Abstract
The tumor suppressor p53 is mutated in approximately half of all cancers, whereas the p53 signaling network is perturbed in almost all cancers. In response to different stress stimuli, p53 selectively activates genes to elicit a cell survival or cell death response. How p53 makes the decision between life and death remains a fascinating question and an exciting field of research. Understanding how this decision is made has major implications in improving cancer treatments, particularly in recently evolved immune checkpoint inhibition therapy. Here, we highlight progress and challenges in understanding the mechanisms governing p53’s life-and-death decision-making process, and discuss how this decision is relevant to immune system regulation. Finally, we discuss how knowledge of the p53 pro-survival and pro-death decision node can be applied to optimize immune checkpoint inhibitor therapy for cancer treatment.
Keywords: p53, cell survival, cell death, cancer, cancer therapeutics, cancer immunotherapy
p53, the Cellular Gatekeeper
Since its discovery and characterization as a tumor suppressor several decades ago, p53 has become one of the most researched molecules in biomedical science. As a cellular gatekeeper, p53 plays a pivotal role in the maintenance of genome stability, which is antithetical to the tendency of cancer cells to accumulate mutations. As a result, somatic mutations in p53 occur in almost every type of cancer, and even in cancers without p53 mutation, the p53 signaling pathway is frequently perturbed [1] (reviewed in [2]).
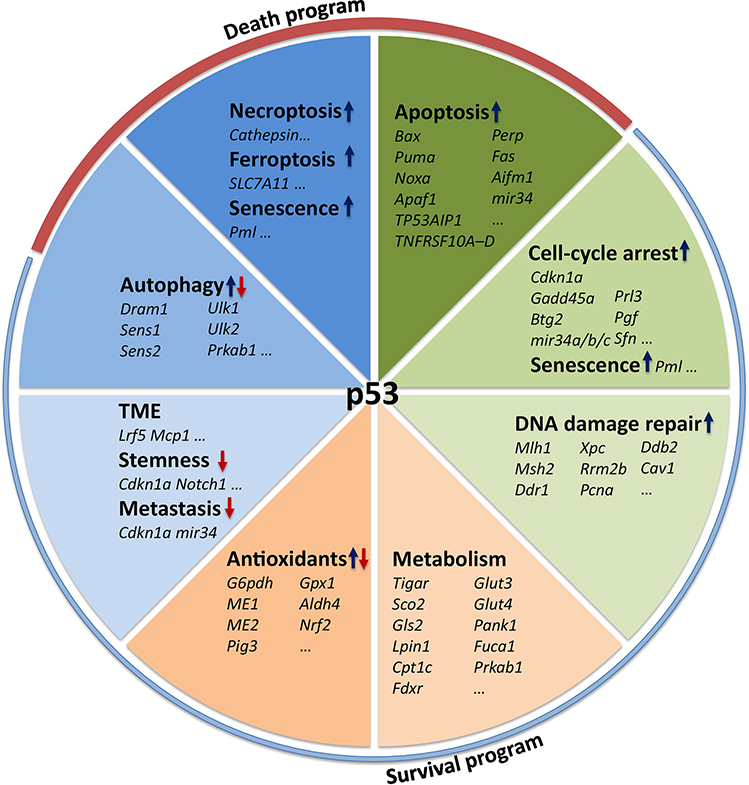
p53 is best known as a transcription factor. Upon activation, p53 forms a symmetric tetramer and binds to its consensus response element in the promoter region of target genes. Under unstressed conditions, p53 protein is maintained at low levels and inactivated through concerted actions of its major negative regulators MDM2 and MDMX. This negative regulation is important to prevent aberrant growth inhibition and cell death. In the presence of stress, such as DNA damage, oncogenic activation, hypoxia, metabolic stress, and osmotic pressure, p53 protein accumulates and becomes activated, which, in turn, transcriptionally up- or down-regulates hundreds of genes and non-coding RNAs involved in several biological processes that can be broadly categorized into cell death and survival programs (Figure 1). Specifically, p53 can trigger palpable changes such as apoptosis (see Glossary), cell cycle arrest, and senescence, as well as more subtle changes such as metabolic reprogramming, autophagy, redox rebalance, and DNA damage repair (reviewed in [3]).
Figure 1. The p53 Transcriptome.
The tumor suppressor p53 regulates hundreds of target genes, through which p53 commits cells to either survival or death programs.
With a role in so many diverse pathways and biological outcomes, p53 is appropriately subjected to tight regulation, as too little p53 activity can result in tumor development, and too much p53 activity can result in indiscriminate cell cycle arrest and cell death. In this review, we highlight recent progress in understanding how p53 differentially regulates pro-survival and pro-death programs. We also explore how this information can be used to improve cancer treatment, particularly with the potential to combine p53-based treatment with newly developed immunotherapies.
p53 as A Decision Node for Cell Survival and Destruction
Upon severe stress, p53 triggers cell death by transcribing genes involved in multiple death pathways. Induction of apoptosis in neoplastic cells is among the first discovered biological functions for p53 [4, 5]. Cellular stressors, such as oncogenic activation and DNA damage, activate p53, which induces apoptosis by either directly driving the expression of pro-apoptotic factors, such as BAX, PUMA, NOXA, FAS, and death receptors (reviewed in [6]), or by translocating itself to the mitochondria, where p53 facilitates mitochondrial outer membrane permeabilization (MOMP) [7]. In addition to apoptosis, p53 also induces ferroptosis in response to reactive oxygen species (ROS) by repressing the expression of SLC7A11, a key component of the cystine/glutamate antiporter [8]. Moreover, in the absence of the apoptotic proteins BAX and BAK, p53 induces cathepsin Q expression, which leads to necroptosis and inhibits tumor growth [9]. Thus, p53 protects the genome through the elimination of extensively damaged cells by activating pro-death programs within those cells.
Despite the ability to induce distinct cellular outcomes, p53 only induces cell death as a last resort. Although some aspects of p53 activity remain speculative, a commonly accepted primordial function is that p53 initially evolved as a checkpoint to stall proliferation to allow the cell a chance to repair damage, which contributes to organismal longevity and adaptation to environmental stress. Indeed, in cells exposed to genotoxic stress, p53 transcriptionally upregulates p21, a cyclin-dependent kinase (CDK) inhibitor that halts the cell cycle at the G1 phase [10]. In a context- and stress-dependent manner, specifically in response to sub-lethal stress, p53 then activates a series of pro-survival response genes to allow cells to recover from the stress. For instance, p53 maintains cell fitness according to the status of the nutrient supply. In the presence of excess nutrient supply, p53 primes energy storage in lipid droplets [11], whereas when the cell experiences nutrient deficiency, p53 immediately drives catabolism and burns fat [12]. Moreover, in response to moderate DNA damage, p53 increases cellular disulfide reduction capacity to remove mutagenic reactive oxygen species (ROS), activates autophagy for the re-allocation of cellular resources, and reprograms metabolism to meet the high demand for energy and nucleotide biosynthesis for DNA damage repair. These effector functions, coordinately and collectively, rescue cells from death [3].
Stressors that activate p53 do not always result in the same outcome. For example, although different doses of a single DNA damaging agent, such as doxorubicin, can activate p53, the outcomes can be completely different. Whereas low-dose doxorubicin-activated p53 results in cell survival, higher doses produce more widespread cell death [13]. Likewise, different p53-activating stresses can also result in variable outcomes, as well as different p53-regulated gene expression patterns. These differences support the role of p53 as a decision node tasked with choosing which of the different gene programs to activate as an appropriate response to the context and severity of a given stress.
How does p53 Make Life-and-Death Decisions?
As the p53 transcription network expands, an interesting question regarding the p53-regulated gene programs concerns the antagonistic effects of some programs towards others. One of the more obvious examples is that during the p53-dependent induction of apoptosis, the simultaneous p53-dependent activation of DNA repair or antioxidant pathways counteract to the intended apoptotic outcome. Therefore, pleiotropic transcription factors such as p53 likely require mechanisms to selectively express some genes but not others according to the specific context. As one of the major mysteries that persist regarding p53 regulation, several models have been proposed to explain mechanistically how p53 decides which programs to activate, many of which have focused on differences in p53 itself.
A p53 Post-Translational Modification Model
p53 is subject to a large and diverse array of posttranslational modifications (PTMs), including but not limited to ubiquitination, phosphorylation, acetylation, and methylation. Among them, the modification of certain residues, particularly by phosphorylation and acetylation, contribute to the ability of p53 to differentially activate cell cycle arrest or apoptosis genes. For example, the phosphorylation of Ser46 in p53 correlates strongly with the induction of apoptotic genes and an apoptotic outcome [14]. p53 acetylation at residue Lys382 occurs subsequent to Ser46 phosphorylation, and this acetylation mark is required for p53 apoptotic gene expression [15]. p53 can also be functionally acetylated at Lys320 and Lys373, which results in the expression of cell cycle arrest and apoptotic genes, respectively [16]. Mechanistically, acetylation at Lys320 prevents the phosphorylation of N-terminal p53 residues thereby restricting p53 to high-affinity promoters such as those of cell cycle arrest gene p21. In contrast, acetylation of Lys373 promotes the phosphorylation of N-terminal p53 residues and allows p53 to bind to low-affinity promoters such as those of the apoptotic target genes. In addition, acetylation of Lys120 also appears to dictate the affinity between p53 and cell cycle arrest and apoptosis gene response elements [17]. Lys120 acetylation is necessary for the induction of p53 apoptotic genes, as mutation of this residue inhibits the ability of p53 to upregulate apoptotic gene expression. Similarly, PRMT5-mediated p53 arginine methylation results in preferential p21 transcription as opposed to apoptotic genes [18]. Collectively, much like the “histone code,” p53 PTMs are thought to provide at least part of the instructive information from which p53 target genes are selectively expressed in response to stress type and severity.
A p53 Response Element Model
Another prevalent model for p53 differential gene regulation is based on the binding affinity between p53 and cell cycle arrest or apoptotic p53 response elements (REs). Through the analysis and confirmation of several dozen authentic p53 REs, these REs deviate, sometimes considerably, from the consensus RE (RRRCWWGYYY). In one study comparing the p53 RE of the cell cycle arrest gene p21 and the apoptotic gene TP53I3 (also called PIG3), the p53 RE for p21 was bound more rapidly than that of TP53I3 in response to DNA damage, suggesting that p53 binds its p53 REs at different rates and affinities [19]. In another study, p53 was shown to bind preferentially to the REs for p21 and the senescence target gene GADD45 in senescent cells [20]. This binding pattern was not observed in cells treated with an acute dose of doxorubicin, which show increased accumulation of p53 at the promoters of apoptotic target genes [20]. Using a biophysical approach, the Fersht group determined the in vitro dissociation constants for 20 known p53 response elements for the p53 tetramer and found that all of the low-affinity response elements corresponded to apoptotic genes, which could also be consistent with the p53 threshold model (see below) [21]. Although these studies provide evidence that at least some of the p53 REs display different affinities for p53 binding, other studies have hinted at the possibility that certain p53 PTMs could be required to enhance its binding to the low-affinity promoters.
A p53 Threshold Model
Multiple studies have suggested that the outcome of a cell in dealing with stress depends on the levels of p53 [22, 23]. A study using a p53-inducible cell model that allows for the precise control of p53 levels in the absence of genotoxic stress showed a strong correlation between the levels of p53 and the expression patterns of cell cycle arrest and apoptosis genes [23]. Additionally, the threshold for p53 transcriptional repression of anti-apoptotic Bcl-2 family genes is higher than the threshold for the transactivation of pro-apoptotic genes [24]. As a result, inhibition of BCL-2 family proteins results in increased apoptosis in the presence of an otherwise sub-lethal level of p53 [25]. These results support a model whereby only high levels of p53 are able to induce apoptosis. While low levels of p53 can activate some apoptotic gene expression, the abundance of apoptotic gene expression does not surpass the threshold required for cell death. This model is further corroborated by a recent finding showing a dynamic, rather than a fixed, p53 threshold needs to be met to execute an apoptotic program [26]. In this study, an increase in the p53 apoptotic threshold results in drug-dependent induction of anti-apoptotic genes, predominantly in the inhibitors of apoptosis (IAP) family. Together, this model fits well with several studies that have shown that to trigger an apoptotic response, a certain threshold of pro-apoptotic gene expression (relative to anti-apoptotic gene expression) must be met [27, 28].
A p53 Dynamic Expression Pattern Model
Researchers have recently found that ionizing radiation (IR) induces a pulsed pattern of p53 expression, whereas ultraviolet (UV) irradiation induces a sustained pattern of p53 activation. The pulsed patterns of p53 induction tend to result in non-lethal and reversible outcomes for the cell, while sustained activation of p53 results in cell death or senescence [29, 30]. The p53-activating molecule nutlin-3a can convert the IR-associated pulsed p53 into a sustained expression pattern, which changes the cell survival outcome associated with the pulsed pattern into a senescence outcome [22]. Moreover, prolonged challenge with low-dose doxorubicin triggers sequential p53 nulses, which, upon exceeding an effective threshold, transactivates pro-apoptotic genes and results in a terminal pulse enacting apoptosis at a comparable rate with that induced by an acute and high-dose treatment [31]. Thus, these studies collectively suggest that p53 expression pattern plays a role in whether a cell commits apoptosis or cell cycle arrest in response to a given stress.
Another fundamental question is whether p53 pulses cause dynamic expression of its target genes and therefore impact cell fate. Hafner and colleagues systematically analyzed the pattern of p53 DNA binding, as well as the transcriptome along with p53 oscillation induced by DNA damage [32]. Surprisingly, p53 DNA binding reveals a pulsatile pattern that is uniform across all genomic loci; however, identical p53 oscillation activates target genes that exhibit multiple distinct dynamics, indicating that post-transcriptional mechanisms are responsible for the differences in gene expression dynamics. Indeed, a recent study demonstrated that p53 oscillation-associated target gene expression dynamics, in the format of either pulsing or sustained p53 levels, result from different mRNA decay rates of target genes [33], indicating that p53 pulses coordinate target gene dynamics to determine cell fate.
A p53 Regulation of miRNA Model
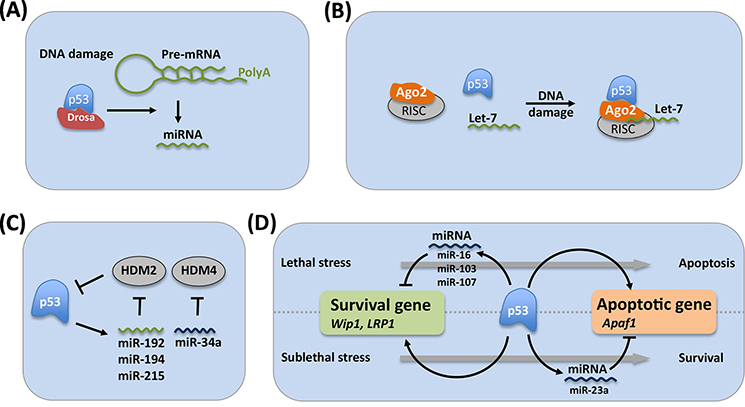
As one of the master transcription factors responsible for sensing stress, p53 regulates the maturation and expression of several miRNAs [34] (Figure 2). p53 interacts with the Drosha processing complex and facilitates the processing of primary miRNAs to precursor miRNAs, thus promoting the post-transcriptional maturation of several antiproliferative miRNAs, including miR-16–1, miR-143, and miR-145, in response to DNA damage [35]. In addition to general control of miRNA maturation, DNA damage induces p53 binding to AGO2 and alters the association of Let-7 family miRNAs with AGO2, leading to cell cycle arrest [36]. Moreover, p53 directly induces the transcription of several miRNAs, including miR-23a, miR-145, miR-107, miR-192, and miR-205 [37], and the miR-34 family [38], all of which contribute to the tumor suppressive function of p53 by targeting oncogenes such as E2F, Bcl-2, and PD-L1 [39, 40]. As an added layer of self-regulation, the p53-regulated genes miR-34a, miR-192, miR-194, and miR-215 repress MDM2 and MDMX expression, creating a positive feedback loop with p53 to accelerate the p53 pro-death outcome (Figure 2) [41, 42].
Figure 2. Involvement of miRNA in p53 Transcriptome.
A. p53 regulates miRNA biogenesis by interacting with the Drosha processing complex through the association with DEAD-Box RNA helicase p68. B. In response to DNA damage, p53 binds to Ago2, an important component of RISC, and leads to increased association of Let-7 family miRNA to Ago2 for further degradation of target mRNA. C. p53 directly regulates the expression of a group of miRNA genes, including miR-34a and miR-192/miR-194/miR215, which repress the expression of HDM2 and HDM4, respectively. D. p53-induced miR-23a directly targets Apaf1, a p53-regulated gene, and protects cells from apoptosis. p53 also induces miR-16 expression, which inhibits Wip1 expression, alleviating cells from cell cycle arrest and causing cell death. Similarly, p53 activates miR-103 and miR-107, which inhibit LRP1 expression and promote apoptosis. Both Wip1 and LRP1 are direct p53 target genes.
Some p53-regulated miRNAs target known p53-regulated coding genes [43]. For example, the p53-regulated gene miR-23a is expressed in response to p53-activating stresses. Upon upregulation, miR-23a down-regulates the expression of the apoptotic p53 target gene Apaf1. Through this mechanism, miR-23a expression can promote apoptotic resistance in some cancer cells [44–46]. Although these studies were performed in cancer cells, it is likely that p53-regulated miRNAs contribute to the regulation of other p53 target genes in normal cells as well. A recent study identified the gene low-density lipoprotein receptor-related protein 1 (LRP1) as a direct p53 target gene that is also targeted by p53-regulated miRNAs miR-103 and miR-107 [30]. In this study, only a sub-lethal dose, but not a lethal dose, of doxorubicin results in an increase in LRP1 protein expression. By contrast, p53 activates miR-103 and miR-107 expression only in response to lethal doses of doxorubicin, and these miRNAs in turn target LRP1 transcript to repress its expression, which results in increased apoptosis. These results suggest that lethal doses of chemotherapeutics induce p53 to switch to a pro-apoptotic response in part through the upregulation of miRNAs that actively suppress the expression of pro-survival p53 target genes.
Collectively, with hundreds, if not thousands, of potential p53 “target genes” being identified through transcriptomic studies and some 350 being validated experimentally [47], different models have been proposed to address differential gene regulation by p53 in the context of determining cell survival and death. Although often proposed as single mechanisms, evidence suggests that these models often overlap and form dynamic networks [31]. Thus, systematic analysis of different models along with different stresses and tumor lineages may shed new light on understanding p53 gene regulation to help guide novel therapeutic strategies.
Implications of p53 in Cancer Immunotherapy
Given the importance of p53 in orchestrating DNA damage responses and making life or death decisions in cancer cells, most conventional chemo- and radiotherapies nemonstrate p53 dependence by introducing general DNA damage in cancer cells. Increasing evidence indicates that the efficiency of some of these therapies largely depends on the host immune response [48, 49], which raises the question of how DNA damage insults relate to the immune microenvironment in tumors and to what extent p53 signaling is involved in this process. If this is the case, it is worthwhile to evaluate the potential of combining p53-targeted therapy with anti-cancer immunotherapy.
Dual Role for p53 in Immune Regulation
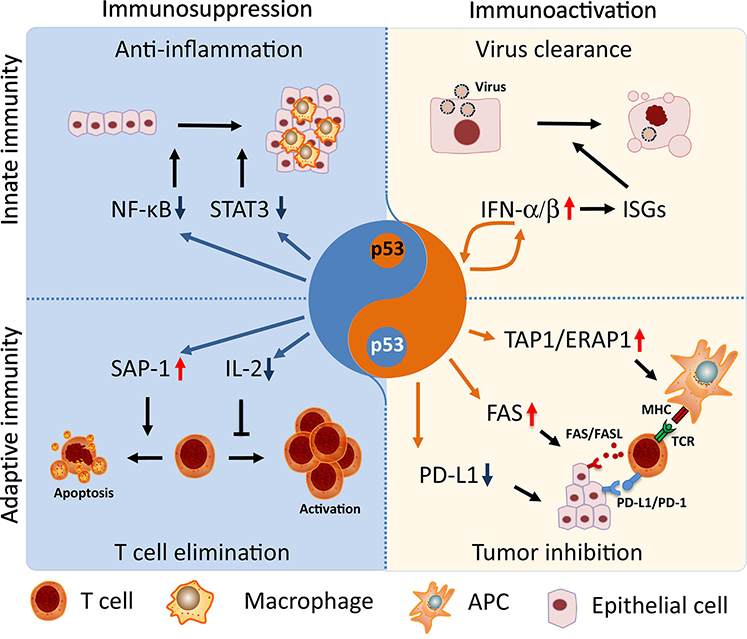
As a master regulator of cellular homeostasis, p53 comprehensively modulates immune integrity by directly transactivating hub genes of immune signaling pathways (Figure 3, Key Figure). p53 activates innate immune responses through the transactivation of regulators involved in pathogen sensing, cytokine expression, and inflammation [50]. For example, Toll-like receptors (TLRs) play important roles in pathogen recognition by immune system. Among them, TLR3, TLR5 and TLR9 are transcriptional targets of p53 that regulate agonist-induced cell death and inflammation [51]. On the other hand, upon virus infection, interferon-alpha/beta induces p53 expression, which in turn boosts IFN signaling through a positive feedback loop [52], subsequently triggering p53-dependent apoptosis to eliminate virally infected cells and prevent virus spread [53, 54]. Consistent with this observation, “super p53” mice, which carry an extra copy of p53, demonstrate latent virus replication and are resistant to viral infection [55]. On the contrary, p53 inhibits immune infiltration by counteracting NF-κB, the pivotal pro-inflammatory pathway, thereby suppressing excessive expression of pro-inflammatory cytokines across cancer types [56]. Indeed, p53 deficiency is associated with increased expression of pro-inflammatory factors such as TNF-α and interleukins, leading to an increased tendency towards inflammatory disease [57]. This phenomenon is phenocopied in mice lacking the p53 pro-survival target genes p21 or GADD45 [58, 59], indicating possible involvement of a p53 pro-survival function in anti-inflammation.
Figure 3. p53 Fine-Tunes Immune Homeostasis.
p53 plays dual roles in the regulation of the immune response to maintain tissue homeostasis. On one hand, p53 counteracts pro-inflammatory pathways, such as NF-κBand JAK-STAT3, thereby inhibiting tumor infiltration of macrophages, neutrophils, and CD4+ T cells. Moreover, p53 transcriptionally induces the expression of apoptotic factor SAP-1 and directly inhibits the expression of T cell growth factor IL-2, resulting in increased apoptosis and compromised proliferation in T cells, thereby counterbalancing T cell activity and avoiding autoimmune diseases. On the other hand, p53 activates type-I interferon signaling through a positive feedback loop, promoting viral eradication through the induction of IFN-stimulated genes (ISGs)-associated apoptosis in infected cells. p53 is able to activate anti-tumor adaptive immunity through multiple pathways. For instance, p53 induces the expression of TAP1 and ERAP1, which mediates the transporting and processing of tumor-associated antigens for further delivery to the membrane of APC cells, so that antigen-associated MHC can be recognized by TCRs from T cells. Moreover, p53 activates the co-stimulating machinery Fas/FasL and interrupts the immune-suppressive complex PD-1/PD-L1, thus bypassing PD-L1-associated immune-escape and enhancing the tumor killing effect of CTLs.
Chronic inflammation has commonly been considered a driving force of tumorigenesis. In a study with mice carrying intestinal epithelial cell (IEC)-specific p53 deletion, p53 deficiency does not cause spontaneous intestinal tumorigenesis but remarkably enhances carcinogen-induced adenoma transition to invasive carcinoma by prompting the formation of an NF-κB-dependent inflammatory microenvironment, which allows neoplastic cells to penetrate the epithelial barrier and further metastasize [60]. Similar p53-dependent anti-inflammatory tumor suppressive effects were shown in colon and breast cancer with gain-of-function p53 mutants activating pro-inflammatory Wnt and STAT3 signaling [61–63]. Alternatively, p53 activates natural killer (NK) cells and triggers anticancer innate immune response through induction of ULBP1 and ULBP2 [64]. Thus, through making life and death decisions, p53 fine-tunes the innate immune response and suppresses tumor development.
Unlike the innate immune system, the adaptive immune response is an antigen-specific process that is not only responsible for the direct elimination of antigens but, also triggers immunological memory for long-term protection from immunogens. During carcinogenesis, the accumulation of mutations changes the composition of cell surface proteins in cancer cells, resulting in the expression of tumor-associated antigens (TAAs). Different TAAs are presented to T cells via major histocompatibility complex (MHC) class I or II on antigen-presenting cells (APCs), such as dendritic cells and macrophages, which in turn triggers T cell activation and clonal expansion. Among T cell lineages, CD8+ cytotoxic T cells (CTLs) are responsible for the ultimate elimination of tumor cells by direct cell-mediated cytotoxicity as well as indirect antibody complement-mediated cytotoxicity. CD4+ T helper cells (Ths) promote a humoral immune response and/or activate CTLs through the secretion of cytokines. As a subset of Ths, regulatory T cells (Tregs), which mediate tolerance to self-antigens and counterbalance the immune response, are commonly hijacked by tumor cells for immunosuppression [65].
Until recently, the function of p53 in anti-tumor immunity has been largely under appreciated. One role of p53 in anti-tumor immunity involves regulating peripheral T cells, which, upon activation, undergo an exquisitely orchestrated self-renewal process as a component of tissue homeostasis and to avoid autoimmune diseases. p53 activation coincides with T cell activation, which leads to T cell elimination through the induction of the lymphocyte-specific pro-apoptotic gene SLAM-associated protein (SAP) [66], as well as by repression of the pro-proliferation gene interleukin-2 (IL-2) [67]. Recently, Watanabe et al. found that T cell receptor (TCR) signaling results in a decrease in both p53 mRNA and protein, which promotes antigen-specific CD4+ T cell proliferation [68]. Consistently, in TCR transgenic mice, p53 deletion leads to hyperactive glycolysis, which correlates with enhanced T cell proliferation and augments anti-melanoma activity, implicating a role for p53 in metabolic microenvironment regulation through its effects in T cells [69].
p53 also plays a direct role in coordinating the anti-tumor response through its regulation of different nodes of the adaptive immune response. For example, once TAAs are phagocytosed by APCs, they are transported from the cytosol to the endoplasmic reticulum (ER) by the Transporter Associated Antigen Processing 1 (TAP1) for processing by the protein Endoplasmic Reticulum Amino Peptidase 1(ERAP1) [70]. p53 transcriptionally upregulates both TAP1 and ERAP1, through which p53 directly facilitates antigen presentation via the major histocompatibility complex (MHC) I pathway [71, 72]. In addition to its direct impact on single cells, p53 promotes an anticancer adaptive immune response by enhancing cancer cell-CTL communication, which is driven by the immunological synapse consisting of MHC-I on cancer cells and TCR on CTLs. One of the mechanisms through which CTLs induce cancer cell apoptosis is by expressing Fas-ligand (FasL), a transmembrane protein that binds Fas receptor on cancer cells, within the synapse during cancer cell-CTL interaction. Notably, p53 promotes CTL-mediated tumor cell killing by upregulating Fas/APO-1 in cancer cells [73]. To escape CTL killing, cancer cells develop negative co-stimulating signals to block the immunological synapse and promote immune tolerance. For example, programmed death-1 (PD-1) and cytotoxic T-lymphocyte associated antigen 4 (CTLA4) are major CTL immune checkpoint receptors that down-regulate immune responses upon activation. Cancer cells directly activate these receptors by overexpressing PD-1 ligand (PD-L1) on their surface, which marks the cancer cells as self-antigens from the perspective of CTLs [74]. p53 down-regulates PD-L1 expression in cancer cells by upregulating PD-L1-targeting miR-34a. In a phase I clinical trial investigating the effects of miR-34a administration in non-small cell lung cancer, miR-34a-loaded liposomes reduced PD-L1 expression in human tumors, which correlated with increased CTL tumor infiltration and reduced immunosuppressive macrophages and Tregs, resulting in marked sensitization of cancer cells to radiotherapy [40]. These data suggest that p53 can suppress tumor growth by recruiting CTLs and activating their cell killing effects on cancer cells.
Although several questions remain to be addressed in future studies, current evidence suggests that p53 regulation of the immune response in tumors shows a Yin-Yang balance. The “dark side” (Yin), which includes T cell development and the inflammatory response, is p53’s effects on immune cells themselves and functions as an immune checkpoint. By controlling the T cell populations and by inactivating pro-inflammatory factors, such as NF-κB, p53 promotes tissue homeostasis and prevents the detrimental effects that originate from prolonged and/or excessive immune responses. The opposite side is the “bright side” (Yang), which mainly responds to viral infection or mutagenesis and promotes the removal of infected non-immune cells [75]. This side is associated with boosted IFN signaling, as well as coordinating a T cell response to recognize the target cell as non-self antigen for destruction and removal. These features provide a distinct opportunity to combine p53-targeted therapies with current immunotherapies to develop more effective cancer treatments.
Potential to Combine p53 with Immunotherapy
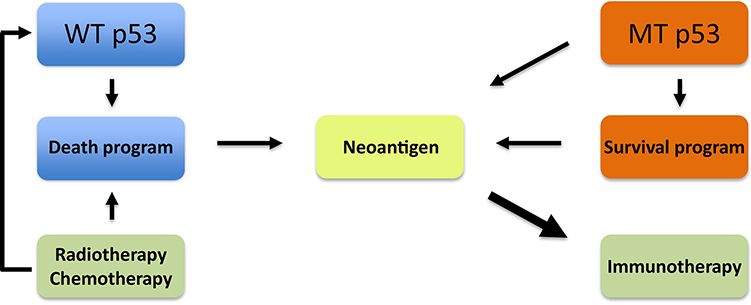
Immunotherapy has emerged as one of the most promising modalities of cancer treatment. Breakthroughs have been made in the clinical application of immune checkpoint inhibitors (ICIs) (see Glossary) and chimeric antigen receptor-T (CAR-T) cell therapy. As a milestone of ICI therapy, drugs that target PD-1/PD-L1 have produced durable responses in patients with many different types of cancers [76]. Although the function of p53 in the regulation of the immune system remains incompletely understood, p53 signaling has recently been shown to exert remarkable immunogenic effects, thus representing a promising strategy in anti-cancer immunotherapy (Figure 4).
Figure 4. p53-Based Neoantigenesis and Corresponding Therapeutic Strategies in Cancer Treatment.
Despite the slow progress in other strategies of p53-based cancer treatments, applying these principles to immunotherapy has major potential as a new direction for cancer therapy. Neoantigen generation has proven indispensible for the success of immunotherapy. Recent studies have revealed that nutlin-3a creates neoantigens through the induction of WT p53-dependent apoptosis, which could be an effective adjuvant in chemoradiation-induced immunogenesis. In addition, mutant p53 is a cancer neoantigen itself, which offers the possibility of using mutant p53 as an indicator for immunotherapy treatment.
p53 Turns Up the Heat for ICI.
Although there has been impressive progress with the nnti-cancer ICI therapy, a bottleneck is that many cancer patients are resistant to ICI treatment. In lung squamous cancer patients treated with ICI, only 20% of patients showed a durable response to anti-PD-1/PD-L1 therapy [77]. This partial response is also observed for many other types of cancers and can be related to the antigenicity of a given tumor [78]. An effective adaptive immune response against tumor cells requires sufficiently activated CTLs that efficiently navigate to the tumor milieu, as well as adequate TAAs that are presented on MHC by APCs [79]. However, many neoantigens enriched in tumors and proven to be immune-active fail to activate the immune response in cancer patients [80], which could partially account for ICI resistance in some patients. Therefore, increasing the neoantigen repertoire remains a promising direction to improve ICI responses.
It has long been known that dendritic cells are highly efficient at activating CTLs [81], indicating a critical role for these cells in neoantigene-driven tumor cell apoptosis. Several DNA damaging agents such as doxorubicin, mitoxantrone, oxaliplatin, cyclophosphamide, and bortezomib induce immunogenic cell death (ICD) (see Glossary) [82], through which dying cells release host molecules and damage-associated molecular patterns (DAMPs) such as HMGB1 [83]. Of note, using clinically relevant ICI-resistant murine models and a combination of immunogenic drugs to trigger T cell infiltration into tumors, Pittet and colleagues found that immunogenic chemotherapy induces tumor sensitivity to anti-PD-1 antibody treatment [84]. p53 adenovirus (Ad-p53) treatment can significantly reverse anti-PD-1 resistance in both ipsilateral and contralateral melanomas where there is no Ad-p53 exposure [85, 86]. This global effect of p53 raises the question about whether p53-dependent apoptosis is favorable for inducing immunogenesis. Indeed, a recent study using a xenograft melanoma model showed that local activation of WT p53 with nutlin-3a overcomes immune suppression and amplifies anti-tumor immunity via the induction of immunogenic cell death [87]. Of note, this p53-dependent tumor regression only occurs in tumor models with present of tumor-infiltrating lymphocytes (TILs), suggesting the participation of the immune system in this process. Moreover, mice that experienced complete tumor regression due to nutlin-3a injection rejected subsequent tumor re-challenges, indicating the establishment of anti-tumor memory. At least part of the initial tumor rejection is due to p53 directly activating miR-34, which represses PD-L1 expression [40]. Thus p53 deficiency protects cancer cells from CD8+ T cell killing by restoring PD-L1 expression in both mouse models and patients [88], which suggests that the reactivation of WT p53 unleashes cytotoxic T cells from tumor cell-mediated inhibition and thus attenuates the effects of cancer-driven immunosuppression. Collectively, these observations imply that chemo-induced p53-dependent apoptosis has the potential to sensitize unresponsive tumors to ICI therapy.
Mutant p53 Augments Neoantigenesis.
Cancer cells are often genetically unstable and produce neoantigens, which provides a basis for distinguishing tumor cells from normal cells. In many clinical studies, tumor types that present with higher rate of genomic mutations are more responsive to anti-PD-1 therapy, presumably due to the expression of foreign proteins associated with the mutations, which serve as potential immune response-inducing antigens, or neoantigens [89]. This concept of using greater mutational burden as a predictive marker of response to immune checkpoint blockade therapy has been demonstrated in colorectal cancer trials [90], and lately has expanded to eleven other cancer types with an overall significantly improved response rate in patients with MMR deficiency [91]. As mutant p53 increases tumor mutational burden (TMB) by interrupting DNA damage repair and deactivating cell cycle checkpoints [2], mutant p53 could be a predictor for ICI treatment efficacy.
In a recent study of 75 patients with NSCLC treated with anti-PD-1 antibody nivolumab plus CTLA-4-targeting ipilimumab, whole exome sequencing revealed that the magnitude of TMB correlates with superior outcomes. Further analysis demonstrated that p53 mutation correlates with TMB and presents as the only enriched gene in responders, suggesting a positive effect of p53 mutation-associated genome instability (see Glossary) in ICI therapy [92]. This finding was further validated by another clinical study that included 245 untreated patients with lung adenocarcinoma (LUAD) and 32 LUAD patients treated with the anti-PD-1 antibody nivolumab. p53 mutation-bearing tumors were again identified with the highest tumor-infiltrated CTL density and PD-L1 expression. In this tumor subtype, pathways related to antigen presentation and T cell activation were up-regulated. Prolonged progression-free survival was observed in nivolumab-treated patients harboring p53-mutant tumors, suggesting a strong connection between p53 mutation and ICI efficacy [93]. Finally, analysis of TCGA data [94] showed similar findings. Overall, these studies show that p53 mutation could be an important “helper” in promoting neoantigenesis that can be leveraged through the use of ICI therapy.
p53 hotspot mutations themselves appear to serve as immuno-active neoantigens in cancer. For example, in a recent clinical trial evaluating T cell responsive neoantigens in metastatic ovarian cancer, p53 hotspot mutations (Y220C and G245S) were enriched in peptides presented to the MHC-I of CTLs and were identified in tumor-infiltrating lymphocytes, whereas other neoantigens recognized by T cells arose exclusively from random and nonsynonymous somatic mutations [95]. This observation was corroborated by a follow-up analysis of a cohort of 140 patients diagnosed with different types of epithelial cancer [96]. Among these patients, p53 neoantigen–specific human leukocyte antigen (HLA) restriction elements and T cell receptors (TCRs) were identified in one-third of patients who carry p53 hotspot mutations. Both TILs and TCR gene-engineered T cells recognize and respond to these p53 neoantigens. Collectively, these results imply that hijacking mutant p53 could be a useful strategy for sensitizing tumors to immunotherapy.
Concluding Remarks
Understanding p53 downstream functions, specifically in the tumor context, offers valuable insight for the development of p53-based anti-cancer therapeutic strategies. Towards this direction, there are many questions remaining to be addressed (see Outstanding Questions). Together with cell cycle arrest and senescence, p53-induced apoptosis has long been considered as the main process through which p53 suppresses tumor development. Until recently, this dogma has been challenged by findings that p53-dependent apoptosis, cell cycle arrest, and senescence are dispensable for tumor suppression in mouse models harboring either p53 gene mutations or deletions of key p53 target genes [97, 98]. It is true that none of the individual effector functions of p53 can suppress tumor development alone. Instead, growing evidence indicates that different processes may operate synchronically, interactively, and synergistically depending on the type and degree of stresses, as well as the lineage of the cancer and cell types [99]. Therefore, it is important to know how p53 makes the choice between activating survival machinery or triggering cell death. It is also necessary to understand how individual effector functions crosstalk and coordinate in response to the environmental triggers that are most relevant for each specific tumor environment.
Outstanding Questions.
Among the hundreds target genes and multiple biological processes that are regulated by p53, what are the major executors to deliver the tumor suppressive function of p53?
How do individual p53 effectors crosstalk and coordinate each other in response to specific tumor environment?
How does p53 regulate immune system under both normal and cancerous conditions?
Within different tumor microenvironment how p53 modulate immune landscape to create a niche for immune response or immune escape?
How can the underlying mechanism of p53 regulation of immune response be explored for cancer therapeutics?
As the p53 network grows ever bigger and more complex, it is not surprising that the p53 regulatory network rarely follows a linear path to a given function. Instead, p53 regulates multiple sides of many, if not all, biological processes, and therefore maintains global tissue homeostasis through its life and death decision-making. When choosing the former, p53 arrests the cell cycle, activates DNA damage repair, activates autophagy, and reprograms metabolism to save cells from DNA damage stress. When choosing the latter, p53 not only triggers apoptosis, but also plays a role in the immune system-mediated clearance of the cells[3]. Similarly, in terms of immune regulation, p53 counterbalances both innate and adaptive immune responses when there is present of prolonged inflammation or hyperactive T cells. On the other hand, p53 boosts both arms of the immune system to eliminate cells that are damaged by viral infection or expressing tumor-associated antigens. Altogether, p53 is a neutralizer of cell stress, functioning as a master stress sensor and response director.
Finally, while the efficacy of immunotherapy has been demonstrated in the treatment of multiple cancers, the potential of p53 as a source of neoantigenesis and as an adjuvant for ICI illustrates the value of new pipelines focused on developing p53-based cancer therapies. Our current knowledge of p53 regulation of the immune system remains limited and fragmented. Moreover, it remains to be discovered how p53 regulates the immune system under both normal and cancerous conditions, as well as how p53 modulates the immune landscape in the tumor microenvironment to create a niche for immunoresponse or immunoescape. While many of these studies are just beginning to scratch the surface, a comprehensive understanding of p53 regulation of cancer immunology looks very promising in developing clinically valuable medicines.
Highlights.
The tumor suppressor p53 plays a critical role in cell fate decision-making. Mild and short-term DNA damage leads to p53 activation of pro-survival mechanisms, allowing the cells to repair and recover from the damages. Intensive and prolonged DNA damage triggers p53-dependent cell death to avoid accumulation of genomic mutations.
The life-and-death decision-making by p53 is influenced by multiple mechanisms, including p53 post-translational modification, different binding affinity to p53 response element, p53 threshold levels and dynamic patterns, and p53 regulation of miRNAs.
WT p53 enhances anti-tumor immunity by sensitizing cells to immunogenic cell death, whereas mutant p53 increases mutational burden and promotes neoantigenesis.
The p53-dependent anti-cancer immunity represents a promising direction for the development of cancer therapeutic strategies.
Acknowledgements
We apologize for not being able to cite all of the relevant papers due to limited space. Y. Z. is support by grants from The National Institute of Health (CA100302, CA127770, CA167637, and CA212407). Y. L. is supported by the National Natural Science Foundation of China (Grant No. 81672618, 81972723) and the Natural Science Foundation of the Jiangsu Higher Education Institutions (Grant No. 18KJA320012).
Glossary
- Apoptosis
a form of programmed cell death indicated by caspase activation, nuclear fragmentation and cell shrinkage. Apoptosis is critically important for embryo development, metamorphosis, and removal of unnecessary or damaged cells.
- Genome instability
an increased tendency of alteration in genome, including mutation, amplification or deletion of nucleic acids, rearrangements of chromosomes, and aneuploidy. Cancer cells are commonly characterized as genomic instable.
- Immunogenic cell death
ICD is a type of cell death through which dying cells release host molecules and damage-associated molecular patterns (DAMPs), which subsequently triggers adaptive immune responses. ICD of cancer cell is an important contributor to some anti-cancer therapeutic approaches to suppress cancer progression, by releasing tumor-associated antigens.
- Immune checkpoint inhibitor
Immune checkpoints are cell surface proteins responsible for the maintenance of immune homeostasis by preventing over-active immune responses, and sometimes keeping T cells from killing cancer cells. Immune checkpoint inhibitor (ICI) is a type of molecule that target and block immune checkpoints.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hollstein M et al. (1991) p53 mutations in human cancers. Science 253 (5015), 49–53. [DOI] [PubMed] [Google Scholar]
- 2.Muller PA and Vousden KH (2014) Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 25 (3), 304–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kruiswijk F et al. (2015) p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol 16 (7), 393–405. [DOI] [PubMed] [Google Scholar]
- 4.Yonish-Rouach E et al. (1991) Wild-type p53 induces apoptosis of myeloid leukaemic cells that is inhibited by interleukin-6. Nature 352 (6333), 345–7. [DOI] [PubMed] [Google Scholar]
- 5.Shaw P et al. (1992) Induction of apoptosis by wild-type p53 in a human colon tumor-derived cell line. Proc Natl Acad Sci U S A 89 (10), 4495–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aubrey BJ et al. (2018) How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ 25 (1), 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaseva AV et al. (2012) p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell 149 (7), 1536–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang L et al. (2015) Ferroptosis as a p53-mediated activity during tumour suppression. Nature 520 (7545), 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tu HC et al. (2009) The p53-cathepsin axis cooperates with ROS to activate programmed necrotic death upon DNA damage. Proc Natl Acad Sci U S A 106 (4), 1093–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.el-Deiry WS et al. (1993) WAF1, a potential mediator of p53 tumor suppression. Cell 75 (4), 817–25. [DOI] [PubMed] [Google Scholar]
- 11.Liu S et al. (2017) Protection against High-Fat-Diet-Induced Obesity in MDM2(C305F) Mice Due to Reduced p53 Activity and Enhanced Energy Expenditure. Cell Rep 18 (4), 1005–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y et al. (2014) Ribosomal protein-Mdm2-p53 pathway coordinates nutrient stress with lipid metabolism by regulating MCD and promoting fatty acid oxidation. Proc Natl Acad Sci U S A 111 (23), E2414–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rinaldo C et al. (2007) MDM2-regulated degradation of HIPK2 prevents p53Ser46 phosphorylation and DNA damage-induced apoptosis. Mol Cell 25 (5), 739–50. [DOI] [PubMed] [Google Scholar]
- 14.Oda E et al. (2000) Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288 (5468), 1053–8. [DOI] [PubMed] [Google Scholar]
- 15.Hofmann TG et al. (2002) Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat Cell Biol 4 (1), 1–10. [DOI] [PubMed] [Google Scholar]
- 16.Knights CD et al. (2006) Distinct p53 acetylation cassettes differentially influence gene-expression patterns and cell fate. J Cell Biol 173 (4), 533–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sykes SM et al. (2006) Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell 24 (6), 841–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jansson M et al. (2008) Arginine methylation regulates the p53 response. Nat Cell Biol 10 (12), 1431–9. [DOI] [PubMed] [Google Scholar]
- 19.Szak ST et al. (2001) Kinetics of p53 binding to promoter sites in vivo. Mol Cell Biol 21 (10), 3375–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson JG and Pereira-Smith OM (2006) p53 is preferentially recruited to the promoters of growth arrest genes p21 and GADD45 during replicative senescence of normal human fibroblasts. Cancer Res 66 (17), 8356–60. [DOI] [PubMed] [Google Scholar]
- 21.Weinberg RL et al. (2005) Comparative binding of p53 to its promoter and DNA recognition elements. J Mol Biol 348 (3), 589–96. [DOI] [PubMed] [Google Scholar]
- 22.Purvis JE et al. (2012) p53 dynamics control cell fate. Science 336 (6087), 1440–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kracikova M et al. (2013) A threshold mechanism mediates p53 cell fate decision between growth arrest and apoptosis. Cell Death Differ 20 (4), 576–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grinkevich VV et al. (2009) Ablation of key oncogenic pathways by RITA-reactivated p53 is required for efficient apoptosis. Cancer Cell 15 (5), 441–53. [DOI] [PubMed] [Google Scholar]
- 25.Pan R et al. (2017) Synthetic Lethality of Combined Bcl-2 Inhibition and p53 Activation in AML: Mechanisms and Superior Antileukemic Efficacy. Cancer Cell 32 (6), 748–760.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paek AL et al. (2016) Cell-to-Cell Variation in p53 Dynamics Leads to Fractional Killing. Cell 165 (3), 631–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spencer SL and Sorger PK (2011) Measuring and modeling apoptosis in single cells. Cell 144 (6), 926–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bentele M et al. (2004) Mathematical modeling reveals threshold mechanism in CD95-induced apoptosis. J Cell Biol 166 (6), 839–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lahav G et al. (2004) Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet 36 (2), 147–50. [DOI] [PubMed] [Google Scholar]
- 30.Leslie PL et al. (2018) p53 Regulates the Expression of LRP1 and Apoptosis through a Stress Intensity-Dependent MicroRNA Feedback Loop. Cell Rep 24 (6), 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu M et al. (2017) p53 dynamics orchestrates with binding affinity to target genes for cell fate decision. Cell Death Dis 8 (10), e3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hafner A et al. (2017) p53 pulses lead to distinct patterns of gene expression albeit similar DNA-binding dynamics. Nat Struct Mol Biol 24 (10), 840–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Porter JR et al. (2016) p53 Pulses Diversify Target Gene Expression Dynamics in an mRNA Half-Life-Dependent Manner and Delineate Co-regulated Target Gene Subnetworks. Cell Syst 2 (4), 272–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng Z et al. (2011) Tumor suppressor p53 meets microRNAs. J Mol Cell Biol 3 (1), 44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki HI et al. (2009) Modulation of microRNA processing by p53. Nature 460 (7254), 529–33. [DOI] [PubMed] [Google Scholar]
- 36.Krell J et al. (2016) TP53 regulates miRNA association with AGO2 to remodel the miRNA-mRNA interaction network. Genome Res 26 (3), 331–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piovan C et al. (2012) Oncosuppressive role of p53-induced miR-205 in triple negative breast cancer. Mol Oncol 6 (4), 458–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang TC et al. (2007) Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 26 (5), 745–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hermeking H (2012) MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer 12 (9), 613–26. [DOI] [PubMed] [Google Scholar]
- 40.Cortez MA et al. (2016) PDL1 Regulation by p53 via miR-34. J Natl Cancer Inst 108 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pichiorri F et al. (2010) Downregulation of p53-inducible microRNAs <em>192</em>, <em>194</em>, and <em>215</em> Impairs the p53/MDM2 Autoregulatory Loop in Multiple Myeloma Development. Cancer Cell 18 (4), 367–381. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Okada N et al. (2014) A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes Dev 28 (5), 438–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J et al. (2017) MicroRNA Control of p53. J Cell Biochem 118 (1), 7–14. [DOI] [PubMed] [Google Scholar]
- 44.Yamakuchi M et al. (2010) P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad Sci U S A 107 (14), 6334–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shang J et al. (2014) MicroRNA-23a antisense enhances 5-fluorouracil chemosensitivity through APAF-1/caspase-9 apoptotic pathway in colorectal cancer cells. J Cell Biochem 115 (4), 772–84. [DOI] [PubMed] [Google Scholar]
- 46.Chen Q et al. (2014) MicroRNA-23a/b and microRNA-27a/b suppress Apaf-1 protein and alleviate hypoxia-induced neuronal apoptosis. Cell Death Dis 5, e1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fischer M (2017) Census and evaluation of p53 target genes. Oncogene 36 (28), 3943–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weichselbaum RR et al. (2017) Radiotherapy and immunotherapy: a beneficial liaison? Nat Rev Clin Oncol 14 (6), 365–379. [DOI] [PubMed] [Google Scholar]
- 49.Xu MM et al. (2016) The Role of Adaptive Immunity in the Efficacy of Targeted Cancer Therapies. Trends Immunol 37 (2), 141–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Munoz-Fontela C et al. (2016) Emerging roles of p53 and other tumour-suppressor genes in immune regulation. Nat Rev Immunol 16 (12), 741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shatz M et al. (2012) The human TLR innate immune gene family is differentially influenced by DNA stress and p53 status in cancer cells. Cancer Res 72 (16), 3948–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Munoz-Fontela C et al. (2008) Transcriptional role of p53 in interferon-mediated antiviral immunity. J Exp Med 205 (8), 1929–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takaoka A et al. (2003) Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature 424 (6948), 516–23. [DOI] [PubMed] [Google Scholar]
- 54.Miciak J and Bunz F (2016) Long story short: p53 mediates innate immunity. Biochim Biophys Acta 1865 (2), 220–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Munoz-Fontela C et al. (2005) Resistance to viral infection of super p53 mice. Oncogene 24 (18), 3059–62. [DOI] [PubMed] [Google Scholar]
- 56.Wellenstein MD and de Visser KE (2018) Cancer-Cell-Intrinsic Mechanisms Shaping the Tumor Immune Landscape. Immunity 48 (3), 399–416. [DOI] [PubMed] [Google Scholar]
- 57.O'Neill LA et al. (2013) The history of Toll-like receptors - redefining innate immunity. Nat Rev Immunol 13 (6), 453–60. [DOI] [PubMed] [Google Scholar]
- 58.Salvador JM et al. (2002) Mice lacking the p53-effector gene Gadd45a develop a lupus-like syndrome. Immunity 16 (4), 499–508. [DOI] [PubMed] [Google Scholar]
- 59.Balomenos D et al. (2000) The cell cycle inhibitor p21 controls T-cell proliferation and sex-linked lupus development. Nat Med 6 (2), 171–6. [DOI] [PubMed] [Google Scholar]
- 60.Schwitalla S et al. (2013) Loss of p53 in enterocytes generates an inflammatory microenvironment enabling invasion and lymph node metastasis of carcinogen-induced colorectal tumors. Cancer Cell 23 (1), 93–106. [DOI] [PubMed] [Google Scholar]
- 61.Wellenstein MD et al. (2019) Loss of p53 triggers WNT-dependent systemic inflammation to drive breast cancer metastasis. Nature 572 (7770), 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Di Minin G et al. (2014) Mutant p53 reprograms TNF signaling in cancer cells through interaction with the tumor suppressor DAB2IP. Mol Cell 56 (5), 617–29. [DOI] [PubMed] [Google Scholar]
- 63.Cooks T et al. (2013) Mutant p53 prolongs NF-kappaB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer Cell 23 (5), 634–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Textor S et al. (2011) Human NK cells are alerted to induction of p53 in cancer cells by upregulation of the NKG2D ligands ULBP1 and ULBP2. Cancer Res 71 (18), 5998–6009. [DOI] [PubMed] [Google Scholar]
- 65.Spurrell EL and Lockley M (2014) Adaptive immunity in cancer immunology and therapeutics. Ecancermedicalscience 8, 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Madapura HS et al. (2012) p53 contributes to T cell homeostasis through the induction of pro-apoptotic SAP. Cell Cycle 11 (24), 4563–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chaudhry S et al. (2002) Cross-regulation of T cell growth factor expression by p53 and the Tax oncogene. J Immunol 169 (12), 6767–78. [DOI] [PubMed] [Google Scholar]
- 68.Watanabe M et al. (2014) Downmodulation of tumor suppressor p53 by T cell receptor signaling is critical for antigen-specific CD4(+) T cell responses. Immunity 40 (5), 681–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Banerjee A et al. (2016) Lack of p53 Augments Antitumor Functions in Cytolytic T Cells. Cancer Res 76 (18), 5229–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leone P et al. (2013) MHC class I antigen processing and presenting machinery: organization, function, and defects in tumor cells. J Natl Cancer Inst 105 (16), 1172–87. [DOI] [PubMed] [Google Scholar]
- 71.Zhu K et al. (1999) p53 induces TAP1 and enhances the transport of MHC class I peptides. Oncogene 18 (54), 7740–7. [DOI] [PubMed] [Google Scholar]
- 72.Wang B et al. (2013) p53 increases MHC class I expression by upregulating the endoplasmic reticulum aminopeptidase ERAP1. Nat Commun 4, 2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Owen-Schaub LB et al. (1995) Wild-type human p53 and a temperature-sensitive mutant induce Fas/APO-1 expression. Mol Cell Biol 15 (6), 3032–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Riley JL (2009) PD-1 signaling in primary T cells. Immunol Rev 229 (1), 114–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Levine AJ (2020) P53 and The Immune Response: 40 Years of Exploration-A Plan for the Future. Int J Mol Sci 21 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Postow MA et al. (2015) Immune Checkpoint Blockade in Cancer Therapy. J Clin Oncol 33 (17), 1974–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gettinger SN et al. (2015) Overall Survival and Long-Term Safety of Nivolumab (Anti-Programmed Death 1 Antibody, BMS-936558, ONO-4538) in Patients With Previously Treated Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 33 (18), 2004–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu D et al. (2018) Mechanisms of Resistance to Immune Checkpoint Blockade. Am J Clin Dermatol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brown JS et al. (2018) Combining DNA damaging therapeutics with immunotherapy: more haste, less speed. Br J Cancer 118 (3), 312–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stronen E et al. (2016) Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science 352 (6291), 1337–41. [DOI] [PubMed] [Google Scholar]
- 81.Albert ML et al. (1998) Dendritic cells acquire antigen from apoptotic cells and induce class I-restricted CTLs. Nature 392 (6671), 86–9. [DOI] [PubMed] [Google Scholar]
- 82.Kroemer G et al. (2013) Immunogenic cell death in cancer therapy. Annu Rev Immunol 31, 51–72. [DOI] [PubMed] [Google Scholar]
- 83.Hernandez C et al. (2016) Damage-associated molecular patterns in cancer: a double-edged sword. Oncogene 35 (46), 5931–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pfirschke C et al. (2016) Immunogenic Chemotherapy Sensitizes Tumors to Checkpoint Blockade Therapy. Immunity 44 (2), 343–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sobol RE et al. (2017) Effect of adenoviral p53 (Ad-p53) tumor suppressor immune gene therapy on checkpoint inhibitor resistance and abscopal therapeutic efficacy. Journal of Clinical Oncology 35 (15_suppl), e14610–e14610. [Google Scholar]
- 86.Chada S et al. (2018) Reversing immune checkpoint inhibitor resistance with adenoviral IL-24 and p53 tumor suppressor immune gene therapy. Journal of Clinical Oncology 36 (5_suppl), 64–64. [Google Scholar]
- 87.Guo G et al. (2017) Local Activation of p53 in the Tumor Microenvironment Overcomes Immune Suppression and Enhances Antitumor Immunity. Cancer Res 77 (9), 2292–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cha YJ et al. (2016) Clinicopathological and prognostic significance of programmed cell death ligand-1 expression in lung adenocarcinoma and its relationship with p53 status. Lung Cancer 97, 73–80. [DOI] [PubMed] [Google Scholar]
- 89.Schumacher TN and Schreiber RD (2015) Neoantigens in cancer immunotherapy. Science 348 (6230), 69–74. [DOI] [PubMed] [Google Scholar]
- 90.Le DT et al. (2015) PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 372 (26), 2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Le DT et al. (2017) Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357 (6349), 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hellmann MD et al. (2018) Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Biton J et al. (2018) TP53, STK11, and EGFR Mutations Predict Tumor Immune Profile and the Response to Anti-PD-1 in Lung Adenocarcinoma. Clin Cancer Res 24 (22), 5710–5723. [DOI] [PubMed] [Google Scholar]
- 94.Dong ZY et al. (2017) Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin Cancer Res 23 (12), 3012–3024. [DOI] [PubMed] [Google Scholar]
- 95.Deniger DC et al. (2018) T-cell Responses to TP53 "Hotspot" Mutations and Unique Neoantigens Expressed by Human Ovarian Cancers. Clin Cancer Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Malekzadeh P et al. (2019) Neoantigen screening identifies broad TP53 mutant immunogenicity in patients with epithelial cancers. J Clin Invest 129 (3), 1109–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li T et al. (2012) Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 149 (6), 1269–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Valente LJ et al. (2013) p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa. Cell Rep 3 (5), 1339–45. [DOI] [PubMed] [Google Scholar]
- 99.Janic A et al. (2018) DNA repair processes are critical mediators of p53-dependent tumor suppression. Nat Med 24 (7), 947–953. [DOI] [PubMed] [Google Scholar]