Abstract
Changeux et al. (Changeux et al. C. R. Biol. 343:33–39.) recently suggested that the SARS-CoV-2 spike protein may interact with nicotinic acetylcholine receptors (nAChRs) and that such interactions may be involved in pathology and infectivity. This hypothesis is based on the fact that the SARS-CoV-2 spike protein contains a sequence motif similar to known nAChR antagonists. Here, we use molecular simulations of validated atomically detailed structures of nAChRs and of the spike to investigate the possible binding of the Y674-R685 region of the spike to nAChRs. We examine the binding of the Y674-R685 loop to three nAChRs, namely the human α4β2 and α7 subtypes and the muscle-like αβγδ receptor from Tetronarce californica. Our results predict that Y674-R685 has affinity for nAChRs. The region of the spike responsible for binding contains a PRRA motif, a four-residue insertion not found in other SARS-like coronaviruses. The conformational behavior of the bound Y674-R685 is highly dependent on the receptor subtype; it adopts extended conformations in the α4β2 and α7 complexes but is more compact when bound to the muscle-like receptor. In the α4β2 and αβγδ complexes, the interaction of Y674-R685 with the receptors forces the loop C region to adopt an open conformation, similar to other known nAChR antagonists. In contrast, in the α7 complex, Y674-R685 penetrates deeply into the binding pocket in which it forms interactions with the residues lining the aromatic box, namely with TrpB, TyrC1, and TyrC2. Estimates of binding energy suggest that Y674-R685 forms stable complexes with all three nAChR subtypes. Analyses of simulations of the glycosylated spike show that the Y674-R685 region is accessible for binding. We suggest a potential binding orientation of the spike protein with nAChRs, in which they are in a nonparallel arrangement to one another.
Significance
It was recently suggested that the SARS-CoV-2 spike protein may interact with nicotinic acetylcholine receptors and that such interactions may be involved in pathology and infectivity. We investigate this hypothesis by molecular dynamics simulations. Our results predict that a viral spike protein peptide (adjacent to the furin cleavage site) exhibits favorable binding affinity to nicotinic acetylcholine receptors and suggest subtype-specific dynamics for the peptide. We show that this peptide is accessible in the fully glycosylated spike. We model how the spike may interact with these receptors and find that interaction is possible with the two proteins in a nonparallel arrangement.
Introduction
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is a novel strain of coronavirus that first appeared in China in late 2019 and causes the potentially fatal disease coronavirus disease 2019 (COVID-19). This virus initially infects respiratory epithelial cells by binding to the angiotensin-converting 2 enzyme (ACE2) receptor (1). Although predominantly recognized as a respiratory disease (2,3), SARS-CoV-2 also causes severe inflammation and damage in other organs (4, 5, 6, 7). Under certain conditions (and as with other coronaviruses (8)), SARS-CoV-2 may enter the central nervous system (CNS) through the bloodstream by disrupting the blood-brain barrier or infecting the peripheral nerves (e.g., (7,9, 10, 11, 12)).
Since it emerged as a human pathogen, SARS-CoV-2 has caused more than 80.8 million confirmed cases of COVID-19 and more than 1.7 million deaths worldwide, as of December 28, 2020 (13). Several major risk factors for the development of COVID-19 have been identified, including age, heart disease, diabetes, and hypertension (14). Given the apparently low prevalence of smokers among hospitalized COVID-19 patients (15, 16, 17), it was proposed that nicotine may offer some protective value to mitigate COVID-19 (the “protection” hypothesis) (15). It has been suggested that medicinal nicotine (either in patches, gum, or electronic delivery systems) should be investigated as a therapeutic option for this disease (e.g., (15,18)). Clinical trials for nicotine are underway (e.g., https://clinicaltrials.gov/ct2/show/NCT04429815). Alternative explanations to the protection hypothesis have been proposed (19); the first relates to the failure in correctly identifying smokers upon hospital admission (19) and the second is that hospitalized COVID-19 patients may be less likely to smoke as their comorbidities motivate them to quit (“smoking cessation” hypothesis) (19).
Based on the early observations of the lower-than-expected smoking prevalence in hospitalized COVID-19 patients, Changeux and colleagues suggested a role for nicotinic acetylcholine receptors (nAChRs) in the pathophysiology of COVID-19 via a direct interaction between these receptors and the viral spike glycoprotein (20). This suggestion was based on the fact that the spike protein contains a sequence motif similar to known nAChR antagonists (20) (Fig. S1), such as α-bungarotoxin from Bungarus multicinctus and glycoprotein from Rabies lyssavirus (formerly Rabies virus). Changeux et al. and others also proposed that COVID-19 might be controlled or mitigated by the use of nicotine, if the latter can compete with the virus for binding to these receptors (e.g., (9,18,20, 21, 22, 23, 24)). If interactions with nAChRs are important, they may be relevant for some of the systemic effects observed in COVID-19.
nAChRs are cation channels that belong to the pentameric ligand-gated ion channel family (25). They are present in both the peripheral (at the skeletal neuromuscular junction and in the autonomic nervous system) and CNS (26). The neuronal receptors have emerged as important targets for the treatment of Alzheimer’s disease, schizophrenia, pain, and nicotine addiction (26,27). Mutations of muscle nAChR can cause congenital myasthenia gravis (27). A large repertoire of nAChR subtypes differ in the homo- or heteromeric assembly of five monomers arranged around a central channel axis (28, 29, 30). Each nAChR subtype shows different selectivity for agonists and antagonists (28, 29, 30). All nAChRs share the same basic architecture (Fig. 1 B), formed of a large N-terminal extracellular domain (ECD), where the agonist-binding site is located; a transmembrane domain (TMD) surrounding the ion channel; an intracellular domain (ICD); and a short extracellular C-terminal domain (CTD) (28, 29, 30) The ligand-binding pocket is located at the interface between two neighboring subunits (Fig. 1 B) and is formed by loops A, B, and C from the principal subunit and D, E, and F from the complementary subunit (Fig. S2). The α4β2 nAChR is the most prevalent heteromeric subtype in the brain; it is implicated in diverse processes such as cognition, mood, and reward and is necessary for nicotine addiction (28, 29, 30,34). The homomeric α7 nAChR is also abundant and widely expressed in the CNS, where it contributes to cognition, sensory processing, and attention (35). The α7 subtype is also expressed on a variety of nonneuronal cells, such as immune cells, astrocytes, microglia, and endothelial cells, where it contributes to anti-inflammatory pathways (36, 37, 38). Because of its role in the downregulation of the production of proinflammatory cytokines (36, 37, 38), it has been suggested that the α7 nAChR may be involved in the hyperinflammation response that can be caused by SARS-CoV-2 (9,18,24,39). The muscle-type receptor derived from the electric organ of Tetronarce californica (formerly Torpedo californica) is one of the most extensively studied nAChRs and has provided significant structural insight into this receptor family. It is formed by two α- and one each of β-, δ-, and γ-subunits and has high sequence similarity (55–80% identity) with its human counterpart (40). For this reason and because its structure is available (33), we used it in this work as a proxy for the human muscle-type nAChRs. Muscle fatigue, myalgia, and arthralgia are common symptoms in COVID-19 patients (e.g., (41, 42, 43)). However, it is still unclear if these symptoms result from direct muscle damage from viral infection or from the body’s inflammatory response (7,41).
Figure 1.
Overview of the three-dimensional structures of the SARS-CoV-2 spike protein and the αβγδ nAChR from T. californica. (A) The model for the complete, fully glycosylated, SARS-CoV-2 spike represents the closed state of the protein after furin cleavage (31). The spike protein is a homotrimer (32); each monomer is shown in different colors, namely green, cyan, and orange, with glycans depicted in pink. Each monomer is formed by three domains: head, stalk, and cytoplasmic tail (CT) (32). The Y674-R685 region is shown in red. (B) Shown is the cryoEM structure of the muscle-type receptor from T. californica (PDB: 6UWZ) (33). This receptor is a heteropentamer formed of two α- (green), one β- (blue), one δ- (yellow), and one γ- (orange) subunits. Each monomer is formed by four domains (28, 29, 30): ECD, transmembrane domain (TMD), intracellular domain (ICD), and C-terminal domain (CTD). The agonist binding site is located in the ECDs at the interface between two neighboring subunits. To see this figure in color, go online.
According to Changeux et al.’s “nicotinic hypothesis,” direct interaction between SARS-CoV-2 and nAChRs occurs via a small region in the viral spike protein (20) (Figs. S3 and S4). The spike is a fusion protein (32,44) found on the surface of the virion that mediates entry into host cells. It is an extensively glycosylated homotrimer, with each monomer formed by three domains (Fig. 1 A): head, stalk, and cytoplasmic tail (32). The head comprises two subunits: S1, which binds the ACE2 receptor on host cells (32), and S2, which facilitates membrane fusion (32). The spike contains two proteolytic cleavage sites (32): one (“furin cleavage” site) at the S1/S2 boundary thought to activate the protein (45) and a second in the S2 subunit that releases the fusion peptide (46). The region suggested by Changeux et al. (20) to be directly involved in the interaction with nAChRs spans from Y674 to R685 and is located in the head region of the protein at the interface between the S1 and S2 domains, immediately preceding the S1/S2 cleavage point (32) (Fig. 1 A; Figs. S3 and S4). Furin cleaves the peptide bond after R685, thus separating it from its neighbor S686 (e.g., before viral exit from the host cell) (45). Cleavage activation of viral glycoproteins is known to be important for infectivity and virulence (32,45).
The Y674-R685 region contains a four-residue, polybasic PRRA insertion not present in other SARS-CoV-related coronaviruses (47) that is homologous to several neurotoxins known to target nAChRs (20). In SARS-CoV-2, abrogation of the PRRA motif moderately affects virus entry into cells (32,45). This motif has recently been shown experimentally to interact with neuropilin-1 receptors (48), T cell receptors (49), and host cell glycans, such as heparin sulfate (50,51). The high sequence similarities between the Y674-R685 region and several known nAChR antagonists (Fig. S1) suggests that this region of the SARS-CoV-2 spike protein may bind to nAChRs, potentially acting as an antagonist (20). Hence, it has been postulated that nicotine may have an effect in COVID-19 by competing and interfering with this binding. Note that an alternative region (G381 to K386 in the S1 subunit) in the spike protein has been hypothesized to interact with nAChRs (52), but glycosylation makes this unlikely.
Here, we use molecular simulations to examine the “nicotinic hypothesis” proposed by Changeux et al. (20), in particular to test whether the SARS-CoV-2 spike protein can bind stably to nAChRs via the Y674-R685 region and identify interactions that may be involved in the stabilization of the complexes. To test this, we have built structural models for the complexes formed by the 12-residue region from the spike (S-peptide) and the ECDs of three different nAChRs, namely the human α4β2, human α7, and muscle-like αβγδ receptor from T. californica (hereafter named αβγδ). These simulations build on our successful previous extensive simulations of nAChRs, which have e.g., identified a general mechanism for signal propagation in this receptor family (53, 54, 55) and simulations of the spike (31,56, 57, 58) and its interactions (48,59).
Results and discussion
Interactions between the SARS-CoV-2 S-peptide and nAChRs
Structural models of the three SARS-CoV-2 S-peptide-nAChR complexes were built based on the cryo-electron microscopy (cryoEM) structure of the αβγδ receptor from T. californica with bungarotoxin (33). α-Bungarotoxin is a neurotoxin that acts as a nAChR antagonist, directly competing with acetylcholine (60), and has high sequence similarity with the Y674-R685 region of the spike protein of SARS-CoV-2 (Fig. S1). Twenty models were generated for each complex, and the one with the lowest Modeler objective function (61) (Fig. 2; Fig. S7) was used as the starting point for molecular dynamics (MD) simulations (see the Supporting materials and methods for more details). Three replicate simulations, each 300-ns long, were performed for each complex to investigate the peptide-receptor conformational behavior and possible induced-fit effects.
Figure 2.
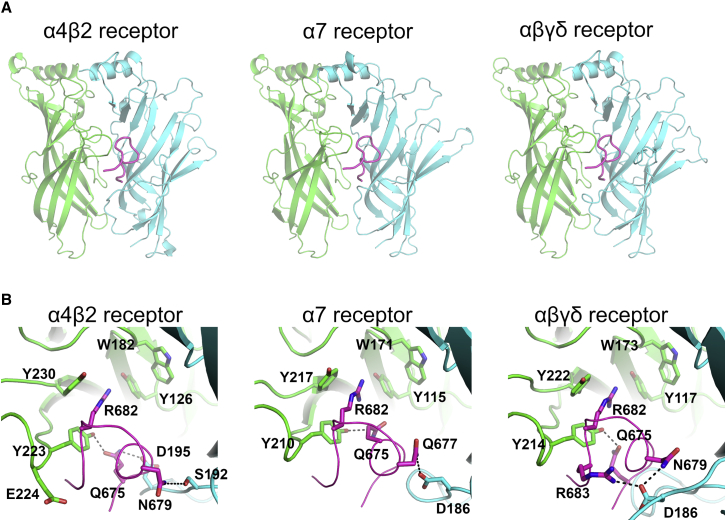
Predicted binding modes of the SARS-CoV-2 S-peptide to different nAChRs. (A) Shown are complexes formed by the S-peptide and three different nAChRs, namely the human α4β2, human α7, and the muscle-like αβγδ receptor from T. californica. The S-peptide (region Y674-R685) is highlighted in magenta, and the principal and complementary subunits of the receptors are colored in green and cyan, respectively. These models show the conformation of the S-peptide bound to the first pocket at the beginning of the simulations. In the human α4β2 receptor, the binding pocket is formed by one α4 and one β2 subunit, whereas in the human α7 nAChR, the pocket is formed by two α7 subunits. In the αβγδ receptor, the two binding pockets are nonequivalent; one is formed by an α and a δ and the second by α- and γ-subunits. (B) Shown is a close-up view of the peptide-receptor interaction region. Residues involved in binding of the S-peptide are shown with sticks. Note that the side chain of R682 in the S-peptide is located inside the aromatic box establishing cation-π interactions with some of the highly conserved aromatic residues lining the pocket. Note also that all residue numbers used in this work, unless stated otherwise, refer to the human α7 (Uniprot: P36544), human α4 (Uniprot: P43681), human β2 (Uniprot: P17787), T. californica α (Uniprot: P02710), T. californica δ (Uniprot: P02718), T. californica γ (Uniprot: P02714), and SARS-CoV-2 spike protein (Uniprot: P0DTC2) sequences. To see this figure in color, go online.
At the beginning of the simulations, the S-peptide was located in the binding pocket, bound by interactions with both the principal and complementary subunits (Fig. 2; Fig. S7). A close-up view of the peptide-receptor interface reveals extensive contacts (Fig. 2 B; Fig. S7 B), mainly with the principal subunit. In all three complexes, the side chain of R682 of the S-peptide binds as the recognized positively charged group, a strictly conserved pharmacophore of all nAChR ligands (62,63). As can be seen in Fig. 2 B, the guanidinium group of R682 is well positioned inside the aromatic box, forming several cation-π interactions with TyrC1 (α4Y223, α7Y210, and αY214 in the human α4β2, human α7, and muscle-like αβγδ receptor from T. californica, respectively), TyrC2 (α4Y230, α7Y217, and αY222), and TyrA (α4Y126, α7Y115, and αY117). Note that these cation-π interactions do not entirely mimic the binding of nicotine, as no interactions with TrpB are present (64). R682 is part of the four-residue PRRA insertion not found in other SARS-like coronaviruses (47), and it forms part of the furin cleavage site located at the boundary between the S1 and S2 subunits (32). Additional binding interactions with the peptide are also observed with different residues depending on the receptor subtype; in the α4β2 nAChR, hydrogen bonds involving the side chains of α4Y223, α4E224, β2S192, and β2D195 in the receptor and Q675, N679, and the main-chain nitrogen of A684 of the S-peptide are observed; in the α7 nAChR, two hydrogen bonds between α7D186 and α7Y210 in the receptor and S-peptide Q675 and Q677 are seen; and in the αβγδ receptor from T. californica, hydrogen bonds involving αY214 and δD186 from the receptor and Q675, N679, R682, and R683 of the peptide are observed.
The simulations show distinct patterns of dynamical behavior for the S-peptide in the different receptor subtypes. In the α4β2 and α7 complexes, the peptide showed high positional and conformational variability, whereas in the αβγδ complex, it generally remained in the same pose throughout the simulation (Figs. S8 and S10). Similar behavior is observed for the peptides in the two binding pockets in each complex. When bound to the α4β2 and α7 nAChR, the peptide adopted many different binding modes inside the pocket, ranging from highly compact to fully extended conformations (Fig. S10). In contrast, in the αβγδ receptor, the peptide was more compact (Fig. S10). The range of the radius of gyration values for the S-peptide in all three complexes is similar to that observed in the simulations of the full-length glycosylated SARS-CoV-2 spike protein embedded in a viral membrane (Fig. S6; (31)). Principal component analysis (PCA) of the peptide dynamics revealed different conformational behavior of the peptide in the three complexes. When bound to the muscle-like receptor, the peptide shows limited dynamical freedom; it explores a restricted conformational space spanned by the first two principal components (Fig. S11).
The number of hydrogen bonds between the peptide and the receptors over the simulations was determined (Fig. S12). Two more H-bonds are observed in the αβγδ complex than in the α4β2 and α7 receptors (Fig. S12). These additional interactions with the complementary subunit (Fig. S12) probably contribute to the increased stability of this complex and the more compact conformation of the peptide in the αβγδ receptor.
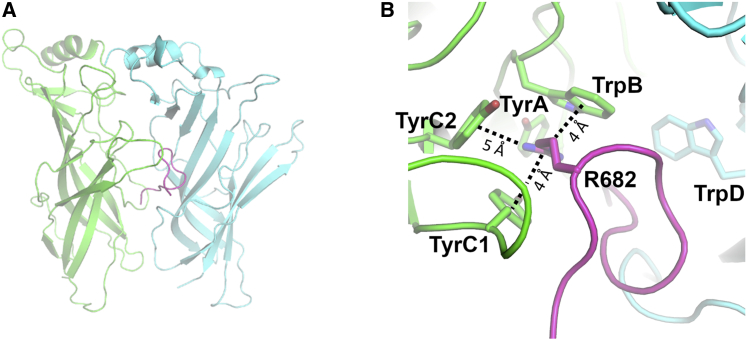
Analysis of the distribution of the distance between the R682 of the peptide and the conserved aromatic residues forming the aromatic box shows the distinctive behavior of the peptide when bound to different receptors (Fig. S13). Interactions with R682, TyrC1, and TyrC2 are quite frequent in all three complexes, being present more than 60% of the time. To examine how deeply into the binding pocket the peptide inserts, we monitored the interactions of R682 with TrpB, a residue lining the back wall of the nAChR aromatic box. TrpB (α4W182, α7W171, and αW173) is highly conserved across the nAChR family, and it makes cation-π and H-bond interactions with the positively charged group on the ligands (62,63). In the α4β2 and αβγδ complexes, the S-peptide does not extend far into the pocket, and interactions between R682 and TrpB are mostly absent (Fig. S13). In contrast, in the α7 complex, the peptide binds more deeply into the hydrophobic cavity, adopting conformations that allow not only for the direct contact between R682 and TrpB (Figs. S14–S15) but also achieve optimal core-binding interactions (Fig. 3). In such configurations, other interactions are present in addition to those with TrpB, namely cation-π interactions with TyrC1 and TyrC2 (Fig. S15). Although no direct contact between R682 and TyrA is observed, both residues are connected through a H-bond network mediated by Q675 from the S-peptide (Fig. S16). This is significant because interactions with TyrA, TrpB, TyrC1, and TyrC2 are known to be critical for ligand binding and to modulate gating in the α7 subtype (65, 66, 67).
Figure 3.
Representative conformation of the α7 complex, in which direct interaction between TrpB and R682 is observed. (A) Shown is an overall view of the S-peptide:α7 complex. (B) Shown is a close-up view of the R682 interaction region within the aromatic box. The principal and complementary subunits of the α7 receptor are colored in green and cyan, respectively. The S-peptide is highlighted in magenta. Interactions between the guanidinium group of R682 and the aromatic rings of TrpB (α7W171), TyrC1 (α7Y210), and TyrC2 (α7Y217) are shown with dashed lines. See also Figs. S13–S16 for more details about the behavior of the S-peptide when bound to α7 nAChR. To see this figure in color, go online.
The binding of a ligand or a peptide can be expected to affect the conformational dynamics of the receptors (e.g., (53, 54, 55,68, 69, 70)). To investigate this, the root mean-square fluctuation (RMSF) profiles of the Cα atoms were determined for all three receptors. Distinct dynamic behaviors are observed for the binding site regions in the different subtypes (Figs. S17–S19). These differences are mostly in loops C and F, two structural motifs important for binding and selectivity (66,71,72). Loop F shows decreased flexibility in the α4β2 complex, whereas loop C dynamics is more restricted in the muscle-like αβγδ receptor compared with the other two subtypes.
At the beginning of the simulations, in all the three complexes, loop C adopted an open conformation because of the steric interference of the peptide. During the simulations, the αβγδ and α4β2 receptors mostly maintained this open conformation. In the α7 complex, as the peptide moved deeper into the binding pocket, loop C rotated inwards, adopting a semiclosed structure. Loop C capping is known to be important for the anchoring of the ligands in the binding pocket (66,71) and has been suggested to be indirectly involved in gating (54,73). A relationship between loop C position and ligand activation has also been proposed (72); agonists are proposed to stabilize more compact loop conformations, whereas antagonists disfavor loop closing. On this basis, our findings suggest that the S-peptide may act as an antagonist in the αβγδ and α4β2 receptors, thus inhibiting gating. However, in the α7 subtype, it is unclear whether the peptide may be an agonist or antagonist and whether it can promote gating. How the S-peptide affects the different nAChRs may be relevant to understanding COVID-19 pathophysiology (9,18,24,39).
A molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) approach (74,75) was used to calculate the free energy of binding of the S-peptide to the different receptors (Table 1; Table S1). MM-PBSA calculations are an efficient and often useful method to estimate binding free energies (74,75) and are widely used to study protein-ligand interactions in medicinal chemistry (76, 77, 78), including in drug design for nAChRs (79,80). The favorable calculated binding energies suggest stable complex formation between the S-peptide and all three nAChRs (Table 1; Table S1), with different binding affinities depending on the subtype.
Table 1.
MM-PBSA relative binding energy values for the S-peptide in the human α4β2, human α7, and muscle-like αβγδ nAChR from T. californica
| Average ΔGbind for the complexes (kJ/mol) |
|||
|---|---|---|---|
| α4β2 | α7 | αβγδ | |
| First pocket | −215.9 (80.4) | −184.5 (24.3) | −374.3 (98.5) |
| Second pocket | −215.7 (55.7) | −114.9 (46.6) | −391.5 (75.8) |
Numbers in brackets represent the SDs. Note that the values reported in this table are averaged over all replicates (see Table S1 for the ΔGbind for the individual replicates) and do not contain the entropic contribution to the binding energy.
In silico alanine-scanning mutagenesis was performed to identify important residues (referred to as “hotspots”) in peptide-receptor association (Figs. S20–S22). Hotspots are residues with high energetic contributions to the thermodynamic stability of a given complex (81). Alanine-scanning provides a detailed energy map of a protein-binding interface (81). Here, we used the fast in silico method, BudeAlaScan (81), in which every residue, for both receptor and peptide, is mutated (singly, in turn) to alanine. Hotspots are determined by the difference between the binding free energies of the alanine mutant and wild-type complexes (ΔΔGbind) (81). Hotspots were identified at the interface of the receptor, some of them common to all three subtypes (Fig. S23; Tables 2; Table S2). In particular, TyrC1 (α4Y223, α7Y210, and αY214) and the negatively charged residues in the upper part of loop F (β2D195, α7D186, δD201, and δE203) strongly stabilize the complex. In the human α7 nAChR, the substitution of several key agonist-binding residues in the aromatic box (namely TyrA (α7Y115), TyrC1 (α7Y210), TrpB (α7W171), and TrpD (α7W77)) by alanine is also predicted to destabilize the interface between the peptide and the receptor. Of the residues in the peptide, Y674, R682, and R685 are the major contributors to stabilizing the interface (Fig. S24). This analysis reinforces the critical role of R682 in binding to nAChRs.
Table 2.
BUDE Alanine-scanning predicted average ΔΔGbind for the hot spots (−3 kJ/mol ≥ residue contribution ≤ 3 kJ/mol) in the first binding pocket of the receptors
| First binding pocket | |||||
|---|---|---|---|---|---|
|
α4β2 receptor |
α7 receptor |
Muscle-like αβγδ receptor |
|||
| Residue | ΔΔGbind (kJ/mol) | Residue | ΔΔGbind (kJ/mol) | Residue | ΔΔGbind (kJ/mol) |
| β2D195 | 9.5 (3.6) | α7Y210 | 7.6 (2.2) | αY214 | 12.1 (2.6) |
| α4Y223 | 7.7 (2.0) | α7W77 | 5.1 (2.0) | δD201 | 6.1 (1.9) |
| α4Y230 | 3.7 (2.2) | α7Y115 | 3.8 (2.9) | δW197 | 4.6 (2.3) |
| β2W32 | 3.3 (1.7) | α7S188 | 3.7 (1.9) | δI199 | 4.0 (0.8) |
| α7D186 | 3.1 (2.1) | δD186 | 3.9 (1.7) | ||
| δE203 | 3.8 (2.0) | ||||
| αT215 | 3.1 (1.5) | ||||
The average value was calculated over the three replicates. Numbers in brackets represent the SDs (calculated over the 303 frames per complex). Note that the ΔΔGbind corresponds to the difference between mutant and wild-type complexes, and as such, positive ΔΔGbind values mean that the mutation to alanine destabilizes the complex.
Accessibility of the SARS-CoV-2 S-peptide in MD simulations of the full-length glycosylated spike
Since the beginning of the pandemic, the computational structural biology/biomolecular simulation community has investigated the SARS-CoV-2 spike protein in different states and conditions and the complexes that it forms (e.g., (31,56, 57, 58, 59,82, 83, 84, 85, 86)). Simulations have revealed the dynamics of the spike and its glycan shield (31,57, 58, 59,82,83,85,86) and the effects of the binding of small molecules (56,84). Here, to further explore the “nicotinic hypothesis,” we show that the Y674-R685 region (corresponding to the S-peptide) is accessible for binding, using the available MD simulations of the fully glycosylated full-length SARS-CoV-2 spike protein in the open and closed states by Casalino et al. (31). We note that in these models, the Y674-R685 region was modeled de novo as it was entirely (open spike) or partially (closed spike) missing in the initial cryoEM structures (32,44) (for more details, see Supporting materials and methods). In these simulations, the Y674-R685 region adopts conformations potentially compatible with binding to nAChRs (Fig. 4 A). Our analysis reveals that the Y674-R685 loop is only weakly shielded by the glycans and is predominantly solvent exposed (Fig. 4; Fig. S5). Especially when the spike is in the closed state (Fig. 4 B), the Y674-R685 loop appears highly accessible to a probe with a radius ranging from 1.4 to 15 Å. In contrast, in the open spike (Fig. 4 C), a larger variability of the accessible area is observed, preventing an unambiguous interpretation of the glycan shield effect on Y674-R685 for probes with a radius larger than 7 Å. The slightly different and less variable accessibility of the Y674-R585 loop observed in the closed spike when compared the open spike protein is in agreement with the sharper distribution of the radius of gyration calculated for this region in the closed spike (Fig. S6). This behavior might indicate different binding propensity of the S-peptide in the open and closed spike states. We hypothesize that it might be linked to a different packing of the three spike monomers in the two states. We note that the accessibility of this region makes it available to bind other receptors that may also bind the PRRA motif, such as neuropilin-1.
Figure 4.
Accessible surface area (ASA) of Y674-R685 region in the context of the fully glycosylated full-length SARS-CoV-2 spike. (A) Shown is a snapshot taken from the simulations by Casalino et al. (31) of the glycosylated full-length SARS-CoV-2 spike in the closed state showing Y674-R685 loop protruding into the solvent. The protein is depicted with a gray surface, whereas 674–685 loop is shown as a cyan ribbon. The glycans are illustrated with blue sticks. (B–C) The ASA of the of residues 674–685 (corresponding to the S-peptide) and the area shielded by glycans at multiple probe radii from 1.4 Å (water molecule) to 15 Å are calculated using the available MD trajectories of the full-length models of the glycosylated SARS-CoV-2 spike protein in the closed (B) and open states (C) from Casalino et al. (31). The area of 674–685 shielded by the glycans is presented in blue, whereas the gray line represents the accessible area of 674–685 in the absence of glycans. Highlighted in cyan is the area of 674–685 that remains accessible in the presence of glycans. The calculated values have been averaged across the three chains and across the different replicas performed for each system by Casalino et al. (31). Error bars correspond to ± SD. To see this figure in color, go online.
Conclusions
In summary, the findings reported here support the hypothesis that the SARS-CoV-2 spike protein can interact with nAChRs. Our calculations indicate stable binding of the spike protein to these receptors through a region adjacent to the furin cleavage site and corresponding to the Y674-R685 region. These calculations also show apparent subtype-specific interactions and dynamics for the Y674-R685 region. COVID-19 is known to cause a range of neurological (87,88), muscular (41), and respiratory (89) symptoms, and these predicted interactions may be relevant to understand the pathophysiology associated with this disease.
Our results predict that the Y674-R685 region of the spike protein has affinity for nAChRs. The region in the spike responsible for binding to nAChRs harbors the PRRA motif and shares high sequence similarity with neurotoxins known to be nAChR antagonists. The guanidinium group of R682 is the key anchoring point to the binding pocket, in which it forms several interactions with the residues that form the aromatic box. Analysis of the structure and dynamics of the full-length glycosylated spike shows that the Y674-R685 region protrudes outside the glycan shield, is solvent accessible (Fig. 4; Fig. S5), and is flexible (Fig. S6), showing that it is accessible to bind to nAChRs (and to other receptors such as neuropilins (48)). Modeling the interaction between the full-length spike and nAChRs indicates that association is possible with the proteins in a nonparallel orientation to one another (Fig. S4). cryoEM and tomography experiments and coarse-grained simulations show considerable bending and tilting of the spike. A tilt angle up to 60° relative to the normal axis of the membrane is observed (57,58,85,90,91). This flexibility of the spike protein would facilitate binding to host nAChRs.
Evidence that the interaction between the Y674-R685 region of the spike and nAChRs is possible comes from the recently characterized interaction of the spike protein with neuropilin-1 (48), which was shown to occur via the same region as the one proposed here. Having explored various possible orientations, we find that only approximately nonparallel arrangements of the spike and receptor allow for their interaction. This nonparallel interaction may not be immediately obvious, but it is consistent with other observations and is possible for two principal reasons: first, membrane curvature and deformation, and second, bending of the stalk of the spike. Experiments (e.g., cryoEM and tomography) and coarse-grained simulations show that a significant degree of stalk bending is possible and that the spike can adopt a wide range of conformations with different degrees of bending (57,85,90,91) given by the three flexible hinges in the spike protein (85).
In the α4β2 and αβγδ complexes, the conformational dynamics of the bound Y674-R685 peptide are compatible with the hypothesis of it acting as an antagonist; it forces loop C to adopt an open conformation and prevents the formation of important interactions within the binding pocket. Intriguingly, in the α7 complexes, the peptide adopts binding modes that allow strong interactions within the aromatic box, raising the question of whether it promotes gating in this subtype. This is important because activation of α7 nAChR triggers anti-inflammatory signaling mechanisms in inflammatory cells, leading to a decrease in cytokine production, which may have relevance in understanding early COVID-19 pathology (9,18,24,39). If nicotine does indeed prove to have any clinical value, it is likely that it would be due to interfering with the association with nAChRs. If so, nicotine analogs (e.g., smoking cessation agents such as varenicline (92), cytisine (93), and, potentially, cytisine variants (55)) could also find a useful application for COVID-19.
Given the promising results presented here, structural, mutational, and single-channel studies will be of interest to test the importance of the interactions of the SARS-CoV-2 spike with nAChRs and the potential relevance of these interactions to pathology and infectivity in COVID-19. To assist with further investigations, we make our simulation files and datasets available and openly accessible, in accordance with the sharing principles agreed to by our community for simulations relevant to COVID-19 (94).
Author contributions
Conceptualization/design of the work: A.S.F.O. and A.J.M. Acquisition and analysis of the data: A.S.F.O., A.A.I., L.C., and Z.G. Writing of the manuscript: A.S.F.O., A.A.I., L.C., and A.J.M. Review and editing of the manuscript: A.S.F.O., A.J.M., I.B., Z.G., T.G., D.K.S., R.B.S., and R.E.A. Funding acquisition: T.M., R.E.A., and A.J.M.
Acknowledgments
A.J.M. and A.S.F.O. thank EPSRC (grant EP/M022609/1) and the Elizabeth Blackwell Institute for Health Research, University of Bristol for financial support (Elizabeth Blackwell Institute Rapid Response Funding Call (COVID-19)). MD simulations were carried out using the computational facilities of the Advanced Computing Research Centre, University of Bristol (http://www.bris.ac.uk/acrc) and using Oracle Public Cloud Infrastructure (https://cloud.oracle.com/en_US/iaas) under an award for COVID-19 research. We thank Drs. Simon Bennie and Jonathan Barnoud for help with the Cluster-in-the-cloud and the creation of a scalable cluster on the Oracle Cloud. A.J.M., A.S.F.O., R.B.S., and D.K.S. also thank EPSRC for provision of ARCHER HPC time through HECBioSim (HECBioSim.ac.uk) under a COVID-19 award. R.E.A. acknowledges support from NIH (GM132826), NSF RAPID (MCB-2032054), an award from the RCSA Research Corp., and a UC San Diego Moore’s Cancer Center 2020 SARS-COV-2 seed grant. R.E.A., L.C., and Z.G. thank the Texas Advanced Computing Center (TACC) Frontera team and acknowledge computer time made available through a Director’s Discretionary Allocation (made possible by the NSF award OAC-1818253).
Editor: Tamar Schlick.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.01.037.
Supporting citations
References (95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123) appear in the Supporting material.
Supporting material
References
- 1.Yan R., Zhang Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lake M.A. What we know so far: COVID-19 current clinical knowledge and research. Clin. Med. (Lond.) 2020;20:124–127. doi: 10.7861/clinmed.2019-coron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puntmann V.O., Carerj M.L., Nagel E. Outcomes of cardiovascular magnetic resonance imaging in patients recently recovered from coronavirus disease 2019 (COVID-19) JAMA Cardiol. 2020;5:1265–1273. doi: 10.1001/jamacardio.2020.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varga Z., Flammer A.J., Moch H. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puelles V.G., Lütgehetmann M., Huber T.B. Multiorgan and renal tropism of SARS-CoV-2. N. Engl. J. Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y., Geng X., Wang H. New understanding of the damage of SARS-CoV-2 infection outside the respiratory system. Biomed. Pharmacother. 2020;127:110195. doi: 10.1016/j.biopha.2020.110195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desforges M., Le Coupanec A., Talbot P.J. Human coronaviruses and other respiratory viruses: underestimated opportunistic pathogens of the central nervous system? Viruses. 2019;12:1–28. doi: 10.3390/v12010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tizabi Y., Getachew B., Aschner M. Nicotine and the nicotinic cholinergic system in COVID-19. FEBS J. 2020;287:3656–3663. doi: 10.1111/febs.15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baig A.M., Khaleeq A., Syeda H. Evidence of the COVID-19 virus targeting the CNS: tissue distribution, host-virus interaction, and proposed neurotropic mechanisms. ACS Chem. Neurosci. 2020;11:995–998. doi: 10.1021/acschemneuro.0c00122. [DOI] [PubMed] [Google Scholar]
- 11.Baig A.M., Sanders E.C. Heralding healthcare professionals: recognition of neurological deficits in COVID-19. ACS Chem. Neurosci. 2020;11:1701–1703. doi: 10.1021/acschemneuro.0c00286. [DOI] [PubMed] [Google Scholar]
- 12.Alam S.B., Willows S., Sandhu J.K. Severe acute respiratory syndrome coronavirus 2 may be an underappreciated pathogen of the central nervous system. Eur. J. Neurol. 2020;27:2348–2360. doi: 10.1111/ene.14442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang C., Wang Y., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farsalinos K., Barbouni A., Niaura R. Systematic review of the prevalence of current smoking among hospitalized COVID-19 patients in China: could nicotine be a therapeutic option? Intern. Emerg. Med. 2020;15:845–852. doi: 10.1007/s11739-020-02355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan W.J., Ni Z.Y., Zhong N.S., China Medical Treatment Expert Group for Covid-19 Clinical characteristics of coronavirus Disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu W., Tao Z.W., Hu Y. Analysis of factors associated with disease outcomes in hospitalized patients with 2019 novel coronavirus disease. Chin. Med. J. (Engl.) 2020;133:1032–1038. doi: 10.1097/CM9.0000000000000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez-Rubio J., Navarro-Lopez C., Najera A. Cytokine release syndrome (CRS) and nicotine in COVID-19 patients: trying to calm the storm. Front. Immunol. 2020;11:1359. doi: 10.3389/fimmu.2020.01359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen B., Nichols A., Lester H. Successful cessation programs that reduce comorbidity may explain surprisingly low smoking rates among hospitalized COVID-19 patients. Qeios. 2020 doi: 10.32388/WURFH0. [DOI] [Google Scholar]
- 20.Changeux J.P., Amoura Z., Miyara M. A nicotinic hypothesis for Covid-19 with preventive and therapeutic implications. C. R. Biol. 2020;343:33–39. doi: 10.5802/crbiol.8. [DOI] [PubMed] [Google Scholar]
- 21.Altable Pérez M., De la Serna J.M. Neuroinvasion and viral reservoir in COVID-19. Cureus. 2020;12:e11014. doi: 10.7759/cureus.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Virgiliis F., Di Giovanni S. Lung innervation in the eye of a cytokine storm: neuroimmune interactions and COVID-19. Nat. Rev. Neurol. 2020;16:645–652. doi: 10.1038/s41582-020-0402-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leitzke M., Stefanovic D., Schönknecht P. Autonomic balance determines the severity of COVID-19 courses. Bioelectron. Med. 2020;6:22. doi: 10.1186/s42234-020-00058-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dratcu L., Boland X. Does nicotine prevent cytokine storms in COVID19? Cureus. 2020;12:e11220. doi: 10.7759/cureus.11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wonnacott S. Presynaptic nicotinic ACh receptors. Trends Neurosci. 1997;20:92–98. doi: 10.1016/s0166-2236(96)10073-4. [DOI] [PubMed] [Google Scholar]
- 26.Gharpure A., Noviello C.M., Hibbs R.E. Progress in nicotinic receptor structural biology. Neuropharmacology. 2020;171:108086. doi: 10.1016/j.neuropharm.2020.108086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cecchini M., Changeux J.P. The nicotinic acetylcholine receptor and its prokaryotic homologues: structure, conformational transitions & allosteric modulation. Neuropharmacology. 2015;96:137–149. doi: 10.1016/j.neuropharm.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 28.Thompson A.J., Lester H.A., Lummis S.C. The structural basis of function in Cys-loop receptors. Q. Rev. Biophys. 2010;43:449–499. doi: 10.1017/S0033583510000168. [DOI] [PubMed] [Google Scholar]
- 29.Nemecz Á., Prevost M.S., Corringer P.J. Emerging molecular mechanisms of signal transduction in pentameric ligand-gated ion channels. Neuron. 2016;90:452–470. doi: 10.1016/j.neuron.2016.03.032. [DOI] [PubMed] [Google Scholar]
- 30.Changeux J.P. The nicotinic acetylcholine receptor: a typical ‘allosteric machine’. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018;373:20170174. doi: 10.1098/rstb.2017.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casalino L., Gaieb Z., Amaro R.E. Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. ACS Cent. Sci. 2020;6:1722–1734. doi: 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walls A.C., Park Y.J., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rahman M.M., Teng J., Hibbs R.E. Structure of the native muscle-type nicotinic receptor and inhibition by snake venom toxins. Neuron. 2020;106:952–962.e5. doi: 10.1016/j.neuron.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dineley K.T., Pandya A.A., Yakel J.L. Nicotinic ACh receptors as therapeutic targets in CNS disorders. Trends Pharmacol. Sci. 2015;36:96–108. doi: 10.1016/j.tips.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haydar S.N., Dunlop J. Neuronal nicotinic acetylcholine receptors - targets for the development of drugs to treat cognitive impairment associated with schizophrenia and Alzheimer’s disease. Curr. Top. Med. Chem. 2010;10:144–152. doi: 10.2174/156802610790410983. [DOI] [PubMed] [Google Scholar]
- 36.Martelli D., Farmer D.G., Yao S.T. The splanchnic anti-inflammatory pathway: could it be the efferent arm of the inflammatory reflex? Exp. Physiol. 2016;101:1245–1252. doi: 10.1113/EP085559. [DOI] [PubMed] [Google Scholar]
- 37.Tracey K.J. The inflammatory reflex. Nature. 2002;420:853–859. doi: 10.1038/nature01321. [DOI] [PubMed] [Google Scholar]
- 38.Maturo M.G., Soligo M., Nardini C. The greater inflammatory pathway-high clinical potential by innovative predictive, preventive, and personalized medical approach. EPMA J. 2019;11:1–16. doi: 10.1007/s13167-019-00195-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manni L., Tieri P., Soligo M. A contribution to the hypothesis of nicotinic challenge as therapeutic option for COVID-19 patients. Qeios. 2020 doi: 10.32388/UJX3KN.2. [DOI] [Google Scholar]
- 40.Unwin N. Nicotinic acetylcholine receptor and the structural basis of neuromuscular transmission: insights from Torpedo postsynaptic membranes. Q. Rev. Biophys. 2013;46:283–322. doi: 10.1017/S0033583513000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cipollaro L., Giordano L., Maffulli N. Musculoskeletal symptoms in SARS-CoV-2 (COVID-19) patients. J. Orthop. Surg. Res. 2020;15:178. doi: 10.1186/s13018-020-01702-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goyal P., Choi J.J., Safford M.M. Clinical characteristics of Covid-19 in New York city. N. Engl. J. Med. 2020;382:2372–2374. doi: 10.1056/NEJMc2010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vetter P., Vu D.L., Jacquerioz F. Clinical features of covid-19. BMJ. 2020;369:m1470. doi: 10.1136/bmj.m1470. [DOI] [PubMed] [Google Scholar]
- 44.Wrapp D., Wang N., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davidson A.D., Williamson M.K., Matthews D.A. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020;12:68. doi: 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Apellániz B., Huarte N., Nieva J. The three lives of viral fusion peptides. Chem Phys Lipids. 2014;181:40–55. doi: 10.1016/j.chemphyslip.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daly J.L., Simonetti B., Yamauchi Y. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng M.H., Zhang S., Bahar I. Superantigenic character of an insert unique to SARS-CoV-2 spike supported by skewed TCR repertoire in patients with hyperinflammation. Proc. Natl. Acad. Sci. USA. 2020;117:25254–25262. doi: 10.1073/pnas.2010722117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Q., Chen C.Z., Ye Y. Heparan sulfate assists SARS-CoV-2 in cell entry and can be targeted by approved drugs in vitro. Cell Discov. 2020;6:80. doi: 10.1038/s41421-020-00222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clausen T.M., Sandoval D.R., Esko J.D. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183:1043–1057.e15. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Farsalinos K., Eliopoulos E., Poulas K. Nicotinic cholinergic system and COVID-19: in silico identification of an interaction between SARS-CoV-2 and nicotinic receptors with potential therapeutic targeting implications. Int. J. Mol. Sci. 2020;21:5807. doi: 10.3390/ijms21165807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oliveira A.S.F., Shoemark D.K., Mulholland A.J. Identification of the initial steps in signal transduction in the α4β2 nicotinic receptor: insights from equilibrium and nonequilibrium simulations. Structure. 2019;27:1171–1183.e3. doi: 10.1016/j.str.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 54.Oliveira A.S.F., Edsall C.J., Mulholland A.J. A general mechanism for signal propagation in the nicotinic acetylcholine receptor family. J. Am. Chem. Soc. 2019;141:19953–19958. doi: 10.1021/jacs.9b09055. [DOI] [PubMed] [Google Scholar]
- 55.Campello H.R., Del Villar S.G., Gallagher T. Unlocking nicotinic selectivity via direct C‒H functionalisation of (‒)-cytisine. Chem. 2018;4:1710–1725. [Google Scholar]
- 56.Toelzer C., Gupta K., Schaffitzel C. Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein. Science. 2020;370:725–730. doi: 10.1126/science.abd3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Casalino L., Dommer A., Amaro R.E. AI-driven multiscale simulations illuminate mechanisms of SARS-CoV-2 spike dynamics. bioRxiv. 2020 doi: 10.1101/2020.11.19.390187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu A., Pak A.J., Voth G.A. A multiscale coarse-grained model of the SARS-CoV-2 virion. Biophys. J. 2020;120:1–8. doi: 10.1016/j.bpj.2020.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barros E.P., Casalino L., Amaro R.E. The flexibility of ACE2 in the context of SARS-CoV-2 infection. Biophys. J. 2020;120:1–13. doi: 10.1016/j.bpj.2020.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang G.K., Schmidt J. Primary structure and binding properties of iodinated derivatives of alpha-bungarotoxin. J. Biol. Chem. 1980;255:11156–11162. [PubMed] [Google Scholar]
- 61.Sali A. Comparative protein modeling by satisfaction of spatial restraints. Mol. Med. Today. 1995;1:270–277. doi: 10.1016/s1357-4310(95)91170-7. [DOI] [PubMed] [Google Scholar]
- 62.Dougherty D.A. Cys-loop neuroreceptors: structure to the rescue? Chem. Rev. 2008;108:1642–1653. doi: 10.1021/cr078207z. [DOI] [PubMed] [Google Scholar]
- 63.Corringer P.J., Poitevin F., Changeux J.P. Structure and pharmacology of pentameric receptor channels: from bacteria to brain. Structure. 2012;20:941–956. doi: 10.1016/j.str.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 64.Morales-Perez C.L., Noviello C.M., Hibbs R.E. X-ray structure of the human α4β2 nicotinic receptor. Nature. 2016;538:411–415. doi: 10.1038/nature19785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams D.K., Stokes C., Papke R.L. Differential regulation of receptor activation and agonist selectivity by highly conserved tryptophans in the nicotinic acetylcholine receptor binding site. J. Pharmacol. Exp. Ther. 2009;330:40–53. doi: 10.1124/jpet.109.151225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Puskar N.L., Xiu X., Dougherty D.A. Two neuronal nicotinic acetylcholine receptors, alpha4beta4 and alpha7, show differential agonist binding modes. J. Biol. Chem. 2011;286:14618–14627. doi: 10.1074/jbc.M110.206565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Arnam E.B., Blythe E.E., Dougherty D.A. An unusual pattern of ligand-receptor interactions for the α7 nicotinic acetylcholine receptor, with implications for the binding of varenicline. Mol. Pharmacol. 2013;84:201–207. doi: 10.1124/mol.113.085795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suresh A., Hung A. Molecular simulation study of the unbinding of α-conotoxin [ϒ4E]GID at the α7 and α4β2 neuronal nicotinic acetylcholine receptors. J. Mol. Graph. Model. 2016;70:109–121. doi: 10.1016/j.jmgm.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 69.Grazioso G., Sgrignani J., Cavalli A. Allosteric modulation of alpha7 nicotinic receptors: mechanistic insight through metadynamics and essential dynamics. J. Chem. Inf. Model. 2015;55:2528–2539. doi: 10.1021/acs.jcim.5b00459. [DOI] [PubMed] [Google Scholar]
- 70.Arias H.R., Feuerbach D., Ortells M. Functional and structural interaction of (-)-lobeline with human α4β2 and α4β4 nicotinic acetylcholine receptor subtypes. Int. J. Biochem. Cell Biol. 2015;64:15–24. doi: 10.1016/j.biocel.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 71.Horenstein N.A., McCormack T.J., Papke R.L. Reversal of agonist selectivity by mutations of conserved amino acids in the binding site of nicotinic acetylcholine receptors. J. Biol. Chem. 2007;282:5899–5909. doi: 10.1074/jbc.M609202200. [DOI] [PubMed] [Google Scholar]
- 72.Nys M., Kesters D., Ulens C. Structural insights into Cys-loop receptor function and ligand recognition. Biochem. Pharmacol. 2013;86:1042–1053. doi: 10.1016/j.bcp.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 73.Purohit P., Auerbach A. Loop C and the mechanism of acetylcholine receptor-channel gating. J. Gen. Physiol. 2013;141:467–478. doi: 10.1085/jgp.201210946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang C., Greene D., Luo R. Recent developments and applications of the MMPBSA method. Front. Mol. Biosci. 2018;4:87. doi: 10.3389/fmolb.2017.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015;10:449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Slynko I., Schmidtkunz K., Sippl W. Identification of highly potent protein kinase C-related kinase 1 inhibitors by virtual screening, binding free energy rescoring, and in vitro testing. ChemMedChem. 2016;11:2084–2094. doi: 10.1002/cmdc.201600284. [DOI] [PubMed] [Google Scholar]
- 77.Evers A., Klabunde T. Structure-based drug discovery using GPCR homology modeling: successful virtual screening for antagonists of the alpha1A adrenergic receptor. J. Med. Chem. 2005;48:1088–1097. doi: 10.1021/jm0491804. [DOI] [PubMed] [Google Scholar]
- 78.Raza S., Ranaghan K.E., Azam S.S. Visualizing protein-ligand binding with chemical energy-wise decomposition (CHEWD): application to ligand binding in the kallikrein-8 S1 Site. J. Comput. Aided Mol. Des. 2019;33:461–475. doi: 10.1007/s10822-019-00200-4. [DOI] [PubMed] [Google Scholar]
- 79.Grazioso G., Pomè D.Y., De Amici M. Design of novel alpha7-subtype-preferring nicotinic acetylcholine receptor agonists: application of docking and MM-PBSA computational approaches, synthetic and pharmacological studies. Bioorg. Med. Chem. Lett. 2009;19:6353–6357. doi: 10.1016/j.bmcl.2009.09.073. [DOI] [PubMed] [Google Scholar]
- 80.Grazioso G., Cavalli A., De Micheli C. Alpha7 nicotinic acetylcholine receptor agonists: prediction of their binding affinity through a molecular mechanics Poisson-Boltzmann surface area approach. J. Comput. Chem. 2008;29:2593–2602. doi: 10.1002/jcc.21019. [DOI] [PubMed] [Google Scholar]
- 81.Ibarra A.A., Bartlett G.J., Wilson A.J. Predicting and experimentally validating hot-spot residues at protein-protein interfaces. ACS Chem. Biol. 2019;14:2252–2263. doi: 10.1021/acschembio.9b00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sikora M., Bulow S.v., Hummer G. Map of SARS-CoV-2 spike epitopes not shielded by glycans. bioRxiv. 2020 doi: 10.1101/2020.07.03.186825. [DOI] [Google Scholar]
- 83.Zimmerman M.I., Porter J., Bowman G.R. SARS-CoV-2 simulations go exascale to capture spike opening and reveal cryptic pockets across the proteome. bioRxiv. 2020 doi: 10.1101/2020.06.27.175430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shoemark D., Colenso C., Mulholland A. Molecular simulations suggest vitamins, retinoids and steroids as ligands binding the free fatty acid pocket of SARS-CoV-2 spike protein. ChemRxiv. 2020 doi: 10.26434/chemrxiv.13143761.v1. [DOI] [Google Scholar]
- 85.Turoňová B., Sikora M., Beck M. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science. 2020;370:203–208. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fallon L., Belfon K., Simmerling C. Free energy landscapes for RBD opening in SARS-CoV-2 spike glycoprotein simulations suggest key interactions and a potentially druggable allosteric pocket. ChemRxiv. 2020 doi: 10.26434/chemrxiv.13502646.v1. [DOI] [Google Scholar]
- 87.Ellul M.A., Benjamin L., Solomon T. Neurological associations of COVID-19. Lancet Neurol. 2020;19:767–783. doi: 10.1016/S1474-4422(20)30221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Paterson R.W., Brown R.L., Zandi M.S. The emerging spectrum of COVID-19 neurology: clinical, radiological and laboratory findings. Brain. 2020;143:3104–3120. doi: 10.1093/brain/awaa240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yuki K., Fujiogi M., Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin. Immunol. 2020;215:108427. doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yao H., Song Y., Li S. Molecular architecture of the SARS-CoV-2 virus. Cell. 2020;183:730–738.e13. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ke Z., Oton J., Briggs J.A.G. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hays J.T., Ebbert J.O. Varenicline for tobacco dependence. N. Engl. J. Med. 2008;359:2018–2024. doi: 10.1056/NEJMct0800146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Etter J.F. Cytisine for smoking cessation: a literature review and a meta-analysis. Arch. Intern. Med. 2006;166:1553–1559. doi: 10.1001/archinte.166.15.1553. [DOI] [PubMed] [Google Scholar]
- 94.Amaro R.E., Mulholland A.J. A community letter regarding sharing biomolecular simulation data for COVID-19. J. Chem. Inf. Model. 2020;60:2653–2656. doi: 10.1021/acs.jcim.0c00319. [DOI] [PubMed] [Google Scholar]
- 95.Walsh R.M., Jr., Roh S.H., Hibbs R.E. Structural principles of distinct assemblies of the human α4β2 nicotinic receptor. Nature. 2018;557:261–265. doi: 10.1038/s41586-018-0081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lester H.A. Blockade of acetylcholine receptors by cobra toxin: electrophysiological studies. Mol. Pharmacol. 1972;8:623–631. [PubMed] [Google Scholar]
- 97.Bateman A., Martin M., Consortium U., The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McWilliam H., Li W., Lopez R. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013;41:W597–W600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sievers F., Higgins D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018;27:135–145. doi: 10.1002/pro.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sali A., Potterton L., Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 101.Laskowski R.A., Macarthur M.W., Thornton J.M. Procheck - a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 102.Søndergaard C.R., Olsson M.H., Jensen J.H. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J. Chem. Theory Comput. 2011;7:2284–2295. doi: 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- 103.Olsson M.H., Søndergaard C.R., Jensen J.H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 104.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 105.Lindorff-Larsen K., Piana S., Shaw D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 107.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 108.Nosé S., Klein M.L. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983;50:1055–1076. [Google Scholar]
- 109.Essmann U., Perera L., Berkowitz M.L. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 110.Allen M.P., Tildesley D.J. Clarendon Press; Oxford, UK: 1987. Computer simulation of liquids. [Google Scholar]
- 111.Abraham M.J., Murtola T., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;2:19–25. [Google Scholar]
- 112.Delano W.L. Delano Scientific LLC, San Carlos, CA; 2003. The Pymol molecular graphics system. version 0.98. [Google Scholar]
- 113.DeLano W.L. Abstracts of Papers of the American Chemical Society. 2009. PyMOL molecular viewer: updates and refinements. [Google Scholar]
- 114.Ng H.W., Laughton C.A., Doughty S.W. Molecular dynamics simulations of the adenosine A2a receptor: structural stability, sampling, and convergence. J. Chem. Inf. Model. 2013;53:1168–1178. doi: 10.1021/ci300610w. [DOI] [PubMed] [Google Scholar]
- 115.Kumari R., Kumar R., Lynn A., Open Source Drug Discovery Consortium g_mmpbsa--a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 116.Baker N.A., Sept D., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wood C.W., Ibarra A.A., Sessions R.B. BAlaS: fast, interactive and accessible computational alanine-scanning using BudeAlaScan. Bioinformatics. 2020;36:2917–2919. doi: 10.1093/bioinformatics/btaa026. [DOI] [PubMed] [Google Scholar]
- 118.Wood C.W., Heal J.W., Woolfson D.N. ISAMBARD: an open-source computational environment for biomolecular analysis, modelling and design. Bioinformatics. 2017;33:3043–3050. doi: 10.1093/bioinformatics/btx352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McIntosh-Smith S., Price J., Ibarra A.A. High performance in silico virtual drug screening on many-core processors. Int. J. High Perform. Comput. Appl. 2015;29:119–134. doi: 10.1177/1094342014528252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lentz T.L., Burrage T.G., Tignor G.H. Is the acetylcholine receptor a rabies virus receptor? Science. 1982;215:182–184. doi: 10.1126/science.7053569. [DOI] [PubMed] [Google Scholar]
- 121.Hueffer K., Khatri S., Schulte M.K. Rabies virus modifies host behaviour through a snake-toxin like region of its glycoprotein that inhibits neurotransmitter receptors in the CNS. Sci. Rep. 2017;7:12818. doi: 10.1038/s41598-017-12726-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Donnelly-Roberts D.L., Lentz T.L. Synthetic peptides of neurotoxins and rabies virus glycoprotein behave as antagonists in a functional assay for the acetylcholine receptor. Pept. Res. 1989;2:221–226. [PubMed] [Google Scholar]
- 123.Lan J., Ge J., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.