Introduction
Since John McCarthy introduced the term ‘artificial intelligence (AI)’ in 19551, AI research has been growing. ‘AI’ is an umbrella term that encompasses a vast degree of computer technologies (e.g., expert systems, computer vision, robotics, and machine learning) (Figure 1A) as well as a concept of a machine imitating human intelligence2,3 (Table 1). Modern AI is defined as a system’s ability to 1) perceive the current world, i.e., data; 2) to employ and compare different approaches to achieve specific goals based on given data; 3) to tune their performance and apply to unseen data, and; 4) to repeat the previous processes multiple times and update the previous learning4. When reviewing results from AI models, it is therefore critical to understand whether they are appropriately developed and validated (Figure 1B).
Figure 1A.
Table 1.
Definition of Artificial Intelligence and Its Subfields
| Artificial intelligence (AI) | • An umbrella term that encompasses a vast degree of computer technologies (e.g., expert systems, computer vision, robotics, and machine learning) as well as the concept of a machine imitating human intelligence • A system with the ability to perceive data, to employ and compare different algorithms to achieve specific goals, to analyze their performance and tune them, and then apply to unseen data, and repeat the previous process and update the previous learning |
| Expert system | • Subset of AI • Rule-based systems built with explicit coding of decision rules |
| Machine learning | • Subset of AI • Training a computer model to solve problems (e.g., prediction) by using statistical theories or identifying specific patterns in the data (e.g., phenotyping) |
| Deep learning | • Subset of machine learning • Algorithms to process multiple layers of information to model intricate relationships among data |
| Decision tree | • Supervised machine learning algorithm • Flowchart structure like a tree that has internal nodes, branches, and leaves ○ Internal nodes contain questions such as whether a patient has a fever >100.4F ○ Branches represent the answer (i.e., yes or no) ○ Leaves represent final class labels • Random forest is an ensemble of decision trees |
| K-nearest neighbor | • Supervised machine learning algorithm • Is used for classification and regression tasks based on similarities (i.e., proximity or distance) between available data and new data |
| Naïve Bayes | • Supervised machine learning algorithm • Probabilistic classifiers based on Bayes’ theorem with an assumption of independence among predictor variables |
| K-means clustering | • Unsupervised machine learning algorithm • Identify similar characteristics in the dataset and partition into subgroups |
AI; artificial intelligence
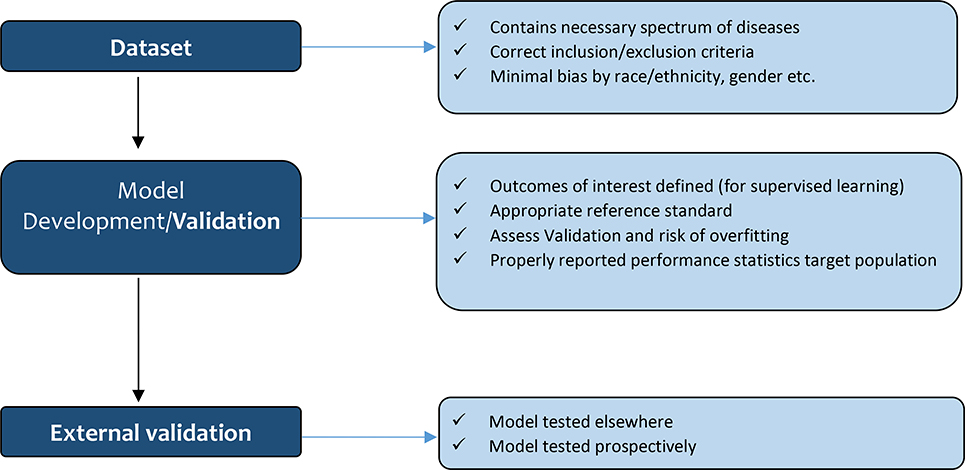
Figure 1B.
The Quality and Quantity of Input data
Like conventional parametric and semi-parametric diagnostic or prognostic models, the quality of the conclusions drawn from the AI-based algorithms relies on the characteristics of the dataset, which is used to train the model. If the model is trained on/based on a biased dataset, the model itself is likely to be biased5. AI might assume that probabilities are static or outcomes within the dataset are optimized, which might not be the case. Hence, similar to conventional methods, it is essential to look at the exclusion and inclusion criteria of the study. For example, a model for post-liver transplant (LT) graft survival, which focuses on matching LT donor and recipient pairs, might not be accurate for recipients with hepatocellular carcinoma (HCC) if the input dataset did not include enough HCC patients. Also, an imbalanced distribution of covariates (e.g., race) or outcomes (e.g., rejection) within a dataset, can affect the usefulness of the algorithm. If black patients are under-represented in the dataset, the model might not accurately provide a diagnostic or prognostic prediction for black patients. The input dataset for a disease diagnostic model needs to have a representative epidemiological spectrum of disease to be predictive6. A recent report on the overall accuracy of diagnosing kidney allograft rejection was reported to be 92.9%7. The authors used a deep learning-based computer-aided diagnostic system using diffusion-weighted magnetic resonance imaging combined with creatinine clearance and serum creatinine7. But the grades of rejection and how rejection was confirmed, was not reported. Thus, for instance, if the input dataset mainly included Banff II or III rejection, the model may not accurately detect Banff I rejection. Similarly, if the outcome variable is not balanced, it is unlikely that the model, which is trained on an imbalanced dataset, truly represents the performance of the outcome of interest.
Model development and validation
Appropriate reference standard and outcomes of interest
There are two broad divisions of machine learning (ML) which have different objectives: supervised and unsupervised learning. Supervised learning trains a model to predict a known status or group5. The outcomes of interest need to be very clearly defined. Many algorithms are readily available such as decision tree, K-nearest neighbor, and Naïve Bayes (Table 1)8. For example, for kidney allograft rejection, the precise definition of rejection is paramount, which many papers have failed to do7,9. Reeves et al. used outcomes confirmed by three pathologists as the reference standard to verify the diagnosis of kidney allograft T cell-mediated rejection and antibody-mediated rejection reliablly10. In contrast, unsupervised learning models do not require clearly defined outcomes5, and the computer will identify similar patterns within the dataset, such as the clustering of subgroups with similar characteristics, e.g., K-means clustering8 (see Table 1). This method can be useful for hypothesis generation and to identify clusters of patients (phenotyping) or events that are more similar to each other.
Model development
We will focus mainly on supervised learning from this point on since most literature in transplantation is based on supervised learning. Many ML models divide data into a training set and a test set during the model development5. After the initial training of the model, the model undergoes internal validation to assess its performance. Sensitivity and regularization parameters can be fine-tuned at this stage to help optimize prediction without overfitting the data. For internal validation, K-fold cross-validation and bootstrapping are commonly used methods5,6. For example, in 10-fold cross-validation, the dataset is split into 10 sets, and then one set is used for validation, and 9 sets are used for training. This provides an average performance of 10 sets to reduce selection bias and to obtain more stable error estimates11. Bootstrapping is a method of random sampling, and it is commonly used for very small datasets to get inferences about the dataset, such as standard error, confidence interval11. If manuscripts about AI or ML models do not describe the internal validation or cross-validation process, the readers should view the results with caution5. Overfitting refers to a condition where the model fits too well to the training set, but then the results likely do not apply/fit well to an external validation set6. For example, a model could simply ‘memorize’ the labels of the input dataset, which would cause the accuracy to be 100%. However, this model would have very poor generalizability when predicting new data5. This can be mitigated partially by regularization or reducing the ‘flexibility’ of the algorithm11, paradoxically reducing the ‘learning’ ability but increasing the potential generalizability to unseen data. Given that the chief benefit of AI is flexibility above traditional models, comparing the final model to traditional methods with an appropriate statistical test is very important.
Discrimination and Calibration
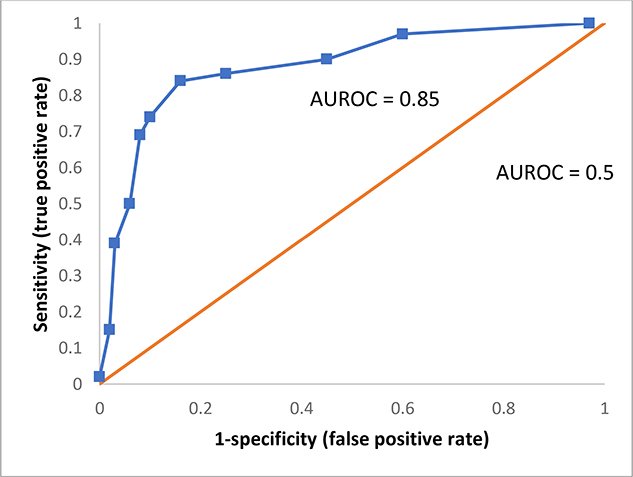
Discrimination is defined as a model’s ability to distinguish different conditions12, such as rejection or not. The C-statistic or the area under the receiver operating characteristic (AUROC) is the most commonly reported model performance metrics for classification problems. The AUROC curve indicates the relationship between false-positive rate (1-specificity) and true positive rate (sensitivity)6(Figure 1–C). A greater AUROC indicates that the model has higher sensitivity with the same false-positive rate and therefore possesses better discriminatory power. In general, an AUROC <0.60 discriminates poorly, an AUROC 0.60 – 0.75 is regarded as possibly helpful discrimination, and an AUROC >0.75 is considered useful12. The AUROC can also assist in choosing optimal cut-off values depending on the desire to optimize sensitivity, specificity, disease prevalence, clinical setting, and cost of misclassification6. However, the use of AUROC does not reflect a model’s performance well when the proportion of negative outcomes is very high13. For example, data in which 95% have no rejection will give the same AUROC as with a balanced data set even though the false positive rate using the unbalance data will be high.
Figure 1C.
Calibration or goodness of fit refers to how accurately a model can predict the absolute risk estimates12. Hence, to adequately interpret the results, discrimination and calibration need to be assessed at the same time12. For example, if a model predicts the risk of dying much lower or higher than the actual risk of dying, then the calibration of that model is poor. The calibration curve and the Hosmer-Lemeshow test are commonly used to assess the degree calibration6.
Other measures
Accuracy, precision (or positive predictive value), recall (or sensitivity), specificity, negative predictive value, and F1 scores are also commonly used to report on model performance. These measures can be easily obtained from a confusion matrix and should all be evaluated (Table 2). The confusion matrix, or error matrix, is a table summarizing the model performance14.
Table 2). Examples of a Confusion Matrix.
In a set of 300 biopsy results there are 180 cases of “rejection” and 120 cases of “no rejection”. Two machine learning models were trained and produced these predictions.
| Actual “rejection” | Actual “no rejection” | ||
|---|---|---|---|
| Model A | Predicted “rejection” | 170 (TP) | 70 (FP) |
| Predicted “no rejection” | 10 (FN) | 50 (TN) | |
| Model B | Predicted “rejection” | 120 (TP) | 10 (FP) |
| Predicted “no rejection” | 60 (FN) | 110 (TN) | |
| Results for Models A and B | |||
| Model A | Model B | ||
| Sensitivity | 0.94 | 0.67 | |
| Specificity | 0.42 | 0.92 | |
| Accuracy | 0.73 | 0.77 | |
| PPV | 0.71 | 0.92 | |
| NPV | 0.83 | 0.65 | |
| F1 Score | 0.81 | 0.77 | |
False negative ~ FN, False positive ~ FP, Negative predictive value ~ NPV, Positive predictive value ~ PPV, True positive ~ TR, True positive rate ~ TPR, True negative ~ TN
Equations and calculations for Model A:
Sensitivity (or recall) = Specificity =
Accuracy =
Precision (or positive predictive value) =
Negative predictive value =
F1 Score =
Accuracy refers to the total fraction of correct predictions14. Model A correctly predicted 170 true rejections and 50 true non-rejections in a cohort of 300, leading to an accuracy of 0.73 (Table 2). While Model A and Model B have similar accuracy (0.73, 0.77), their abilities to detect rejection vary. For example, Model A can predict 170 cases as rejection out of 180 total rejection, while Model B predicted only 120 cases (sensitivity 94% vs. 67%). Hence, accuracy alone can give a false impression of a model, especially if the dataset is imbalanced. In datasets with very high true negatives rejection (and therefore low true positives rejection), accuracy can be high, yet it is not useful to detect rejection.
Positive predictive value (PPV) or Precision (true positives/ [true positives + false positives]) refers to the ratio of correctly predicted positive cases to all predicted positive cases. It estimates the probability that a positive result implies the actual presence of the disease or outcomes15. When a model has a high PPV, the positive result indicates the patient almost certainly has the disease. Unlike sensitivity, PPV is affected by the prevalence of the disease or the outcome15. For example, the PPV in Model A and B is 0.71 and 0.92, respectively (Table 2). If the prevalence of rejection is very high, Model B is useful to detect a disease even with low sensitivity due to the high PPV15.
Negative predictive value (true negatives / [false negatives + true negatives])14 is defined as the correct negative predictions from the total of negative predictions (Table 2). Similar to PPV, a high negative predictive value (NPV) indicates that the patient with a negative predicted outcome is highly likely not to have a disease. Unlike specificity, NPV is influenced by the prevalence15.
F1 score accounts for both precision and recall (or sensitivity) but is not influence by true negative rates14,16.
Therefore the F1 score is a more suitable performance metric when the data is imbalanced with a large number of true negatives16. As mentioned above, high accuracy does not reflect the model’s performance adequately if the input dataset is imbalanced. An F1 score ranges from 0 to 1 and a higher value denotes better classification performance14(Table 2).
Imbalanced data should be accounted for in model development. When the positive (rejection) and negative (no rejection) outcomes are not properly balanced (usually less than 10% positive outcomes) in training data, the model performance may suffer. This can be mitigated by synthetically balancing the data using algorithms such as Synthetic Minority Over-Sampling Technique, using measures such as balanced accuracy which is defined as an average of sensitivity and specificity17, or a Bayesian modeling approach (i.e., adding weight to the smaller subset to balance out the dominant subset)18. In any case, the algorithm trained and then tested on an imbalanced dataset needs to be re-trained and re-tested after balancing.
External Validation/Generalization
Ideally, AI or ML models should be validated in a different patient population, at another site or in prospective cohorts, much like in traditional statistical models. Because the model performance could drop in a new data environment such as different computed tomography (CT) image resolution with different CT scanner, different electronic medical health records system. It is, therefore, essential to pay attention to whether a model is externally validated.
AI in transplantation
The impact of AI in transplantation has not been extensively studied but has gained more attention in recent years. We will introduce some recently published AI-related papers in transplantation and use the above-mentioned quality tests to describe the papers.
One-year post-heart transplant survival was investigated using the United Network of Organ Sharing data from 1987 to 201419. The authors tested multiple ML algorithms (e.g., neural network) and traditional statistical models (e.g., logistic regression)19. Overall, the study was well designed and followed proper methodology. For example, 80% of the data was used for training, and 20% was used for validation. Serial bootstrapping was performed to examine the stability of the C statistics. Calibration was assessed with calibration curves and the Hosmer - Lemeshow goodness of fit. The neural network showed similar C statistics (0.66) compared with logistic regression C statistics (0.65). However, the calibration of the neural network was lower than the traditional statistical models. The authors concluded that the ML algorithms trained on this dataset did not outperform traditional methods. However, they suggested that ML might outperform traditional statistical methods if augmented by data from electronic health records.
Reeve et al. reported on ensembles ML models providing automated kidney biopsy reports in conjunction with molecular measurement for allograft rejection10. Their random forest-based algorithms demonstrated a similar level of balanced accuracy compared to pathologists (92% for T cell-mediated rejection and 94% antibody-mediated rejection). The study has multiple strengths: a very large dataset with a diverse population and a gold-standard reference such as confirmed rejection by pathologists. Also, the paper reported on the accuracy, sensitivity, specificity, PPV, NPV, and balanced accuracy for the readers to understand the model’s performance. This study would be strengthened by external validation.
Bertsimas et al. developed an optimized prediction of mortality model (OPOM) to predict LT candidate’s 3-month waitlist mortality or removal from the waitlist20. During the model development, the authors used a very large dataset (1,618,966 observations) from the Standard Transplant Analysis dataset in patients 12 years or older. The impact of OPOM was assessed with the Liver Simulated Allocation Model and compared with the Model for End-Stage Liver Disease (MELD). Allocation based on the OPOM model showed 417.96 fewer deaths per year compared to MELD, and more LT in female patients. The AUROC of OPOM was highest at 0.859 (0.841 MELD-Na). Overall, the study was well designed and improved on the current system, such as MELD. Interestingly, the authors included the pediatric population, which might impact the prediction results for adults. The authors carefully designed the study to be useful for HCC and non-HCC patients. The OPOM should be tested in a new patient group for external validation.
Conclusion
The use of AI in transplantation to answer pertinent questions is increasing and undoubtedly will continue to provide valuable information. As with any method or tool, it is essential to understand its limits and to be able to evaluate its application critically. Is the input dataset appropriate to answer the question? Is the internal and external validation of the model appropriate? How well did the algorithm perform discrimination, accuracy, calibration, F1 score, NPV, precision? Users should know the strength and limitations of the AI to avoid overreliance, which can lead to spurious conclusions or neglect clinical warning signs. Since AI algorithms may struggle to detect very rare diseases or unusual cases, clinicians’ discretion is important for those cases. Therefore, having a basic understanding of how to evaluate AI/ML papers will ensure that this promising methodology is appropriately applied.
Acknowledgments
3. Funding and acknowledgment
1. Sookhyeon Park acknowledged the supports from the National Institutes of Health (NIH) training grant T32 DK-108738 and the George M. O’Brien Kidney Research Core Center (P30) funded by the National Institutes of Diabetes and Digestive and Kidney Diseases (NIDDK).
2. Nikhilesh R Mazumder acknowledged the supports from the National Institutes of Health (NIH) training grant T32DK077662-11.
3. Sanjay Mehrotra acknowledged the grant 1R01DK118425-01A1 from the National Institute of Diabetes, Digestive and Kidney Diseases (NIDDK)
4. Daniela Ladner acknowledged the grant 1R01DK118425-01A1 from the National Institute of Diabetes, Digestive and Kidney Diseases (NIDDK).
Abbreviations
- AI
artificial intelligence
- AUROC
the area under the receiver operating characteristic
- CT
computed tomography
- FN
false negative
- FP
false positive
- HCC
hepatocellular carcinoma
- LT
liver transplant
- MELD
the Model for End-Stage Liver Disease
- ML
machine learning
- NLP
natural language processing
- NPV
negative predictive value
- OPOM
an optimized prediction of mortality model
- PPV
positive predictive value
- TN
true negative
- TP
true positive
- TPR
true positive rate
Footnotes
2. Disclosure/ conflict of interest
Sookhyeon Park, Nikhilesh R Mazumder, Sanjay Mehrotra, Bing Ho, Bruce Kaplan, and Daniela Ladner do not have any conflict of interest.
Contributor Information
Sookhyeon Park, Northwestern University Transplant Outcomes Research Collaborative, Comprehensive Transplant Center, Feinberg School of Medicine, Northwestern University; Division of Nephrology, Department of Medicine, Northwestern Medicine, Chicago, Illinois.
Nikhilesh R Mazumder, Northwestern University Transplant Outcomes Research Collaborative, Comprehensive Transplant Center, Feinberg School of Medicine, Northwestern University; Division of Hepatology, Department of Medicine, Northwestern Medicine, Chicago, Illinois.
Sanjay Mehrotra, Northwestern University Transplant Outcomes Research Collaborative, Comprehensive Transplant Center, Feinberg School of Medicine, Northwestern University; Industrial Engineering and Management Sciences, Northwestern University, Evanston, IL; Center for Engineering and Health, Institute for Public Health and Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL.
Bing Ho, Northwestern University Transplant Outcomes Research Collaborative, Comprehensive Transplant Center, Feinberg School of Medicine, Northwestern University; Division of Nephrology, Department of Medicine, Northwestern Medicine, Chicago, Illinois.
Bruce Kaplan, Baylor Scott and White Health System, Temple/ Dallas, Office of Vice President.
Daniela P Ladner, Northwestern University Transplant Outcomes Research Collaborative, Comprehensive Transplant Center, Feinberg School of Medicine, Northwestern University; Division of Transplantation, Department of Surgery, Northwestern Medicine, Chicago, Illinois.
References
- 1.John M, Marvin LM, Nathaniel R, Claude ES. A Proposal for the Dartmouth Summer Research Project on Artificial Intelligence, August 31, 1955. AI Magazine. 2006;27(4). [Google Scholar]
- 2.Korf RE. Proceedings of the 4th AAAI Conference on Deep Blue Versus Kasparov: The Significance for Artificial Intelligence. 1997. [Google Scholar]
- 3.Yu K-H, Beam AL, Kohane IS. Artificial intelligence in healthcare. Nature Biomedical Engineering. 2018;2(10):719–731. [DOI] [PubMed] [Google Scholar]
- 4.Kaplan A, Haenlein M. Siri, Siri, in my hand: Who’s the fairest in the land? On the interpretations, illustrations, and implications of artificial intelligence. Business Horizons. 2019;62(1):15–25. [Google Scholar]
- 5.Liu Y, Chen PC, Krause J, Peng L. How to Read Articles That Use Machine Learning: Users’ Guides to the Medical Literature. Jama. 2019;322(18):1806–1816. [DOI] [PubMed] [Google Scholar]
- 6.Park SH, Han K. Methodologic Guide for Evaluating Clinical Performance and Effect of Artificial Intelligence Technology for Medical Diagnosis and Prediction. Radiology. 2018;286(3):800–809. [DOI] [PubMed] [Google Scholar]
- 7.Abdeltawab H, Shehata M, Shalaby A, et al. A Novel CNN-Based CAD System for Early Assessment of Transplanted Kidney Dysfunction. Scientific reports. 2019;9(1):5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rashidi HH, Tran NK, Betts EV, Howell LP, Green R. Artificial Intelligence and Machine Learning in Pathology: The Present Landscape of Supervised Methods. Academic pathology. 2019;6:2374289519873088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tapak L, Hamidi O, Amini P, Poorolajal J. Prediction of Kidney Graft Rejection Using Artificial Neural Network. Healthcare informatics research. 2017;23(4):277–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reeve J, Madill-Thomsen KS, Halloran PF. Using ensembles of machine learning classifers to maximize the accuracy and stability of molecular biopsy interpretation. American Journal of Transplantation. 2019;19:452–453. [DOI] [PubMed] [Google Scholar]
- 11.Alpaydin E Introduction to Machine Learning. MIT Press; 2014. [Google Scholar]
- 12.Alba AC, Agoritsas T, Walsh M, et al. Discrimination and Calibration of Clinical Prediction Models: Users’ Guides to the Medical Literature. Jama. 2017;318(14):1377–1384. [DOI] [PubMed] [Google Scholar]
- 13.Saito T, Rehmsmeier M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PloS one. 2015;10(3):e0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tharwat A Classification assessment methods. Applied Computing and Informatics. 2018. [Google Scholar]
- 15.Sadrzadeh H, Baskin L, Kline G. Chapter 1 - Variables affecting endocrine tests results, errors prevention and mitigation. In: Sadrzadeh H, Kline G, eds. Endocrine Biomarkers. Elsevier; 2017:1–40. [Google Scholar]
- 16.Waegeman W, Dembczynski K, Jachnik A, Cheng W, Hüllermeier E. On the bayes-optimality of F-measure maximizers. J Mach Learn Res. 2014;15(1):3333–3388. [Google Scholar]
- 17.Iram S, Vialatte F-B, Qamar MI. Chapter 1 - Early Diagnosis of Neurodegenerative Diseases from Gait Discrimination to Neural Synchronization. In: Al-Jumeily D, Hussain A, Mallucci C, Oliver C, eds. Applied Computing in Medicine and Health. Boston: Morgan Kaufmann; 2016:1–26. [Google Scholar]
- 18.Klein K, Hennig S, Paul SK. A Bayesian Modelling Approach with Balancing Informative Prior for Analysing Imbalanced Data. PloS one. 2016;11(4):e0152700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller PE, Pawar S, Vaccaro B, et al. Predictive Abilities of Machine Learning Techniques May Be Limited by Dataset Characteristics: Insights From the UNOS Database. Journal of cardiac failure. 2019;25(6):479–483. [DOI] [PubMed] [Google Scholar]
- 20.Bertsimas D, Kung J, Trichakis N, Wang Y, Hirose R, Vagefi PA. Development and validation of an optimized prediction of mortality for candidates awaiting liver transplantation. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2019;19(4):1109–1118. [DOI] [PubMed] [Google Scholar]