Abstract
Changes in mitochondrial DNA (mt-DNA) copy number in blood/tissue have been linked to increased risk of several cancers; however, studies on their association in breast cancer is still lacking. In this pilot study, we investigated mt-DNA copy number variation in peripheral blood and tissue samples from metastatic breast cancer patients and compared their differences. For the study, peripheral blood samples from non-cancer individuals (control) and breast cancer patients, along with resected tissues from adjacent and tumor sites from same breast cancer patients were collected. Total genomic DNA was isolated and changes in mt-DNA copy number were measured by relative quantification using SYBR green based quantitative real time PCR method. Our results indicated a significant reduction in mt-DNA copy number in blood samples of breast cancer patients compared to control. However, a significantly higher mt-DNA copy number was observed in tumor tissue when compared with paired non tumor tissue. There was no significant difference in mt-DNA copy number between blood and adjacent tumor tissue samples of the breast cancer patients. Overall, our study reports for the first time a comparison of mt-DNA copy number in blood and paired tissue together and suggested that mt-DNA copy number is differentially regulated in blood and tumor tissues in breast cancer.
Keywords: Mitochondria, Mitochondrial DNA copy Number, Breast cancer, Metastasis
Highlights
-
•
Mitochondrial DNA (mt-DNA) copy number is differentially regulated in blood and tumor tissues of breast cancer.
-
•
A significant decrease in mt-DNA was observed in the blood samples of breast cancer patients compared with non-cancerous control individuals.
-
•
An increase in the mt-DNA copy number was observed in tumor tissue compared with adjacent non-tumor tissue from breast cancer patients.
-
•
Comparison of mt-DNA copy number in blood and tissue samples together is suggested to be a better indicator of mitochondrial changes..
1. Introduction
Breast cancer is one of the most commonly diagnosed cancers in females and a leading cause of cancer deaths contributing to 11.6 % death among all cancers worldwide [1]. In recent years, India has seen a tremendous increase in breast cancer incidences, which has surpassed the cervical cancer to reach to the top of the cancer list [2]. Although due to advancement in diagnostic and treatment strategies, the mortality rate has also decreased specially in western countries, however, it has little effect in developing countries like India. In fact, mortality rate is higher in rural area despite of lower incidences of breast cancer compared to urban cities in India, indicating variation in disease susceptibility and clinical outcome [2]. Therefore analysis of risk factors and their role in disease progression are required to develop effective diagnostic and therapeutic approaches.
In recent years, alterations in mitochondrial mechanisms have been reported in various cancers, and have been linked to carcinogenesis [3]. Specifically, mitochondrial DNA (mt-DNA) alterations such as mt-DNA copy number variations, mt-DNA rearrangements, deletions, and point mutations have been reported in various cancers, and have been investigated for their use as risk factors or early diagnostic markers [4], however role of such alterations in disease progression is still not clear. Mitochondrion contains multiple copies of its DNA (~16.5 kb in size), and due to its close proximity to respiratory chains (RC) complexes and absence of sophisticated DNA repair mechanism, mt-DNA is susceptible to oxidative damage, commonly observed in cancer cells [5]. Therefore alterations in mt-DNA copy number have been reported to be associated with increased cancer risk [6]. However, their positive or inverse relation with cancer incidence is highly variable and depends on various factors including origin of sample and type of cancer. In case of breast cancer, while several studies have reported somatic and germline mutations in mt-DNA, a very few studies have investigated the alterations in mt-DNA copy number in patient samples [[7], [8], [9], [10], [11], [12]]. In these studies, copy number variations have been investigated in either peripheral blood samples or paired tissue samples of breast cancer patients. In addition, some of these reports showed contrasting results indicating the complexity of mt-DNA copy number variations in breast cancer [9,12]. Major reason of such discrepancies are the lack of understanding on their tissue specific changes even in same individuals for understanding the effect of mt-DNA copy number on mitochondrial functions [13]. Therefore, in this pilot study, we investigated the variations in mt-DNA copy number in blood and tissue samples. Mt-DNA copy number was measured in peripheral blood samples from healthy and breast cancer patients, and also from paired non-cancerous adjacent and tumor tissue. Results were compared to see the overall changes in mt-DNA copy number in blood and tissue samples of breast cancer patients.
2. Materials and methods
2.1. Patient samples and DNA isolation
Female breast cancer patient samples; such as peripheral blood and tissue were obtained from Mahaveer Cancer Sansthan, Patna Bihar after approval from Ethics Committee Mahavir Cancer Sansthan to collaborator Dr. Ashok Kumar Ghosh (Ref N. MCS/Admin/2017-18/707). Samples were from those individuals having diagnosed with breast cancer as detailed in Table 1. Pre-surgical blood samples and surgically resected non-cancerous and tumor tissues were collected from same subjects (n = 14). Standard guidelines from American Joint Committee on Cancer (AJCC) were followed for staging of cancer [14]. Briefly, TNM status of tumor was determined considering tumor extent (T), regional lymph nodal spread (N) and distant metastasis (M). It is an indicator of extent of spread of disease and an important prognostic factor for both, cancer free survival and overall survival. Similarly, grade indicates the degree of aggressiveness of the disease and determined by growth of the cells, in which low grade number (grade 1) indicates slow growing cells and less likely to spread, while grade 2 or above reflects fast growing cells and most likely to spread. Lymph Node (LN) status reflects if lymph nodes were involved by tumor, in which N1 reflects that cancer has spread to 1 to 3 axillary lymph nodes, N2 reflects the spread to 4 to 9 axillary lymph nodes and N3 for 10 or more axillary lymph nodes. Tumor diameter is the ‘T’ component of TNM staging depends on tumor size (cm3) and helps in deciding the cancer staging. Peripheral blood from female volunteer free form cancer were collected for age matched controls (n = 14). DNA was extracted from 500μl blood samples using a QIAamp Blood Kit (Qiagen, Frederick, MD, USA) according to the manufacturers’ instructions. DNA quantity was measured using Nano-drop spectrophotometer (Thermo scientific, Fitchburg, WI, USA).
Table-1.
Clinicopathological information of breast cancer patients.
| S.N. | Patient Group | Age (Y) | Stage | LN Status | Tumor Grading | Tumor diameter (cm3) |
|---|---|---|---|---|---|---|
| 1 | P-1 | 42 | 3B | N1 | 2 | 7 |
| 2 | P-2 | 52 | 3B | N2 | 2 | 8 |
| 3 | P-3 | 55 | 3A | N1 | 1 | 4 |
| 4 | P-4 | 64 | 3A | N1 | 2 | 3 |
| 5 | P-5 | 41 | 3B | N2 | 2 | 7 |
| 6 | P-6 | 45 | 3B | N1 | 2 | 8 |
| 7 | P-10 | 59 | 3A | N2 | 2 | 4 |
| 8 | P-11 | 56 | 3A | N1 | 2 | 3 |
| 9 | P-12 | 44 | 3C | N2 | 2 | 12 |
| 10 | P-15 | 39 | 3A | N1 | 2 | 4 |
| 11 | P-16 | 40 | 2B | N0 | 1 | 3 |
| 12 | P-17 | 51 | 3B | N1 | 2 | 7 |
| 13 | P-19 | 45 | 3A | N1 | 2 | 5 |
| 14 | P-20 | 41 | 3A | N1 | 2 | 4 |
|
Mean Age: 48.14 ± 2.133 Median Age: 45 |
||||||
2.2. Mitochondrial DNA copy number measurement
Relative mt-DNA copy number was measured by quantitative PCR (qPCR) through simultaneous amplification of mt-DNA and nuclear DNA, and calculated after normalizing mt-DNA amplification from nuclear amplification. The mt-DNA specific primers for Human t-RNA leucine 1 gene (forward primer: CACCCAAGAACAGGGTTT GT and reverse primer: TGGCCATGGGTATGTTGTTAA) and nuclear DNA primers for 18s r-RNA gene (forward primer: TAGAGGGACAAGTGGCGTTC and reverse primer: CGCTGAGCCAGTCAGTGT) were used for quantitative PCR to determine the mt-DNA copy number following previously published protocols [15,16]. The q-PCR reaction was set up according to the manufacturer’s instruction and optimized for more than 95% amplification efficiency (Fast SYBR Green Master Mix, Applied Biosystems) and run on 7900HT Fast Real-Time PCR System (Applied Biosystems). Relative fold changes were calculated by 2-ΔCT (ΔCT = CT mt-DNA - CT n-DNA) method following previously published [16].
2.3. Statistical analysis
Statistical analysis was done using Graph Pad Prism software (Version 5.0, San Diego, CA, USA). The continuous variables were compared using Student-t-test between two groups, and more than two groups were compared with ANOVA/Bonferroni's multiple comparison test when appropriate. The differences between groups with P < 0.05 were considered statistically significant.
3. Results
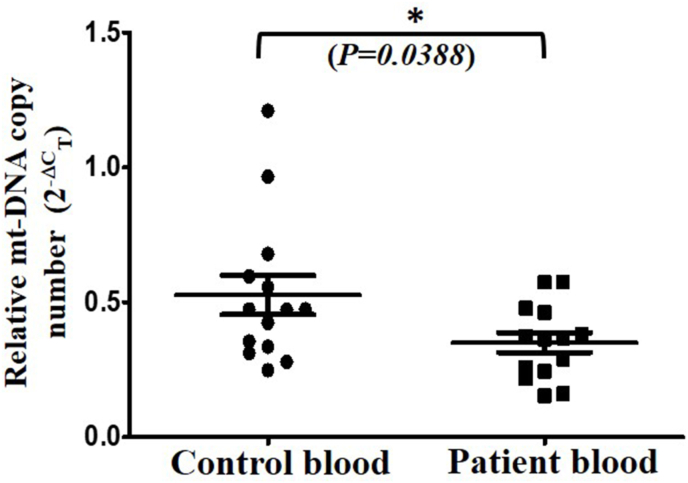
The major aim of this study was to investigate the differences in mt-DNA copy number from blood and tissue compartments of breast cancer patients. Therefore, mt-DNA copy number was determined in 1) blood samples from healthy and breast cancer patients, and 2) paired non-cancerous and tumor tissue from same breast cancer patients who were undergone surgery. The detail of breast cancer patients and age information of control group in this study is summarized in Table-1, Table 2 respectively. The mean age of patients and control subjects (n = 14) at the time of study were 48.14 ± 2.133 and 41.07 ± 3.489 respectively and difference between the age of both groups were found non-significant. Almost all the patients in this study were diagnosed with invasive breast cancer of stage 3A-C, and metastatic type of cancer (N1 or more), with an exception of P-16. Out of fourteen, 12 patients had grade 2 or moderately differentiated tumor mass as confirmed through pathological investigations, and overall tumor size varied from 3-12 cm3 in these patients (Table 1). Total genomic DNA was isolated from peripheral blood samples of control and patient groups and used for analysis of mt-DNA copy number by performing q-RT-PCR through optimized protocol as published earlier [15,16]. The comparison of blood samples showed approximately 34% decrease in mt-DNA copy number in patient groups compared to control group (p<0.0388) (Fig. 1).
Table 2.
Age details of the control group.
| S.N. | Control Group | Age (Y) |
|---|---|---|
| 1 | C-1 | 31 |
| 2 | C-3 | 65 |
| 3 | C-5 | 55 |
| 4 | C-6 | 50 |
| 5 | C-7 | 30 |
| 6 | C-9 | 52 |
| 7 | C-10 | 32 |
| 8 | C-12 | 60 |
| 9 | C-19 | 25 |
| 10 | C-16 | 39 |
| 11 | C-18 | 35 |
| 12 | C-20 | 40 |
| 13 | C-21 | 36 |
| 14 | C-14 | 25 |
| Mean Age = 41.07 ± 3.489 Median Age: 37.5 |
||
Fig. 1.
Mitochondrial DNA copy number variations in peripheral blood samples. Genomic DNA was isolated and used for q-RT-PCR as described in methods. Relative mt-DNA copy number was determined by 2-ΔCT method and compared between values (Mean ± SD) of patient blood samples from control blood samples. Student’s t-test (unpaired, two tailed) was performed to analyze the significance. *(P<0.05).
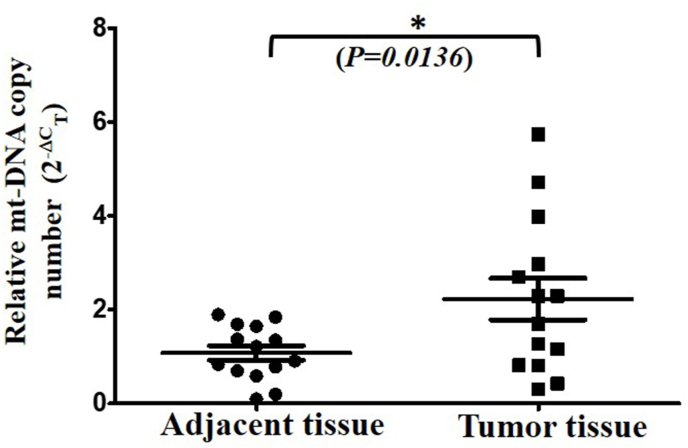
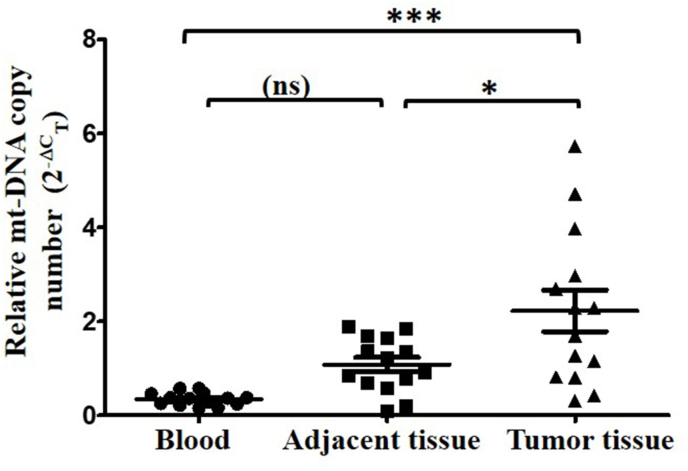
To further investigate the tissue specific changes in mt-DNA copy number, we isolated genomic DNA from resected paired non-cancerous and tumor tissues and measured the mt-DNA copy number as discussed previously. Interestingly, we found an opposite pattern from blood investigations and observed a significant 2 fold increase in mt-DNA copy number in tumor tissue than paired non-cancerous adjacent tissue (p<0.0136) (Fig. 2). In order to investigate the changes in mt-DNA copy number across the samples in breast cancer, we compared the fold changes in matched blood, adjacent and tumor tissues of the patients. Relative to blood mt-DNA copy number, a significant 6.36 fold increase was observed in tumor tissue (Fig. 3). However compared to blood, increase in mt-DNA copy number in adjacent tissue was not significant.
Fig. 2.
Variation in mt-DNA copy number in paired breast cancer tissue. Mt-DNA copy number from paired adjacent non tumor and tumor tissue was determined using q-RT-PCR method. The relative fold change (2-ΔCT) of adjacent and tumor tissue (Mean ± SD) is shown. Two tailed paired Student’s t-test was used to calculate the significance.*(P<0.05).
Fig. 3.
Comparison of mt-DNA copy number in blood and tissue of breast cancer patients. The relative fold change (2-ΔCT) of blood, adjacent and tumor tissue (Mean ± SD) is shown. One way ANOVA and Bonferroni's Multiple Comparison Test was used to calculate the significance.***(P<0001), *(P<0.05), ns-non significant.
Altogether, results on mt-DNA copy number variations indicated that mt-DNA copy number was decreased in blood samples of patients compared to healthy individuals. However, when paired tissue samples were compared, it resulted in an increase in mt-DNA copy number in tumor tissue. Together with paired analysis of mt-DNA copy number in blood and tissue samples of patients, no significant difference was observed in blood and adjacent tissue, while mt-DNA copy number in tumor tissue was significantly elevated.
4. Discussion
Mitochondrial DNA alterations have long been investigated in different cancers to identify their relation to cancer risk; however mt-DNA copy number changes are highly variable and dependent on various factors including contribution of mitochondrial functions in different cancer types [6,17]. In case of breast cancer, only a handful of studies have been reported which have investigated mt-DNA copy number changes, either in blood samples or paired tissue samples with contrasting findings [[7], [8], [9], [10], [11], [12],18]. For example, some of the studies focusing on blood samples reported a significant increase in mt-DNA copy number [8,10,11]. Conversely, other studies on mt-DNA copy number measurements in paired tissue samples, reported a decrease in mt-DNA copy number [9,12,[18], [19], [20]]. In addition, combined with mt-DNA copy number variations with other cancers, several reports suggested variability in blood vs tissue analysis across different cancers [6,17,21]. For example, in an analysis from The Cancer Genome Atlas (TCGA) data, a decrease in mt-DNA copy number in breast cancer tissue samples has been observed [17]. However in a separate study on meta-analysis, increased mt-DNA copy number was associated with breast cancer combining blood and tissue analysis [6]. There is no clarity on predictive value of mt-DNA copy number in breast cancer, as some indicates low mt-DNA copy number for better prognosis for metastasis [22], while other suggests the contrary [21]. In addition, no data is available on comparison in blood and tissue samples together in breast cancer, which may indicate if such differences co-exist in individual patients, and would allow better understanding of mitochondrial contribution in breast cancer for a given set of conditions. In our study, we selected only metastatic breast cancer cases and measured the mt-DNA copy number in blood and paired tissue samples. Although our study is in line with previous reports on showing inverse relation in blood vs tissue samples, while individual comparisons in blood or tissue samples for mt-DNA copy number resulted in opposite trend from previous reports. In our study, the cancer blood samples were first compared with heathy controls and we found a decrease in the mt-DNA copy number (Fig. 1). Since previous studies on measurement of mt-DNA copy number in blood samples were reported in patients above 50 years of age [8,10,11,23], this difference could be possibly due to variation in the age group in our study (median age 45). Further, we observed a significant increase in mt-DNA copy number in tumor tissue when compared with its paired non-cancerous tissue (Fig. 2). This is in line with previous reports in which elevated mt-DNA copy number is associated with invasive breast carcinoma along with higher frequency of D-loop region mutations and over expression of serval hormone receptors and cancer biomarkers [9]. Since mt-DNA copy number varies with pathological characteristics, the increase in mt-DNA copy number has also been associated with increasing grade (from grade II to III) and very large size of tumor (>50cm3) as reported earlier [19]. Similarity, in an early study by Yu et al., although an overall decrease in mt-DNA copy number was observed in paired breast cancer tissue, a significant upregulation was also observed to be associated in patients below 50 years of age (66.7%) and patients with histological grade 2 tumor [12]. In our study, most of the cases had metastatic breast cancer (LN status: N1 or N2, 13/14) and grade 2 (12/14), and mean age below 50 years, corroborating with these reports of higher mt-DNA copy number in paired breast cancer tissue. However, elevation of mt-DNA copy number increase in our case differ from other studies, in which low mt-DNA copy number has been observed in breast cancer tissue samples [12,18,20]. Such discrepancies have also been observed in other cancers and partly account for metabolic flexibility of different cell types in cancer tissue, genetic diversity, tumor purity and functional status of mitochondria [17]. For example, functional analysis of mitochondria in invasive breast cancer patients suggested that epithelial breast cancer cells show upregulation of mitochondrial biogenesis and transcription and may contribute to fueling the invasiveness of breast cancer [24]. However, this upregulation is absent in adjacent stromal cells that have mitochondrial dysfunction and highly dependent on glycolysis supporting Warburg’s hypothesis [25]. Similarly, several OXPHOS inhibitors are now in clinical trials for certain cancers, which specifically target upregulated mitochondrial functions indicating an increased dependency on mitochondrial functions in carcinogenesis [26]. Comparison of blood and tissue samples together from breast cancer patients in our study resulted in a consistence increase in mt-DNA copy number in tumor tissue; however, there was no significant difference in the mt-DNA copy number in patients blood to adjacent non-cancerous tissue (Fig. 3). Our study also indicated that changes in mt-DNA copy number from tumor tissues were not necessarily reflected in blood samples of cancer patients, as we found a decrease not increase in the blood samples of patients compared to non-cancer individuals. Since no significant difference was observed in blood and adjacent non-tumor tissue samples of patients; it further suggest that; to obtain a more clear idea on mtDNA copy number variations in cancer, analysis of tissue and blood samples together would be more appropriate choice than analyzing in blood or in paired tissue only. However, with this small sample size and lack of functional studies, it is difficult to conclude whether this increase in mtDNA copy number in tumor tissues indicates functional requirement or compensation for impaired mitochondrial functions.
Therefore, it remains crucial to understand if alterations in mt-DNA copy number has any functional association with mitochondria: whether increased mt-DNA copy number reflects increase compensation for mt-DNA mutations/dysfunction or increased demand for mitochondrial energy/metabolites for growth and invasiveness. Although, from our study, this issue remained unsolved, however based on these preliminary data, future studies are planned in a larger sample size to reconfirm our finding on elevated mt-DNA copy number as well as investigating mitochondrial functions. It will help to understand functional role of mt-DNA copy number variations in breast carcinogenesis and would be useful for breast cancer risk assessment or prognosis.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author LKS upon reasonable request.
Authors statement
NKR planned and performed the experiments, complied and analyzed the data. GP and AKG and were involved in sample collection, histopathological reporting, tabulation of data. RH helped in analyzing and critically proofreading the manuscript. LKS wrote the manuscript. Manuscript was reviewed by all the contributing authors.
Declaration of competing interest
None.
Acknowledgements
Author’s laboratory is supported with funding from Indian Council of Medical Research grant (EMR-2019-665/CMB/Adhoc/BMS to LKS). NKR was supported by junior research fellowship [grant no. 16-6(Dec.2017)/2018] from CSIR-UGC.
References
- 1.Bray F., Ferlay J., Soerjomataram I. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Malvia S., Bagadi S.A., Dubey U.S. Epidemiology of breast cancer in Indian women. Asia Pac. J. Clin. Oncol. 2017;13:289–295. doi: 10.1111/ajco.12661. [DOI] [PubMed] [Google Scholar]
- 3.Kumar Sharma L. Mitochondrial alteration: a major player in carcinogenesis. Cell Biol. 2015;3:8. [Google Scholar]
- 4.Lu J., Sharma L.K., Bai Y. Implications of mitochondrial DNA mutations and mitochondrial dysfunction in tumorigenesis. Cell Res. 2009;19:802–815. doi: 10.1038/cr.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yakes F.M., Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu L., Yao X., Shen Y. Altered mitochondrial DNA copy number contributes to human cancer risk: evidence from an updated meta-analysis. Sci. Rep. 2016;6:35859. doi: 10.1038/srep35859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campa D., Barrdahl M., Santoro A. Mitochondrial DNA copy number variation, leukocyte telomere length, and breast cancer risk in the European Prospective Investigation into Cancer and Nutrition (EPIC) study. Breast Cancer Res. 2018;20:29. doi: 10.1186/s13058-018-0955-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lemnrau A., Brook M.N., Fletcher O. Mitochondrial DNA copy number in peripheral blood cells and risk of developing breast cancer. Canc. Res. 2015;75:2844–2850. doi: 10.1158/0008-5472.CAN-14-1692. [DOI] [PubMed] [Google Scholar]
- 9.Lin C.S., Chang S.C., Ou L.H. Mitochondrial DNA alterations correlate with the pathological status and the immunological ER, PR, HER-2/neu, p53 and Ki-67 expression in breast invasive ductal carcinoma. Oncol. Rep. 2015;33:2924–2934. doi: 10.3892/or.2015.3887. [DOI] [PubMed] [Google Scholar]
- 10.Shen J., Platek M., Mahasneh A. Mitochondrial copy number and risk of breast cancer: a pilot study. Mitochondrion. 2010;10:62–68. doi: 10.1016/j.mito.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thyagarajan B., Wang R., Nelson H. Mitochondrial DNA copy number is associated with breast cancer risk. PloS One. 2013;8 doi: 10.1371/journal.pone.0065968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu M., Zhou Y., Shi Y. Reduced mitochondrial DNA copy number is correlated with tumor progression and prognosis in Chinese breast cancer patients. IUBMB Life. 2007;59:450–457. doi: 10.1080/15216540701509955. [DOI] [PubMed] [Google Scholar]
- 13.Clay Montier L.L., Deng J.J., Bai Y. Number matters: control of mammalian mitochondrial DNA copy number. J Genet Genomics. 2009;36:125–131. doi: 10.1016/S1673-8527(08)60099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edge S.B., Compton C.C. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann. Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 15.Fang H., Liu X., Shen L. Role of mtDNA haplogroups in the prevalence of knee osteoarthritis in a southern Chinese population. Int. J. Mol. Sci. 2014;15:2646–2659. doi: 10.3390/ijms15022646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 17.Reznik E., Miller M.L., Senbabaoglu Y. Mitochondrial DNA copy number variation across human cancers. Elife. 2016;5 doi: 10.7554/eLife.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mambo E., Chatterjee A., Xing M. Tumor-specific changes in mtDNA content in human cancer. Int. J. Canc. 2005;116:920–924. doi: 10.1002/ijc.21110. [DOI] [PubMed] [Google Scholar]
- 19.Bai R.K., Chang J., Yeh K.T. Mitochondrial DNA content varies with pathological characteristics of breast cancer. J Oncol. 2011;2011:496189. doi: 10.1155/2011/496189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan A.X., Radpour R., Haghighi M.M. Mitochondrial DNA content in paired normal and cancerous breast tissue samples from patients with breast cancer. J. Canc. Res. Clin. Oncol. 2009;135:983–989. doi: 10.1007/s00432-008-0533-9. [DOI] [PubMed] [Google Scholar]
- 21.Chen N., Wen S., Sun X. Elevated mitochondrial DNA copy number in peripheral blood and tissue predict the opposite outcome of cancer: a meta-analysis. Sci. Rep. 2016;6:37404. doi: 10.1038/srep37404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weerts M.J., Sieuwerts A.M., Smid M. Mitochondrial DNA content in breast cancer: impact on in vitro and in vivo phenotype and patient prognosis. Oncotarget. 2016;7:29166–29176. doi: 10.18632/oncotarget.8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen J., Wan J., Song R. Peripheral blood mitochondrial DNA copy number, length heteroplasmy and breast cancer risk: a replication study. Carcinogenesis. 2015;36:1307–1313. doi: 10.1093/carcin/bgv130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sotgia F., Whitaker-Menezes D., Martinez-Outschoorn U.E. Mitochondria "fuel" breast cancer metabolism: fifteen markers of mitochondrial biogenesis label epithelial cancer cells, but are excluded from adjacent stromal cells. Cell Cycle. 2012;11:4390–4401. doi: 10.4161/cc.22777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 26.Ashton T.M., McKenna W.G., Kunz-Schughart L.A. Oxidative phosphorylation as an emerging target in cancer therapy. Clin. Canc. Res. 2018;24:2482–2490. doi: 10.1158/1078-0432.CCR-17-3070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author LKS upon reasonable request.