Abstract
Glioma is a highly heritable disease with a strong genetic component. The N6-methyladenosine (m6A) modification core genes play important roles in the context of cancer. However, the effects of polymorphisms in the m6A modification core genes on the risk of pediatric glioma remain undefined. Here, we intended to demonstrate the relationship between 24 functional single-nucleotide polymorphisms (SNPs) in eight m6A modification core genes and glioma risk. Case-control design and multinomial logistic regression were used to develop models to estimate the risk of glioma while accounting for the subtypes of glioma. A total of 171 glioma cases and 228 controls from South China were genotyped using a TaqMan assay. The WTAP rs7766006, YTHDF2 rs3738067, and FTO rs9939609 variants conferred a statistically significant increased risk of glioma, respectively. YTHDC1 rs2293595, YTHDC1 rs3813832, and FTO rs8047395 were associated with a significant inverse association with risk of glioma, respectively. The significant associations were more predominant in stratification analyses of certain subgroups. Functional annotations revealed that WTAP rs7766006 and YTHDF2 rs3738067 could be potential functional variants by increasing expression of WTAP and YTHDF2 mRNA, respectively. Overall, these findings implicate variants in the m6A modification core genes as playing a role in pediatric glioma etiology.
Keywords: glioma, m6A, polymorphism, susceptibility, Chinese
Graphical Abstract
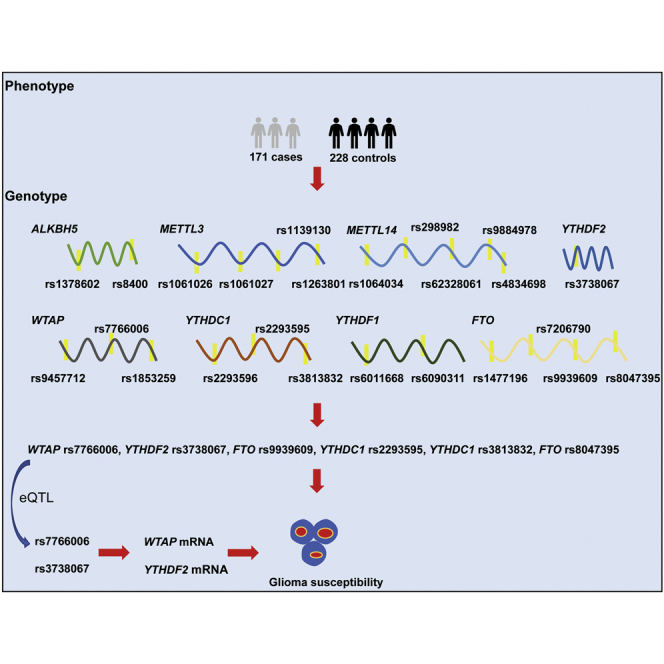
The role of m6A modification core gene polymorphisms in glioma remains to be elucidated. He and colleagues showed that m6A gene polymorphisms are associated with glioma risk, possibly by increasing their own mRNA expression. Their work highlights the critical roles of m6A modification gene polymorphisms in the etiology of glioma.
Introduction
Brain tumors are characterized by high incidence and mortality owing to their notoriously invasive nature.1 Glioma is the most prevalent primary malignant brain tumor and accounts for almost 30% of all primary brain tumors.2,3 Gliomas are mainly derived from neuroglial stem or progenitor cells.4,5 The majority of pediatric gliomas are benign and thus classified as grade I or II by the WHO classification. These pediatric low-grade gliomas (LGGs) rarely undergo malignant transformation and present favorable prognosis under current treatment strategies. However, a significant portion of gliomas are malignant and progress rapidly and are therefore classified as grade III or IV.6 Despite all therapeutic efforts, patients with high-grade gliomas (HGGs) retain a limited prognosis, with the most aggressive forms being lethal within months.7
Many environmental factors have been explored, yet only one definite factor (ionizing radiation) is recognized as a causative agent.8, 9, 10 Hereditary factors, lifestyle, and diet are suggested to confer risk of glioma, but causal relationships should be solidified.11 Evidence for a genetic component to glioma risk has been growing. Several genome-wide association studies (GWASs) have identified a dozen glioma risk-associated single-nucleotide polymorphisms (SNPs), which are located in genes CCDC26, PHLDB1, TP53, EGFR, and CDKN2A-CDKN2B.12, 13, 14, 15 Collectively, however, these variants still account for only a small portion of glioma risk, and additional predisposition loci likely remain to be discovered. Exploration of other causative genetic variation is warranted to better understand the etiology of glioma.
N6-methyladenosine (m6A) is the most distributed mRNA post-transcriptional modification in eukaryotic cells.16 The m6A modification process is accomplished by a series of proteins. According to the different roles of proteins in the RNA methylation process, m6A core proteins are currently mainly divided into writers, erasers, and readers.17 The m6A methyltransferase complexes methyltransferase-like 3 (METTL3), METTL14, and Wilms tumor 1-associated protein (WTAP) act as m6A writers, mainly mediating the m6A methylation of mRNA.18 m6A demethylase FTO and ALKBH5 act as erasers, mainly mediating m6A demethylation of mRNA. A series of m6A binding proteins, YT521-B homology domain family (YTHDF)1/2/3, YTHDC1/2, IGF2BP1/2/3, and eIF3, act as readers and are involved in determining the fate of m6A-modified target mRNA transcripts.19 The effect of abnormal levels of m6A methylated core proteins on cancer progression has also been investigated. Compelling evidence has pointed to the participation of ALKBH5, METTL3, METTL14, WTAP, YTHDC1, YTHDF1, YTHDF2, and FTO in human cancers, including glioma.20, 21, 22, 23, 24, 25, 26, 27 Xi et al.28 found that WTAP expression predicts poor prognosis in malignant glioma patients. Zhang et al.27 revealed that m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program.
Nevertheless, the role of SNPs in m6A modification core genes on glioma risk has been poorly unraveled. Given the evidence that cells modulated by m6A contribute to tumorigenesis, we hypothesize that SNPs of m6A modification core genes may predispose to the risk of glioma. To test this hypothesis, here we conducted a case-control study to investigate the association of SNPs with susceptibility to glioma among children of Chinese ancestry.
Results
Characteristics of the participants
Detailed frequency distributions of demographic and clinical characteristics of glioma cases (n = 171) and cancer-free controls (n = 228) are presented in Table S1. Controls were frequency matched to cases by age (p = 0.623) and gender (p = 0.190). Among these cases, the astrocytic tumors accounted for 125 (73.10%), the ependymoma for 24 (14.62%), the neuronal and mixed neutonal-glial tumors for 14 (8.19%), and embryonal tumors for 7 (4.09%). According to the WHO grades, 103 glioma cases (60.23%) were classified into grade I, 28 (16.37%) into grade II, 15 (8.77%) into grade III, and 25 (14.62%) into grade IV.
Association of m6A modification core genes and glioma risk
Our case-control study successfully genotyped 24 SNPs in the 8 m6A modification core genes. The single-locus analysis was applied to estimate the associations between each selected SNP and glioma risk (Table 1). None of the 24 SNPs violated the Hardy-Weinberg equilibrium (HWE) in control populations (all p values > 0.05). The WTAP rs7766006 (adjusted odds ratio [OR] = 1.58, 95% confidence interval [CI] = 1.04–2.40, p = 0.034) and YTHDF2 rs3738067 (adjusted OR = 1.86, 95% CI = 1.24–2.80, p = 0.003) variants were associated with a statistically significant increased risk of glioma, respectively, in dominant model. YTHDC1 rs2293595 (adjusted OR = 0.59, 95% CI = 0.39–0.90, p = 0.013), YTHDC1 rs3813832 (adjusted OR = 0.61, 95% CI = 0.40–0.91, p = 0.016), and FTO rs8047395 (adjusted OR = 0.63, 95% CI = 0.41–0.95, p = 0.026) were associated with a significant inverse association with risk of glioma, respectively, in dominant model. Under recessive model, only FTO rs9939609 (adjusted OR = 7.39, 95% CI = 1.56–35.11, p = 0.012) was associated with risk of glioma.
Table 1.
Association of m6A modification core genes and glioma risk in Southern Chinese children
| Gene | Polymorphism | Allele | Case (n = 171) | Control (n = 228) | AOR (95% CI)a | pa | AOR (95% CI)b | pb | HWE | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | AA | AB | BB | AA | AB | BB | |||||||
| ALKBH5 | rs1378602 | G | A | 139 | 30 | 2 | 182 | 43 | 2 | 0.97 (0.58–1.63) | 0.920 | 1.27 (0.17–9.21) | 0.815 | 0.757 |
| ALKBH5 | rs8400 | G | A | 51 | 85 | 35 | 71 | 118 | 38 | 1.14 (0.74–1.77) | 0.548 | 1.37 (0.82–2.31) | 0.233 | 0.350 |
| METTL3 | rs1061026 | T | G | 141 | 28 | 2 | 187 | 37 | 4 | 0.98 (0.58–1.65) | 0.935 | 0.66 (0.12–3.74) | 0.641 | 0.185 |
| METTL3 | rs1061027 | C | A | 112 | 55 | 4 | 139 | 79 | 10 | 0.80 (0.53–1.22) | 0.295 | 0.55 (0.17–1.79) | 0.317 | 0.771 |
| METTL3 | rs1139130 | A | G | 62 | 85 | 24 | 91 | 102 | 35 | 1.13 (0.75–1.71) | 0.559 | 0.88 (0.50–1.56) | 0.665 | 0.470 |
| METTL3 | rs1263801 | G | C | 91 | 67 | 13 | 112 | 90 | 26 | 0.83 (0.55–1.23) | 0.349 | 0.66 (0.33–1.33) | 0.241 | 0.230 |
| METTL14 | rs1064034 | T | A | 77 | 83 | 11 | 112 | 97 | 19 | 1.23 (0.82–1.84) | 0.316 | 0.82 (0.38–1.77) | 0.605 | 0.755 |
| METTL14 | rs298982 | G | A | 135 | 36 | 0 | 179 | 46 | 3 | 1.02 (0.63–1.67) | 0.930 | – | – | 0.982 |
| METTL14 | rs62328061 | A | G | 106 | 63 | 2 | 154 | 66 | 8 | 1.29 (0.85–1.97) | 0.232 | 0.33 (0.07–1.59) | 0.168 | 0.778 |
| METTL14 | rs9884978 | G | A | 110 | 53 | 8 | 155 | 62 | 11 | 1.14 (0.75–1.75) | 0.534 | 0.91 (0.35–2.35) | 0.848 | 0.150 |
| METTL14 | rs4834698 | T | C | 45 | 81 | 45 | 51 | 118 | 59 | 0.79 (0.49–1.25) | 0.313 | 1.03 (0.65–1.63) | 0.892 | 0.583 |
| WTAP | rs9457712 | G | A | 116 | 51 | 4 | 148 | 71 | 9 | 0.86 (0.56–1.32) | 0.487 | 0.58 (0.17–1.95) | 0.381 | 0.894 |
| WTAP | rs1853259 | A | G | 57 | 85 | 29 | 62 | 120 | 46 | 0.74 (0.48–1.14) | 0.168 | 0.77 (0.45–1.29) | 0.314 | 0.382 |
| WTAP | rs7766006 | G | T | 57 | 88 | 26 | 97 | 106 | 25 | 1.58 (1.04–2.40)c | 0.034c | 1.46 (0.81–2.65) | 0.211 | 0.620 |
| YTHDC1 | rs2293596 | T | C | 108 | 55 | 8 | 140 | 76 | 12 | 0.91 (0.60–1.38) | 0.668 | 0.84 (0.33–2.12) | 0.708 | 0.689 |
| YTHDC1 | rs2293595 | T | C | 77 | 74 | 20 | 74 | 112 | 42 | 0.59 (0.39–0.90)c | 0.013c | 0.61 (0.34–1.08) | 0.091 | 0.974 |
| YTHDC1 | rs3813832 | T | C | 109 | 53 | 9 | 115 | 96 | 17 | 0.61 (0.40–0.91)c | 0.016c | 0.72 (0.31–1.67) | 0.448 | 0.619 |
| YTHDF1 | rs6011668 | C | T | 132 | 32 | 7 | 168 | 56 | 4 | 0.81 (0.50–1.29) | 0.369 | 2.66 (0.75–9.43) | 0.131 | 0.787 |
| YTHDF1 | rs6090311 | A | G | 61 | 75 | 35 | 92 | 103 | 33 | 1.20 (0.79–1.81) | 0.400 | 1.58 (0.93–2.68) | 0.093 | 0.633 |
| YTHDF2 | rs3738067 | A | G | 83 | 78 | 10 | 144 | 74 | 10 | 1.86 (1.24–2.80)c | 0.003c | 1.31 (0.53–3.25) | 0.560 | 0.900 |
| FTO | rs1477196 | G | A | 105 | 58 | 8 | 127 | 87 | 14 | 0.76 (0.51–1.15) | 0.193 | 0.80 (0.32–1.96) | 0.617 | 0.860 |
| FTO | rs9939609 | T | A | 129 | 33 | 9 | 175 | 51 | 2 | 1.07 (0.67–1.72) | 0.757 | 7.39 (1.56–35.11)c | 0.012c | 0.411 |
| FTO | rs7206790 | C | G | 110 | 53 | 8 | 165 | 59 | 4 | 1.45 (0.94–2.24) | 0.090 | 2.73 (0.80–9.38) | 0.110 | 0.626 |
| FTO | rs8047395 | A | G | 75 | 78 | 18 | 75 | 116 | 37 | 0.63 (0.41–0.95)c | 0.026c | 0.63 (0.34–1.16) | 0.139 | 0.481 |
AOR, adjusted odds ratio; CI, confidence interval; HWE, Hardy-Weinberg equilibrium.
Adjusted for age and gender for dominant model.
Adjusted for age and gender for recessive model.
Significant results.
Stratification analysis
Stratification analysis was further performed for those significant SNPs based on age, gender, subtypes, and tumor grades. For WTAP genotypes (Table 2), rs7766006 GT/TT increased glioma risk in children aged ≥ 60 months (adjusted OR = 2.10, 95% CI = 1.17–3.78, p = 0.013), subtype of embryonal tumors (adjusted OR = 17.00, 95% CI = 1.03–281.79, p = 0.048), and patients with tumors in the tumor grades I+II (adjusted OR = 1.61, 95% CI = 1.02–2.54, p = 0.041). We then treated rs9457712 GG/GA, rs1853259 AA/AG, and rs7766006 GT/TT as risk genotypes. After combining the risk genotypes, we observed that patients with 3 risk genotypes were more likely to develop glioma in children aged ≥60 months (adjusted OR = 2.10, 95% CI = 1.17–3.78, p = 0.013), subtype of embryonal tumors (adjusted OR = 17.00, 95% CI = 1.03–281.79, p = 0.048), and patients with tumors in the tumor grades I+II (adjusted OR = 1.61, 95% CI = 1.02–2.54, p = 0.041).
Table 2.
Stratification analysis between WTAP genotypes and glioma risk
| Variables | rs7766006 (cases/controls) | AOR (95% CI)a | pa | Risk genotypesb (cases/controls) | AOR (95% CI)a | pa | ||
|---|---|---|---|---|---|---|---|---|
| GG | GT/TT | 0–2 | 3 | |||||
| Age, months | ||||||||
| <60 | 27/40 | 58/79 | 1.09 (0.60–1.98) | 0.775 | 27/40 | 58/79 | 1.09 (0.60–1.98) | 0.775 |
| ≥60 | 30/57 | 56/52 | 2.10 (1.17–3.78)c | 0.013c | 30/57 | 56/52 | 2.10 (1.17–3.78)c | 0.013c |
| Gender | ||||||||
| Females | 30/40 | 51/53 | 1.39 (0.75–2.60) | 0.300 | 30/40 | 51/53 | 1.39 (0.75–2.60) | 0.300 |
| Males | 27/57 | 63/78 | 1.76 (1.00–3.12) | 0.052 | 27/57 | 63/78 | 1.76 (1.00–3.12) | 0.052 |
| Subtypes | ||||||||
| Astrocytic tumors | 43/97 | 82/131 | 1.54 (0.96–2.45) | 0.071 | 43/97 | 82/131 | 1.54 (0.96–2.45) | 0.071 |
| Ependymoma | 10/97 | 15/131 | 0.98 (0.42–2.32) | 0.971 | 10/97 | 15/131 | 0.98 (0.42–2.32) | 0.971 |
| Neuronal and mixed | 3/97 | 11/131 | 2.48 (0.66–9.24) | 0.177 | 3/97 | 11/131 | 2.48 (0.66–9.24) | 0.177 |
| Embryonal tumors | 1/97 | 6/131 | 17.00 (1.03–281.79)c | 0.048c | 1/97c | 6/131c | 17.00 (1.03–281.79)c | 0.048c |
| Tumor grades | ||||||||
| I | 31/97 | 72/131 | 1.86 (1.12–3.08) | 0.017 | 31/97 | 72/131 | 1.86 (1.12–3.08) | 0.017 |
| II | 12/97 | 16/131 | 0.98 (0.44–2.19) | 0.964 | 12/97 | 16/131 | 0.98 (0.44–2.19) | 0.964 |
| III | 5/97 | 10/131 | 1.31 (0.43–4.02) | 0.632 | 5/97 | 10/131 | 1.31 (0.43–4.02) | 0.632 |
| IV | 9/97 | 16/131 | 1.71 (0.67–4.33) | 0.259 | 9/97 | 16/131 | 1.71 (0.67–4.33) | 0.259 |
| I+II | 43/97 | 88/131 | 1.61 (1.02–2.54)c | 0.041c | 43/97 | 88/131 | 1.61 (1.02–2.54)c | 0.041c |
| III+IV | 14/97 | 26/131 | 1.56 (0.76–3.22) | 0.226 | 14/97 | 26/131 | 1.56 (0.76–3.22) | 0.226 |
AOR, adjusted odds ratio; CI, confidence interval.
Adjusted for age and gender, omitting the corresponding stratify factor.
Risk genotypes were carriers with rs9457712 GG/GA, rs1853259 AA/AG, and rs7766006 GT/TT genotypes.
Significant results.
For YTHDC1 genotypes (Table 3), rs2293595 TC/CC was associated with decreased glioma risk in children aged ≥60 months, females, subtype of astrocytic tumors, patients with tumors in tumor grade I, patients with tumors in tumor grade II, and patients with tumors in tumor grade I+II. rs3813832 TC/CC was associated with decreased glioma risk in children aged ≥ 60 months, females, subtype of astrocytic tumors, patients with tumors in tumor grade II, and patients with tumors in tumor grade I+II. rs2293596 TC/CC, rs2293595 TC/CC, and rs3813832 TC/CC were further referred to as protective genotypes. Compared to 0–1 protective genotypes, those with 2–3 protective genotypes were less likely to develop glioma in children aged ≥60 months, females, subtype of astrocytic tumors, patients with tumors in tumor grade I, patients with tumors in tumor grade II, and patients with tumors in tumor grade I+II.
Table 3.
Stratification analysis between YTHDC1 genotypes and glioma risk
| Variables | rs2293595 | AOR (95% CI)a | pa | rs3813832 | AOR (95% CI)a | pa | Protective genotypesb | AOR (95% CI)a | pa | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (cases/controls) | (cases/controls) | (cases/controls) | ||||||||||
| TT | TC/CC | TT | TC/CC | 0–1 | 2–3 | |||||||
| Age, months | ||||||||||||
| <60 | 34/40 | 51/79 | 0.76 (0.43–1.35) | 0.348 | 51/58 | 34/61 | 0.63 (0.36–1.11) | 0.111 | 45/53 | 40/66 | 0.71 (0.4–1.24) | 0.229 |
| ≥60 | 43/34 | 43/75 | 0.46 (0.25–0.82)c | 0.009c | 58/57 | 28/52 | 0.55 (0.30–0.99)c | 0.044c | 55/48 | 31/61 | 0.46 (0.26–0.82)c | 0.008c |
| Gender | ||||||||||||
| Females | 40/27 | 41/66 | 0.42 (0.22–0.80)c | 0.008c | 56/46 | 25/47 | 0.47 (0.25–0.88)c | 0.018c | 52/39 | 29/54 | 0.44 (0.24–0.82)c | 0.010c |
| Males | 37/47 | 53/88 | 0.77 (0.44–1.34) | 0.358 | 53/69 | 37/66 | 0.74 (0.43–1.27) | 0.275 | 48/62 | 42/73 | 0.74 (0.43–1.27) | 0.271 |
| Subtypes | ||||||||||||
| Astrocytic tumors | 59/74 | 66/154 | 0.54 (0.34–0.86)c | 0.009c | 83/115 | 42/113 | 0.54 (0.34–0.86)c | 0.009c | 76/101 | 49/127 | 0.53 (0.34–0.84)c | 0.007c |
| Ependymoma | 10/74 | 15/154 | 0.76 (0.32–1.78) | 0.520 | 14/115 | 11/113 | 0.77 (0.33–1.79) | 0.546 | 12/101 | 13/127 | 0.87 (0.38–2.01) | 0.744 |
| Neuronal and mixed | 6/74 | 8/154 | 0.68 (0.22–2.04) | 0.489 | 7/115 | 7/113 | 1.01 (0.34–3.00) | 0.984 | 7/101 | 7/127 | 0.83 (0.28–2.46) | 0.732 |
| Embryonal tumors | 2/74 | 5/154 | 0.80 (0.13–4.84) | 0.806 | 5/115 | 2/113 | 0.48 (0.08–2.85) | 0.415 | 5/101 | 2/127 | 0.36 (0.06–2.15) | 0.264 |
| Tumor grades | ||||||||||||
| I | 46/74 | 57/154 | 0.60 (0.37–0.97)c | 0.037c | 63/115 | 40/113 | 0.68 (0.42–1.10) | 0.112 | 59/101 | 44/127 | 0.61 (0.38–0.98)c | 0.042c |
| II | 15/74 | 13/154 | 0.42 (0.19–0.92)c | 0.031c | 21/115 | 7/113 | 0.34 (0.14–0.83)c | 0.017c | 19/101 | 9/127 | 0.38 (0.16–0.87)c | 0.022c |
| III | 6/74 | 9/154 | 0.74 (0.25–2.17) | 0.577 | 8/115 | 7/113 | 0.86 (0.30–2.47) | 0.777 | 7/101 | 8/127 | 0.91 (0.32–2.62) | 0.859 |
| IV | 10/74 | 15/154 | 0.72 (0.29–1.78) | 0.479 | 17/115 | 8/113 | 0.58 (0.23–1.47) | 0.250 | 15/101 | 10/127 | 0.66 (0.27–1.61) | 0.355 |
| I+II | 61/74 | 70/154 | 0.55 (0.35–0.86)c | 0.009c | 84/115 | 47/113 | 0.59 (0.38–0.92)c | 0.019c | 78/101 | 53/127 | 0.55 (0.36–0.85)c | 0.008c |
| III+IV | 16/74 | 24/154 | 0.73 (0.36–1.47) | 0.377 | 25/115 | 15/113 | 0.67 (0.33–1.36) | 0.265 | 22/101 | 18/127 | 0.72 (0.36–1.43) | 0.342 |
AOR, adjusted odds ratio; CI, confidence interval.
Adjusted for age and gender, omitting the corresponding stratify factor.
Protective genotypes were carriers with rs2293596 TC/CC, rs2293595 TC/CC, and rs3813832 TC/CC genotypes.
Significant results.
For YTHDF2 rs3738067 A > G polymorphism (Table 4), AG/GG was associated with increased glioma risk in children aged <60 months, children aged ≥60 months, females, subtype of astrocytic tumors, patients with tumors in tumor grade I, patients with tumors in tumor grade IV, patients with tumors in tumor grade I+II, and patients with tumors in tumor grade III+IV.
Table 4.
Stratification analysis between YTHDF2 rs3738067 A > G polymorphism and glioma risk
| Variables | rs3738067 (cases/controls) | Crude OR | p | AOR (95% CI)a | pa | |
|---|---|---|---|---|---|---|
| AA | AG/GG | (95% CI) | ||||
| Age, months | ||||||
| <60 | 39/73 | 46/46 | 1.87 (1.07–3.29)b | 0.029b | 1.87 (1.06–3.29)b | 0.030b |
| ≥60 | 44/71 | 42/38 | 1.78 (1.00–3.18) | 0.050 | 1.81 (1.01–3.23)b | 0.046b |
| Gender | ||||||
| Females | 38/61 | 43/32 | 2.16 (1.17–3.97)b | 0.014b | 2.27 (1.22–4.25)b | 0.010b |
| Males | 45/83 | 45/52 | 1.60 (0.93–2.74) | 0.089 | 1.61 (0.93–2.77) | 0.087 |
| Subtypes | ||||||
| Astrocytic tumors | 61/144 | 64/84 | 1.80 (1.16–2.80)b | 0.009b | 1.90 (1.20–2.99)b | 0.006b |
| Ependymoma | 13/144 | 12/84 | 1.58 (0.69–3.63) | 0.278 | 1.58 (0.68–3.64) | 0.287 |
| Neuronal and mixed | 7/144 | 7/84 | 1.71 (0.58–5.06) | 0.329 | 1.63 (0.55–4.84) | 0.381 |
| Embryonal tumors | 2/144 | 5/84 | 4.29 (0.81–22.58) | 0.086 | 6.82 (1.00–46.76) | 0.051 |
| Tumor grades | ||||||
| I | 52/144 | 51/84 | 1.68 (1.05–2.69)b | 0.031b | 1.78 (1.10–2.89)b | 0.018b |
| II | 14/144 | 14/84 | 1.71 (0.78–3.77) | 0.180 | 1.71 (0.78–3.77) | 0.182 |
| III | 8/144 | 7/84 | 1.50 (0.53–4.29) | 0.449 | 1.47 (0.51–4.23) | 0.475 |
| IV | 9/144 | 16/84 | 3.05 (1.29–7.20)b | 0.011b | 3.08 (1.23–7.71)b | 0.016b |
| I+II | 66/144 | 65/84 | 1.69 (1.09–2.61)b | 0.018b | 1.77 (1.14–2.75)b | 0.012b |
| III+IV | 17/144 | 23/84 | 2.32 (1.17–4.59)b | 0.016b | 2.29 (1.14–4.60)b | 0.019b |
AOR, adjusted odds ratio; CI, confidence interval.
Adjusted for age and gender, omitting the corresponding stratify factor.
Significant results.
For FTO genotypes (Table 5), rs8047395 AG/GG was associated with decreased glioma risk in subtype of astrocytic tumors, patients with tumors in tumor grade I, and patients with tumors in tumor grade I+II. rs1477196 GA/AA, rs9939609 TT/TA, rs7206790 CC/CG, and rs8047395 GT/TT were further referred to as protective genotypes. Compared to 0–2 protective genotypes, those with 3–4 protective genotypes were less likely to develop glioma in patients with tumors in tumor grade I and patients with tumors in tumor grade I+II.
Table 5.
Stratification analysis between FTO genotypes and glioma risk
| Variables | rs8047395 (cases/controls) | AOR (95% CI)a | pa | Protective genotypesb (cases/controls) | AOR (95% CI)a | pa | ||
|---|---|---|---|---|---|---|---|---|
| AA | AG/GG | 0–2 | 3–4 | |||||
| Age, months | ||||||||
| <60 | 39/41 | 46/78 | 0.62 (0.35–1.09) | 0.098 | 38/41 | 47/78 | 0.65 (0.37–1.15) | 0.138 |
| ≥60 | 36/34 | 50/75 | 0.64 (0.35–1.15) | 0.133 | 35/33 | 51/76 | 0.64 (0.35–1.16) | 0.139 |
| Gender | ||||||||
| Females | 36/30 | 45/63 | 0.61 (0.32–1.13) | 0.116 | 35/30 | 46/63 | 0.64 (0.34–1.19) | 0.159 |
| Males | 39/45 | 51/90 | 0.65 (0.37–1.12) | 0.120 | 38/44 | 52/91 | 0.65 (0.37–1.13) | 0.128 |
| Subtypes | ||||||||
| Astrocytic tumors | 55/75 | 70/153 | 0.62 (0.39–0.98)c | 0.039c | 53/74 | 72/154 | 0.64 (0.41–1.02) | 0.060 |
| Ependymoma | 12/75 | 13/153 | 0.53 (0.23–1.23) | 0.141 | 12/74 | 13/154 | 0.53 (0.23–1.22) | 0.137 |
| Neuronal and mixed | 6/75 | 8/153 | 0.66 (0.22–1.99) | 0.459 | 6/74 | 8/154 | 0.65 (0.22–1.86) | 0.444 |
| Embryonal tumors | 2/75 | 5/153 | 1.31 (0.22–7.85) | 0.767 | 2/74 | 5/154 | 1.23 (0.21–7.26) | 0.821 |
| Tumor grades | ||||||||
| I | 48/75 | 55/153 | 0.58 (0.36–0.93)c | 0.025c | 47/74 | 56/154 | 0.58 (0.36–0.95)c | 0.029c |
| II | 11/75 | 17/153 | 0.76 (0.34–1.70) | 0.504 | 11/74 | 17/154 | 0.74 (0.33–1.67) | 0.473 |
| III | 8/75 | 7/153 | 0.44 (0.15–1.26) | 0.124 | 8/74 | 7/154 | 0.43 (0.15–1.25) | 0.120 |
| IV | 8/75 | 17/153 | 1.07 (0.42–2.73) | 0.892 | 7/74 | 18/154 | 1.27 (0.48–3.34) | 0.635 |
| I+II | 59/75 | 72/153 | 0.60 (0.39–0.94)c | 0.026c | 58/74 | 73/154 | 0.61 (0.39–0.95)c | 0.028c |
| III+IV | 16/75 | 24/153 | 0.72 (0.36–1.45) | 0.355 | 15/74 | 25/154 | 0.78 (0.38–1.58) | 0.489 |
AOR, adjusted odds ratio; CI, confidence interval.
Adjusted for age and gender, omitting the corresponding stratify factor.
Protective genotypes were carriers with rs1477196 GA/AA, rs9939609 TT/TA, rs7206790 CC/CG, and rs8047395 GT/TT genotypes.
Significant results.
Expression quantitative trait loci (eQTL) analyses
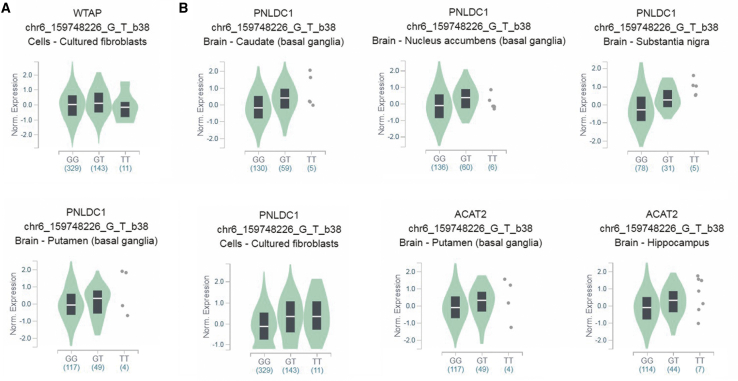
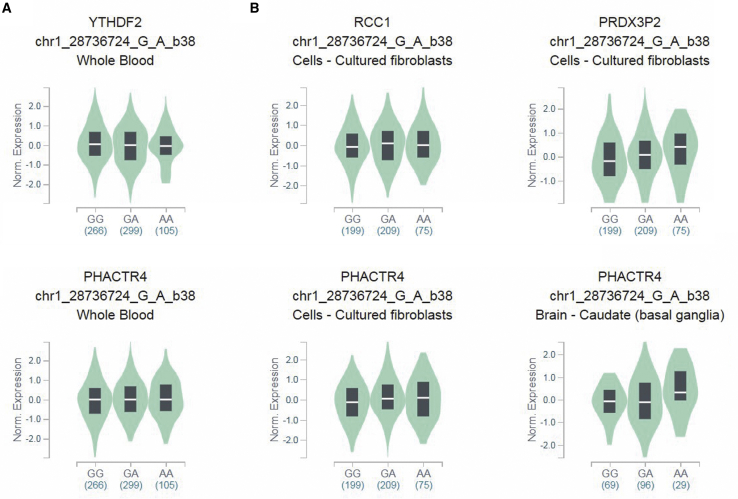
We further assessed the putative functional relevance of WTAP rs7766006 and YTHDF2 rs3738067 using released data from GTEx. Samples with rs7766006 T genotype had significantly higher WTAP mRNA levels in the cell-cultured fibroblasts than samples with rs7766006 G genotype (Figure 1A). We also found that rs7766006 T genotype confers to higher mRNA level of neighboring genes, including PNLDC1 and ACAT2 (Figure 1B). Samples with rs3738067 A genotype had significantly higher YTHDF2 mRNA levels in the whole blood than samples with rs3738067 G genotype (Figure 2A). Our cis-eQTL analysis also detected an association between rs3738067 A and increased expression of genes PHACTR4, RCC1, and PRDX3P2 (Figure 2B).
Figure 1.
Functional implication of WTAP gene rs7766006 polymorphism based on the public database GTEx portal
(A) The genotype of rs7766006 and expression of WTAP gene in cell-cultured fibroblasts. (B) The genotype of rs7766006 and expression of its neighboring genes PNLDC1 and ACAT2 in different tissues.
Figure 2.
Functional implication of YTHDF2 gene rs3738067 polymorphism based on the public database GTEx portal
(A) The genotype of rs3738067 and expression of YTHDF2 gene in whole blood. (B) The genotype of rs3738067 and expression of its neighboring genes PHACTR4, RCC1, and PRDX3P2 in different tissues.
Functional annotation of WTAP and YTHDF2
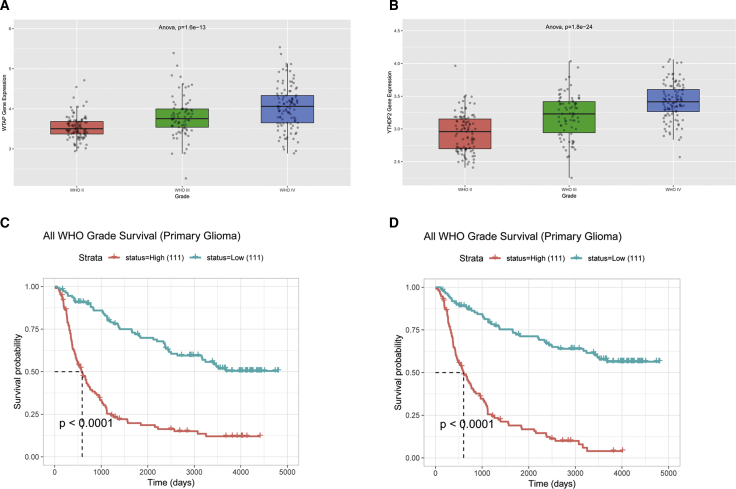
Functional annotation of WTAP and YTHDF2 expression was determined in public data deposited in the Chinese Glioma Genome Atlas (CGGA) database (mRNaseq_325). WTAP (Figure 3A) and YTHDF2 (Figure 3B) expression were significantly higher in grade III and IV samples than in grade II samples. We also performed survival analysis to investigate the clinical relevance of WTAP and YTHDF2 expression in patient survival. The results demonstrated that elevated WTAP (Figure 3C) and YTHDF2 (Figure 3D) expression was clinically correlated with unfavorable outcomes of glioma patients.
Figure 3.
Correlation of WTAP and YTHDF2 with progression and overall survival of glioma based on the CGGA database
(A) WTAP expression increased in grades III and IV compared to grade II. (B) YTHDF2 expression increased in grades III and IV compared to grade II. (C) Correlation between WTAP expression and the survival of glioma patients. (D) Correlation between YTHDF2 expression and the survival of glioma patients.
Discussion
Emerging evidence has been growing regarding the significant contributions of m6A modification core genes to the initiation and development of various cancers. However, no reports have been found in analyzing the impact of these critical gene SNPs on risk of glioma. Discovering genetic variants that distinguish glioma is of critical clinical importance for disease prevention and treatment. In this analysis of Asian children from China, we first identified several glioma risk variants: WTAP rs7766006, YTHDF2 rs3738067, FTO rs9939609, YTHDC1 rs2293595, YTHDC1 rs3813832, and FTO rs8047395.
Regarding the epidemiology assessment of m6A modification core gene SNPs on cancer, only two available studies have been conducted. In 2019, Meng et al.29 performed a first case-control study on m6A modification core gene SNPs and colorectal cancer risk. Their study applied a two-stage design: discovery stage (1,150 cases and 1,342 controls) and replication stage (932 cases and 966 controls). A total of 240 SNPs in 20 m6A modification-related genes were genotyped. Surprisingly, only one variant, SND1 gene rs118049207, was associated with colorectal cancer risk. SND1 gene rs118049207 might impact colorectal cancer risk by changing the mRNA expression of the SND1 gene and then lead to alteration of m6A level. More recently, the group of Yang et al.30 performed another epidemiology study regarding m6A genes SNPs and cancer risk. They carried out the first study to examine the association between genetic variants in m6A modification genes and esophageal squamous cell carcinoma (ESCC) risk. They observed that one potentially functional SNP located upstream of YTHDC2, rs2416282, predisposes to ESCC risk in Chinese population by altering the expression of YTHDC2. Given the important role of m6A modification of core gene SNPs in cancer, we aim to investigate the association between m6A modification core gene SNPs and the risk of glioma.
Our study found that WTAP rs7766006, YTHDF2 rs3738067, and FTO rs9939609 variants could contribute to glioma risk, respectively. However, we detected a significant inverse association between YTHDC1 rs2293595, YTHDC1 rs3813832, FTO rs8047395, and susceptibility to glioma, respectively. We then proceed to determine the possible mechanisms for the conferring risk role of WTAP rs7766006 and YTHDF2 rs3738067. The eQTL results indicated that the increased glioma risk was linked to the upregulated expression levels of the WTAP and the YTHDF2 gene. WTAP and its partner Wilms tumor 1 (WT1) protein are present together throughout the nucleoplasm as well as in nuclear speckles and partially colocalize with splicing factors.31 WTAP is ubiquitously expressed in diverse tissues, rather than the tissue-specific expression pattern of WT1.31 Substantial evidence supports the implication of WTAP in various cellular processes, including m6A methylation modification,18 alternative splicing,32 X chromosome inactivation,33 and cell cycle regulation.34 Moreover, WTAP has also been reported to be extensively involved in several cancers. WTAP can be treated as a marker for predicting poor prognosis in malignant glioma patients.28 Jin et al.22 suggested that WTAP may play an oncogenic role in glioma. This research provides further support for our result that WTAP rs7766006 contributed to increased glioma risk by upregulating expression levels of the WTAP gene. The YTHDF proteins are cytoplasmic m6A readers that specifically recognize and bind to m6A within the consensus RR(m6A)CH sequence.35 Human YTHDF proteins include three members, YTHDF1–3, each of which comprises a highly conserved single-stranded RNA-binding domain located at the carboxy terminus (the YTH domain) and a less conserved amino-terminal region.36 YTHDF2 specifically recognizes m6A by its aromatic cage. Chen et al.37 showed that METTL3 promotes liver cancer progression through YTHDF2-dependent post-transcriptional silencing of SOCS2. Recently, Chai and his colleagues38 carried out an observational study to examine if m6A modification genes can be used in a purely prognostic perspective for glioma. They reported a direct correlation between the expression of WTAP, YTHDF2, and the WHO grade of glioma, respectively. In other words, WTAP and YTHDF2 act as “risky” genes in glioma. These data combined with our results shed light on the biological mechanisms of how WTAP rs7766006 and YTHDF2 rs3738067 function to enhance glioma risk. We also applied CGGA data to explore whether these genes were associated with glioma progression and prognosis. The result showed that higher expression of WTAP and YTHDF2 were positively correlated to glioma progression and unfavorable overall survival. Taken together, these results suggest the potential value of WTAP and YTHDF2 as markers in the outcome prediction of glioma patients. Of note, specific data regarding the role of WTAP and YTHDF2 in pediatric glioma remains to be explored.
Our study has several limitations. First, while this sample does represent the largest dataset of genotyped Chinese glioma cases, small numbers of subjects in some of the subgroup analysis may have limited the ability to detect associations with certain SNPs. Expansion of sample size is necessary to confirm the associations detected in this analysis, which necessitates additional multicenter collaborations. Second, all the participants were Han Chinese descents. Conclusion obtained from the single population here might not be generalized to overall ethnic groups. Third, environmental factors and environmental-genetic interactions contributed to the glioma, yet these factors were not interrogated in the risk models. Of note, plenty of critical genetic events such as H3K27M mutation, Isocitrate dehydrogenase (IDH) mutation, and v-raf murine sarcoma viral oncogene homolog B1 (BRAF) fusions play an important role in glioma development.39 However, the impact of these critical genetic events on the risk of glioma remains to be elucidated. Therefore, many other additional variables that likely to impact the glioma phenotype should be considered. Thus, the significant SNPs obtained herein all require additional validation, particularly involving environmental factors and environmental-genetic interaction analysis.
This is the first epidemiology study to assess common variants within m6A modification core genes in relation to different glioma subgroups. In conclusion, we have identified common variants within m6A modification core genes that are associated with glioma risk. Further functional study of m6A modification core gene SNPs in glioma is warranted and may lead to unearthing the genetic and epigenetic mechanisms underlying this disease.
Materials and methods
Study subjects
In brief, we conducted a hospital-based case-control study in China. 171 children with primary glioma and 228 children who were free of glioma were enrolled. Epidemiological data were collected using structured questionnaires. The inclusion criterion for case subjects was biopsy confirmed or histologically verified glioma. Control subjects, who were recruited concurrently with case subjects, were randomly selected from the volunteers visiting the hospital and matched according to the expected age and gender distribution of cases. All participants gave written informed consent to use their samples for research purposes. The institutional review board of Guangzhou Women and Children’s Medical Center approved the current study. A more detailed relevant sample selection could be found in our previous work.40
SNP selection and genotyping
Eight m6A modification core genes were contained: Wilms tumor 1-associated protein (WTAP), methyltransferase like 3 (METTL3), methyltransferase like 14 (METTL14), alpha-ketoglutarate dependent dioxygenase (FTO), alkB homolog 5 (ALKBH5), YTH m6A RNA binding protein 1 (YTHDF1), YTH m6A RNA binding protein 2 (YTHDF2), and YTH domain containing 1 (YTHDC1). The potentially functional SNPs were selected by using the NCBI dbSNP database and SNPinfo (https://snpinfo.niehs.nih.gov/). SNPs that fulfilled the following selection criteria were chosen: (1) the minor allele frequency (MAF) reported in HapMap was > 5% for Chinese Han subjects; (2) SNPs located in the 5′ flanking region, exon, 5′ untranslated region (5′ UTR), and 3′ UTR, which might affect transcription activity or binding capacity of the microRNA binding site; and (3) SNPs in low linkage disequilibrium with each other (R2 < 0.8). A total of 24 SNPs were selected. Specifically, 2 ALKBH5, 4 METTL3, 5 METTL14, 3 WTAP, 3 YTHDC1, 2 YTHDF1, 1 YTHDF2, and 4 FTO SNPs were genotyped. Genomic DNA was prepared from peripheral blood samples using a QIAamp DNA blood mini kit (QIAGEN, Valencia, CA, USA). The TaqMan genotyping for the SNP was performed on an ABI 7900 (Applied Biosystems, Foster City, CA, USA). All case/control status was carried out blind to the laboratory personnel. Genotyping of the proposed SNPs was all performed in the laboratory of Guangzhou. The conditions of reactions were set as follows: pre-read stage at 60°C for 30 s, holding stage at 95°C for 10 min, repeated 45 cycles each of denaturation at 95°C for 15 s, annealing and extension at 60°C for 1 min. 10% duplicate test samples and water samples (negative controls) were included in each 96-well plate. A 100% concordance rate of the duplicated samples was achieved. Detailed information about SNPs selection and genotyping have been described previously.41, 42, 43
Statistical analysis
The compliance of genotypes with HWE among controls was appraised by a χ2 test. Differences in demographic characteristics between the cases and the controls were evaluated using χ2 test or t test as appropriate. The age- and gender-adjusted ORs and 95% CIs for the relationships between the SNPs and glioma risk were determined by multivariate logistic regression analysis. We also conducted stratified analyses by reclassifying cases with different subgroups, including age, gender, subtypes, and tumor grade. Further functional annotation of the significant SNPs was performed using the Genotype-Tissue Expression (GTEx) (https://gtexportal.org).44 Gene expression and glioma patient survival data were obtained from the CGGA database http://www.cgga.org.cn/. All tests for statistical significance used a two-sided alpha of 0.05. All statistical analyses were conducted using the SAS v10.0 (SAS Institute, Cary, NC, USA).
Acknowledgments
This study was supported by grants from the Natural Science Foundation of Guangdong Province (nos. 2019A1515010360 and 2020A1515010188); the National Natural Science Foundation of China (nos. 81802346 and 81672496); the Science and Technology Project of Guangzhou (no. 201804010042); Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease (no. 2019B030301004); and the China Postdoctoral Science Foundation (nos. 2020T130132 and 2020M682668).
Author contributions
J.H., L.Y., and H.L. designed and performed the study and wrote the manuscript; A. Lin, H.C., A. Luo, and Z.Z. collected the samples and information; J.H. and L.Y. participated in analyzing data; J.H., Z.Z., and X.L. coordinated the study over the entire time. All authors reviewed the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2020.12.013.
Contributor Information
Zhenjian Zhuo, Email: zhenjianzhuo@163.com.
Xiaoping Liu, Email: liu_xiaoping@gwcmc.org.
Supplemental Information
References
- 1.Udaka Y.T., Packer R.J. Pediatric Brain Tumors. Neurol. Clin. 2018;36:533–556. doi: 10.1016/j.ncl.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 2.Ostrom Q.T., Gittleman H., Liao P., Rouse C., Chen Y., Dowling J., Wolinsky Y., Kruchko C., Barnholtz-Sloan J. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007-2011. Neuro-oncol. 2014;16(Suppl 4):iv1–iv63. doi: 10.1093/neuonc/nou223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neglia J.P., Robison L.L., Stovall M., Liu Y., Packer R.J., Hammond S., Yasui Y., Kasper C.E., Mertens A.C., Donaldson S.S. New primary neoplasms of the central nervous system in survivors of childhood cancer: a report from the Childhood Cancer Survivor Study. J. Natl. Cancer Inst. 2006;98:1528–1537. doi: 10.1093/jnci/djj411. [DOI] [PubMed] [Google Scholar]
- 4.Weller M., Wick W., Aldape K., Brada M., Berger M., Pfister S.M., Nishikawa R., Rosenthal M., Wen P.Y., Stupp R., Reifenberger G. Glioma. Nat. Rev. Dis. Primers. 2015;1:15017. doi: 10.1038/nrdp.2015.17. [DOI] [PubMed] [Google Scholar]
- 5.Jones D.T.W., Kieran M.W., Bouffet E., Alexandrescu S., Bandopadhayay P., Bornhorst M., Ellison D., Fangusaro J., Fisher M.J., Foreman N. Pediatric low-grade gliomas: next biologically driven steps. Neuro-oncol. 2018;20:160–173. doi: 10.1093/neuonc/nox141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 7.Tang J.H., Yang L., Chen J.X., Li Q.R., Zhu L.R., Xu Q.F., Huang G.H., Zhang Z.X., Xiang Y., Du L. Bortezomib inhibits growth and sensitizes glioma to temozolomide (TMZ) via down-regulating the FOXM1-Survivin axis. Cancer Commun. (Lond.) 2019;39:81. doi: 10.1186/s40880-019-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bondy M.L., Scheurer M.E., Malmer B., Barnholtz-Sloan J.S., Davis F.G., Il’yasova D., Kruchko C., McCarthy B.J., Rajaraman P., Schwartzbaum J.A., Brain Tumor Epidemiology Consortium Brain tumor epidemiology: consensus from the Brain Tumor Epidemiology Consortium. Cancer. 2008;113(7, Suppl):1953–1968. doi: 10.1002/cncr.23741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vienne-Jumeau A., Tafani C., Ricard D. Environmental risk factors of primary brain tumors: A review. Rev. Neurol. (Paris) 2019;175:664–678. doi: 10.1016/j.neurol.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Cordier D., Gerber M., Brand S. Effects of two types of exercise training on psychological well-being, sleep, quality of life and physical fitness in patients with high-grade glioma (WHO III and IV): study protocol for a randomized controlled trial. Cancer Commun. (Lond.) 2019;39:46. doi: 10.1186/s40880-019-0390-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis M.E. Epidemiology and Overview of Gliomas. Semin. Oncol. Nurs. 2018;34:420–429. doi: 10.1016/j.soncn.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Wrensch M., Jenkins R.B., Chang J.S., Yeh R.F., Xiao Y., Decker P.A., Ballman K.V., Berger M., Buckner J.C., Chang S. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat. Genet. 2009;41:905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shete S., Hosking F.J., Robertson L.B., Dobbins S.E., Sanson M., Malmer B., Simon M., Marie Y., Boisselier B., Delattre J.Y. Genome-wide association study identifies five susceptibility loci for glioma. Nat. Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melin B.S., Barnholtz-Sloan J.S., Wrensch M.R., Johansen C., Il’yasova D., Kinnersley B., Ostrom Q.T., Labreche K., Chen Y., Armstrong G., GliomaScan Consortium Genome-wide association study of glioma subtypes identifies specific differences in genetic susceptibility to glioblastoma and non-glioblastoma tumors. Nat. Genet. 2017;49:789–794. doi: 10.1038/ng.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kinnersley B., Labussière M., Holroyd A., Di Stefano A.L., Broderick P., Vijayakrishnan J., Mokhtari K., Delattre J.Y., Gousias K., Schramm J. Genome-wide association study identifies multiple susceptibility loci for glioma. Nat. Commun. 2015;6:8559. doi: 10.1038/ncomms9559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balacco D.L., Soller M. The m6A Writer: Rise of a Machine for Growing Tasks. Biochemistry. 2019;58:363–378. doi: 10.1021/acs.biochem.8b01166. [DOI] [PubMed] [Google Scholar]
- 17.Chen X.Y., Zhang J., Zhu J.S. The role of m6A RNA methylation in human cancer. Mol. Cancer. 2019;18:103. doi: 10.1186/s12943-019-1033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ping X.L., Sun B.F., Wang L., Xiao W., Yang X., Wang W.J., Adhikari S., Shi Y., Lv Y., Chen Y.S. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan Q., Liu P.Y., Haase J., Bell J.L., Hüttelmaier S., Liu T. The Critical Role of RNA m6A Methylation in Cancer. Cancer Res. 2019;79:1285–1292. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 20.Cui Q., Shi H., Ye P., Li L., Qu Q., Sun G., Sun G., Lu Z., Huang Y., Yang C.G. m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 2017;18:2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galardi S., Michienzi A., Ciafrè S.A. Insights into the Regulatory Role of m6A Epitranscriptome in Glioblastoma. Int. J. Mol. Sci. 2020;21:2816. doi: 10.3390/ijms21082816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin D.I., Lee S.W., Han M.E., Kim H.J., Seo S.A., Hur G.Y., Jung S., Kim B.S., Oh S.O. Expression and roles of Wilms’ tumor 1-associating protein in glioblastoma. Cancer Sci. 2012;103:2102–2109. doi: 10.1111/cas.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F., Yi Y., Miao Y., Long W., Long T., Chen S., Cheng W., Zou C., Zheng Y., Wu X. N6-Methyladenosine Modulates Nonsense-Mediated mRNA Decay in Human Glioblastoma. Cancer Res. 2019;79:5785–5798. doi: 10.1158/0008-5472.CAN-18-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li F., Zhang C., Zhang G. m6A RNA Methylation Controls Proliferation of Human Glioma Cells by Influencing Cell Apoptosis. Cytogenet. Genome Res. 2019;159:119–125. doi: 10.1159/000499062. [DOI] [PubMed] [Google Scholar]
- 25.Visvanathan A., Patil V., Arora A., Hegde A.S., Arivazhagan A., Santosh V., Somasundaram K. Essential role of METTL3-mediated m6A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37:522–533. doi: 10.1038/onc.2017.351. [DOI] [PubMed] [Google Scholar]
- 26.Xu C., Yuan B., He T., Ding B., Li S. Prognostic values of YTHDF1 regulated negatively by mir-3436 in Glioma. J. Cell. Mol. Med. 2020;24:7538–7549. doi: 10.1111/jcmm.15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang S., Zhao B.S., Zhou A., Lin K., Zheng S., Lu Z., Chen Y., Sulman E.P., Xie K., Bögler O. m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell. 2017;31:591–606.e6. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xi Z., Xue Y., Zheng J., Liu X., Ma J., Liu Y. WTAP Expression Predicts Poor Prognosis in Malignant Glioma Patients. J. Mol. Neurosci. 2016;60:131–136. doi: 10.1007/s12031-016-0788-6. [DOI] [PubMed] [Google Scholar]
- 29.Meng Y., Li S., Gu D., Xu K., Du M., Zhu L., Chu H., Zhang Z., Wu Y., Fu Z., Wang M. Genetic variants in m6A modification genes are associated with colorectal cancer risk. Carcinogenesis. 2020;41:8–17. doi: 10.1093/carcin/bgz165. [DOI] [PubMed] [Google Scholar]
- 30.Yang N., Ying P., Tian J., Wang X., Mei S., Zou D., Peng X., Gong Y., Yang Y., Zhu Y. Genetic variants in m6A modification genes are associated with esophageal squamous-cell carcinoma in the Chinese population. Carcinogenesis. 2020;41:761–768. doi: 10.1093/carcin/bgaa012. [DOI] [PubMed] [Google Scholar]
- 31.Little N.A., Hastie N.D., Davies R.C. Identification of WTAP, a novel Wilms’ tumour 1-associating protein. Hum. Mol. Genet. 2000;9:2231–2239. doi: 10.1093/oxfordjournals.hmg.a018914. [DOI] [PubMed] [Google Scholar]
- 32.Haussmann I.U., Bodi Z., Sanchez-Moran E., Mongan N.P., Archer N., Fray R.G., Soller M. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540:301–304. doi: 10.1038/nature20577. [DOI] [PubMed] [Google Scholar]
- 33.Moindrot B., Cerase A., Coker H., Masui O., Grijzenhout A., Pintacuda G., Schermelleh L., Nesterova T.B., Brockdorff N. A Pooled shRNA Screen Identifies Rbm15, Spen, and Wtap as Factors Required for Xist RNA-Mediated Silencing. Cell Rep. 2015;12:562–572. doi: 10.1016/j.celrep.2015.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horiuchi K., Umetani M., Minami T., Okayama H., Takada S., Yamamoto M., Aburatani H., Reid P.C., Housman D.E., Hamakubo T., Kodama T. Wilms’ tumor 1-associating protein regulates G2/M transition through stabilization of cyclin A2 mRNA. Proc. Natl. Acad. Sci. USA. 2006;103:17278–17283. doi: 10.1073/pnas.0608357103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z., Theler D., Kaminska K.H., Hiller M., de la Grange P., Pudimat R., Rafalska I., Heinrich B., Bujnicki J.M., Allain F.H., Stamm S. The YTH domain is a novel RNA binding domain. J. Biol. Chem. 2010;285:14701–14710. doi: 10.1074/jbc.M110.104711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen M., Wei L., Law C.T., Tsang F.H., Shen J., Cheng C.L., Tsang L.H., Ho D.W., Chiu D.K., Lee J.M. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 38.Chai R.C., Wu F., Wang Q.X., Zhang S., Zhang K.N., Liu Y.Q., Zhao Z., Jiang T., Wang Y.Z., Kang C.S. m6A RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gliomas. Aging (Albany NY) 2019;11:1204–1225. doi: 10.18632/aging.101829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cahill D., Turcan S. Origin of Gliomas. Semin. Neurol. 2018;38:5–10. doi: 10.1055/s-0037-1620238. [DOI] [PubMed] [Google Scholar]
- 40.Zhuo Z., Lu H., Zhu J., Hua R.X., Li Y., Yang Z., Zhang J., Cheng J., Zhou H., Li S. METTL14 Gene Polymorphisms Confer Neuroblastoma Susceptibility: An Eight-Center Case-Control Study. Mol. Ther. Nucleic Acids. 2020;22:17–26. doi: 10.1016/j.omtn.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhuo Z.J., Liu W., Zhang J., Zhu J., Zhang R., Tang J., Yang T., Zou Y., He J., Xia H. Functional Polymorphisms at ERCC1/XPF Genes Confer Neuroblastoma Risk in Chinese Children. EBioMedicine. 2018;30:113–119. doi: 10.1016/j.ebiom.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hua R.X., Zhuo Z.J., Zhu J., Jiang D.H., Xue W.Q., Zhang S.D., Zhang J.B., Li X.Z., Zhang P.F., Jia W.H. Association between genetic variants in the XPG gene and gastric cancer risk in a Southern Chinese population. Aging (Albany NY) 2016;8:3311–3320. doi: 10.18632/aging.101119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhuo Z., Zhou C., Fang Y., Zhu J., Lu H., Zhou H., Wu H., Wang Y., He J. Correlation between the genetic variants of base excision repair (BER) pathway genes and neuroblastoma susceptibility in eastern Chinese children. Cancer Commun. (Lond.) 2020;40:641–646. doi: 10.1002/cac2.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Consortium G.T., GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.