Ursi D., Goossens H., and Ieven M., J. Clin. Microbiol. 45, 421 (2007). 10.1128/JCM.00894-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang S.-Y., Lien K.-Y., Huang K.-J., Lei H.-Y., and Lee G.-B., Biosens. Bioelectron. 24, 855 (2008). 10.1016/j.bios.2008.07.019 [DOI] [PubMed] [Google Scholar]
- 45.Yoon J.-Y., Introduction to Biosensors, 2nd ed. (Springer, 2016), pp. 257–297. 10.1007/978-3-319-27413-3_14 [DOI] [Google Scholar]
- 46.van Heirstraeten L., Spang P., Schwind C., Drese K. S., Ritzi-Lehnert M., Nieto B., Camps M., Landgraf B., Guasch F., Corbera A. H., Samitier J., Goossens H., Malhotra-Kumar S., and Roeser T., Lab Chip 14, 1519 (2014). 10.1039/C3LC51339D [DOI] [PubMed] [Google Scholar]
- 47.Strohmeier O., Keil S., Kanat B., Patel P., Niedrig M., Weidmann M., Hufert F., Drexler J., Zengerle R., and von Stetten F., RSC Adv. 5, 32144 (2015). 10.1039/C5RA03399C [DOI] [Google Scholar]
- 48.Li L., Miao B., Li Z., Sun Z., and Peng N., ACS Sens. 4, 2738 (2019). 10.1021/acssensors.9b01270 [DOI] [PubMed] [Google Scholar]
- 49.Akin D., Li H., and Bashir R., Nano Lett. 4, 257 (2004). 10.1021/nl034987p [DOI] [Google Scholar]
- 50.Lien K.-Y., Hung L.-Y., Huang T.-B., Tsai Y.-C., Lei H.-Y., and Lee G.-B., Biosens. Bioelectron. 26, 3900 (2011). 10.1016/j.bios.2011.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho Y.-K., Lee J.-G., Park J.-M., Lee B.-S., Lee Y., and Ko C., Lab Chip 7, 565 (2007). 10.1039/b616115d [DOI] [PubMed] [Google Scholar]
- 52.Lien K.-Y., Lin J.-L., Liu C.-Y., Lei H.-Y., and Lee G.-B., Lab Chip 7, 868 (2007). 10.1039/b700516d [DOI] [PubMed] [Google Scholar]
- 53.Han N., Shin J. H., and Han K.-H., RSC Adv. 4, 9160 (2014). 10.1039/c3ra47980c [DOI] [Google Scholar]
- 54.Chen G. D., Alberts C. J., Rodriguez W., and Toner M., Anal. Chem. 82, 723 (2010). 10.1021/ac9024522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chaipan C., Pryszlak A., Dean H., Poignard P., Benes V., Griffiths A. D., and Merten C. A., Cell Chem. Biol. 24, 751 (2017). 10.1016/j.chembiol.2017.05.009 [DOI] [PubMed] [Google Scholar]
- 56.Ng E. X., Sun G., Wei S.-C., Miller M. A., DasGupta R., Lam P. Y. P., and Chen C.-H., Anal. Chem. 91, 1277 (2019). 10.1021/acs.analchem.8b02576 [DOI] [PubMed] [Google Scholar]
- 57.Davaji B., Jeong Bak H., Chang W.-J., and Hoon Lee C., Biomicrofluidics 8, 034101 (2014). 10.1063/1.4875656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hsu M. N., Wei S.-C., Guo S., Phan D.-T., Zhang Y., and Chen C.-H., Small 14, 1802918 (2018). 10.1002/smll.201802918 [DOI] [PubMed] [Google Scholar]
- 59.Jing T., Lai Z., Wu L., Han J., Lim C. T., and Chen C.-H., Anal. Chem. 88, 11750 (2016). 10.1021/acs.analchem.6b03370 [DOI] [PubMed] [Google Scholar]
- 60.Agresti J. J., Antipov E., Abate A. R., Ahn K., Rowat A. C., Baret J.-C., Marquez M., Klibanov A. M., Griffiths A. D., and Weitz D. A., Proc. Natl. Acad. Sci. U.S.A. 107, 4004 (2010). 10.1073/pnas.0910781107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen T. N., Gupta A., Zalavadia M. D., and Streets A., Lab Chip 20, 3899 (2020). 10.1039/D0LC00169D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei S.-C., Hsu M. N., and Chen C.-H., Biosens. Bioelectron. 144, 111639 (2019). 10.1016/j.bios.2019.111639 [DOI] [PubMed] [Google Scholar]
- 63.Bhattacharyya A. and Klapperich C. M., Sens. Actuators B Chem. 129, 693 (2008). 10.1016/j.snb.2007.09.057 [DOI] [Google Scholar]
- 64.Gimenez T. D., Bailão A. M., de Almeida Soares C. M., Fiaccadori F. S., Borges de Lima Dias e Souza M., and Duarte G. R. M., Anal. Methods 9, 2116 (2017). 10.1039/C6AY03481K [DOI] [Google Scholar]
- 65.Niimi M., Masuda T., Kaihatsu K., Kato N., Nakamura S., Nakaya T., and Arai F., Sens. Actuators B Chem. 201, 185 (2014). 10.1016/j.snb.2014.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hui W. C., Yobas L., Samper V. D., Heng C.-K., Liw S., Ji H., Chen Y., Cong L., Li J., and Lim T. M., Sens. Actuators A Phys. 133, 335 (2007). 10.1016/j.sna.2006.06.031 [DOI] [Google Scholar]
- 67.Kim S., Dehlinger D., Peña J., Seol H., Shusteff M., Collette N. M., Elsheikh M., Davenport M., Naraghi-Arani P., and Wheeler E., Microfluid. Nanofluid. 23, 9 (2019). 10.1007/s10404-018-2173-y [DOI] [Google Scholar]
- 68.Xia Y., Tang Y., Yu X., Wan Y., Chen Y., Lu H., and Zheng S.-Y., Small 13, 1603135 (2017). 10.1002/smll.201603135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Surawathanawises K., Kundrod K., and Cheng X., Analyst 141, 1669 (2016). 10.1039/C5AN02282G [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang J., Mahalanabis M., Liu L., Chang J., Pollock N., and Klapperich C., Diagnostics 3, 155 (2013). 10.3390/diagnostics3010155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jacinto M. J., Soares R. R. G., Azevedo A. M., Chu V., Tover A., Conde J. P., and Aires-Barros M. R., Sep. Purif. Technol. 154, 27 (2015). 10.1016/j.seppur.2015.09.006 [DOI] [Google Scholar]
- 72.Negrete A., Ling T. C., and Lyddiatt A., J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 854, 13 (2007). 10.1016/j.jchromb.2007.03.041 [DOI] [PubMed] [Google Scholar]
- 73.Jin C. E., Lee T. Y., Koo B., Sung H., Kim S.-H., and Shin Y., Sens. Actuators B Chem. 255, 2399 (2018). 10.1016/j.snb.2017.08.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hagan K. A., Reedy C. R., Uchimoto M. L., Basu D., Engel D. A., and Landers J. P., Lab Chip 11, 957 (2011). 10.1039/C0LC00136H [DOI] [PubMed] [Google Scholar]
- 75.Ferguson B. S., Buchsbaum S. F., Wu T.-T., Hsieh K., Xiao Y., Sun R., and Soh H. T., J. Am. Chem. Soc. 133, 9129 (2011). 10.1021/ja203981w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Du K., Cai H., Park M., Wall T. A., Stott M. A., Alfson K. J., Griffiths A., Carrion R., Patterson J. L., Hawkins A. R., Schmidt H., and Mathies R. A., Biosens. Bioelectron. 91, 489 (2017). 10.1016/j.bios.2016.12.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.