Abstract
Background
The epidemiology of human urinary schistosomiasis caused by Schistosoma haematobium can be complicated by the presence of ruminant schistosomiasis caused, primarily by S. bovis. The two schistosome species may be transmitted by the same Bulinus species, they may occur sympatrically in the same habitat, and their cercariae are very similar in morphology and therefore, difficult to tell them apart. Screening of snails collected from freshwater habitats for schistosome infections is often used to identify transmission sites or to evaluate success or failure of interventions. However, pin-pointing sites involved in S. haematobium transmission can be complicated by the presence of other mammalian schistosomes such as the bovine schistosome, which is a fairly common parasite. A PCR-RFLP method targeting a unique segment of the second internal transcribed spacer (ITS2) region of the ribosomal DNA (rDNA) in the schistosomes was used to identify mammalian schistosome cercariae shed by bulinid snails collected from endemic freshwater habitats located within Machakos county in south-eastern Kenya, with the aim to identify the transmission sites and assess the distribution each of the parasite species in the study area.
Results
A total of 5,034 bulinid snails were collected from 41 different sites and screened for schistosome infections, and out of these, 43 (<1%) were found to be shedding mammalian schistosome cercariae. On analysis using the Polymerase chain reaction- Restriction Fragment Length Polymorphisms (PCR-RFLP) assay, cercariae from 32 snails were identified as S. haematobium while cercariae from 11 snails turned out to be S. bovis. Only two sites out of 40 namely Kisukioni and Katiwa, were active transmission sites. Both sites were active transmission sites for both S. haematobium and S. bovis. The assay reliably identified and distinguished between S. haematobium and S. bovis cercariae, even when only a few cercariae (5–10) were present in the sample, or when the parasite DNA concentrations were as low as five pico grammes (5pg). The FTA® paper offered a more reliable way of collecting, transporting and storing DNA material, and the samples.
Conclusion
The PCR-based assay can potentially be used to support schistosomiasis control efforts, in epidemiological studies of urinary schistosomiasis, or in transmission ecology studies of S. haematobium and S. bovis.
Keywords: Schistosoma haematobium, Schistosoma bovis, PCR-RFLP, Schistosomiasis, Athi River Basin, Kenya, Transmission sites
Schistosoma haematobium, Schistosoma bovis, PCR-RFLP, schistosomiasis, Athi River Basin, Kenya, transmission sites.
1. Introduction
Schistosomiasis (or bilharziasis) remains one of the most significant health burdens for people living in vulnerable parts of the world [1, 2, 3]. Schistosomiasis is a debilitating disease caused by blood flukes of the genus Schistosoma, and worldwide, over 207 million people are known to harbour schistosome parasites with over 90% of the case occurring in Sub-Saharan Africa [4]. In 2000, it was estimated that 70 million people had hematuria, 32 million had dysuria associated with Schistosoma haematobium, 18 million had major bladder wall pathology, 10 million people had S. haematobium related renal failure; and schistosomiasis related bladder cancer, resulting in an estimated mortality of 150 000 people per year in sub- Saharan Africa [5, 6]. Of the schistosome species related to S. haematobium, Schistosoma bovis is the most widespread, with reported distribution in the Middle East, the Mediterranean basin, and most parts of Africa. In Kenya, both human urinary schistosomiasis caused by S. haematobium and the ruminant schistosomiasis caused, primarily by S. bovis, are common. The two species share some fundamental life stage aspects in their biology namely, they can utilize the same Bulinus snail species as intermediate hosts, and in Kenya, both parasite species are transmitted by members of Bulinus africanus group (B. africanus, B. globosus and B. nasutus), may occur sympatrically, and their cercarial larval stages are very similar morphologically. As a result it may be difficult to determine which schistosome species is being transmitted in a particular habitat or locality, at any given time. Snail collection from freshwater habitats and screening them for schistosome infections is routinely used to identify potential transmission sites or to monitor success or failure of intervention measures [7]. Screening field-collected snails for the presence of cercariae is a simple way of determining risk of schistosomiasis transmission in a particular water body or locality. The presence of schistosome cercariae in a snail population present in a water body or locality would suggest risk of schistosomiasis transmission. However, knowing whether the cercariae shed are S. haematobium or S. bovis, and the proportion of snails shedding a particular cercariae species will help determine the level of the risk. This information will be valuable for control programs as control efforts targeting transmission sites can be more accurately directed to the most risky sites thus, help economize on scarce resources available for intervention.
We evaluated and further refined a previously described PCR-based assay that targets the amplification of the second internal transcribed spacer (ITS2) region of genomic DNA of the schistosomes, followed by restriction fragment length polymorphism (RFLP) analysis of the PCR products using Taq1 restriction enzyme, in the identification of mammalian schistosome cercariae shed by Bulinus snails collected from localities in the Machakos-Kitui county in south-eastern Kenya, within the Athi River Basin, an area known to be endemic for urinary schistosomiasis.
2. Materials and methods
2.1. Ethical approval and consent to participants
This study was approved by the Scientific and Ethics Review Unit (SERU), SSC NO. 2145, of the Kenya Medical Research Institute. Written consent were obtained from the parents/guardians of the school children who provided urines for isolating S. haematobium eggs for raising worms used as parasite reference materials. In addition, the children who agree to provide a urine sample assented to participate. All children found infected with S. haematobium were given a standard dose of praziquantel (40 mg/kg body weight) under the supervision of a qualified clinician, free of charge. Approvals were also obtained from the KEMRI Animal care and use committee (ACUC) for the use of laboratory animals.
2.2. Snail collection sites
Buliniid snails were collected from various freshwater habitats in the Machakos-Kitui area, within the Athi River Basin over a period of 12 months, between March 2012 to January 2013, and were screened for schistosomes by “shedding” for mammalian schistosome These localities were selected for the present study because previous studies had indicated that B. africanus snails were common here and urinary schistosomiasis was being transmitted in some of the water bodies in the area.
Snail collection from habitats was done during morning hours, and each habitat was sampled for 30 min by 2 persons, each using a standard snail scoop, and the snails collected were counted and sorted out into species, and were then transported to the laboratory where they were screened for mammalian schistosomes. The snails were individually isolated into wells of a 24-well culture plate in 1ml of double distilled water, and exposed to artificial light to stimulate cercarial release from the snails. The schistosome cercariae shed by the individual snails were then transferred individually, into a labeled 0.2 ml PCR reaction tube, under a dissecting microscope, using an elongated Pasteur pipette, and then preserved in 95% ethanol or the cercariae were blotted directly onto an FTA paper (Whatman Inc., Clifton, NJ) and stored at 4OC until the samples were used for DNA amplification.
2.3. Authentic parasite reference materials for the assay
Authentic adult S. haematobium and S. bovis were used as sources of reference DNA for the PCR-RFLP assay. S. haematobium adult worms were raised in laboratory hamsters exposed to cercariae released from laboratory-bred Bulinus africanus snails, which had previously been exposed to miracidia hatched from S. haematobium ova isolated from pooled urine samples collected from naturally infected school children from Ng'alalia area, Kangundo, in Machakos county. Consent was obtained from parents/guardians of the children who provided urine samples, and all the children who tested positive for urinary schistosomiasis were treated with a single dose of praziquantel (40 mg/kg) under the supervision of a qualified and competent clinician. The urine samples collected from the individual children between 10:00hr and 12:00hr were then screened for S. haematobium infection after filtration using the filtration technique described by Mott et al. [8], which involves use of Millipore filters (12–14 μm pore size). The filters were then examined under a microscope for parasite ova. The S. haematobium ova positive urine samples were pooled, transferred into a conical flask, and topped up with normal saline to slow down ova hatching. After the sample was allowed to settle down, the supernatant was decanted, and the flask filled with distilled water. The flask was then covered with a piece of black cloth and left to sit on the bench for 30–60 min to allow the ova to hatch into miracidia, and the phototropic miracidia to move up the water column to the water surface, where they were then collected with a pipette and transferred into a Petri dish. The miracidia were then used to expose lab bred Bulinus africanus snails, in which the cercariae for hamster infections developed. Approximately, 4 weeks after exposure of the snails to miracidia, the snails were screened using the “shedding” method, for cercariae (the schistosome larval form that infects humans or other susceptible mammalian hosts). The hamsters were exposed to cercariae using the procedure described by Smithers and Terry [9].
Approximately 12 weeks after exposure of hamsters to S. haematobium cercarie, the hamsters were perfused for recovery of adult worms as described by Smithers and Terry, [9]. The worms recovered were washed in normal saline, and stored in 95% ethanol until DNA was extracted.
The adult S. bovis worms were recovered from the mesenteric veinules of naturally infected cattle slaughtered at an abbatoir located in the outskirts of the city of Nairobi using fine tipped forceps. The worms were transferred into normal saline, were rinsed, and then preserved in 95% ethanol until used for DNA extraction.
2.4. Primers
One set of internal transcribed spacer (ITS) region primers previously designed by Barber et al, [10] was used to amplify the ITS2 subunit (including most of the 5.8S gene and 40 bases of the 5′ of the 28S gene). The primer sequences are as follows: Forward, 5′- GCA TCG ATG AAG AAC GCA GC-3′ and Reverse 5′- TCC TCC GCT TAT TGA TAT GC-3’.
2.5. DNA extraction, polymerase chain reaction and gel electrophoresis
Schistosome DNA was extracted using the HotShot method described by Truett et al. [11]. Briefly, an alkaline lysis reagent containing 25mM NaOH, 0.2mM disodium ethylenediaminetetraacetic acid (EDTA) at a pH 12 was prepared by dissolving the salts in water without adjusting the pH. A neutralising reagent made up of 40mM Tris–HCl at pH 5 was also prepared by dissolving Tris-HCl in water without adjusting the pH. Cercariae were also directly blotted on WHATMAN paper and stored at -4 °C until used for DNA amplification.
For the unknown parasite samples (cercariae) or the parasite reference materials (the adult S. haematobium and S. bovis worms) previously preserved in 95% ethanol were socked in distilled water for at least 1 h. Distilled water-soaked individual adult worms were thereafter each transferred into a 0.2 ml PCR tube using a fine pair of forceps. A 75μl volume of the alkaline lysis reagent was then added into each tube and the samples heated to 95 °C for 1 h with occasional vortexing every 15 min to break the worms. After 1 h of heating, the sample was cooled to 4 °C and 75 μl of the neutralising reagent added into each tube. Thirteen microlitre of the DNA extract was used in a PCR reaction in a final volume of 50 μl. Freshly shed cercariae were also, used in this study and the procedure for DNA extraction was the same as for adult worms with the only exception being that less volume (10μL for both the alkaline lysis and the neutralizing reagent) of the reaction reagents were used.
PCR was performed in a total volume of 50 μl containing 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 0.1% Triton® X-100, 200 μM dNTP (Promega, Madison, WI, USA), 2.5mM MgCl2, 0.2 μM of each primer, 1 unit of Taq polymerase (Promega, Madison, WI, USA), and approximately 75 ng of schistosome genomic DNA. The thermal cycling profile included an initial denaturation step at 94 °C for 5 min, followed by 35 cycles of 45 s at 94 °C, 45 s at 58 °C, 30 s at 72 °C and a final step of 7 min at 72 °C using a GeneAmp 2400 (Applied Biosystems, Foster City, CA, USA) thermal cycler. The PCR products were separated on a 2% agarose gel using TAE buffer and 100bp DNA ladder (Promega, Madison, WI, USA), used to determine the product sizes.
2.6. Restriction fragment length polymorphism (RFLP) analysis of the ITS2 region
The ITS2 PCR products of the two schistosome species were used to set up restriction digest with each of the restriction enzyme Taq 1. Taq 1 has restriction site at 200 and 380bps for S. haematobium and 200 and 240 bps for Schistosoma bovis. Each RFLP reaction included 2μL of restriction enzyme specific buffer, 1 μL each of DNass, RNass-free water, Bovine serum albumin, and restriction enzyme and 15 μL of the PCR product. A mineral oil overlay was added to each reaction tube to prevent evaporation of reaction mix within the tube. This was then incubated at restriction enzyme specific temperature (65 °C for Taq 1) initially for 4 h after which an additional 1μL of restriction enzyme was added and the reaction incubated over night to ensure complete digestion. The entire digest reaction mix was thereafter loaded onto a 2% agarose gel and subjected to electrophoresis at 80mV for one and a half hours. Restriction fragments were sized using 100 bp molecular marker (Promega) and Polaroid films used for photography.
3. Results
3.1. Snail species collected, schistosome infections, and parasite identification by PCR-RFLP analysis
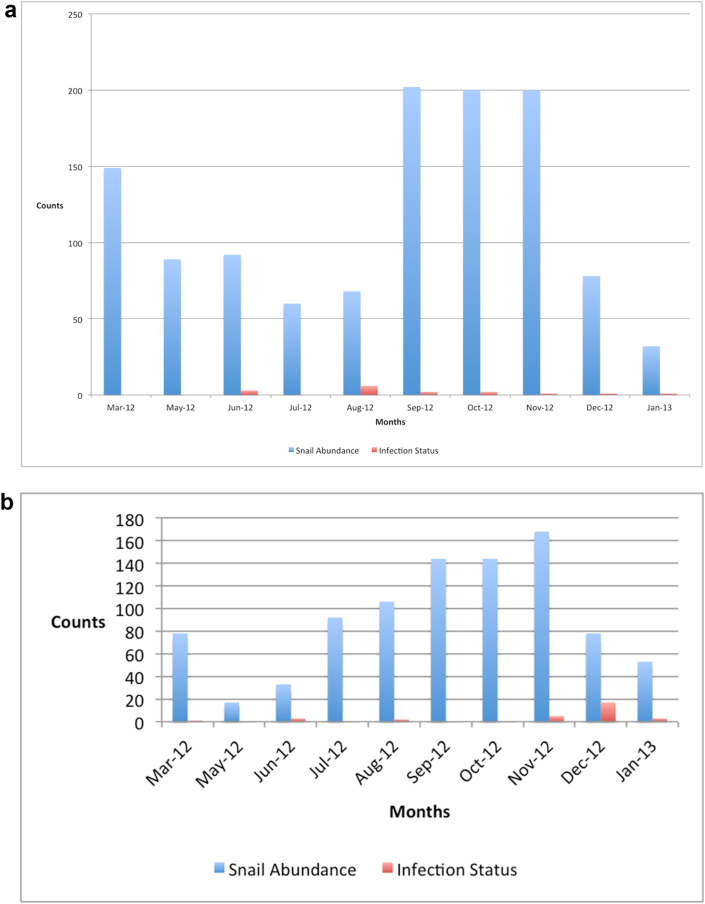
A total of 6472 Bulinus species snails, representing species in the B. africanus, B. tropicus and B. forskalii were collected during the sampling period. Bulinus snails were identified by their shell morphological chacteristics using standard keys [12, 13]. Of the buliniid snails collected, 43 (0.66%) shed mammalian schistosome cercariae. Thirty-two of these shed S. haematobium cercariae and eleven shed S. bovis bases on the PCR-RFLP assay. Table 1 shows the individuals collected for each snail species group, collection habitats, and the habitats from which mammalian schistosome cercariae were collected. Out of the 41 habitats sampled on a monthly basis over a period of 10 months, only two habitats (Kwa Katiwa and Kisukioni) seemed to be active transmission sites for both S. haematobium and S. bovis throughout the study area as shown in Table 1. Of the snails shedding mammalian schistosome cercariae at Kwa Katiwa, majority (approximately 67%) shed S. bovis as determined by PCR-RFLP analysis. At Kisukioni, however, most of the snails shedding schistosomes (90%) shed S. haematobium cercariae. These particular habitats retained water throughout the sampling period, and snails were collected regularly from these habitats, with peak abundance during the period March–May 2012 (Figure 1). The snails shedding mammalian schistosome cercariae were collected throughout the sampling period except in March and May in the case of Kwa Katiwa (Figure 2a) and May, September and October 2012, in the case of Kisukioni (Figure 2b). A distinct peak in abundance of infected snails is observed for kisukioni in december. None of the snails infected with schistosomes, however, was found to harbor mixed infections of both S. haematobium and S. bovis based on the PCR-RFLP analysis. The largest number of snails was collected from the Kwa Katiwa habitat, with a peak abundance being during the period September–December 2012 (see Figure 2a). Sampling was however not done in April 2012 due to logistical reasons.
Table 1.
Sample collection sites and their coordinates, habitat type, number of snails collected, number of Bulinid snails positive for S. haematobium and number of snails positive for S. bovis.
| S.No. | Locality | Latitude | Longitude | Altitude (m) | Habitat type | No. of Bulinid snails collected | No. of Snail + ve S. haem | No. of Snails + S. bovis |
|---|---|---|---|---|---|---|---|---|
| 1 | Nzagathi | 01030′63″S | 038005′56″E | 973 | Dam | 403 | 0 | 0 |
| 2 | Kyalele | 01030″63″S | 038O06′19″E | 981 | Dam | 923 | 0 | 0 |
| 3 | Kwa kasoa | 01O35′29″S | 038O06′54″E | 910 | Dam | 0 | 0 | 0 |
| 4 | Kalundu | 01O22′54″S | 037O59′49″E | 1065 | Stream | 67 | 0 | 0 |
| 5 | Mbusyani | 01O23′16″S | 037O58′16″E | 1115 | Dam | 0 | 0 | 0 |
| 6 | Energy | 01O22′99″S | 037O59′96″E | 1086 | Dam | 0 | 0 | 0 |
| 7 | Masaani | 01O25′48″S | 037O50′22″E | 1101 | Dam | 0 | 0 | 0 |
| 8 | Kyua | 01O24′80″S | 037O45′50″E | 1065 | Dam | 2 | 0 | 0 |
| 9 | Mekilingi | 01O23′48″S | 037O45′43″E | 1040 | Dam | 0 | 0 | 0 |
| 10 | Katangi | 01O24′50″S | 037O41′41″E | 1287 | Dam | 522 | 0 | 0 |
| 11 | Msalani | 01O19′03″S | 037O27′63″E | 1439 | Dam | 22 | 0 | 0 |
| 12 | Masii | 01O20′02″S | 037O26′41″E | 1648 | Dam | 13 | 0 | 0 |
| 13 | Kwa Kivukui | 01O19′85″S | 037O26′12″E | 1290 | Dam | 1 | 0 | 0 |
| 14 | Kwa Matolo | 01O18′44″S | 037O25′78″E | 1258 | Dam | 60 | 0 | 0 |
| 15 | Quarry ponds | 01O19′66″S | 037O17′25″E | 1258 | Ponds/pits | 12 | 0 | 0 |
| 16 | Kwa Katiwa | 01O12′00″S | 037O16′41″E | 1510 | Dam | 1,452 | 4 | 8 |
| 17 | Ngalalia | 01O13′96″S | 037O12′87″E | 1508 | Dam | 61 | 0 | 0 |
| 18 | Matuu dam | 01O13′82″S | 037O14′66″E | 1541 | Dam | 5 | 0 | 0 |
| 19 | Kwa Mweta | 01O15′07″S | 037O15′24″E | 1518 | Dam | 14 | 0 | 0 |
| 20 | Kisukioni | 01O13′49″S | 037O16′93″E | 1521 | Dam | 1013 | 28 | 3 |
| 21 | Maluva | 01O14′49″S | 037O18′62″E | 1506 | Dam | 66 | 0 | 0 |
| 22 | Kwa Kitoi | 01O12′87″S | 037O20′23″E | 1517 | Dam | 15 | 0 | 0 |
| 23 | Kwa Maundu | 01O12′75″S | 037O20′10″E | 1520 | Dam | 6 | 0 | 0 |
| 24 | Kakoo dam | 01O36′63″S | 038O05′50″E | 1012 | Dam | 20 | 0 | 0 |
| 25 | Tala township | 01O36′21″S | 037O19′12″E | 1606 | Dam | 706 | 0 | 0 |
| 26 | Energy dam 3 | 01O22′68″S | 037O59′99″E | 1138 | Dam | 26 | 0 | 0 |
| 27 | Ndivuni | 01O24′54″S | 037O41′85″E | 1224 | Dam | 0 | 0 | 0 |
| 28 | Katsung'wa | 01O36′61″S | 038O07′67″E | 901 | Dam | 0 | 0 | 0 |
| 29 | Kwa Musila | 01O24′43″S | 037O28′91″E | 1327 | Dam | 0 | 0 | 0 |
| 30 | Kakoo stream | 01O24′83″S | 037O28′27″E | 1314 | Dam/River | 48 | 0 | 0 |
| 31 | Maithya dam | 01O28′78″S | 037O27′35″E | 1248 | Dam | 0 | 0 | 0 |
| 32 | Kwa Ndove | 01O27′92″S | 037O26′64″E | 1344 | Dam | 137 | 0 | 0 |
| 33 | Muthale dam | 01O11′07″S | 038O00′47″E | 1319 | Dam | 204 | 0 | 0 |
| 34 | Syokimau dam | 01O07′01″S | 037O55′49″E | 1183 | Dam | 0 | 0 | 0 |
| 35 | Kwa Mutonga | 01O17′97″S | 037O51′32″E | 1151 | Dam | 0 | 0 | 0 |
| 36 | Kiseveni | 01O15′52″S | 037O51′24″E | 1185 | Dam | 34 | 0 | 0 |
| 37 | Kwa Nyika | 01O33′89″S | 037O33′89″E | 1216 | Dam | 526 | 0 | 0 |
| 38 | Kaamita | 01O23′25″S | 037O35′02″E | 1146 | Dam | 56 | 0 | 0 |
| 39 | Muvuti | 01O22′51″S | 037O37′88″E | 1168 | Dam | 1 | 0 | 0 |
| 40 | Kwa Mwinzi | 01O23′82″S | 037O33′33″E | 1239 | Dam | 57 | 0 | 0 |
| 41 | Kwa Musango | 01O24′54″S | 037O33′72″E | 1439 | Dam | 0 | 0 | 0 |
| Total | 6472 | 32 | 11 | |||||
Figure 1.
Bar graph Shows monthly abundance of snails (Data from all study sites).
Figure 2.
(a) Shows the abundance of snails collected within the Katiwa study site by month for the 10 months that sampling was carried out. (b): Shows the abundance of snails and the infection status of Kisukioni sampling site. Kisukioni recorded the highest number of infected snails collected during the 10 months sampling period between March 2012 and Janary 2013.
3.2. Comparison between WHATMAN FTA paper and Hotshot DNA extraction methods
The quality of DNA extracted using either the Whatman FTA paper or the Hotshot methods were comparable (Figure 3), and also, both methods yielded DNA that could easily be amplified.
Figure 3.
Shows PCR product of DNA amplification following extraction using the FTA paper and the HotShot methods. Lane 1 and 10 is 100bp marker, lane 2–4 are PCR product from DNA extracted from 5, 10, 20 cercariae extracted using the FTA paper, and Lane 5–8 PCR product from DNA extracted from 5, 10, 20 cercariae extracted using Hotshot method. Lane 9 is the negative control.
3.3. Sensitivity of the PCR assay
Amplification of the DNA of both S. haematobium and S. bovis by the ITS primers was consistently successful (Figure 4). It was possible to amplify the DNA of S. haematobium or S. bovis at a lower concentration of 5pg, although S. haematobium gave a better quality PCR product (Figure 4).
Figure 4.
Showing the banding pattern of the PCR products following amplification with the ITS2 primers. Lane 1 and 6 are the 100bp molecular markers, lane 2 and 4 are DNA amplified from control S. bovis worms (5fg and 10fg concentrations for lane 2 and 4 respectively) and Lane 3 and 5 are DNA amplified from control DNA extracted from S. haematobium worms (5fg and 10fg concentrations for lane 2 and 4 respectively).
When a variable number of cercariae was used to test the lowest limit that could yield a detectable product, it was possible to amplify DNA from 5 cercariae (Figure 4). The intensity of the resultant band (corresponding to DNA concentration) increased with an increase in the number of cercariae use as the starting material (Figure 4).
3.4. Specificity of the PCR assay
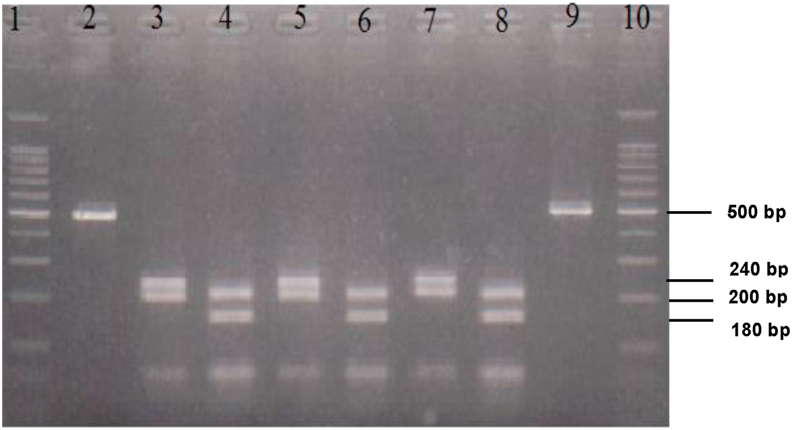
When the control PCR products were subjected to digestion using the Taq 1 restriction enzymes, clear banding patterns were detected and it was possible to distiguish between S. haematobium and S. bovis DNA (see Figure 5). Both S. haematobium and S. bovis shared a band at 200bp. However, S. haematobium had a distinct band at 180 bp while S. bovis had a distinct band at 240 bp.
Figure 5.
Shows the RFLP product following digest of the PCR product with Taq 1 resetriction enzyme. Lane 1 and 10 shows 100 bp marker, lane 2 is the control undigested S. bovis control, lane 9 is the control undigested S. haematobium control, lanes 3–6 are the control digested S. bovis DNA, lane 7 and 8 are the control digested S. haematobium DNA.
When cercariae obtained from field collected Bulinus sp. snails were subjected to the PCR-RFLP assay, we were able to reliably distinguish S. haematobium and S. bovis cerariae as illustrated in Figure 6. The results of this assay were confirmed from the egg morphology of the two species obtained from infected mice.
Figure 6.
Shows the banding patterns of RFLP product of unknown DNA extracted from cercariae shed by Bulinus sp snails collected from snail habitats, following digestion with Taq 1 restriction enzyme. Lane 1 and 10 shows the 100bp marker, lane 2 is control undigested S. bovis, lane 3–6 are the digested unknown mammalian schistosome cercarial DNA, lane 9 is control undigested S. haematobium DNA.
4. Discussion
In this study, the PCR-RFLP assay previously described by Barber el al., [10], was evaluated further for its reliability in the identification of S. haematobium and S. bovis cercariae. We have validated the usefulness of this assay for epidemiological studies of S. haematobium and S. bovis using samples collected from habitats in the Athi Basin in south-eastern Kenya. The FTA papers offered a more reliable way of collecting, transporting and storing DNA as they were convenient for transporting samples from the field and samples were safely stored at room temperature without loss of DNA quality. Taq 1 restriction enzyme used was able to reliably distinguish between the DNA of S. haematobium and that of S. bovis. This was also observed in the study by Barber et al., 2000. The sensitivity of this assay was demonstrated by the fact that schistosome DNA in the range of 5 fg or schistosome cercariae as few as 5 cercariae could be detected in the assay. S. haematobium was found to be the most widespread of the two species in the River Athi Basin although its occurrence has declined over time (REF). This we hypothesize is due to changes in climatic conditions that have led to reduction in the number of suitable habitats. Several methods have been developed for epidemiological studies of S. haematobium. For example, DraI PCR, previously developed by Hamburger et al. [14] has been used in coastal Kenya. The DraI, a repeated sequence, consisting of tandemly arranged 121-bp-long units is highly abundant in the S. haematobium genome and was found to be useful for large scale monitoring of S. haematobium transmission in an endemic locality in coastal Kenya. It was also found to be cheaper and sensitive with a detection limit of less than 10 fg of Schistosoma DNA [15, 16, 17]. However, because the Dra I repeat can cross-hybridized with DNA from S. bovis, S.magrebowiei, S. mattheei, S. curassoni, and S. intercalatum, it cannot distinguish between the various species that are normally co-endemic with S. haematobium. To overcome this problem, Amarir et al. [18], combined Dra1 method with another molecular method known as the Sh110 SmSl which is specific for S. haematobium. This is a 525bp Sh 110 novel repeat sequence of S. haematobium. Although DraI PCR and Sh110 SmSl were highly sensitive and specific for S. haematobium detection, the cost of doing the assay remained high due to the fact that one had to do two sets of PCR for the amplification of the two target markers. Hence there is still need to develop novel primers that can differentiate human and animal schistosomes simultaneously so as to decrease the cost of the assays.
Recently, Abbasi et al. [19], developed a simple and more sensitive PCR assay that enables direct discrimination of S. haematobium from related animal schistosomes, by the primer combination of DraI reverse primer and Sh73 direct primer (73d). The sensitivity of S. haematobium detection was found to be 1 pg, whereas S. bovis detection was 10 pg. However, such assays still required further validation using larger numbers of field snails for large scale monitoring of post-intervention residual transmission.
A more promising, rapid, ‘multiplex’ one-step polymerase chain reaction (RD-PCR) method has also recently been developed by Webster et al., [20]. It utilizes cytochrome oxidase subunit 1 (COX1) mitochondrial DNA. This method is able to discriminate between S. haematobium and S. bovis using a single forward primer and two species-specific reverse primers. However, this method has not been validated for its field applicability and also due to the fact that it is mitochondrial based, it might require the use of nuclear diagnostic markers alongside for it to be used for epidemiological studies.
More recently, Akinwale et al. [21], have developed a new PCR assay using a pair of primers, ShND-1/ShND-2, to amplify a target sequence of 1117 bp from S. haematobium mitochondrion complete genome. The assay was found to be sensitive, specific and was able to successfully differentiate S. haematobium from S. magrebowiei, in addition to its other closely related animal infective schistosome species. However, further validation of the assay for its field applicability is required for field studies since the authors only examined a few field-collected snails. A more promising technique that uses molecular xenomonitoring, DNA-based method that has previously been developed to monitor the transmission of several vector-borne diseases was recently developed by Pennance et al 2020. However, the investigators identified two limiting factor to their assay namely; First, the laborious nature of testing each individual snail that adds time and cost. Second, the need for a secondary screening of the Schistosoma amplicon, via sequencing, to confirm species [22].
The PCR-based method we have evaluated for cercariae identification offers improved sensitivity and specificity and can potentially be used to support schistosomiasis control efforts, and may be suitable for use in epidemiological studies of urinary schistosomiasis or in studies on the transmission ecology of S. haematobium and S. bovis.
Declarations
Author contribution statement
Eric L. Agola: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Ibrahim N. Mwangi, Geoffrey M. Maina, Joseph M. Kinuthia, Martin W. Mutuku: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was supported by an internal research grant (Grant Ref. No. SSC 2145 awarded to ELA) by the Kenya Medical Research Institute (KEMRI), and is published with the approval of the Director, KEMRI.
Data availability statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We acknowledge the help and administrative support of Dr. Kimani Gachuhi, Director, Center for Biotechnology Research and Development (CBRD), Kenya Medical Research Institute (KEMRI) in this project. We thank Mr. Stephen Kamau, for logistical support during field collection of samples.
References
- 1.Steinmann P., Keiser J., Bos R., Tanner M., Utzinger J. Schistosomiasis and water resources development: systematic review, meta-analysis, and estimates of people at risk. Lancet Infect. Dis. 2006;6:411–425. doi: 10.1016/S1473-3099(06)70521-7. [DOI] [PubMed] [Google Scholar]
- 2.Chitsulo L., Engels D., Montresor A., Savioli L. The global status of schistosomiasis and its control. Acta Trop. 2000;77:41–51. doi: 10.1016/s0001-706x(00)00122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Werf M.J., de Vlas S.J., Brooker S., Looman C.W., Nagelkerke N.J., Habbema J.D. Quantification of clinical morbidity associated with schistosome infection in sub-Saharan Africa. Acta Trop. 2003;86:125–139. doi: 10.1016/s0001-706x(03)00029-9. [DOI] [PubMed] [Google Scholar]
- 4.Murray C.J., Vos T., Lozano R., Naghavi M., Flaxman A.D., Michaud C. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990-2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2197–2223. doi: 10.1016/S0140-6736(12)61689-4. [DOI] [PubMed] [Google Scholar]
- 5.King C.H., Dangerfield-Cha M. The unacknowledged impact of chronic schistosomiasis. Chron. Illness. 2008;4:65–79. doi: 10.1177/1742395307084407. [DOI] [PubMed] [Google Scholar]
- 6.Hotez P.J., Brindley P.J., Bethony J.M., King C.H., Pearce E.J., Jacobson J. Helminth infections: the great neglected tropical diseases. J. Clin. Invest. 2008;118:1311–1321. doi: 10.1172/JCI34261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sturrock R.F., Klumpp R.K., Ouma J.H., Butterworth A.E., Fulford A.J., Kariuki H.C. Observations on the effects of different chemotherapy strategies on the transmission of Schistosoma mansoni in Machakos District, Kenya, measured by long-term snail sampling and cercariometry. Parasitology. 1994;109(Pt 4):443–453. doi: 10.1017/s0031182000080690. [DOI] [PubMed] [Google Scholar]
- 8.Mott K.E., Baltes R., Bambagha J., Baldassini B. Field studies of a reusable polyamide filter for detection of Schistosoma haematobium eggs by urine filtration. Tropenmed. Parasitol. 1982;33:227–228. [PubMed] [Google Scholar]
- 9.Smithers S.R., Terry R.J. The infection of laboratory hosts with cercariae of Schistosoma mansoni and the recovery of the adult worms. Parasitology. 1965;55:695–700. doi: 10.1017/s0031182000086248. [DOI] [PubMed] [Google Scholar]
- 10.Barber K.E., Mkoji G.M., Loker E.S. PCR-RFLP analysis of the ITS2 region to identify Schistosoma haematobium and S. bovis from Kenya. Am. J. Trop. Med. Hyg. 2000;62:434–440. doi: 10.4269/ajtmh.2000.62.434. [DOI] [PubMed] [Google Scholar]
- 11.Truett G.E., Heeger P., Mynatt R.L., Truett A.A., Walker J.A., Warman M.L. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT) Biotechniques. 2000;29(52):54. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- 12.Brown D.S. 2 edition. Taylor and Francis; London: 1994. Freshwater Snails of Africa and Their Medical Importance. [Google Scholar]
- 13.DBL-WHO . Danish Bilharziasis Laboratory. WHO collaborating Centre for Applied Malacology; Charlottenlund, Denmark: 1998. A Field Guide to African Freshwater Snails. [Google Scholar]
- 14.Hamburger J., He N., Abbasi I., Ramzy R.M., Jourdane J., Ruppel A. Polymerase chain reaction assay based on a highly repeated sequence of Schistosoma haematobium: a potential tool for monitoring schistosome-infested water. Am. J. Trop. Med. Hyg. 2001;65:907–911. doi: 10.4269/ajtmh.2001.65.907. [DOI] [PubMed] [Google Scholar]
- 15.Hamburger J., Hoffman O., Kariuki H.C., Muchiri E.M., Ouma J.H., Koech D.K. Large-scale, polymerase chain reaction-based surveillance of Schistosoma haematobium DNA in snails from transmission sites in coastal Kenya: a new tool for studying the dynamics of snail infection. Am. J. Trop. Med. Hyg. 2004;71:765–773. [PubMed] [Google Scholar]
- 16.Abbasi I., King C.H., Sturrock R.F., Kariuki C., Muchiri E., Hamburger J. Differentiation of Schistosoma haematobium from related schistosomes by PCR amplifying an inter-repeat sequence. Am. J. Trop. Med. Hyg. 2007;76:950–955. [PMC free article] [PubMed] [Google Scholar]
- 17.Melo F.L., Gomes A.L., Barbosa C.S., Werkhauser R.P., Abath F.G. Development of molecular approaches for the identification of transmission sites of schistosomiasis. Trans. R. Soc. Trop. Med. Hyg. 2006;100:1049–1055. doi: 10.1016/j.trstmh.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 18.Amarir F., Sebti F., Abbasi I., Sadak A., Fellah H., Nhammi H. Schistosoma haematobium detection in snails by DraI PCR and Sh110/Sm-Sl PCR: further evidence of the interruption of schistosomiasis transmission in Morocco. Parasites Vectors. 2014;7:288. doi: 10.1186/1756-3305-7-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abbasi I., King C.H., Muchiri E.M., Hamburger J. Detection of Schistosoma mansoni and Schistosoma haematobium DNA by loop-mediated isothermal amplification: identification of infected snails from early prepatency. Am. J. Trop. Med. Hyg. 2010;83:427–432. doi: 10.4269/ajtmh.2010.09-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webster B.L., Rollinson D., Stothard J.R., Huyse T. Rapid diagnostic multiplex PCR (RD-PCR) to discriminate Schistosoma haematobium and S. bovis. J. Helminthol. 2010;84:107–114. doi: 10.1017/S0022149X09990447. [DOI] [PubMed] [Google Scholar]
- 21.Akinwale O.P., Hock T.T., Chia-Kwung F., Zheng Q., Haimo S., Ezeh C. Differentiating Schistosoma haematobium from Schistosoma magrebowiei and other closely related schistosomes by polymerase chain reaction amplification of a species specific mitochondrial gene. Tropenmed. Parasitol. 2014;4:38–42. doi: 10.4103/2229-5070.129163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pennance Tom, Archer John, Lugli Elena Birgitta, Rostron Penny, Llanwarne Felix, Ali Said Mohammed, Amour Amour Khamis, Suleiman Khamis Rashid, Li Sarah, Rollinson David. Development of a molecular snail xenomonitoring assay to detect schistosoma haematobium and schistosoma bovis infections in their Bulinus snail hosts. Molecules. 2020;25(17):4011. doi: 10.3390/molecules25174011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.