Abstract
The outbreak of coronavirus disease 2019 (COVID-19), caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) infection, has emerged in China in December 2019 and rapidly spread to more than 196 countries worldwide.
The physiopathology of human SARS-CoV-2 has not been completely understood, but its pathogenesis has been linked to a disproportionate response of the immune system. Just as described for SARS and MERS, an uncontrolled systemic inflammatory response, known as cytokine release syndrome (CRS) was observed in severe COVID-19 patients. It results from the release by immune and non-immune effector cells of substantial amounts of pro-inflammatory cytokines and appears to contribute to SARS-CoV-2 pulmonary inflammation and extensive lung damage. In addition, hyper-coagulation and thrombosis resulted from the important release of pro-inflammatory cytokines contribute to the lethality of subjects severely infected with SARS-CoV-2. It is therefore essential to have a deep understanding of the various cytokines involved in this exacerbated immune response, and that could be targeted by potential immunological treatments.
The aim of this review was to gather the current knowledge about the role of pro-inflammatory cytokines, namely IL-1β, IL-6, IL-8, IL-17 and TNFα in SARS-CoV-2 CRS, the probable causes and clinical outcomes of this phenomenon in severe cases of COVID-19.
Keywords: SARS-CoV-2, COVID-19, Inflammation, Cytokines
1. Introduction
In December 2019, a new corona virus appeared, the SARS-CoV-2 which belongs to the Coronaviridae family, (from the latin corona = crown, which is due to their morphology as spherical virions with a core shell and surface projections resembling a solar corona). These viruses are enveloped, with a positive sense single-stranded RNA genome (26–32 kb) [1]. The virus has spread so rapidly worldwide, and the growing cases were so alarming that the WHO Emergency Committee declared it a global health emergency. It was not the first-time countries have been affected by this family of viruses, as in the past two decades, Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and Middle East Respiratory Syndrome Coronavirus (MERS-CoV), caused two large-scale aggressive pandemics known for their silent spreading [2]. It is showing symptoms slowly over an incubation period of about 2 weeks, necessary for the virus to replicate in the upper and lower respiratory tract and forming lesions [3]. The disease outcome depends on several factors, including the host immune status. Indeed, most people infected with the SARS-CoV-2 will experience mild to moderate respiratory illnesses and recover without requiring special treatment. However, older people, and especially the ones with medical history like cardiovascular disease, diabetes, chronic respiratory disease and cancer, are more likely to develop serious illness [2]. It was reported that up to 17% of patients infected with the virus, develop an acute respiratory distress syndrome (ARDS) which was associated with organ injury like acute renal injury, acute respiratory injury and septic shock, which can eventually lead to organ failure [4].
Accumulating evidence suggests that a subgroup of patients with severe COVID-19 might have a cytokine release syndrome. Such patients have high levels of pro-inflammatory cytokines and chemokines that are associated with pulmonary inflammation and extensive lung damage [5]. Recently, many studies showed that SARS-CoV-2 promotes coagulopathy, through either the pro-inflammatory cytokines or the activation of the immune cells upon alarm signals such as tissue damage, the complement activation and/or the formation of autoantibodies and immune complexes, which can exacerbate the coagulation process. All these mechanisms can lead to thrombosis, pulmonary embolism and eventually death. The understanding of CRS mechanism(s) and its profile is crucial to developing effective therapeutic approaches in COVID-19.
Here, we review the current understanding of pro-inflammatory cytokines associated with COVID-19 outcome and their relationship with clinical manifestations involved in the severity of this disease.
2. The interaction of the virus with host cells receptors
The virus has four important structural proteins, the envelope protein (E), the membrane protein (M), the spike protein (S), and the nucleocapsid protein (N). These proteins are important to regulate the virus structure and function. The spike protein (S), present on the outer surface of the virion, is a crucial component for its attachment and entry to the host cells. The virus entry requires the priming of the S protein by cellular proteases, which cleave it into a S1/S2 sites, with the subunit S2 possessing the S2’ site that allows the fusion of viral and cellular membranes.
The Angiotensin-Converting Enzyme 2 (ACE2), a negative regulator of the renin-angiotensin system, is a protein expressed in various organs of the human body, but mainly in the lungs, kidney and intestine [6]. The study of Zhou et al. ascertained that the SARS-CoV-2 uses the ACE2 as an entry receptor in the cells expressing this receptor [7]. The priming of S protein by the serine TMPRSS2 is indispensable for the infection of lung cells [8]. Recently, other receptors appeared to play important roles in this process such as GRP78, a chaperon protein belonging to the Heat Shock Protein 70 (HSP70) family and an immunoglobulin heavy chain binding protein BIP. It is located in Endoplasmic Reticulum (ER) membranes of all eukaryotes and its main function is to correct the folding, the assembly and prevent the transport of proteins. Recent studies suggested that the GRP78 protein is also localized on the cell surface at multiple regions of human lung tissues, and can carry out the virus entry. Indeed, the binding is more favorable between different regions of the model of virus spike protein and GRP78 [9,10]. Otherwise, a recent study confirmed the CD147 and SARS-CoV-2 S protein interaction, and that such interaction enhances the viral invasion into host cells. CD147 is a transmembrane glycoprotein that belongs to the immunoglobulin superfamily and is involved in virus’ invasion [11].
Overall, the cell entry of the virus SARS-CoV-2 requires not only an interacting receptor but different indispensable catalytic proteases as well. This variety of processes could explain the complexity and the fast expansion of the virus.
3. Pro-inflammatory cytokines in COVID-19
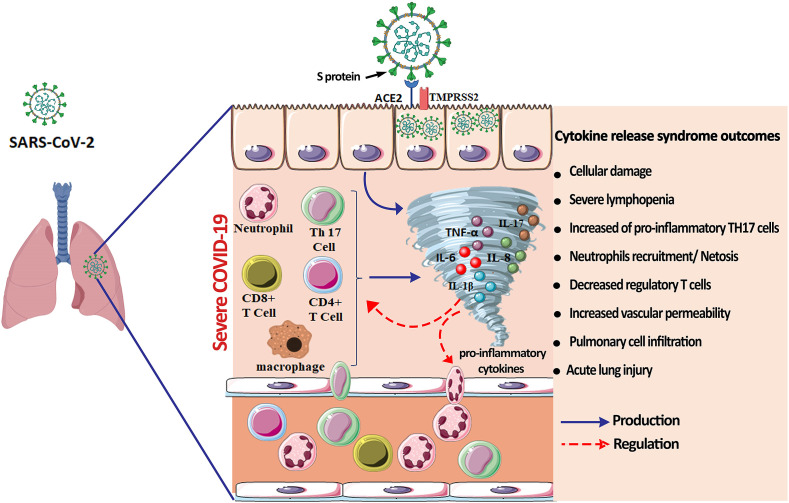
Although the physiopathology of human coronaviruses (H-CoVs) such as SARS-CoV, MERS and SARS-CoV-2, has not been completely understood, they have all been linked to a disrupted and disproportionate response of the immune system particularly cytokine production. Cytokines are membrane-bound or secreted glycoproteins, responsible for the regulation of a large number of biological processes including the balance between the innate and adaptive immune responses, which play a key role in the control of infections or chronic diseases [12]. Therefore, cytokines are believed to play a major role during SARS-CoV-2 infection with the control and resolution of coronavirus infections. However, uncontrolled and exaggerated cytokines production may lead to immunopathogenesis by causing tissue damage in the entire human body. Most COVID-19 patients will develop mild to moderate symptoms, while some infected people may face a hyper-inflammation induced by massive cytokines/chemokines production, known as cytokine release syndrome (CRS), which may lead to fatal pneumonia and acute respiratory distress syndrome [13]. At first, the virus invades the respiratory mucosa, and then infects other cells, triggering a series of immune responses and the generation of a systemic CRS (Fig. 1 ). CRS is characterized by an exacerbated pro-inflammatory cytokines release, usually leading to a systemic inflammation with high inflammation parameters and multiple organ failure [14]. The CRS can occur directly as a result of viral damage, or indirectly by overactivation of the immune system that triggers the infiltration of immune cells such as neutrophils and macrophages into the tissue. This is, in principle, a protective response to limit virus spread, but it ends up being more deleterious than beneficial.
Fig. 1.
The cytokine release syndrome and clinical outcomes in COVID-19 Pathogenesis. Numerous pro-inflammatory cytokines including TNF-α, IL-17, IL-6, IL-8 and IL-1β participate to the CRS onset. These cytokines are secreted by numerous cells like epithelial cells, macrophages, T lymphocytes, neutrophils, and Th17 cells, and exacerbate the SARS-CoV-2 infection. The excessive cytokine release has tremendous consequences such as acute lung injuries, cellular damage, microbiota alteration, severe lymphopenia and a decrease in regulatory T cells.
Clinical studies have shown a significant rise in levels of different cytokines and chemokines in COVID-19. Among key cytokines, we focused on TNF-α, IL-6, IL-17, IL-8 and IL-1β production and their relationship with COVID-19 pathogenesis. Therefore, IL-6 has attracted high levels of interest and its high level seems to be significantly related to the clinical manifestation of severe type patients.
Interleukin 6 (IL-6), also called “hepatocyte stimulating factor”, “hybridoma growth factor” or “B cell stimulating factor”, is a glycoprotein expressed in many cell types such as T and B lymphocytes, monocytes/macrophages, dendritic cells, fibroblasts, and endothelial cells. Its plasma concentration rises during septic shock and various attacks such as trauma and burns [15]. As a pleiotropic cytokine, IL-6 stimulates the growth and differentiation of B lymphocytes and increases the generation of platelets. It also activates the hepatocytes and induces the secretion of inflammation proteins such as reactive protein C (CRP) and fibrinogen [16]. Therefore, IL-6 is involved in the regulation of the immune system, hematopoiesis, inflammation and is a key player in the CRS. The plasma level of IL-6 in COVID-19 patients surpasses the superior limit of normal value in both groups for which the oxygen saturation (SpO2); SpO2 ≥ 90% and the SpO2 < 90% [17]. In many studies, the higher level of IL-6 in serum was positively correlated with the COVID-19 severity [18,19]. A study showed that serum level of IL-6 protein was increased in 2 out of 3 COVID-19 patients. However, the transcription levels of IL-6 did not significantly change in peripheral blood mononuclear cells specimens (PBMC) of COVID-19 patients, suggesting that the secreted IL-6 protein in serum might originate from lung epithelial cells [20]. Statistical analysis showed a negative correlation between T cells number and the serum concentration of IL-6 [21]. Previous studies indicated that in addition to being quantitatively decreased, T cells exhibit elevated exhaustion levels and reduced functional diversity in COVID-19 patients [4]. Despite that certain studies confirmed that SARS-CoV-2 doesn't infect lymphocytes, the lymphocyte cell count was reduced in COVID-19 patients' blood. Among these patients, an important apoptosis gene, TP53, was highly expressed in PBMCs, indicating that apoptosis may be responsible of T lymphocytes number reduction [20]. The literature has highlighted that a decrease of lymphocyte count as well as IL-6 increase may be used as prognostic markers to anticipate severe and fatal COVID-19 patients. In view of this IL-6 increase, antibodies that block the IL-6 receptor are currently under clinical trials for a potential treatment of COVID-19 [22]. Tocilizumab (TCZ) is a recombinant human IL-6 monoclonal antibody, which specifically binds to soluble and membrane-bound IL-6 receptors (IL-6R), thus blocking IL-6 signaling and its mediated inflammatory response [23]. Several clinical trials proved the efficacy of TCZ in the treatment of severe COVID-19. TCZ was associated with an increase of survival, improvement of ARDS, and peripheral lymphocytes count as well as a decline in inflammatory markers [[24], [25], [26], [27]]. According to the Italian guidelines, TCZ can only be used at the end of the initial high viral load phase of COVID-19 (e.g. in patients who have been apyretic for more than 72 h or seven days after the onset of symptoms) in patients with interstitial pneumonia and severe respiratory insufficiency [28].
Tumor necrosis factor (TNF-α), the most studied cytokine in the TNF superfamily, is another key factor in the COVID-19 CRS pathophysiology. It is produced by several cell types, including macrophages, mast cells, T cells, epithelial cells, and smooth muscle cells in the airways. Its synthesis is mainly stimulated by PAMPs and IL-1, via NF-κB activation, and also IL-17 [29]. In healthy subjects, inhaling TNF-α triggers bronchial hyper-responsiveness and inflammation of the airways induced by increased neutrophils recruitment. This cytokine can also cause T cells apoptosis [30]. TNF-α promotes the production of other cytokines such as the IL-1β and IL-6 [31]. Many studies have found that severe COVID-19 cases display increased plasma levels of TNF-α [[32], [33], [34]]. Such increase was also associated with disease severity and inversely correlated with the reduction of T lymphocytes [21]. It has been reported that the prevalence of severe and complicated cases of COVID-19 was lower in patients with anti-TNF-α therapy compared to patients taking steroids. Only 15% of patients with COVID-19 treated with anti-TNF-α had to be hospitalized, and very few of them required an intensive care unit/ventilator or died (3%) compared to patients treated with oral or parenteral steroids and, who had to be hospitalized in 67% of the cases and admitted to the intensive care unit, or died in 25% of the cases [35]. Two additional case reports supported this evidence: one patient suffering from ileal Crohn's disease affected by COVID-19 and was under steroid therapy, and another patient with Crohn's ileitis treated with Adalimumab, a human monoclonal antibody that blocks the interaction between TNF-α and its soluble, membrane-bound receptors [36,37]. To date, a single randomized controlled trial of Adalimumab has been registered (ChiCTR2000030089).
Interleukin 1β (IL-1β) is one of the most important inflammatory pleiotropic cytokines that plays an important role in inflammatory diseases and seems to be involved in COVID-19 CRS. The principal sources of this cytokine are macrophages, monocytes, dendritic cells, B lymphocytes, neutrophils and synovial fibroblasts [38]. The IL-1β effects are quite similar to those of TNF-α, and promote the production of several hematopoietic factors, in particular IL-6 [39]. In addition to its immune functions, IL-1β induces the synthesis of type 2 cyclooxygenase (COX-2), phospholipase A (PLA) and inducible nitric oxide synthase (iNOS). These enzymes are responsible for the formation of nitric oxide (NO), prostaglandin E2 (PGE2) and the platelet activating factor (PAF), involved in fever, pain, vasodilation, hypotension and inflammation components. Moreover, IL-1β increases the expression of chemokines and adhesion molecules, especially in mesenchymal and endothelial cells; thus, promoting the infiltration of immunocompetent cells into injured tissue [40].
IL-1β is secreted during the inflammasome activation in response to several pathogens including viruses such as Coronaviruses. Indeed, a previous study showed that the SARS-CoV Viroporin 3a was sufficient to induce the NLRP3 inflammasome activation and IL-1β production in macrophages stimulated by LPS [41]. A recent investigation showed that IL-1β is highly produced in plasma from both severe and non-severe COVID-19 cases compared with non-infected subjects [33]. Furthermore, another study showed that IL-1β transcripts were significantly up-regulated in whole blood from COVID-19 patients [42]. The increase of IL-1β production is the downstream indicator of cell pyroptosis; this latter is a highly inflammatory form of programmed cell death commonly seen with cytopathic viruses. Therefore, pyroptosis plays a major role in SARS-CoV-2 pathogenesis and is likely a trigger for the subsequent uncontrolled inflammatory response. Moreover, we previously demonstrated that early rapid viral replication may cause big scale cell death leading to the release of pro-inflammatory cytokines [43]. Viral infection and replication in airway epithelial cells could cause high levels of virus-linked pyroptosis with associated vascular leakage, as previously seen in patients with SARS-CoV. Otherwise, high exhaled NO levels are currently used as a marker for eosinophilic airway inflammation in asthmatic subjects, and some authors propose that NO may contribute to tissue damage during airway inflammation [44]. In addition, it is well established that NO production is regulated by cytokines like IL-1β, so this could be one of the mechanisms of the downside effect of IL1-β. Studies based on the inhibition of IL-1β to reduce the massive cytokine release have attracted most attention. Anakinra, a drug that inhibits the biological effect of IL-1β by blocking the IL-1 receptor appears to be a promising treatment. Recent studies showed that the use of Anakinra in patients with COVID-19 is associated with clinical improvement [45]. Canakinumab, a monoclonal antibody targeting IL-1β, is currently the subject of an observational study in a group of patients with COVID-19 (NCT04348448). Furthermore, some drug such as Chloroquine has multiple modes of action including the reduction of IL-1β expression [46].
Interleukin 17 (IL-17), the first biological activity of human IL-17, was demonstrated in 1996 by showing the production of IL-6 and IL-8 by synoviocytes of patients suffering from rheumatoid arthritis in response to IL-17. This indicated the link between IL-17 and inflammation through IL-6 production and the IL-8-induced neutrophils activation [47]. This cytokine is produced in the lung by TH17 cells in response to viruses and leads to the induction of several chemokines that recruit immune cells to the inflammation site [48]. Besides these local effects, IL-17 has also systemic effects. Indeed, the combination of IL-17 with TNF-α induces the expression of pro-coagulation factors, which promotes thrombosis and inhibits endothelial anticoagulatory pathway. These effects account for the increase of the cardiovascular risk associated with inflammatory diseases [49]. In addition to CRS mediation, IL-17 seems to increase the replication of some viruses and leads to viral persistence by inhibiting apoptosis of infected cells in synergy with IL-6 [50]. The serum level of IL-17 was shown to increase in severe cases of COVID-19 patients. Such increase combined to other cytokines production in some COVID-19 patients, was positively correlated with the aggravation of lung lesions [5]. There is recent evidence that TH17 is involved in acute respiratory distress syndrome. The biopsies of patient who died of severe infection induced by SARS-CoV-2, showed a TH17 increase and high CD8+ T cells cytotoxicity. This partly explains the extensive immune damage occurring in this patient [51]. Similarly, another study reported a high density of TH17 cells in the alveolar space of one patient diagnosed as severe case of COVID-19 [33]. This IL-17 association with the COVID-19 severity suggests that this cytokine could also be a plausible target. Moreover, an elderly psoriasis patient affected with COVID-19 presented a favorable outcome while she was treated with Secukinumab, an anti-interleukin-17A monoclonal antibody, approved for psoriasis treatment [52]. In addition, Fedratinib, a JAK2 inhibitor which suppress the TH17 cell induction, could prevent deteriorating outcomes of COVID-19 CRS associated with TH17 [53].
Chemokines are chemo-attractants cytokines that recruit inflammatory cells to migrate from the intravascular space across the endothelium and epithelium into the inflammation site [54]. The relationship between COVID-19 and chemokines dysregulation is not well established. However, some pro-inflammatory chemokines, including CXCL10, CXCL8, CCL2 have been identified at higher levels in the plasma of COVID-19 patients compared to healthy controls. Interestingly, the circulating levels of CXCL10 and CCL2 were found to be significantly higher in patients needing admission to intensive care units compared to patients with a less severe clinical course [54]. According to the transcriptome sequencing on bronchoalveolar lavage fluid (BALF), the excessive release of these two chemokines led to the recruitment of neutrophils, monocytes, and T cells in the lungs [55]. The increased CXCL10 and CCL2 circulating levels was correlated with pulmonary inflammation and pulmonary involvement in SARS patients [54]. Furthermore, other studies showed that the CXCL10 levels were positively and significantly associated with the viral load [56].
Through undertaking single-cell RNA sequencing on nasopharyngeal and bronchial samples from 19 patients with mild or severe COVID-19, Chua et al., showed that in critical cases, the expression of chemokines and chemokine receptors of different cell populations was markedly increased (CCL2, CCL3, CCL20, CXCL1, CXCL3, CXCL10) [57]. Such chemokines increase was also detected in COVID-19 dead patients’ plasma [33].
CXCL8, also called IL-8, has a direct chemotactic and priming effect on neutrophils and is an important mediator of inflammation. It stimulates the neutrophils pro-inflammatory molecules release and has a bactericidal effect by activation of reactive oxygen species (ROS) production [58]. In addition, the clinical and laboratory characteristics of COVID-19 patients showed that IL-8 was strongly produced in severe, rather than non-severe cases [33]. IL-8 is also known to induce Netosis (Neutrophil extracellular traps/NETs). Interestingly, COVID-19 patients showed high levels of cell-free DNA, myeloperoxidase (MPO)-DNA, and citrullinated histone H3 (Cit-H3); those two are highly specific markers of NETs, that may induce organ damage and explain the mortality in COVID-19 patients [59].
Several studies showed an exacerbated inflammatory response during SARS-CoV-2 infection which is characterized by the production of multiple cytokines involved in viral pathology. These cytokines have intertwined relationships where each one contributes to the other's regulation, and often leading to the immune cells hyper-activation. Overall, this hyper-activation has detrimental consequence in the body; and in the absence of appropriate regulation becomes unstoppable, leading to organ failure and death in some cases.
4. How does SARS-CoV-2 initiate cytokine release and what are the potential signaling pathways involved in this process?
The binding of SARS-Cov2 to ACE2 receptor expressed on epithelial cells membrane of the respiratory tract, activates the pro-inflammatory cascade. ACE2 is a major regulator of the renin-angiotensin system (RAS) that acts by converting angiotensin (Ang) II to Ang 1–7, a key regulator of the blood pressure homeostasis and in the prevention of lung injury [60]. The Ang II hormone is vasoconstrictor that directly activates pro-inflammatory responses and the release of cytokines such as IL-6, IL-8 and TNFα by cells through NF-κβ up-regulation, whereas, the Ang 1–7 is a vasodilating hormone, responsible for the anti-inflammatory responses and thereby acting to prevent lungs damage. Interestingly, ACE2 is down-regulated once SARS-CoV-2 enters the host cells; a process that is mediated by ADAM Metallopeptidase Domain 17 (ADAM17), resulting in the release of membrane-bound ACE2 in a soluble form (sACE2). Almost all types of tissues express ADAM17 but its activity is strictly regulated. By shedding ACE2, ADAM17 inhibits Ang 1–7 production, and increases the level of Ang II in COVID-19 patients. The ACE2 down-regulation is associated with RAS imbalance, increased pro-inflammatory response and multi-organ damage from SARS-CoV-2 infection. The mechanism of ACE2 shedding triggered by ADAM17 stimulation is not fully elucidated; however, lymphopenia and the presence of Ang II type 1 receptor (AT1R) are potent activators of such process [60]. In addition, SARS-CoV-2's association with pattern recognition receptors (PRRs) is instrumental in inducing cytokine, chemokine responses to build innate and adaptive immune responses. Nonetheless, the sensors which may recognize the genomics of SARS-CoV are still elusive [61].
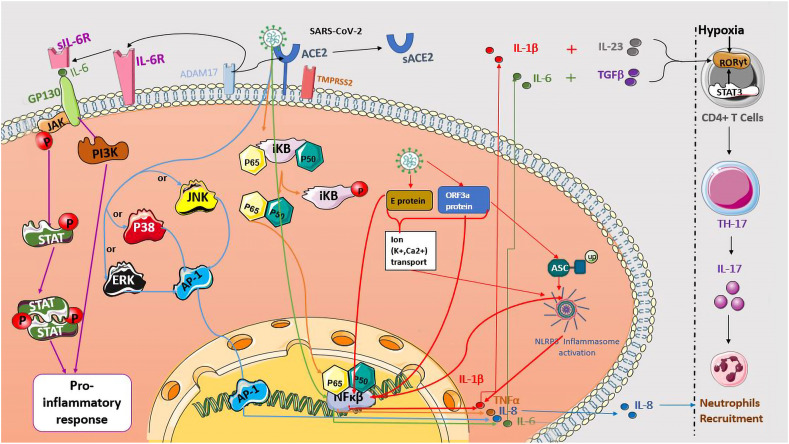
Uncontrolled production of pro-inflammatory cytokines in severe COVID-19 patients is a commonly reported feature of the cytokine release syndrome. Two distinct IL-6 signaling pathways have been identified, the classical and the trans-signaling pathway. IL-6R is produced in two separate forms, IL-6R and its soluble form sIL-6R that is cleaved by ADAM17. In the trans-signaling pathway, IL-6 binds to its soluble receptor SIL-6R and the complex IL-6/SIL-6R activates gp130. Therefore, many intracellular signaling pathways are activated including the Janus kinase (JAK)/signal transducer, transcription activation (STAT) and the phosphatidylinositol 3-kinase (PI3K) pathway, leading to the pro-inflammatory cytokines release by all human cells [62] (Fig. 2 ).
Fig. 2.
Potential signaling pathways involved in COVID-19 cytokine release syndrome. SARS-Cov-2 binds to ACE2 receptor and activates many signaling pathways promoting inflammatory cytokine production. The IL-6 release is initiated by viral proteins, which leads to the transcription factor NF-κβ activation (green arrows). The interaction of IL-6/sIL-6R complex with gp130 protein induces many intracellular signaling pathways, including the Janus kinase (JAK)/(STAT) and (PI3K) leading to pro-inflammatory cytokines release (purple arrows). The phosphorylation of NF-κβ inhibitor proteins (IκB) results in the P65 and P50 proteins release and translocation to the nucleus, inducing NF-κβ transcription and TNFα production (orange arrows). The E protein and ORF3a viral protein activates the NLRP3 inflammasome, which generates IL-1β release (red arrows). The association of the IL-1β, IL-23, IL-6 and TGB-β induce the transcription of RORγt, leading to the differentiation of TCD4+ cells into TH-17 and IL-17 release. The hypoxia can also favor this differentiation. The activation of JNK, P38 or ERK proteins induces the activation and translocation of AP-1 on IL-8 promoter, leading to IL-8 production (blue arrows). Both cytokines IL-8 and IL-17 act by attracting neutrophils to the infectious site. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
TNFα, another CRS member, is synthesized as a membrane-protein, and also cleaved to release its soluble form by ADAM17. TNF soluble and membrane-bound forms are both active although their affinities with TNF family receptors (TNFR) are variable. TNFα signaling is complex, involving as many as 29 different family members of the TNFR. Similarly to IL-6, the TNFR1 broad-based expression gives rise to pleiotropic systemic effects [63]. Recently, the SARS-CoV-2 virus was shown to use the NF-κβ pathway for the release of the pro-inflammatory cytokines such as TNFα and IL-6 (Fig. 2) [64]. A previous study on SARS-CoV reported that S protein significantly increases NF-κβ activity and that the NF-κβ inhibition, leads to a strong inhibition of TNFα and IL-6 release [65]. Moreover, the SARS-CoV N protein directly binds to NF-κβ on IL-6 promoter which induces the activation of IL-6 expression (Fig. 2) [66].
IL-1β is majorly produced by the inflammasome; an innate immune complex which requires two activating signals: the first one is mediated by the NF-κβ pathway which leads to the transcription of the NLRP3 inflammasome and the pro-IL-1β. The second signal leads to the assembly and NLRP3 inflammasome activation [67]. Viruses are able to induce the NLRP3 inflammasome activation and IL-1β production, but the induction of such activation by SARS-CoV-2 remains unclear. Otherwise, SARS-CoV encodes three ion-channel (IC) proteins called Viroporins, the protein E, open reading frame 3a (ORF3a), and ORF8a. SARS-CoV E protein forms a calcium ion (Ca2+) channel in the Endoplasmic Reticulum Golgi Apparatus Intermediate Compartment (ERGIC)/Golgi membranes and leads to the NLRP3 inflammasome activation through Ca2+ transport [68]. SARS-CoV E protein also activates the NF-κβ as a first signal of the NLRP3 inflammasome activation [69]. The ORF3a protein, a potassium (K+) ion channel Viroporin, activates the NLRP3 inflammasome by inducing the K+ efflux and promoting TRAF3-dependent ubiquitination of ASC (Fig. 2) [41,70]. In addition, ORF3a protein induces NF-κβ-mediated up-regulation of the pro-IL-1β transcription and pyroptotic cell death [69].
IL-1 signaling orchestrates the acute response phase to infections, and influences TH17 lymphocyte differentiation which contributes to the pro-inflammatory cytokine's secretion such as IL-17. The IL-17/IL-23 axis promotes inflammatory cytokine secretion such as IL-6 which, in the presence of TGF-β, skews naïve CD4 T cells towards the Th17 phenotype [63]. It has been shown that both IL-1β and IL-23 lead to Th17 differentiation in the presence of TGF-β and IL-6 (Fig. 2). In mice, IL-6 and TGF-β increase Th17 cells differentiation cells through STAT3 activation which subsequently activates RORγt; the “master regulator” of Th17 cell differentiation [71]. An IL-17 signaling modulation was suggested via the JAK/STAT inhibitor Fedratinib [56]. Some studies suggest that reduced ACE2 levels and the extremely hypoxic environment associated with severe COVID-19 can potentiate the Th17 cell differentiation. Indeed, hypoxia-inducible (HIF-1α) may induce the transcription of RORγt leading to an imbalance of the Th17/Treg differentiation, favoring Th17 activity (Fig. 2) [71]. Similar to IL-17, IL-8 is also associated with increased number of alveolar neutrophils (Fig. 2). The signaling pathway of IL-8 in SARS-CoV-2 infection has not been completely understood. SARS-CoV S1 protein activates Extracellular signal-Regulated kinases 1/2, the protein P38, or J c-Jun N-terminal kinases (JNK), initiating the activation of the Activator protein 1 (AP-1) transcription factor on the IL-8 promoter, and then the expression of IL-8 mRNA and IL8 release (Fig. 2) [72]. In addition, IL-6 trans-signaling increases IL-8 production in SARS-COV-2 infection but the precise signaling pathway is still unknown [73].
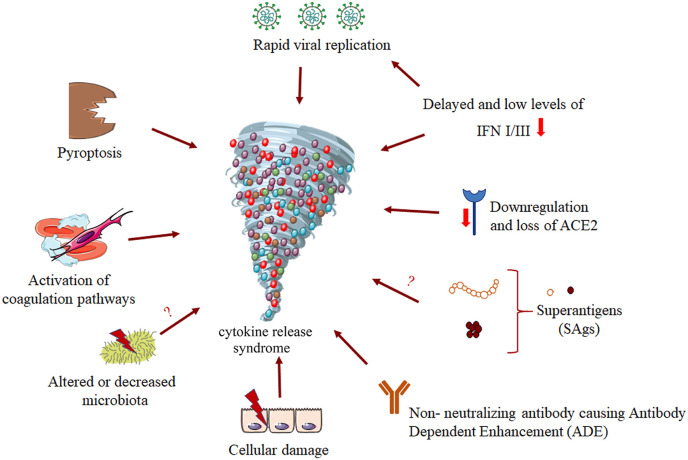
Several factors including, rapid viral replication, cell damage, ACE2 down-regulation, and finally antibody dependent enhancement (ADE) have been linked to the aggressive inflammation caused by SARS-CoV-2 (Fig. 3 ) [74]. The alteration of a protective immune response leads to virus spread and massive destruction of affected tissues. Regarding the innate antiviral immune response, interferons (IFNs) such as IFN type I and type III play a central role in protection against viral infections; they lead to the clearance of the virus and induce tissue repair [75]. Blanco-Melo et al. showed that SARS-CoV-2 infection drives lower antiviral transcriptional response that is marked with an inappropriate inflammation defined by delayed and low levels of type I and III IFN, and loss of viral control in the early phases [76]. The innate immune response may be a critical factor for disease outcome. Indeed, a delayed interferon response and early viral replication within the lungs in conjunction with an increased influx of innate immune cells, mediates tissue damage and may fuel an auto-amplification inflammatory loop, potentially driven by NFκB-induced uncontrolled inflammatory cytokine burst associated with COVID-19 (Fig. 3). Interestingly, some investigations reported that 3.5% of patients (23 of 659) with life-threatening COVID-19 pneumonia had mutations in 8 genes involved in the induction and amplification of type I IFNs, and belong to the TLR3 and IFN-I signaling pathways. These results were confirmed by in vitro studies showing an undetectable IFN-α levels in the blood plasma of people carrying these mutations [77]. Hadjaj et al. found that the immune analysis of a cohort of 50 severe and critical patients, showed a highly impaired interferon (IFN) type I response with no production of IFN-β and low IFN-α. This IFN pathway impairment was associated with a persistent blood viral load as well as an amplified NF-κB-driven inflammatory response, leading to increased cytokines production such as TNF-α and IL-6 [42]. These studies provide proofs that IFN-I pathways impairment may contribute to the COVID-19 severity.
Fig. 3.
Potential causes of COVID-19 cytokine release syndrome. Several factors or pathways may lead to the induction of CRS. For example, the direct rapid replication of the virus inside the organism, the down-regulation of INF type I and III, the down-regulation and loss of the receptor ACE2, non-neutralizing antibodies which are produced by B cells can exacerbate COVID19 through antibody-dependent enhancement (ADE), the alteration or decrease in microbiota, and also the activation of coagulation pathways. Moreover, there are some hypotheses surrounding other mechanisms that may also lead to CRS, like the action of pyroptosis due to the activation of the inflammasome or the Super Antigens (SAgs).
Adaptive immune cells play a central role in defense against viral infections. CD4 T cells stimulate B cells to produce specific antibodies against the virus, and CD8 T cells directly kill virus-infected cells [78]. SARS-CoV-2 may escape these defense mechanisms either by the induction of T lymphocytes apoptosis or the exhaustion of these cells due to the expression of pro-inflammatory cytokines [79]. Severe lymphopenia, decreased regulatory T cells and increased of pro-inflammatory TH17 cells were frequently reported in critically ill COVID-19 patients, suggesting a deregulated adaptive immune response. Increased levels of neutrophils also correlate with disease severity and death; IL-17 and IL-8 are known as cytokines that strongly participate in neutrophils recruitment [33,34]. Pulmonary inflammation is considered to be the main cause of life-threatening respiratory diseases in the severe stage of COVID-19 [51].
Thrombosis is acknowledged as a pathological disruption of homeostasis involving coagulation, platelet activation, and characterized by clot formation and vessel occlusion. The activation of both inflammation and coagulation and the excessive activation of immune cells along with intravascular thrombus formation during systemic infections, have deleterious consequences for host tissues [80]. Previous immunological studies have shown that cytokines like IL-6, TNF-α and, IL-1β are critically involved in the hyper-activation of platelets which may lead to abnormal clot formation and can also play an important role in the down-regulation of important physiological coagulant pathways [81]. These hyperactive platelets are present in both severe and non-severe COVID-19 patients and form aggregates with leucocytes. It has been observed that Extra Vesicules (EV) released by platelets are significantly increased in the circulation. These EV can participate in both inflammation and coagulation processes. A recent study demonstrated that platelets are hyper-responsive, sensitized to release inflammatory cytokines and they aggregate and adhere to a collagen surface more efficiently when originating from COVID-19 patients [82]. This cytokine production shows the dual role of platelets in worsening both inflammation and coagulation.
Some cytokines such as TNF-α, IL-1β, and IL-6 can induce the generation of thrombin in humans. Moreover, TNF-α and IL-1β can also trigger the subsequent release of Plasminogen Activator Inhibitor-1 (PAI-1), and may diminish the release of tissue Plasminogen Activator (tPA), which inhibits the physiological process that degrades blood clots [83]. IL-6, a cytokine that is significantly elevated in severe COVID-19 infection, is a key activator of coagulopathy by inducing tissue factor expression and increasing fibrinogen production. Similarly, IL-6 can mainly enhance the proliferation, differentiation, and activation of megakaryocytes through increasing the production of thrombopoietin [84]. Megakaryocytes along with platelet-derived fibrin characterize multi-organ thrombosis in COVID-19 patients [85]. IL-1β down-regulates thrombomodulin and consequently causes defects in anticoagulant proteins. In a study by J. Bester et al. it was shown that IL-8 was the cytokine that caused the most significant changes at all levels of coagulation, including fibrinogen, thrombin, and cellular interactions [86]. Recent studies have highlighted the link between abnormal coagulation parameters like D-dimers and fibrin degradation products and COVID-19 mortality. The pro-inflammatory cytokines overproduction in COVID-19, mainly IL-6, stimulates mononuclear cells to express TF, which promotes the transformation of prothrombin into thrombin, which then converts circulating fibrinogen into fibrin [87]. In all severe SARS-CoV-2 cases, high values of D-dimers, fibrin/fibrinogen degradation products (FDP), and fibrinogen (FIB) were reported compared to patients with milder forms of COVID-19 or healthy controls [88].
The release of cytokines and chemokines like TNFα, IL-1, IL-6, and IL-8 induces macrophage activation syndrome which triggers the endothelial cells, macrophages, and neutrophils to express TF within the lungs which in turn initiates and further augments pulmonary coagulopathy and microvascular thrombosis [84,89]. These small fibrin thrombi were observed in small pulmonary arterioles, in 8 COVID-19 cases out of 10, in areas of both damaged and preserved lung parenchyma [90]. Thrombin can increase inflammation via Proteinase-Activated Receptors (PARs) and these two pathways regulate each other, as a vicious circle takes place, leading to disastrous consequences like multiorgan injuries and failure [91] (Fig. 3).
Besides the inflammatory/thrombotic damages, gastrointestinal symptoms have also been associated with this disease and were reported to be higher in severe COVID-19 patients compared with non-severe patients [92]. Few studies highlighted the effect of COVID-19 on gut microbiota and showed microbiome alterations in COVID-19 patients [93,94]. There is an enrichment of opportunistic pathogens and the exhaustion of beneficial commensals that were associated with COVID-19 severity. In addition, in elders in whom COVID-19 infection appears to be more deadly, the variety of the intestinal microbiota was decreased. Another study using a machine learning model reported that the disruption of gut microbiota significantly correlated with pro-inflammatory cytokines and may predispose normal individuals to severe COVID-19 [95] (Fig. 3). Indeed, one study found that in host myeloid progenitors, commensal microbiota regulates the systemic TLR-driven immune response by priming JAK signaling which underlines its role in CRS pathogenesis [96]. In addition, COVID-19 microbiota dysbiosis interferes with mitochondrial homeostasis through the production of metabolites. Mitochondrial oxidative stress may affect the coagulation pathways and fuel the inflammatory/oxidative response [97].
Moreover, the pro-inflammatory cytokine release, observed in COVID-19 patients recalls ones induced by superantigens. Indeed, Staphylococcal and Streptococcal Superantigens (SAgs) are responsible of toxic shock syndrome, which is caused by a rapid hyper-inflammatory response characterized by a massive cytokine release [98]. Therefore, SAgs can lead to cytokine and chemokine inflammatory responses, which could be associated with SARS-Cov2 pathology (Fig. 3).
5. How does SARS-CoV-2 relate to SARS-CoV and MERS-CoV in terms of cytokine release syndrome?
Before the COVID-19 pandemic, SARS-CoV and MERS-CoV, two pathogenic zoonotic coronaviruses were also responsible of major outbreaks. In 2003, SARS-CoV caused 8096 confirmed cases with 774 deaths in 29 countries. As for MERS-CoV, the number of confirmed cases that has been reported by January 2020 is 2519, with 866 confirmed deaths. The case fatality rate of MERS-CoV is much higher than that of SARS-CoV. Bats were identified as the reservoir of SARS-CoV, and dromedary camels are MERS-CoV reservoir. However, inter-human transmission of SARS-CoV and MERS-CoV occurs predominantly via nosocomial transmission, and this might be explained by the fact that shedding of the virus happens after symptoms apparition, and patients are at that time already in healthcare settings [99].
SARS-CoV and MERS-CoV are both β-coronaviruses, and have large positive-sense RNA genomes. Genomic analysis comparing SARS-CoV-2 and SARS-CoV by zpicture showed that there is a homology between proteins of both viruses, namely the envelope protein E and the nucleocapsid phosphoprotein N, showing 94.74% and 90.52% of identity, respectively [100]. To infect cells, envelope spike proteins of SARS-CoV and MERS-CoV bind to two different types of receptors, respectively ACE2 (found in cells of the heart, kidneys, testes and also broadly expressed in the lungs, liver, intestine, and brain) [101] and DPP4 (widely expressed on epithelial cells, in the kidney, alveoli, small intestine, liver, prostate, and on activated leukocytes) [102]. Symptoms of SARS-CoV and MERS-CoV infections are diverse including fever, cough, myalgia, shortness of breath, sore throat, and in few cases diarrhea, and vomiting. Thus, clinical manifestations range from symptoms resembling flu to severe acute respiratory distress syndrome (ARDS) [103].
Even though the pathophysiology of SARS-CoV and MERS-CoV viruses is not yet fully understood, they both have been linked to an abnormal and disproportionate cytokine production. During the SARS-CoV outbreak, a CRS was observed and patients had high levels of IFN‐γ IL‐1, IL‐6, IL‐12, IL‐8, MCP‐1 and IP‐10. MERS-CoV also slowly induces the secretion of high levels of pro-inflammatory cytokines, such as IFN-γ, TNF-α, IL-15 and IL-17. A contrast exists between the cytokine profiles of these two human coronaviruses. SARS-CoV induces the activation of TH1 cell‐mediated immunity by the stimulation of NK, cytotoxic T lymphocytes and a hyper-innate inflammatory response, whereas MERS-CoV infection leads to an important pro-inflammatory Th1 and Th17 response [104,105]. Regarding IFNs secreted very early in viral infection, it was shown that there is an unusual lack of expression of type I IFNs in SARS-CoV, whereas MERS-CoV induced a significant increase in the levels of IFN-α2 and IFN-γ. It was also found that delayed release of IFNs in the early stages of SARS-CoV and MERS-CoV infections hampers the antiviral immune response [54,106].
Similarly to SARS-CoV and MERS-CoV, SARS-CoV-2 symptoms range from mild to severe ones, although it usually induces less severe symptoms, allowing it to spread easily among the population. The affinity of SARS-CoV-2 for ACE2 is higher than that of SARS-CoV [54]. In terms of cytokines production by the three human coronaviruses, it has been shown that, like for SARS-CoV, high levels of IL-6 were found in patients with COVID-19. Regarding IFN production, SARS-CoV-2 exhibited more efficient replication but induced significantly less host interferon response than SARS-CoV. Moreover, as opposed to SARS-CoV, SARS-CoV-2 induces the secretion of Th2 anti-inflammatory cytokines IL-4 and IL-10, and TH1 pro-inflammatory cytokines [33]. Additionally, patients with severe COVID-19 had high number of CCR6+ TH17 cells in their PBMC, indicating that like for MERS-CoV, a TH17 type response is also induced during SARS-COV-2 infection [79].
6. Conclusion and perspectives
COVID-19 is a viral-induced illness with a clinical outcome determined by the extent of the host immune system imbalance. The primary immune response is a positive response that leads to viral clearance in the majority of cases. However, for reasons that are still unclear, the secondary immune response may be exaggerated and challenge tissue integrity, eventually leading to multiple organ failure, ARDS and death. This exaggerated response is demonstrated through a cytokine release syndrome for which there is still no specific or effective treatment. Earlier SARS-CoV and MERS-CoV therapeutic strategies have shown that it was possible to improve prognosis, through the reduction of viral load by conducting early-stage interventions and controlling the exacerbated inflammatory responses using immunomodulators. Although there is no specific antiviral therapy for COVID-19, a deep understanding of CRS mechanisms in this disease can help design possible therapeutic strategies.
It would also be relevant to elucidate the kinetics of cytokine activation and expression during SARS-CoV-2 infection. It would be useful to explore whether direct virus-induced tissue damage, systemic inflammatory cytokine release or the synergistic effects of both, contribute to the multiple organ dysfunction associated with severe COVID-19. Moreover, in the clinical context, approaches should be developed in order to prevent transition from mild to severe cases, and more effective anti-virus agents should be unveiled. There is an urgent need to better understand the host–pathogen biology of COVID-19 as this will provide important insights into treatment and management of the disease, including identification of new therapies.
Authors’ contributions
Dounia DARIF: Conceptualization, Methodology, Writing-Original Draft. Ikram HAMMI: Conceptualization, Methodology, Writing-Original Draft. Ayyoub KIHEL: Conceptualization, Methodology, Writing-Original Draft, Imane EL IDRISSI SAIK : Writing-Original Draft. Fadila GUESSOUS: Conceptualization, Writing - Review & Editing. Khadija AKARID: Conceptualization, Validation, Writing-Original Draft.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Li X., Geng M., Peng Y., Meng L., Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm. Anal. 2020;10:102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO Coronavirus. 2020. https://www.who.int/health-topics/coronavirus#tab=tab_1 (accessed June 26, 2020)
- 3.Chan J.F.W., Yuan S., Kok K.H., To K.K.W., Chu H., Yang J., Xing F., Liu J., Yip C.C.Y., Poon R.W.S., Tsoi H.W., Lo S.K.F., Chan K.H., Poon V.K.M., Chan W.M., Ip J.D., Cai J.P., Cheng V.C.C., Chen H., Hui C.K.M., Yuen K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y., Zhang C., Huang F., Yang Y., Wang F., Yuan J., Zhang Z., Qin Y., Li X., Zhao D., Li S., Tan S., Wang Z., Li J., Shen C., Li J., Peng L., Weibo W., Cao M., Xing L., Xu Z., Chen L., Zhou C., Liu W., Liu L., Jiang C. ChinaXiv; 2020. 2019-novel Coronavirus (2019-nCoV) Infections Trigger an Exaggerated Cytokine Response Aggravating Lung Injury. [Google Scholar]
- 6.Warner F.J., Smith A.I., Hooper N.M., Turner A.J. Angiotensin-converting enzyme-2: a molecular and cellular perspective. Cell. Mol. Life Sci. 2004;61:2704–2713. doi: 10.1007/s00018-004-4240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou P., Lou Yang X., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Di Jiang R., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ibrahim I.M., Abdelmalek D.H., Elshahat M.E., Elfiky A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 2020;80:554–562. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu H., Chan C.M., Zhang X., Wang Y., Yuan S., Zhou J., Au-Yeung R.K.H., Sze K.H., Yang D., Shuai H., Hou Y., Li C., Zhao X., Poon V.K.M., Leung S.P., Yeung M.L., Yan J., Lu G., Jin D.Y., Gao G.F., Chan J.F.W., Yuen K.Y. Middle East respiratory syndrome coronavirus and bat coronavirus HKU9 both can utilize GRP78 for attachment onto host cells. J. Biol. Chem. 2018;293:11709–11726. doi: 10.1074/jbc.RA118.001897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K., Chen W., Zhou Y.-S., Lian J.-Q., Zhang Z., Du P., Gong L., Zhang Y., Cui H.-Y., Geng J.-J., Wang B., Sun X.-X., Wang C.-F., Yang X., Lin P., Deng Y.-Q., Wei D., Yang X.-M., Zhu Y.-M., Zhang K., Zheng Z.-H., Miao J.-L., Guo T., Shi Y., Zhang J., Fu L., Wang Q.-Y., Bian H., Zhu P., Chen Z.-N. BioRxiv; 2020. SARS-CoV-2 Invades Host Cells via a Novel Route: CD147-Spike Protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartee E., McFadden G. Cytokine synergy: an underappreciated contributor to innate anti-viral immunity. Cytokine. 2013;63:237–240. doi: 10.1016/j.cyto.2013.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., Across Speciality Collaboration H. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C., Wu Z., Li J.W., Zhao H., Wang G.Q. Cytokine release syndrome in severe COVID-19: interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int. J. Antimicrob. Agents. 2020;55:105954. doi: 10.1016/j.ijantimicag.2020.105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones S.A., Jenkins B.J. Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer. Nat. Rev. Immunol. 2018;18:773–789. doi: 10.1038/s41577-018-0066-7. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka T., Narazaki M., Kishimoto T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb Perspect Biol. 2014;6:a016295. doi: 10.1101/cshperspect.a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z., Yang B., Li Q., Wen L., Zhang R. Clinical features of 69 cases with coronavirus disease 2019 in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X., Zhao B., Qu Y., Chen Y., Xiong J., Feng Y., Men D., Huang Q., Liu Y., Yang B., Ding J., Li F. Detectable serum SARS-CoV-2 viral load (RNAaemia) is closely associated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients. Clin. Infect. Dis. 2020 doi: 10.1101/2020.02.29.20029520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herold T., Jurinovic V., Arnreich C., Lipworth B.J., Hellmuth J.C., von Bergwelt-Baildon M., Klein M., Weinberger T. Elevated levels of IL-6 and CRP predict the need for mechanical ventilation in COVID-19. J. Allergy Clin. Immunol. 2020 doi: 10.1016/j.jaci.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z., Wu H., Lin Y., Zhang M., Zhang Q., Shi M., Liu Y., Zhou Y., Lan K., Chen Y. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microb. Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G., Yuan Z., Feng Z., Zhang Y., Wu Y., Chen Y. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henriksen Marius. Anti-il6 treatment of serious COVID-19 disease with threatening respiratory failure - full text view - ClinicalTrials.gov. 2020. https://www.clinicaltrials.gov/ct2/show/NCT04322773 (accessed June 19, 2020)
- 23.Zhang W., Zhao Y., Zhang F., Wang Q., Li T., Liu Z., Wang J., Qin Y., Zhang X., Yan X., Zeng X., Zhang S. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19) Clin. Immunol. 2020;214:108393. doi: 10.1016/j.clim.2020.108393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu X., Han M., Li T., Sun W., Wang D., Fu B., Zhou Y., Zheng X., Yang Y., Li X., Zhang X., Pan A., Wei H. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl. Acad. Sci. U.S.A. 2020;117:10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toniati P., Piva S., Cattalini M., Garrafa E., Regola F., Castelli F., Franceschini F., Airò P., Bazzani C., Beindorf E.A., Berlendis M., Bezzi M., Bossini N., Castellano M., Cattaneo S., Cavazzana I., Contessi G.B., Crippa M., Delbarba A., De Peri E., Faletti A., Filippini M., Frassi M., Gaggiotti M., Gorla R., Lanspa M., Lorenzotti S., Marino R., Maroldi R., Metra M., Matteelli A., Modina D., Moioli G., Montani G., Muiesan M.L., Odolini S., Peli E., Pesenti S., Pezzoli M.C., Pirola I., Pozzi A., Proto A., Rasulo F.A., Renisi G., Ricci C., Rizzoni D., Romanelli G., Rossi M., Salvetti M., Scolari F., Signorini L., Taglietti M., Tomasoni G., Tomasoni L.R., Turla F., Valsecchi A., Zani D., Zuccalà F., Zunica F., Focà E., Andreoli L., Latronico N. Tocilizumab for the treatment of severe COVID-19 pneumonia with hyperinflammatory syndrome and acute respiratory failure: a single center study of 100 patients in Brescia, Italy. Autoimmun. Rev. 2020;19:102568. doi: 10.1016/j.autrev.2020.102568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sciascia S., Aprà F., Baffa A., Baldovino S., Boaro D., Boero R., Bonora S., Calcagno A., Cecchi I., Cinnirella G., Converso M., Cozzi M., Crosasso P., De Iaco F., Di Perri G., Eandi M., Fenoglio R., Giusti M., Imperiale D., Imperiale G., Livigni S., Manno E., Massara C., Milone V., Natale G., Navarra M., Oddone V., Osella S., Piccioni P., Radin M., Roccatello D., Rossi D. Pilot prospective open, single-arm multicentre study on off-label use of tocilizumab in patients with severe COVID-19. Clin. Exp. Rheumatol. 2020;38:529–532. [PubMed] [Google Scholar]
- 27.Alattar R., Ibrahim T.B.H., Shaar S.H., Abdalla S., Shukri K., Daghfal J.N., Khatib M.Y., Aboukamar M., Abukhattab M., Alsoub H.A., Almaslamani M.A., Omrani A.S. Tocilizumab for the treatment of severe coronavirus disease 2019. J. Med. Virol. 2020 doi: 10.1002/jmv.25964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lombardy Section Italian Society Infectious And Tropical Diseases Vademecum for the treatment of people with COVID-19. Edition 2.0, 13 March 2020. Le Infez. Med. 2020;28:143–152. http://www.ncbi.nlm.nih.gov/pubmed/32275256 [PubMed] [Google Scholar]
- 29.Luis Muñoz-Carrillo J., Francisco Contreras-Cordero J., Gutiérrez-Coronado O., Trinidad Villalobos-Gutiérrez P., Guillermo Ramos-Gracia L., Elizabeth Hernández-Reyes V. IntechOpen; 2019. Cytokine Profiling Plays a Crucial Role in Activating Immune System to Clear Infectious Pathogens. [DOI] [Google Scholar]
- 30.Makwana R., Gozzard N., Spina D., Page C. TNF-α-induces airway hyperresponsiveness to cholinergic stimulation in Guinea pig airways. Br. J. Pharmacol. 2012;165 doi: 10.1111/j.1476-5381.2011.01675.x. 1978–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peiris J.S.M., Lai S.T., Poon L.L.M., Guan Y., Yam L.Y.C., Lim W., Nicholls J., Yee W.K.S., Yan W.W., Cheung M.T., Cheng V.C.C., Chan K.H., Tsang D.N.C., Yung R.W.H., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., Zhang X., Zhang M., Wu S., Song J., Chen T., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenner E., Ungaro R., Colommbel J., Kappelman M. SECURE-IBD Database; 2020. Current Summary Data.https://covidibd.org/current-data/ 2020. (accessed June 20, 2019) [Google Scholar]
- 36.Mazza S., Sorce A., Peyvandi F., Vecchi M., Caprioli F. A fatal case of COVID-19 pneumonia occurring in a patient with severe acute ulcerative colitis. Gut. 2020;69:1148–1149. doi: 10.1136/gutjnl-2020-321183. [DOI] [PubMed] [Google Scholar]
- 37.Tursi A., Angarano G., Monno L., Saracino A., Signorile F., Ricciardi A., Papa A. COVID-19 infection in Crohn's disease under treatment with adalimumab. Gut. 2020;69:1364–1365. doi: 10.1136/gutjnl-2020-321240. [DOI] [PubMed] [Google Scholar]
- 38.Dinarello C.A. Immunological and inflammatory functions of the interleukin-1 family. Annu. Rev. Immunol. 2009;27:519–550. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- 39.Sironi M., Breviario F., Proserpio P., Biondi A., Vecchi A., Van Damme J., Dejana E., Mantovani A. IL-1 stimulates IL-6 production in endothelial cells. J. Immunol. 1989;142:549–553. http://www.ncbi.nlm.nih.gov/pubmed/2783442 (accessed June 19, 2020) [PubMed] [Google Scholar]
- 40.Dinarello C.A. Interleukin-1β. Crit. Care Med. 2005;33:460–462. doi: 10.1097/01.CCM.0000185500.11080.91. [DOI] [PubMed] [Google Scholar]
- 41.Chen I.-Y., Moriyama M., Chang M.-F., Ichinohe T. Severe acute respiratory syndrome coronavirus Viroporin 3a activates the NLRP3 inflammasome. Front. Microbiol. 2019;10:50. doi: 10.3389/fmicb.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P., Carlier N., Gauzit R., Morbieu C., Pène F., Marin N., Roche N., Szwebel T.A., Merkling S.H., Treluyer J.M., Veyer D., Mouthon L., Blanc C., Tharaux P.L., Rozenberg F., Fischer A., Duffy D., Rieux-Laucat F., Kernéis S., Terrier B. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science (80-. ) 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.da Costa L.S., Outlioua A., Anginot A., Akarid K., Arnoult D. RNA viruses promote activation of the NLRP3 inflammasome through cytopathogenic effect-induced potassium efflux. Cell Death Dis. 2019;10:346. doi: 10.1038/s41419-019-1579-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warke T.J., Fitch P.S., Shields M.D., Brown V., Ennis M., Taylor R., Lyons J.D.M. Exhaled nitric oxide correlates with airway eosinophils in childhood asthma. Thorax. 2002;57:383–387. doi: 10.1136/thorax.57.5.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cavalli G., De Luca G., Campochiaro C., Della-Torre E., Ripa M., Canetti D., Oltolini C., Castiglioni B., Tassan Din C., Boffini N., Tomelleri A., Farina N., Ruggeri A., Rovere-Querini P., Di Lucca G., Martinenghi S., Scotti R., Tresoldi M., Ciceri F., Landoni G., Zangrillo A., Scarpellini P., Dagna L. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: a retrospective cohort study. Lancet Rheumatol. 2020;2:e325–e331. doi: 10.1016/S2665-9913(20)30127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Devaux C.A., Rolain J.M., Colson P., Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int. J. Antimicrob. Agents. 2020;55:105938. doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fossiez F., Djossou O., Chomarat P., Flores-Romo L., Ait-Yahia S., Maat C., Pin J.J., Garrone P., Garcia E., Saeland S., Blanchard D., Gaillard C., Das Mahapatra B., Rouvier E., Golstein P., Banchereau J., Lebecque S. T cell interleukin-17 induces stromal cells to produce proinflammatory and hematopoietic cytokines. J. Exp. Med. 1996;183:2593–2603. doi: 10.1084/jem.183.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryzhakov G., Lai C.C., Blazek K., To K., Hussell T., Udalova I. IL-17 boosts proinflammatory outcome of antiviral response in human cells. J. Immunol. 2011;187:5357–5362. doi: 10.4049/jimmunol.1100917. [DOI] [PubMed] [Google Scholar]
- 49.Hot A., Lenief V., Miossec P. Combination of IL-17 and TNFα induces a pro-inflammatory, pro-coagulant and pro-thrombotic phenotype in human endothelial cells. Ann. Rheum. Dis. 2012;71:768–776. doi: 10.1136/annrheumdis-2011-200468. [DOI] [PubMed] [Google Scholar]
- 50.Hou W., Jin Y.-H., Kang H.S., Kim B.S. Interleukin-6 (IL-6) and IL-17 synergistically promote viral persistence by inhibiting cellular apoptosis and cytotoxic T cell function. J. Virol. 2014;88:8479–8489. doi: 10.1128/jvi.00724-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J., Xia P., Dong J., Zhao J., Wang F.S. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Di Lernia V., Bombonato C., Motolese A. COVID-19 in an elderly patient treated with secukinumab. Dermatol. Ther. 2020 doi: 10.1111/dth.13580. [DOI] [PubMed] [Google Scholar]
- 53.Wu D., Yang X.O. TH17 responses in cytokine storm of COVID-19: an emerging target of JAK2 inhibitor Fedratinib. J. Microbiol. Immunol. Infect. 2020;53:368–370. doi: 10.1016/j.jmii.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang Y., Liu J., Zhang D., Xu Z., Ji J., Wen C. Cytokine storm in COVID-19: the current evidence and treatment strategies. Front. Immunol. 2020;11:1708. doi: 10.3389/fimmu.2020.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Y., Shen C., Li J., Yuan J., Yang M., Wang F., Li G., Li Y., Xing L., Peng L., Wei J., Cao M., Zheng H., Wu W., Zou R., Li D., Xu Z., Wang H., Zhang M., Zhang Z., Liu L., Liu Y. MedRxiv; 2020. Exuberant Elevation of IP-10, MCP-3 and IL-1ra during SARS-CoV-2 Infection Is Associated with Disease Severity and Fatal Outcome. [DOI] [Google Scholar]
- 57.Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F., Debnath O., Thürmann L., Kurth F., Völker M.T., Kazmierski J., Timmermann B., Twardziok S., Schneider S., Machleidt F., Müller-Redetzky H., Maier M., Krannich A., Schmidt S., Balzer F., Liebig J., Loske J., Suttorp N., Eils J., Ishaque N., Liebert U.G., von Kalle C., Hocke A., Witzenrath M., Goffinet C., Drosten C., Laudi S., Lehmann I., Conrad C., Sander L.E., Eils R. COVID-19 severity correlates with airway epithelium–immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020;38:970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 58.Kobayashi Y. The role of chemokines in neutrophil biology. 2008. [DOI] [PubMed]
- 59.Zuo Y., Yalavarthi S., Shi H., Gockman K., Zuo M., Madison J.A., Blair C., Weber A., Barnes B.J., Egeblad M., Woods R.J., Kanthi Y., Knight J.S. Neutrophil extracellular traps in COVID-19. JCI Insight. 2020;5 doi: 10.1172/jci.insight.138999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mahmud-Al-Rafat A., Muzammal Haque Asim M., Taylor-Robinson A.W., Majumder A., Muktadir A., Muktadir H., Karim M., Khan I., Mainul Ahasan M., Morsaline Billah M. A combinational approach to restore cytokine balance and to inhibit virus growth may promote patient recovery in severe COVID-19 cases. Cytokine. 2020;136:155228. doi: 10.1016/j.cyto.2020.155228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sallenave J.M., Guillot L. Innate immune signaling and proteolytic pathways in the resolution or exacerbation of SARS-CoV-2 in Covid-19: key therapeutic targets? Front. Immunol. 2020;11:1229. doi: 10.3389/fimmu.2020.01229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ebihara N., Matsuda A., Nakamura S., Matsuda H., Murakami A. Role of the IL-6 classic-and trans-signaling pathways in corneal sterile inflammation and wound healing. Investig. Ophthalmol. Vis. Sci. 2011;52:8549–8557. doi: 10.1167/iovs.11-7956. [DOI] [PubMed] [Google Scholar]
- 63.Arnaldez F.I., O'Day S.J., Drake C.G., Fox B.A., Fu B., Urba W.J., Montesarchio V., Weber J.S., Wei H., Wigginton J.M., Ascierto P.A. The Society for Immunotherapy of Cancer perspective on regulation of interleukin-6 signaling in COVID-19-related systemic inflammatory response. J. Immunother. Cancer. 2020;8 doi: 10.1136/jitc-2020-000930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma Q., Li R., Pan W., Huang W., Liu B., Xie Y., Wang Z., Li C., Jiang H., Huang J., Shi Y., Dai J., Zheng K., Li X., Hui M., Fu L., Yang Z. Phillyrin (KD-1) exerts anti-viral and anti-inflammatory activities against novel coronavirus (SARS-CoV-2) and human coronavirus 229E (HCoV-229E) by suppressing the nuclear factor kappa B (NF-κB) signaling pathway. Phytomedicine. 2020;78:153296. doi: 10.1016/j.phymed.2020.153296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang W., Ye L., Ye L., Li B., Gao B., Zeng Y., Kong L., Fang X., Zheng H., Wu Z., She Y. Up-regulation of IL-6 and TNF-α induced by SARS-coronavirus spike protein in murine macrophages via NF-κB pathway. Virus Res. 2007;128:1–8. doi: 10.1016/j.virusres.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang X., Wu K., Wang D., Yue X., Song D., Zhu Y., Wu J. Nucleocapsid protein of SARS-CoV activates interleukin-6 expression through cellular transcription factor NF-κB. Virology. 2007;365:324–335. doi: 10.1016/j.virol.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kihel A., Hammi I., Darif D., Lemrani M., Riyad M., Guessous F., Akarid K. The Different Faces of the NLRP3 Inflammasome in Cutaneous Leishmaniasis: A Review. Cytokine. 2020:155248. doi: 10.1016/j.cyto.2020.155248. [DOI] [PubMed] [Google Scholar]
- 68.Nieto-Torres J.L., Verdiá-Báguena C., Jimenez-Guardeño J.M., Regla-Nava J.A., Castaño-Rodriguez C., Fernandez-Delgado R., Torres J., Aguilella V.M., Enjuanes L. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology. 2015;485:330–339. doi: 10.1016/j.virol.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shah A. Novel coronavirus-induced NLRP3 inflammasome activation: a potential drug target in the treatment of COVID-19. Front. Immunol. 2020;11:1021. doi: 10.3389/fimmu.2020.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Siu K., Yuen K., Castano‐Rodriguez C., Ye Z., Yeung M., Fung S., Yuan S., Chan C., Yuen K., Enjuanes L., Jin D. Severe acute respiratory syndrome Coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3‐dependent ubiquitination of ASC. Faseb. J. 2019;33:8865–8877. doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Orlov M., Wander P.L., Morrell E.D., Mikacenic C., Wurfel M.M. A case for targeting Th17 cells and IL-17A in SARS-CoV-2 infections. J. Immunol. 2020;205:892–898. doi: 10.4049/jimmunol.2000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chang Y.-J., Liu C.Y.-Y., Chiang B.-L., Chao Y.-C., Chen C.-C. Induction of IL-8 release in lung cells via activator protein-1 by recombinant baculovirus displaying severe acute respiratory syndrome-coronavirus spike proteins: identification of two functional regions. J. Immunol. 2004;173:7602–7614. doi: 10.4049/jimmunol.173.12.7602. [DOI] [PubMed] [Google Scholar]
- 73.Kang S., Tanaka T., Inoue H., Ono C., Hashimoto S., Kioi Y., Matsumoto H., Matsuura H., Matsubara T., Shimizu K., Ogura H., Matsuura Y., Kishimoto T. IL-6 trans-signaling induces plasminogen activator inhibitor-1 from vascular endothelial cells in cytokine release syndrome. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:22351–22356. doi: 10.1073/pnas.2010229117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fu Y., Cheng Y., Wu Y. Understanding SARS-CoV-2-mediated inflammatory responses: from mechanisms to potential therapeutic tools. Virol. Sin. 2020 doi: 10.1007/s12250-020-00207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Makris S., Paulsen M., Johansson C. Type I interferons as regulators of lung inflammation. Front. Immunol. 2017;8:259. doi: 10.3389/fimmu.2017.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., TenOever B.R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., Rosain J., Bilguvar K., Ye J., Bolze A., Bigio B., Yang R., Arias A.A., Zhou Q., Zhang Y., Onodi F., Korniotis S., Karpf L., Philippot Q., Chbihi M., Bonnet-Madin L., Dorgham K., Smith N., Schneider W.M., Razooky B.S., Hoffmann H.H., Michailidis E., Moens L., Han J.E., Lorenzo L., Bizien L., Meade P., Neehus A.L., Ugurbil A.C., Corneau A., Kerner G., Zhang P., Rapaport F., Seeleuthner Y., Manry J., Masson C., Schmitt Y., Schlüter A., Le Voyer T., Khan T., Li J., Fellay J., Roussel L., Shahrooei M., Alosaimi M.F., Mansouri D., Al-Saud H., Al-Mulla F., Almourfi F., Al-Muhsen S.Z., Alsohime F., Al Turki S., Hasanato R., Van De Beek D., Biondi A., Bettini L.R., D'Angio’ M., Bonfanti P., Imberti L., Sottini A., Paghera S., Quiros-Roldan E., Rossi C., Oler A.J., Tompkins M.F., Alba C., Vandernoot I., Goffard J.C., Smits G., Migeotte I., Haerynck F., Soler-Palacin P., Martin-Nalda A., Colobran R., Morange P.E., Keles S., Çölkesen F., Ozcelik T., Yasar K.K., Senoglu S., Karabela Ş.N., Rodríguez-Gallego C., Novelli G., Hraiech S., Tandjaoui-Lambiotte Y., Duval X., Laouénan C., Snow A.L., Dalgard C.L., Milner J.D., Vinh D.C., Mogensen T.H., Marr N., Spaan A.N., Boisson B., Boisson-Dupuis S., Bustamante J., Puel A., Ciancanelli M.J., Meyts I., Maniatis T., Soumelis V., Amara A., Nussenzweig M., García-Sastre A., Krammer F., Pujol A., Duffy D., Lifton R.P., Zhang S.Y., Gorochov G., Béziat V., Jouanguy E., Sancho-Shimizu V., Rice C.M., Abel L., Notarangelo L.D., Cobat A., Su H.C., Casanova J.L. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;80–:370. doi: 10.1126/SCIENCE.ABD4570. eabd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Swain S.L., McKinstry K.K., Strutt T.M. Expanding roles for CD4 + T cells in immunity to viruses. Nat. Rev. Immunol. 2012;12:136–148. doi: 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yi Y., Lagniton P.N.P., Ye S., Li E., Xu R.H. COVID-19: what has been learned and to be learned about the novel coronavirus disease. Int. J. Biol. Sci. 2020;16:1753–1766. doi: 10.7150/ijbs.45134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Engelmann B., Massberg S. Thrombosis as an intravascular effector of innate immunity. Nat. Rev. Immunol. 2013;13:34–45. doi: 10.1038/nri3345. [DOI] [PubMed] [Google Scholar]
- 81.Miesbach W., Makris M. COVID-19: coagulopathy, risk of thrombosis, and the rationale for anticoagulation. Clin. Appl. Thromb. Hemost. 2020;26 doi: 10.1177/1076029620938149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zaid Y., Puhm F., Allaeys I., Naya A., Oudghiri M., Khalki L., Limami Y., Zaid N., Sadki K., Ben El Haj R., Mahir W., Belayachi L., Belefquih B., Benouda A., Cheikh A., Langlois M.-A., Cherrah Y., Flamand L., Guessous F., Boilard E. Platelets can associate with SARS-CoV-2 RNA and are hyperactivated in COVID-19. Circ. Res. 2020;127:1404–1418. doi: 10.1161/CIRCRESAHA.120.317703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van der Poll T., de Jonge E., an H. ten C. 2013. Cytokines as Regulators of Coagulation.https://www.ncbi.nlm.nih.gov/books/NBK6207/ (accessed June 24, 2020) [Google Scholar]
- 84.Hadid T., Kafri Z., Al-Katib A. Coagulation and anticoagulation in COVID-19. Blood Rev. 2020:100761. doi: 10.1016/j.blre.2020.100761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rapkiewicz A.V., Mai X., Carsons S.E., Pittaluga S., Kleiner D.E., Berger J.S., Thomas S., Adler N.M., Charytan D.M., Gasmi B., Hochman J.S., Reynolds H.R. Megakaryocytes and platelet-fibrin thrombi characterize multi-organ thrombosis at autopsy in COVID-19: a case series. EClinicalMedicine. 2020;24 doi: 10.1016/j.eclinm.2020.100434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bester J., Pretorius E. Effects of IL-1β, IL-6 and IL-8 on erythrocytes, platelets and clot viscoelasticity. Sci. Rep. 2016;6:32188. doi: 10.1038/srep32188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Han H., Yang L., Liu R., Liu F., Wu K.L., Li J., Liu X.H., Zhu C.L. Prominent changes in blood coagulation of patients with SARS-CoV-2 infection. Clin. Chem. Lab. Med. 2020;58:1116–1120. doi: 10.1515/cclm-2020-0188. [DOI] [PubMed] [Google Scholar]
- 89.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dolhnikoff M., Duarte-Neto A.N., de Almeida Monteiro R.A., Ferraz da Silva L.F., Pierre de Oliveira E., Nascimento Saldiva P.H., Mauad T., Marcia Negri E. Pathological evidence of pulmonary thrombotic phenomena in severe COVID-19. J. Thromb. Haemostasis. 2020;18:1517–1519. doi: 10.1111/jth.14844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jose R.J., Manuel A. COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir. 2020;395:e46–e47. doi: 10.1016/S2213-2600(20)30216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tian Y., Rong L., Nian W., He Y. Review article: gastrointestinal features in COVID-19 and the possibility of faecal transmission, Aliment. Pharmacol. Ther. 2020;51:843–851. doi: 10.1111/apt.15731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zuo T., Zhang F., Lui G.C.Y., Yeoh Y.K., Li A.Y.L., Zhan H., Wan Y., Chung A., Cheung C.P., Chen N., Lai C.K.C., Chen Z., Tso E.Y.K., Fung K.S.C., Chan V., Ling L., Joynt G., Hui D.S.C., Chan F.K.L., Chan P.K.S., Ng S.C. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xu K., Cai H., Shen Y., Ni Q., Chen Y., Hu S., Li J., Wang H., Yu L., Huang H., Qiu Y., Wei G., Fang Q., Zhou J., Sheng J., Liang T., Li L. Management of corona virus disease-19 (COVID-19): the Zhejiang experience, Zhejiang da Xue Xue bao. Yi Xue Ban. 2020;49:147–157. doi: 10.3785/j.issn.1008-9292.2020.02.02. https://covid19.elsevierpure.com/tr/publications/management-of-corona-virus-disease-19-covid-19-the-zhejiang-exper (accessed June 24, 2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gou W., Fu Y., Yue L., Chen G., Cai X., Shuai M., Xu F., Yi X., Chen H., Zhu Y.J., Xiao M., Jiang Z., Miao Z., Xiao C., Shen B., Wu X., Zhao H., Ling W., Wang J., Chen Y., Guo T., Zheng J.-S. Cold Spring Harbor Laboratory Press; 2020. Gut Microbiota May Underlie the Predisposition of Healthy Individuals to COVID-19. [DOI] [Google Scholar]
- 96.Weaver L.K., Minichino D., Biswas C., Chu N., Lee J.J., Bittinger K., Albeituni S., Nichols K.E., Behrens E.M. Microbiota-dependent signals are required to sustain TLR-mediated immune responses. JCI Insight. 2019;4 doi: 10.1172/jci.insight.124370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Saleh J., Peyssonnaux C., Singh K.K., Edeas M. Mitochondria and microbiota dysfunction in COVID-19 pathogenesis. Mitochondrion. 2020;54:1–7. doi: 10.1016/j.mito.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Joan P., Davis J.P., Purdy W.K., Wand P.J., Chesney R.W. Clinical manifestations of toxic shock syndrome. JAMA, J. Am. Med. Assoc. 1981;246:741–748. doi: 10.1001/jama.1981.03320070025019. [DOI] [PubMed] [Google Scholar]
- 99.De Wit E., Van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xu J., Zhao S., Teng T., Abdalla A.E., Zhu W., Xie L., Wang Y., Guo X. Systematic Comparison of two Animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020;12:244. doi: 10.3390/v12020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Müller M.A., Dijkman R., Muth D., Demmers J.A.A., Zaki A., Fouchier R.A.M., Thiel V., Drosten C., Rottier P.J.M., Osterhaus A.D.M.E., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yin Y., Wunderink R.G. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23:130–137. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wong C.K., Lam C.W.K., Wu A.K.L., Ip W.K., Lee N.L.S., Chan I.H.S., Lit L.C.W., Hui D.S.C., Chan M.H.M., Chung S.S.C., Sung J.J.Y. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mahallawi W.H., Khabour O.F., Zhang Q., Makhdoum H.M., Suliman B.A. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine. 2018;104:8–13. doi: 10.1016/j.cyto.2018.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Channappanavar R., Fehr A.R., Zheng J., Wohlford-Lenane C., Abrahante J.E., Mack M., Sompallae R., McCray P.B., Meyerholz D.K., Perlman S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 2019;129:3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]