Abstract
Biofoundries integrate high-throughput software and hardware platforms with synthetic biology approaches to enable the design, execution and analyses of large-scale experiments. The unique and powerful combination of laboratory infrastructure and expertise in molecular biology and automation programming, provide flexible resources for a wide range of workflows and research areas. Here, we demonstrate the applicability of biofoundries to molecular microbiology, describing the development and application of automated workflows to identify the genetic basis of growth inhibition of the plant pathogen Streptomyces scabies by a Pseudomonas strain isolated from a potato field. Combining transposon mutagenesis with automated high-throughput antagonistic assays, the workflow accelerated the screening of 2880 mutants to correlate growth inhibition with a biosynthetic gene cluster within 2 weeks.
Keywords: biofoundry, automated testing workflow, Pseudomonas, biocontrol, Streptomyces scabies
1. Introduction
Biofoundries are integrated automation platforms for the design, construction and characterization of organisms for biotechnology applications and research. They combine experimental approaches, such as mathematical and statistical modeling (i.e. design of experiments, DoE), computer-aided design (CAD) and analytical software, together with automated experimental platforms to enable high-throughput automated workflows. Over the last decade, many research institutions have established biofoundries to accelerate both fundamental and applied research (1). Previous to this, the use of automation in research labs was sporadic with uptake mainly limited to service laboratories performing repetitive workflows, such as diagnostics or genotyping.
The era of synthetic biology has applied engineering principles, such as standardization and predictive models to inform experimental design (2). This has enabled experiments to be scaled, making automated workflows not only attractive but necessary. Additional benefits of automation include improvements in the accuracy and reproducibility of experimentation (1, 3). A further tenet of engineering prescribed by synthetic biology is the progression of design-build-test-learn (DBTL) cycles, around which biofoundry workflows are typically centered. In the ‘design’ phase of the cycles, CAD tools are often used to assemble complex experimental designs, e.g. Ref. (4–6) and statistical techniques, such as DoE, are used to identify critical influencing factors and define the scale of the design space (7). Following this, automated workflows are used to assemble or ‘build’ the designs, typically from DNA. In the ‘test’ phase, specific analytic measurements are taken using techniques relevant to the phenotype or characteristic of interest. Finally, data are analyzed in the ‘learn’ phase and conclusions are used to inform the design phase of the subsequent cycle (1).
While the DBTL has clear applications in metabolic engineering (8), the applicability of biofoundries to the wider bioscience research community is not always obvious. The significant costs of establishing and maintaining biofoundries require an active user base. Barriers identified for offsetting the cost of newly established biofoundries include a limited awareness of the capabilities among potential users (1). The basic software and hardware infrastructure of biofoundries, combined with the associated expertise in large-scale experimental design can, however, be applied to a wide range of molecular and microbial workflows enabling biofoundries to become central research infrastructures.
Here, we demonstrate the broader applicability of biofoundries to established molecular and microbial methods through the development and application of automated workflows for the rapid and scalable creation and phenotypic screening of mutant libraries. The ability to identify single-gene determinants of inhibitory activity in random mutagenesis studies is limited only by library size, which presents a significant limitation in manual screening efforts. In this example, we describe workflows for the identification of genes associated with the inhibition of microbial growth. This leads to the identification of a biosynthetic gene cluster (BGC) in the genome of Pseudomonas sp. Ps652, recovered from a commercial potato field in Norfolk (UK). This isolate had previously been shown to display strong in vitro activity against Streptomyces scabies 87-22, a commercially relevant potato pathogen (9, 10). The determinants of Pseudomonas sp. Ps652 inhibition of S. scabies 87-22 were not evident from the Ps652 genome sequence or rational gene deletions and, given the large zones of inhibition observed on plates, it was not possible to use a standard 96 solid pin replicator to assay transposon mutants on the same plate alongside the pathogen. To overcome this constraint, we developed biofoundry workflows to generate and assay a library of random transposon mutants. This enabled the identification of a BGC associated with the production of a compound able to inhibit the growth of S. scabies 87-22. There is substantial interest in the role of microbial antagonism in the health of humans (11), animals (12) and plants (13), but it can be difficult to elucidate the genetic determinants of this antagonism, which can be complex and manifold. The workflow described here provides a template for the automated identification of these genes.
2. Materials and methods
2.1 Bacterial strains and media
Pseudomonas sp. Ps652ΔHCN, a cyanide null mutant of an environmental isolate from a commercial potato field in Norfolk, UK (GenBank Accession GCA_902497775.1) was used as the parent strain for transposon mutagenesis. Pseudomonas sp. Ps652ΔHCN was stored at −80°C in 925 µl L medium with 75 µl DMSO. Streptomyces coelicolor M145 and S. scabies 87-22 were stored as spore stocks that were grown to sporulation on potato dextrose agar, harvested in 20% glycerol and stored at −80°C. The same spore stock was used for the entire workflow.
All media used in this study were adjusted to pH 7.2. Rye Sucrose Agar (RSA): 60 g/l rye grains, 20 g/l sucrose, 20 g/l Agar was prepared by soaking rye grains in a 1/40 dilution of 10% chlorite solution for 4 min before rinsing with reverse osmosis purified water and germinating overnight. Germinated rye grains were then ground at high speed for 2 min with a hand blender, before being transferred to a water bath for 3 h at 50°C in Milli-Q water. This was then filtered through a sieve and muslin cloth before addition of sucrose and agar and sterilization by autoclaving. Malt Extract-Yeast Extract Maltose Medium (MYM): 10 g/l malt extract, 4 g/l yeast extract, 4 g/l maltose, 18 g/l bacteriological agar (Sigma-Aldrich, UK), made up to 1 l with tap water. Lennox Broth and Lennox Agar (L): 10 g/l tryptone, 5 g/l yeast extract, 5 g/l NaCl, 1 g/l D-glucose, 10 g/l Formedium agar (Lennox Agar), made up to 1 l with Milli-Q water. Lysogeny Broth (LB): 10 g/l tryptone, 5 g/l yeast extract, 10 g/l NaCl, made up to 1 l with Milli-Q water. Potato Dextrose Agar: 41 g/l potato dextrose agar (Formedium PDA0102S), made up to 1 l with Milli-Q water.
2.2 Production and selection of a Pseudomonas mutant library
Pseudomonas sp. Ps652ΔHCN was transformed by electroporation in 300 mM sucrose with the mariner transposon-containing plasmid pALMAR3 (14) and 100 µl was plated directly in single-well NuncTM OmniTrayTM plates (242811, Thermo Scientific, UK) containing L agar + 25 µg/ml tetracycline with a plating density that enabled automated colony picking. Plates were incubated overnight at 28°C for 16 h and resulting colonies were selected using the Hamilton Microlab STARplus (Hamilton, Bonaduz AG, Switzerland) equipped with one 96-channel pipetting head and eight individual single channels capable of liquid level detection, a light table and camera for colony picking, and grippers for plate movement steps (Supplementary Data S1). Methods were programmed using the Venus Three software (version 4.4.07740, Hamilton). Details of scripts, deck layout and associated Hamilton libraries are provided in Supplementary Data S2.
The detection and selection of the maximum number of viable colonies were enabled by a modified picking method using the EasypickII software (Version 1.0.2, Hamilton). A maximum number of 500 colonies from each plate were selected to generate the library. Criteria for colony picking were established to avoid cross-contamination and aberrant phenotypes. Colonies were defined with a minimal circularity factor of 0.05 and an area of 0.4–15 mm2. The liquid transfer and aspiration process were initially hampered by the presence of air bubbles formed by successive pipetting that hampered the autosensing of liquid levels, leading to inaccurate transfers. To overcome this, the pipette position was altered during aspiration and ejection of media. Suitable colonies were transferred using disposable 300 µl tips into liquid media (200 µl L + 25 µg/ml tetracycline) in 96-well microplates (4ti-0117, Brooks Life Science, UK) (Supplementary Data S2). Cultures were grown at 28°C, 250 rpm for 16 h. 15 µl DMSO was added to each well manually using a multichannel pipette, and plates were stored at −80°C until required.
2.3 Automated plating of S. coelicolor M145
Spores of S. coelicolor M145 were resuspended into Milli-Q water at a concentration of 35 μl spore stock per 80 ml H2O. A Hamilton Microlab STARplus, as described above, was used to dispense 20 µl of spore suspension into the center of each well of 120 24-well plates (Greiner Bio-One, 662102) containing 1 ml RSA per well. Spores were distributed using the on-deck shaker at 1500 rpm for 5 s. The plates were dried for 1 h before use in automated bioassays. Supplementary Data S3 provides the link to the automated spore plating method scripts, deck layout and associated Hamilton libraries.
2.4 Automated bioassays against S. coelicolor M145
The library of Pseudomonas sp. Ps652ΔHCN mutants were spiked onto the S. coelicolor M145 plates using a Hamilton Microlab STARplus as described above. Pipette tips were used to transfer droplets by submersion without aspiration followed by a brief touch of the agar surface using a liquid level setting of 0 mm. Plates were incubated at 28°C for 5 days before photographic imaging (Canon Ixus 175, Canon, Tokyo, Japan). Supplementary Data S4 provides the link to the bioassay method scripts, deck layout and associated Hamilton libraries.
2.5 Growth curves
Mutants of interest were streaked by hand to single colonies and grown overnight in 10 ml LB. Growth curves were performed by manually inoculating 5 µl of overnight culture diluted to an optical density at 600 nm (OD600) = 0.01 into 150 µl of LB medium in 96-well plates. Plates were incubated at 30°C with shaking at 200 rpm for 48 h with absorbance readings taken every 30 min. Data were acquired on a SPECTROstar Nano UV/Vis microplate reader (BMG Labtech, Ortenberg, Germany), and processed using Microsoft Excel and R (15). Three replicates were used per mutant.
2.6 Cross-streak assays
Cross-streak assays against S. scabies 87-22 were performed manually on MYM tap agar. Streptomyces spores were streaked in two parallel lines on the plate using a sterile toothpick, which were then ‘cross streaked’ at a 90° angle with overnight cultures of Pseudomonas sp. Ps652ΔHCN mutants of interest. All mutants were assayed in triplicate. Plates were incubated for 5 days at 28°C before photographic imaging (Canon Ixus 175, Canon, Tokyo, Japan).
2.7 Insertion site sequencing
Sites of transposon insertion were determined by polymerase chain reaction (PCR) as described in Ref. (16). DNA flanking the insertion sites was enriched in two rounds of amplification using primers specific to the ends of the mariner transposon element (Arb1b 5'-GGCCAGCGAGCTAACGAGACNNNNGATAT-3' and Arb-PCR 5′-CGCAAACCAACCCTTGGCAG-3′) with an initial denaturation at 95°C for 3 min followed by 30 cycles of: 95°C for 15 s, 38°C for 30 s and 72°C for 90 s, followed by a final extension of 72°C for 3 min. Products were purified, eluted in 30 µl sterile water and used as template for a second round of PCR with degenerate primers that anneal to chromosomal sequences flanking the transposon (Arb1 5′-GGCCAGCGAGCTAACGAGAC-3′ and Almar3-seq 5′-ACATATCCATCGCGTCCGCC-3′) with an initial denaturation of 95°C for 3 min followed by 30 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 90 s, followed by a final extension of 72°C for 3 min. Amplicons were purified using a QIAquick Spin PCR Purification Kit (Qiagen, Hilden, Germany) as per the manufacturer’s instructions and sequenced by Sanger sequencing (Eurofins Genomics, Luxembourg City, Luxembourg) using the Almar3-seq primer (5′-ACATATCCATCGCGTCCGCC-3′).
3. Results
3.1 Automated production of a Pseudomonas sp. Ps652ΔHCN mutant library
Prior analysis of the Pseudomonas sp. Ps652 genome determined that it features no BGCs for known antimicrobial compounds (10), with the exception of a hydrogen cyanide (HCN) BGC (17). However, in preliminary experiments (unpublished data), we determined that HCN production in Pseudomonas sp. Ps652 only partly explained inhibition of S. scabies growth and that a non-volatile compound was contributing to inhibition. Therefore, by using a cyanide null mutant, Ps652ΔHCN, as the parent strain for the generation of a transposon mutant library, we predicted the screen would identify new genes relevant to the inhibitory phenotype.
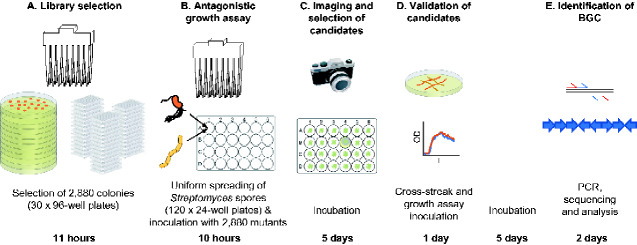
The primary bottleneck in the production of microbial mutant libraries is the selection of colonies and their subsequent curation. Manual colony-picking and subsequent screening of the resultant library is a tedious and time-consuming task with a consequential risk of human error and repetitive strain injury. We, therefore, automated the selection, culture and screening of a library of Pseudomonas sp. Ps652ΔHCN mutants to which the mariner transposon-containing plasmid pALMAR3 (14) had been delivered. Mariner type-transposons consist of a transposase gene flanked by inverted tandem repeats that insert their DNA cargo into the target genome at TA sites (18); the Ps652 genome contains 228 924 TA sites meaning that insertions of the transposase cargo DNA occur approximately randomly across the genome. The use of automation enabled the identification and selection of 2880 colonies (30 × 96-well plates) in 11 h with the added benefits of colony traceability (a digital record that links the original position of colonies to plate position for verification) and walk-away time (Figure 1A, Supplementary Data S2). The workflow was optimized to avoid colonies with aberrant morphologies and to prevent cross-contamination from neighboring colonies, facilitating scalability. The selection of 2880 mutants was based on the number of accessory coding sequences typically found in environmental Pseudomonas strains (19), however, the experiment is easily scalable.
Figure 1.
A Biofoundry workflow for the automated screening of microbial interactions. A mutant library of Pseudomonas sp. Ps652ΔHCN was created by automated selection of 2880 colonies (A). These were used in an automated bioassay with S. coelicolor M145 (B) and image analyzed after 5 days of incubation (C). Candidate strains were validated in growth assays and against the target pathogen, S. scabies, in a licensed laboratory (D) before the insertion site was identified by PCR and sequencing (E).
3.2 Automated identification of Pseudomonas sp. Ps652 mutants unable to inhibit growth of S. coelicolor M145
Streptomyces coelicolor M145 was used in automated bioassays as a non-pathogenic proxy for S. scabies 87-22, which we were unable to work with in the biofoundry due to plant health license limitations. Streptomyces scabies and S. coelicolor both belong to clade II of a genome-based phylogeny for the Streptomyces genus (20), and preliminary work indicated that both were inhibited by Ps652ΔHCN (data not shown). Given the large zones of inhibition observed in pilot experiments, it was not possible to assay the mutant library on plates containing pathogen inoculum using a standard 96-pin replicator. We, therefore, developed an automated assay to assess the ability of 2880 Pseudomonas sp. Ps652ΔHCN mutants to affect the growth of S. coelicolor M145 using 24-well plates. This automated workflow consists of two steps. Firstly, the uniform spread of Streptomyces spores to each well (Figure 1B; Supplementary Data S3) and, secondly, inoculation with Pseudomonas sp. Ps652ΔHCN (Figure 1B; Supplementary Data S4). The automated assay required the optimization of (i) pipetting settings to minimize the time taken for the transfer of spore solution and plate inoculation, (ii) inoculation position (spiking) settings to enable the use of different volumes of agar in 24-well plates, (iii) lid-gripping and plate-gripping positions for accurate and rapid transfer of plates to stacks, and (iv) the speed (rpm) and time (s) for which plates were shaken to achieve a reproducibly homogenous spread of spores in every individual well. We also noted that it was essential to allow the spore solution to dry on the plates for 60 min before spiking the mutant library, to prevent Ps652 mutants from colonizing the entire well.
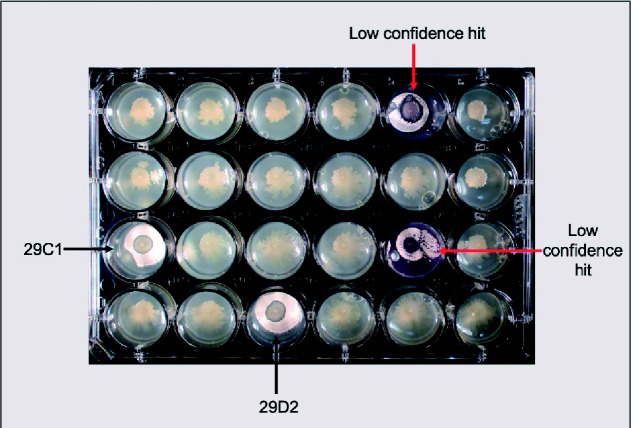
The optimized bioassay was performed across 120 24-well plates and took ∼10 h to set up. Following incubation at 28°C for 5 days, the plates were imaged to identify mutants unable to inhibit S. coelicolor M145 growth (Figure 1C). Mutants of interest we defined as those in which S. coelicolor M145 grew immediately adjacent to the pseudomonad. High-confidence mutants were those in which the growth of the mutants themselves was uncompromised. Low-confidence mutants were defined as those in which Pseudomonas sp. Ps652ΔHCN growth appeared abnormal or in which a minimal zone of inhibition was visible (Figure 2). We identified 23 mutants of interest, which were further investigated in low-throughput screens.
Figure 2.
Representative image of a 24-well plate showing both low-confidence and high-confidence candidates, including 29C1 and 29D2, which were selected for further analysis.
3.3 Validation of candidates in cross-streak assays with S. scabies
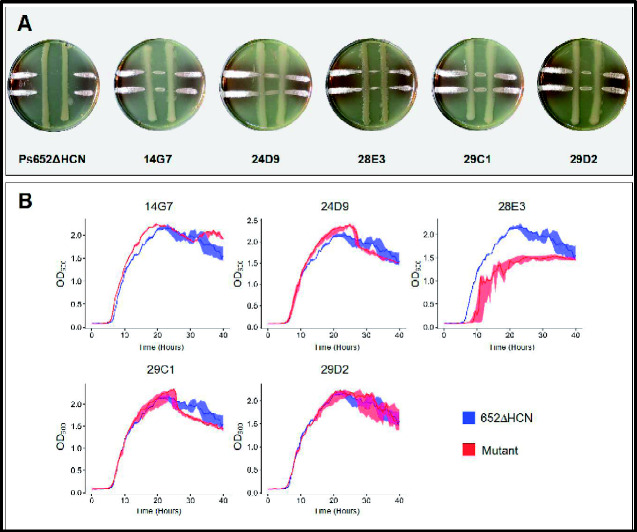
In order to validate the high-throughput screen, the 23 mutants identified in the high-throughput screen against S. coelicolor M145 were streaked to single colonies and used to inoculate cross-streak assays (see Section 2) with the target strain, S. scabies 87-22 (Figure 1D). Five mutant strains, 14G7, 24D9, 28E3, 29C1 and 29D2 were also unable to inhibit the growth of S. scabies 87-22 (Figure 3A). To rule out mutants that simply caused a Ps652 growth defect, growth curves were performed in liquid medium. Under these conditions, mutant 28E3 displayed a significant growth defect when compared with the parent strain and was excluded from further consideration (Figure 3B). Sequencing revealed that 28E3 had a transposon insertion in the rfbA gene, which encodes RmlA, a thymidylyltransferase involved in the biosynthesis of the cell wall component deoxythymidine diphosphate-L-rhamnose (21), highlighting the potential for off-target hits and false positives in this type of screen. All 22 remaining initial hits retained normal growth characteristics, including 14G7, 24D9, 29C1 and 29D2 (Figure 3B). Sites of transposon insertion for all 23 mutants are available in Supplementary Data S5.
Figure 3.
(A) Cross-streak assays of five Pseudomonas sp. Ps652ΔHCN mutants (vertical lines) against S. scabies 87-22 (horizontal lines). Images are representative of 3 replicates after incubation for five days. (B) Growth curves of Pseudomonas sp. Ps652ΔHCN (control) and five transposon mutants. Solid lines represent the median at each time point and the shaded area represent the maximum and minimum values; n = 3.
3.4 Sequencing of transposon insertion sites identifies a candidate gene cluster
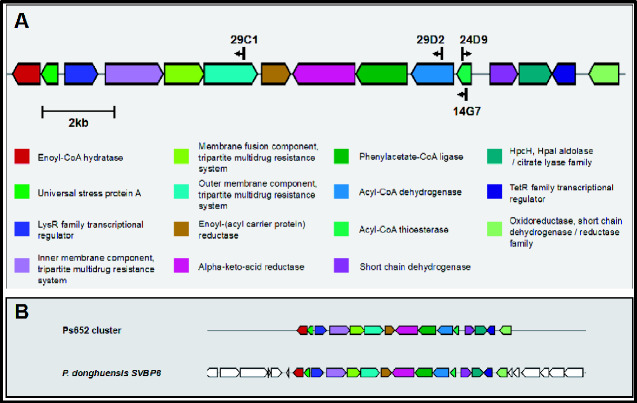
The sites of transposon insertion were determined by PCR, and the four Ps652 mutants that were unable to inhibit the growth of both S. coelicolor M145 and S. scabies 87-22 all harbored transposon insertions within the same 8 kbp chromosomal region (Figure 4A). This region had not been identified as a BGC via antiSMASH 5.0 (22) or PRISM (23) analysis of the Ps652 genome, but did match a putative BGC that had been identified in Pseudomonas donghuensis SVBP6 in parallel with our experimental work. This was associated with antifungal activity and 7-hydroxytropolone biosynthesis in P. donghuensis (24). Comparative sequence analyses using MultiGeneBlast (25) indicated that the BGCs in Pseudomonas sp. Ps652 and P. donghuensis SVBP6 are highly similar, where they share the same genetic architecture (Figure 4B), as well as high identity/coverage scores for all genes (Supplementary Data S6).
Figure 4.
(A) Schematic of the Pseudomonas sp. Ps652 BGC indicating the location of transposon insertions (black arrows) and the conserved domains of the proteins encoded in this BGC. (B) Comparison of the cluster identified in Pseudomonas sp. Ps652 to a recently reported cluster linked to 7-hydroxytropolone production in P. donghuensis SVBP6.
4. Discussion
High-throughput screening is an emerging approach in many scientific fields and can be a useful tool in investigating microbial interactions (26–29). However, many researchers lack access to automation equipment or to programming expertise to enable existing equipment to be applied to their experimental workflow. As a result, high-throughput approaches often bypassed in favor of simple or low-throughput methodologies. This is particularly pertinent in microbial interactions, where the ability to scale bioassays holds immense promise but is impractical without the application of automation (27). In this study, we exemplified the relevance to biofoundries to the study of microbial interactions in the soil interactions, by conducting an automated screen that identified a BGC implicated in growth inhibition in just a few days of laboratory time. Such microbial interactions are of interest to agriculture, particularly the identification and characterization of plant-associated or rhizospheric soil bacteria capable of inhibiting the growth of specific plant pathogens (13, 30).
Advances in bioinformatic approaches to natural product discovery can be applied to such problems, aiding discovery of novel antimicrobial compounds in plant-associated bacteria (31). However, many existing bioinformatic approaches rely on homology to characteristic enzymes, such as polyketide synthases and non-ribosomal peptide synthetases, which represent roughly 65% of microbial metabolites used commercially (32). At present, it is likely that such approaches may be less reliable in identifying novel BGCs that display unusual and unprecedented genetic architectures, or that lack unifying features shared across the whole molecular class. Similarly, currently available bioinformatics tools can be of limited use in uncovering specialized metabolites where the biosynthesis is performed by a single protein, as has been shown previously for a volatile compound that is involved in biocontrol, 1-undecene (33). Consequently, novel compounds with societally relevant activities are still being discovered through bioactivity guided screening methods (34). As such, interesting phenotypes from select bacterial isolates still warrant investigation through screening against pathogens of interest.
To develop biofoundry workflows for studying microbial interactions, we focused on a Pseudomonas strain for which previous investigations had not identified BGCs of known antimicrobials (10, 24). The utility of our automated workflow was initially validated by growth curves and cross-streak assays (Figure 3), and subsequently by the identification of a BGC highly likely to produce the tropolonoid antimicrobial 7-hydroxytropolone. In parallel with our work, this BGC was identified through a transposon mutant screen of P. donghuensis SVBP6 (24) (Figure 4B). In that study, the authors associated the BGC with inhibitory activity of fungi and oomycetes, but not Bacillus subtilis or Escherichia coli. Our screen identified this BGC as being implicated in the inhibition of the filamentous Gram-positive bacterium S. scabies 87-22, a commercially important potato pathogen (35). Lacking the natural product class-defining proteins and being considerably different to published non-Pseudomonas tropolone BGCs (36), this cluster was not readily identified by genome mining tools.
The development of the workflow identified a number of important considerations and advantages of automated processes. The screening of bacterial libraries often relies on 96-solid pin replicators (37); however, some phenotypes are affected by the genotype or phenotype of the surrounding colonies on the plate. In the case of Pseudomonas sp. Ps652, mutant colonies that had lost the ability to produce a diffusible natural product were undetectable due to the production of the inhibitory natural product by neighboring colonies. Conducting the assays in individual wells of 24-well plates, an approach only practicable with automation, overcame this complication. Automating the assay also enabled the uniform distribution of Streptomyces spores, as well as the exact positioning and depth of the Pseudomonas inoculum. This used the liquid level detection feature and conductive tips to avoid the challenges of inconsistent agar depth. An additional advantage of automation is the ability to screen very large libraries. This is particularly important for single-gene determinants of phenotypes. Finally, the selection of false positives can be a potent obfuscating factor. These can increase due to inconsistent assay set-up as well as human errors or bias in visual screening. The ability to accurately and consistently inoculate assays and to program defined parameters for the automated identification of colony size and shape aided the robustness and reproducibility of the assay. Through the application of biofoundry workflows and approaches to experimentation, we were able to rapidly identify and prioritize a manageable number of high-confidence candidates for further characterization using manual approaches, ultimately implicating a BGC proposed to produce 7-hydroxytropolone in the biological control of the important potato pathogen S. scabies 87-22.
Most biofoundries are already engaged in the reconstruction and optimization of biosynthetic pathways to enable natural product biosynthesis. This study highlights their potential in the upstream experimental processes of gene discovery. In the workflow described here, the DBTL cycle consists of the selection of appropriate tools for the production of libraries, the construction and arraying of such libraries, phenotypic screens and genotyping of candidates, and, finally, data analysis to provide new knowledge of gene function. The outcomes may lead to either iterative cycles in which either library complexity or the robustness of the phenotypic screen are improved, or excitingly, link directly though the provision of novel genes to interconnected DBTL cycles in which the aim is biosynthesis. To aid this, critical comparisons of library production methods and the integration of high-throughput computer-aided quantitative phenotyping into biofoundry platforms would be highly beneficial.
SUPPLEMENTARY DATA
Supplementary Data are available at SYNBIO Online.
Supplementary Material
Acknowledgments
We thank James Titchmarch (Hamilton Company) for the original Hamilton picking method, which was adapted by A.E. We are grateful to Jacob Malone (John Innes Centre) for providing the pALMAR3 mariner transposon plasmid, and to David Widdick (John Innes Centre) for advice on transposon mutagenesis screening.
Funding
The authors would like to acknowledge support from the UK Research and Innovation (UKRI) Biotechnology and Biological Sciences Research Council (BBSRC) Core Capability Grant BB/CCG1720/1 for the Earlham Institute and the National Capability BBS/E/T/000PR9815. Additional funding was provided via the BBSRC MfN Institute Strategic Programme grant [BBS/E/J/000PR9790] for the John Innes Centre, a Norwich Research Park BBSRC Doctoral Training Partnership grant [BB/M011216/1] for A.D.M. and a Royal Society University Research Fellowship for A.W.T.
Data availability
All relevant data (scripts packages for the automated colony picking, spore spreading and automated inoculation of Pseudomonas sp. Ps652ΔHCN mutant library) are available via the GitHub repository of the Earlham Institute Biofoundry (https://github.com/eibiofoundry/A-biofoundry-workflow-for-the-identification-of-genetic-determinants-of-microbial-growth-inhibition). All videos depicting the automated workflows are available via the YouTube channel of the Earlham Institute Biofoundry (Automated colony picking: https://youtu.be/cLQZDdMr0l0, automated distribution of S. coelicolor M145 spores on 24-well plates https://youtu.be/3TEFXuciNu8, and automated inoculation of 24-well plates containing S. coelicolor M145 with the Pseudomonas sp. Ps652ΔHCN mutant library: https://youtu.be/nlIDy0Vf5ww).
References
- 1. Hillson N., Caddick M., Cai Y., Carrasco J.A., Chang,M.W., Curach,N.C., Bell,D.J., Le Feuvre,R., Friedman,D.C., Fu,X. et al. (2019) Building a global alliance of biofoundries. Nat. Commun., 10, 2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jessop-Fabre M.M., Sonnenschein N. (2019) Improving reproducibility in synthetic biology. Front. Bioeng. Biotechnol., 7, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chao R., Mishra S., Si T., Zhao H. (2017) Engineering biological systems using automated biofoundries. Metab. Eng., 42, 98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Czar M.J., Cai Y., Peccoud J. (2009) Writing DNA with genoCADTM. Nucleic Acids Res., 37, W40–W47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hillson N.J., Rosengarten R.D., Keasling J.D. (2012) j5 DNA assembly design automation software. ACS Synth. Biol., 1, 14–21. [DOI] [PubMed] [Google Scholar]
- 6. Nielsen A.A.K., Der B.S., Shin J., Vaidyanathan,P., Paralanov,V., Strychalski,E.A., Ross,D., Densmore,D. and Voigt,C.A. (2016) Genetic circuit design automation. Science, 352, aac7341. [DOI] [PubMed] [Google Scholar]
- 7. Braniff N., Ingalls B. (2018) New opportunities for optimal design of dynamic experiments in systems and synthetic biology. Curr. Opin. Syst. Biol., 9, 42–48. [Google Scholar]
- 8. Carbonell P., Jervis A.J., Robinson C.J., Yan,C., Dunstan,M., Swainston,N., Vinaixa,M., Hollywood,K.A., Currin,A., Rattray,N.J.W. et al. (2018) An automated Design-Build-Test- Learn pipeline for enhanced microbial production of fine chemicals. Commun. Biol., 1, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Seipke R.F., Loria R. (2008) Streptomyces scabies 87-22 possesses a functional tomatinase. J. Bacteriol., 190, 7684–7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stefanato F.L., Trippel C., Uszkoreit S., Ferrafiat L., Grenga L., Dickens R., Kelly N., Kingdon A.D.H., Ambrosetti L., Findlay K.C., et al. (2019) Pan-genome analysis identifies intersecting roles for Pseudomonas specialized metabolites in potato pathogen inhibition. bioRxiv, doi: 10.1101/783258 [DOI] [PMC free article] [PubMed]
- 11. Waldman A.J. and Balskus E.P., (2018) The human microbiota, infectious disease, and global health: challenges and opportunities. ACS Infect. Dis., 4, 14–26. [DOI] [PubMed] [Google Scholar]
- 12. Mueller U.G., Sachs J.L. (2015) Engineering microbiomes to improve plant and animal health. Trends Microbiol., 23, 606–617. [DOI] [PubMed] [Google Scholar]
- 13. Kim Y.C., Leveau J., McSpadden Gardener B.B., Pierson,E.A., Pierson,L.S. 3rd and Ryu,C.M. (2011) The multifactorial basis for plant health promotion by plant-associated bacteria. Appl. Environ. Microbiol., 77, 1548–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Malone J.G., Jaeger T., Spangler C., Ritz,D., Spang,A., Arrieumerlou,C., Kaever,V., Landmann,R. and Jenal,U. (2010) YfiBNR mediates cyclic di-GMP dependent small colony variant formation and persistence in Pseudomonas aeruginosa. PLoS Pathog., 6, e1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. R Core Team. (2019) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- 16. O'Toole G.A., Kolter R. (1998) Initiation of biofilm formation in Pseudomonas fluorescens wcs365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol., 28, 449–461. [DOI] [PubMed] [Google Scholar]
- 17. Pessi G., Haas D. (2000) Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the Quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J. Bacteriol., 182, 6940–6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lampe D.J., Grant T.E., Robertson H.M. (1998) Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics, 149, 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. De Vrieze M., Varadarajan A.R., Schneeberger K., Bailly,A., Rohr,R.P., Ahrens,C.H. and Weisskopf,L. (2020) Linking comparative genomics of nine potato-associated Pseudomonas isolates with their differing biocontrol potential against late blight. Front. Microbiol., 11, 857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McDonald B.R. and Currie C.R., (2017) Lateral gene transfer dynamics in the ancient bacterial genus Streptomyces. mBio., 8, e00644-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blankenfeldt W., Asuncion M., Lam J.S. et al. (2000) The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA). EMBO J., 19, 6652–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Blin K., Shaw S., Steinke K., Villebro,R., Ziemert,N., Lee,S.Y., Medema,M.H. and Weber,T. (2019) AntiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res., 47, W81–W87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Skinnider M.A., Merwin N.J., Johnston C.W., Magarvey N.A. (2017) PRISM 3: expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res., 45, W49–W54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Muzio F.M., Agaras B.C., Masi M., Tuzi,A., Evidente,A. and Valverde,C. (2020) 7-hydroxytropolone is the main metabolite responsible for the fungal antagonism of Pseudomonas donghuensis strain SVBP6. Environ. Microbiol., 22, 2550–2563. [DOI] [PubMed] [Google Scholar]
- 25. Medema M.H., Takano E., Breitling R. (2013) Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol. Biol. Evol., 30, 1218–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Geng H., Bruhn J.B., Nielsen K.F., Gram,L. and Belas,R. (2008) Genetic dissection of tropodithietic acid biosynthesis by marine roseobacters. Appl. Environ. Microbiol., 74, 1535–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang R., Gallant É., Seyedsayamdost M.R. (2016) Investigation of the genetics and biochemistry of roseobacticide production in the Roseobacter clade bacterium Phaeobacter inhibens. mBio., 7, e02118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Santos Kron A., Zengerer V., Bieri M., Dreyfuss,V., Sostizzo,T., Schmid,M., Lutz,M., Remus-Emsermann,M.N.P. and Pelludat,C. (2020) Pseudomonas orientalis F9 pyoverdine, safracin, and phenazine mutants remain effective antagonists against Erwinia amylovora in apple flowers. Appl. Environ. Microbiol., 86, e02620–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Agaras B.C., Iriarte A., Valverde C.F. (2018) Genomic insights into the broad antifungal activity, plant-probiotic properties, and their regulation, in Pseudomonas donghuensis strain SVBP6. PLoS One, 13, e0194088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Philippot L., Raaijmakers J.M., Lemanceau P., van der Putten W.H. (2013) Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol., 11, 789–799. [DOI] [PubMed] [Google Scholar]
- 31. Paterson J., Jahanshah G., Li Y., Wang,Q., Mehnaz,S. and Gross,H. (2017) The contribution of genome mining strategies to the understanding of active principles of PGPR strains. FEMS Microbiol. Ecol., 93, fiw249. [DOI] [PubMed] [Google Scholar]
- 32. Katz L., Baltz R.H. (2016) Natural product discovery: past, present, and future. J. Ind. Microbiol. Biotechnol., 43, 155–176. [DOI] [PubMed] [Google Scholar]
- 33. Rui Z., Li X., Zhu X., Liu,J., Domigan,B., Barr,I., Cate,J.H.D. and Zhang,W. (2014) Microbial biosynthesis of medium-chain 1-alkenes by a nonheme iron oxidase. Proc. Natl. Acad. Sci. U S A, 111, 18237–18242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ling L.L., Schneider T., Peoples A.J., Spoering,A.L., Engels,I., Conlon,B.P., Mueller,A., Schäberle,T.F., Hughes,D.E., Epstein,S. et al. (2015) A new antibiotic kills pathogens without detectable resistance. Nature, 517, 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lerat S., Simao-Beaunoir A.M., Beaulieu C. (2009) Genetic and physiological determinants of Streptomyces scabies pathogenicity. Mol. Plant Pathol., 10, 579–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen X., Xu M., Lü J., Xu J., Wang Y., Lin S., Deng Z., Tao M. (2018) Biosynthesis of tropolones in Streptomyces spp.: Interweaving biosynthesis and degradation of phenylacetic acid and hydroxylations on the tropone ring. Appl. Environ. Microbiol., 84, e00349–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Drenkard E., Hibbler R.M., Gutu D.A., Eaton A.D., Silverio A.L., Ausubel F.M., Hurley B.P., Yonker L.M. (2018) Replication of the ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. J. Vis. Exp., 4, 57298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data (scripts packages for the automated colony picking, spore spreading and automated inoculation of Pseudomonas sp. Ps652ΔHCN mutant library) are available via the GitHub repository of the Earlham Institute Biofoundry (https://github.com/eibiofoundry/A-biofoundry-workflow-for-the-identification-of-genetic-determinants-of-microbial-growth-inhibition). All videos depicting the automated workflows are available via the YouTube channel of the Earlham Institute Biofoundry (Automated colony picking: https://youtu.be/cLQZDdMr0l0, automated distribution of S. coelicolor M145 spores on 24-well plates https://youtu.be/3TEFXuciNu8, and automated inoculation of 24-well plates containing S. coelicolor M145 with the Pseudomonas sp. Ps652ΔHCN mutant library: https://youtu.be/nlIDy0Vf5ww).