Abstract
Listeria monocytogenes is a pathogen contaminated food, it is the cause of listeriosis worldwide. The aims of this study were to investigate the occurrence, antimicrobial resistance, serotyping and virulence genes of L. monocytogenes isolated from foods in Meknes city of Morocco. From June 2017 to May 2018, 520 food samples were randomly collected from a traditional market and two overcrowded popular neighborhoods (Lahdim and Hamria) and subjected to the detection of L. monocytogenes. Then, the antimicrobial susceptibility of the isolated strains were evaluated using the standard disk diffusion method and the determination of serotypes and virulence genes was performed by PCR. The results showed the detection of L. monocytogenes in fifteen (2.9%) of 520 samples, including three (5.7%) isolates in traditional whey, raw minced meat and raw sausage, two (3.8%) in raw milk and one (1.9%) in smen (traditional butter), raw bovine meat, raw poultry meat and raw fish, while salads and rayeb (traditional coagulated milk) were not contaminated. Among the fifteen isolated L. monocytogenes, nine (60%) belonged to the serogroup (1/2a, 1/2c, 3a and 3c), two (13.3%) belonged to the serogroup (1/2b, 3b, 4b and 4d) and four (26.6%) do not belong to any studied serogroup. Furthermore, fifteen (100%) isolates showed the presence of actA gene, fourteen (93.3%) harbored hlyA, prfA and plcB genes, thirteen (86.7%) carried inlA and inlC genes and twelve (80%) showed inlJ gene. The antimicrobial susceptibility analysis showed that the isolated strains were more resistant to amoxicillin/clavulanic acid (67.0%), erythromycin (60.0%), sulphamethoxazole (40.0%), ampicillin and sulphamethoxazole/trimethoprim (33.0%) and tetracycline (20.0%). Furthermore, 66.7% (10/15) were multidrug-resistant. From this study, we can conclude that foods marketed in Meknes city were contaminated by multidrug-resistant strains of L. monocytogenes harboring virulence genes, which may cause a serious risk to public health.
Keywords: Foods, Listeria monocytogenes, Serotypes, Virulence genes, Multidrug-resistance
Foods; Listeria monocytogenes; serotypes; virulence genes; multidrug-resistance.
1. Introduction
Listeria monocytogenes is a Gram-positive bacilli, facultative anaerobic, and can grow in a wide range of pH (4.3–9.4), temperatures (from 0 to 45 °C), at a high salt concentration (of up 14%) and water activity (higher than 0.92) [1,2,3]. These particular physicochemical factors are in favor of the survival and proliferation of L. monocytogenes in a wide variety of foodstuffs, including seafood, meat and meat products, milk and dairy products, and vegetables [4].
Invasion of host cells by L. monocytogenes involves many virulence factors. The hly gene encodes an extracellular listeriolysin O (LLO) which has a role in the regulation of the host cell by L. monocytogenes. The ActA protein is essential for actin polymerization and intracytoplasmic movement of L. monocytogenes [5], plcA and plcB are involved in the lysis of the double membrane vacuole formed during cell-to-cell propagation [6]. PrfA is a positive regulatory factor for hly, plcA, mpl, actA and plcB, it regulates the expression of factors necessary for cell invasion (InlA and InlB) and intracellular proliferation (Hpt) [6]. InlA is implicated in the invasion of L. monocytogenes into intestinal epithelial cells by expressing the E-cadherin receptor. The InlB gene induces hepatocyte invasion via the c-Met receptor [6]. For somatic (O) and flagellar (H) antigens, they are used as monoclonal and polyclonal antibodies. There are 15 somatic (O) (I-XV) and 4 flagellar (H) (A-D) antigens [7]. Based on the characteristics of somatic (O) and flagellar (H) antigens agglutination, thirteen serotypes (1/2a, 1/2b, 1/2c, 3a, 3b, 3c, 4a, 4ab, 4b, 4c, 4d, 4e and 7) have been identified in L. monocytogenes [7]. Five other serogroups have been determined in L. monocytogenes such as IIa (1/2a-3a), IIb (1/2b-3b-7), IIc (1/2c-3c), IVa (4a-4c) and IVb (4ab- 4b, 4d-4e) [8], the serotypes (1/2a, 1/2b and 4b) are responsible for human listeriosis at a rate of almost 95%, of which 1/2a, 1/2b are mainly isolated from food and 4b from clinical cases [7].
L. monocytogenes is responsible for listeriosis with a high fatality rate of 20%–30% [7]. This pathogen can cause, meningoencephalitis, cerebral abscesses, cerebritis, bacteremia, meningitis, and sepsis, especially in the immunocompromised individuals and pregnant women [9,10]. On the other hand, antimicrobial resistance spread rapidly worldwide, which cause a health threat and economic burden, owing to the excessive use of antibiotic in human and veterinary medicine [11,12]. The resistance of L. monocytogenes to antibiotics which are commonly used in the treatment of human and animal diseases is worrisome [8].
Listeriosis causes severe damages to public health. In the European Union, the incidence of listeriosis is about 0.47 cases per 100, 000 population [4] and 0.24 cases per 100, 000 population in the United States [13]. In Morocco, the actual incidence of listeriosis remains unknown due to the lack of epidemiological surveillance, in fact only one case of neonatal listeriosis has been reported [14]. However, several studies showed the prevalence of L. monocytogenes in raw and processing foods [15,16].
The objectives of this study were (i) to evaluate the occurrence of L. monocytogenes in food samples collected from Meknes city of Morocco, (ii) to determine their susceptibility profiles of antibiotics and serotypes (iii) and to study their virulence by the amplification of the targeted virulence genes.
2. Material and methods
2.1. Sample collections
From June 2017 to May 2018, a total of 520 samples including raw milk, whey, rayeb, smen, raw bovine meat, raw poultry meat, raw minced meat, raw sausage, raw fish and salads (52 samples per each) were randomly collected from street traders, butcheries and restaurants with forty-three samples per month. Sampling was carried out in a traditional market and two overcrowded neighborhoods popular (Lahdim and Hamria). Then, samples were transported to the Laboratory of Microbiology at the Faculty of Science in Meknes. The microbiological analyses were performed on the same day of sampling.
2.2. Isolation and identification of L. monocytogenes
The protocol was made according to the Moroccan standard method [17]. Briefly, 10 g of each sample was aseptically homogenized with 90 mL of half Fraser broth in a stomacher 400 Circulator (Seward, West Sussex, UK) for 3 min at 260 rpm and incubated at 30 °C for 24 h. After incubation, 0.1 mL was transferred to the tube containing 10 mL of Fraser broth (Biokar, Beauvais, France) and incubated at 37 °C for 48 h. From it, a streak culture was performed on Agar Listeria acc. to Ottaviani & Agosti (ALOA, Biolife, Milan, Italy) and incubated at 37 °C for 48 h. A maximum of 5 colonies presumed to be L. monocytogenes were purified onto tryptone soya yeast extract agar (Biokar, Beauvais, France) and incubated at 30 °C for 24 h. Gram staining, catalase, oxidase, β-hemolysis, CAMP test and Listeria api have been used to confirme L. monocytogenes strains.
2.3. PCR-serorogroups analysis and virulence genes determination
DNA extraction of L. monocytogenes isolates was performed using heating method. Multiplex PCR was used for the determination of serotypes [18] and the amplification of virulence genes of the isolated strins [19], using the specific amorces described in Tables 1 and 2. PCR assays were performed in final volume of 25 μL, which containing 13.9 μL of ddH2O, 2.5 μL of buffer (10×), 2 μL of 25 mM MgCl2, 2.5 μL of 1 μM dNTP mix (KAPA Biosystems), 0.1 μL of Taq DNA polymerase (1 U/μL KAPA Biosystems), 2 μL of template DNA, 1 μL of each 10 μM primer. The PCR program was set as follows for all studied primers: initial denaturation at 5 min for 95 °C, 35 cycles of denaturation at 94 °C for 80 s, annealing at 58 °C for 90 s, elongation at 72 °C for 1 min and final elongation at 72 °C for 5 min primer. The amplified PCR products were visualized by ethidium bromide in 2 % agarose gel under UV light. L. monocytogenes strain ATCC19112 considered as positif control.
Table 1.
Primer used for the amplification of virulence genes of L. monocytogenes.
| Gene | Sequences (5′-3′) | Length (bp) |
|---|---|---|
| inlA | F-CCTAGCAGGTCTAACCGCAC R-TCGCTAATTTGGTTATGCCC |
256 |
| inlC | F-AATTCCCACAGGACACAACC R-CGGGAATGCAATTTTTCACTA |
517 |
| inlJ | F-TGTAACCCCGCTTACACAGTT R-AGCGGCTTGGCAGTCTAATA |
238 |
| actA | F-CCAAGCGAGGTAAATACGGGA R-GTCCGAAGCATTTACCTCTTC |
650 |
| prfA | F-ACCAATGGGATCCACAAGA R-CAGCTGAGCTATGTGCGAT |
467 |
| hlyA | F-ATCATCGACGGCAACCTCGGAGAC R-CACCATTCCCAAGCTAAACCAGTGC |
404 |
| plcB | F-AATATTTCAATCAATCGGTGGCTGA R-GGGTAGTCCGCTTTCGCTCTT |
289 |
Table 2.
Primer used for serotype determination of L. monocytogenes.
| Primer | Sequences (5′-3′) | Product size (bp) | Serotype specificity |
|---|---|---|---|
| lmo0737 | F-AGGGCTTCAAGGACTTACCC R-ACGATTTCTGCTTGCCATTC |
691 | 1/2a, 1/2c, 3a and 3c |
| ORF2819 | F-AGCAAAATGCCAAAACTCGT R-CATCACTAAAGCCTCCCATTG |
471 | 1/2b, 3b, 4b and 4d |
2.4. Antimicrobial susceptibility
The determination of antimicrobial susceptibility profile was carried out by disk diffusion test with reference to the recommendations of the Clinical and Laboratory Standards Institute (CLSI) [18], and European Committee on Antimicrobial Susceptibility Testing (EUCAST) [20]. Fifteen antimicrobials were selected for this study on basis of their uses in the treatment of diseases in humans and veterinary medicine [8]: amoxicillin/clavulanic acid (30 μg), ampicillin (10 μg), penicillin (10 μg), amikacin (30 μg), gentamicin (30 μg), streptomycin (10 μg), imipenem (10 μg), erythromycin (15 μg), tetracycline (30 μg), vancomycin (30 μg), chloramphenicol (30 μg), sulphamethoxazole/trimethoprim (25 μg), sulphamethoxazole (200 μg), ciprofloxacin (5 μg), and kanamycin (30 μg). In this study, the isolated strains showing a decrease in susceptibility (intermediate) were considered as resistant, and L. monocytogenes ATCC19112 was used as a reference strain. Afterward, the Multiple Antibiotic Resistance (MAR) index was assessed as, the number of antimicrobials to which the isolated strains were resistant divided by the total number of antimicrobials to which the isolated strains were tested [21,22].
2.5. Statistical analysis
Statistical analysis was performed using SPSS software (SPSS version 20, IBM Corp, Armonk, NY, USA), The Chi-squared test was performed to assess the relationship between the variables of interest. P-value ˂ 0.05 was used in testing the statistical significance of all experimental data.
3. Results
3.1. Isolation and identification of L. monocytogenes
From a total of 520 analyzed food samples, 15 (2.9%) were positive for L. monocytogenes (Table 3). The highest value was detected in traditional whey, raw minced and raw sausage with 5.7%, followed by raw milk (3.8%), and finally smen, raw bovine meat, poultry meat and raw fish with 1.9%. However, L. monocytogenes was not detected in salads and rayeb (Table 3). The statistical analysis showed that the occurrence of L. monocytogenes do not depends on the food matrice (p = 0.47), seasons (p = 0.52) and sites (p = 0.82).
Table 3.
Rate of occurrence of L. monocytogenes in different food products.
| Food type | No. of samples | No. of positive isolates (%) |
|---|---|---|
| Raw milk | 52 | 2 (3.8) |
| Traditional whey | 52 | 3 (5.7) |
| Rayeb | 52 | 0 (0) |
| Smen | 52 | 1 (1.9) |
| Raw bovine meat | 52 | 1 (1.9) |
| Raw poultry meat | 52 | 1 (1.9) |
| Raw minced meat | 52 | 3 (5.7) |
| raw sausage | 52 | 3 (5.7) |
| raw fish | 52 | 1 (1.9) |
| Salads | 52 | 0 (0) |
| Total | 520 | 15 (2.9) |
The bold indicates the total number of samples studied and the percentage of positive isolates.
3.2. PCR analysis of virulence genes and serotyping of L. monocytogenes
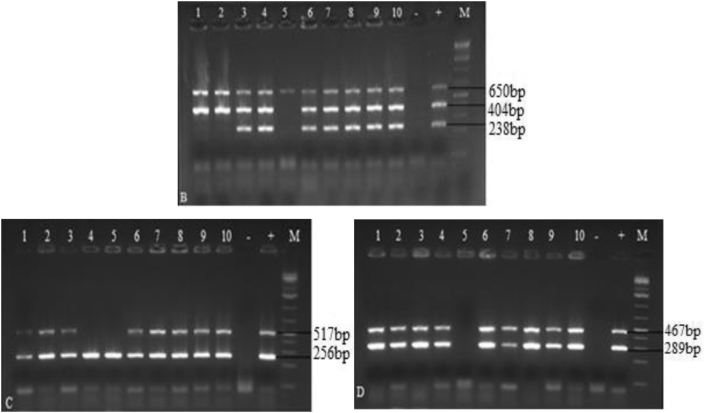
The serotyping analysis showed that among the 15 tested L. monocytogenes strains, nine isolates (60%) belonged to serogroup (1/2a, 1/2c, 3a and 3c) and two (13.3%) belonged to serogroup (1/2b, 3b, 4b and 4d), while four (26.6%) did not belong to any of the studied serogroup (Figure 1 and Table 4). Seven virulence-associated genes (inlA, inlC, inlJ, prfA, plcB, hlyA and actA) were identified. All strains carried actA gene. Whereas, hlyA, prfA and plcB were detected in 14 (93.3%) strains, inlA and inlC in 13 (86.7%) strains and inlJ in 12 (80%) strains. Furthermore, the genes (actA, inlA and inlC), (actA, hlyA, plcB and prfA) and (actA, hlyA, plcB and prfA) were detected in L. monocytogenes strains from raw milk (strain MK9), raw bovine meat (B510) and raw minced meat (M23), respectively (Figure. 2 and Table 4).
Figure 1.
Serotypes identified in L. monocytogenes. A: ORF2819 (471 bp), lmo0737 (691bp), M: Size marker (1 kb), +: L. monocytogenes strain ATCC19112, -: Negative control, From one to ten: L. monocytogenes isolates tested.
Table 4.
Source, antimicrobial resistance profiles, serotypes and virulence genes of L. monocytogenes isolated from foods.
| Source | L. monocytogenes isolate code | Antimicrobial resistance profile | MAR index | Virulence genes | Serotypes |
|---|---|---|---|---|---|
| Traditional whey | W500 | AMC,AMP, E, SMX, SXT, VA, AK, C, CN, K, S | 0.73 | actA, hlyA, inlJ, inlA, inlC, plcB, prfA | 1/2a, 1/2c, 3a and 3c |
| Smen | S220 | AMC, AMP, E, SMX, SXT, CIP | 0.40 | actA, hlyA, inlJ, inlA, inlC, plcB, prfA | 1/2b, 3b, 4b and 4d |
| Raw poultry meat | P2 | AMC, E, SMX, TE | 0.26 | actA, hlyA, inlJ, inlA, inlC, plcB, prfA | 1/2a, 1/2c, 3a and 3c |
| Raw minced meat | M80 | AMC, E, SMX, TE | 0.26 | actA, hlyA, inlJ, inlA, inlC, plcB, prfA | I1/2a, 1/2c, 3a and 3c |
| Traditional whey | W77 | AMC, SMX, SXT | 0.20 | actA, hlyA, inlJ, inlA, inlC, plcB, prfA | 1/2a, 1/2c, 3a and 3c |
| Raw sausage | Sg 90 | AMC, SMX, SXT | 0.20 | actA, hlyA, inlJ, inlA, inlC, plcB, prfA | 1/2a, 1/2c, 3a and 3c |
| Raw milk | Mk 60 | AMC, AMP, E | 0.20 | actA, hlyA, inlJ, inlA, inlC, plcB, prfA | 1/2b, 3b, 4b and 4d |
| Raw milk | Mk 9 | AMC, AMP, E | 0.20 | actA, inlA, inlC, | - |
| Raw fish | F300 | E, SMX, SXT | 0.20 | actA, hlyA, inlJ, inlA, inlC, plcB, prfA | 1/2a, 1/2c, 3a and 3c |
| Traditional whey | W5 | AMC, E, TE | 0.20 | actA, hlyA, inlJ, inlA, inlC, plcB, prfA | 1/2a, 1/2c, 3a and 3c |
| Raw bovine meat | B510 | AMC, AMP | 0.13 | actA, hlyA, plcB, prfA | - |
| Raw sausage | Sg44 | AMC, SMX | 0.13 | actA, hlyA, inlJ, inlA, inlC, plcB, prfA | 1/2a, 1/2c, 3a and 3c |
| Raw minced meat | M23 | AMC, E, | 0.13 | actA, hlyA, plcB, prfA | - |
| Raw minced meat | M140 | E, TE | 0.13 | actA, hlyA, inlJ, inlA, inlC, plcB, prfA | - |
| Raw sausage | Sg310 | - | 0.00 | actA, hlyA, inlJ, inlA, inlC, plcB, prfA | 1/2a, 1/2c, 3a and 3c |
AK: amikacin; AMC: amoxicillin/clavulanic acid; AMP: ampicillin; C: chloramphenicol; CIP: ciprofloxacin; CN: gentamycin; E: erythromycin; IPM: imipenem; K: kanamycin; MAR: multiple antimicrobial resistance; P: penicillin; S: streptomycin; SMX: sulphamethoxazole; SXT: sulphamethoxazole/trimethoprim; TE: tetracycline; VA: vancomycin; MAR: Multiple Antibiotic Resistance.
Figure 2.
Virulence genes of L. monocytogenes. B: actA (650 bp), hlyA (404 bp), inlJ (238 bp), C: inlA (256 bp), inlC (517 bp), D: plcB (289 bp), prfA (467 bp), M: Size marker (1 kb), +: L. monocytogenes strain ATCC19112, -: Negative control, From one to ten: L. monocytogenes strains tested.
3.3. Antimicrobial susceptibility
The results of this study showed that L. monocytogenes isolated from food present a high resistance to amoxicillin/clavulanic acid (67.0%), followed by erythromycin (60.0%), sulphamethoxazole (40.0%), ampicillin and sulphamethoxazole/trimethoprim (33.0%), tetracycline (20.0%), chloramphenicol, gentamicin, ciprofloxacin, amikacin, streptomycin, and vancomycin (17.0%). However, they were susceptible to penicillin and imipenem (Table 5). In addition, the MAR index value of the isolated L. monocytogenes ranged from 0.00 to 0.73 (Table 4).
Table 5.
Antimicrobial resistance percentages of L. monocytogenes isolated from foods.
| Antimicrobial agent | No. of L. monocytogenes isolates (n = 15) |
|
|---|---|---|
| S | R | |
| Amoxicillin/clavulanic acid (30μg) | 5 (33) | 10 (67) |
| Erythromycin (15 μg) | 6 (40) | 9 (60) |
| Sulphamethoxazole (200 μg) | 9 (60) | 6 (40) |
| Ampicillin (10 μg) | 10 (67) | 5 (33) |
| Sulphamethoxazole/trimethoprim(25 μg) | 10 (67) | 5 (33) |
| Tetracycline (30 μg) | 12 (80) | 3 (20) |
| Chloramphenicol (30 μg) | 14 (93) | 1 (7) |
| Gentamicin (30 μg) | 14 (93) | 1 (7) |
| Ciprofloxacin (5 μg) | 14 (93) | 1 (7) |
| Amikacine (30 μg) | 14 (93) | 1 (7) |
| Streptomycin (10 μg) | 14 (93) | 1 (7) |
| Vancomycin (30 μg) | 14 (93) | 1 (7) |
| Kanamycin (30 μg) | 14 (93) | 1 (7) |
| Penicillin G (10 μg) | 15 (100) | 0 (0) |
| Imipenem (10 μg) | 15 (100) | 0 (0) |
4. Discussion
The present study showed that the rate of occurrence of L. monocytogenes in foods consumed in Meknes city of Morocco was 2.9%. This value is in agreement with those reported in Iran (2.99%) [18], Estonia (2.6%) [1] and Algeria (2.6%) [23]. However, it's higher than that found previously in Tetouan city of Morocco (1.5%) [24], in Japan (1.7%) [25] and India (1.5%) [26], and lower than that reported previously in Casablanca city of Morocco (23.3%) [27], Ireland (5.8 %) [28], Uruguay (11.2%) [29], China (21.7%) [30], Chile (25%) [31], Greece (14.3%) [32] and Spain (6.2%) [33]. Furthermore, traditional whey, minced mead, and raw sausage were the most contaminated foods (5.7%), followed by raw milk (3.8%), smen, bovine meat, poultry meat and raw fish (1.9%). These results were higher than those reported in other regions of Morocco for poultry meat (6.6%), red meat products (6.5%), salads (6%) and seafoods (0%) [34], ground meat and sausage (3.3%) and raw poultry (1.3%) [27], raw milk (0.83%) [35], dairy products (0.74%) and poultry meat (0%) [24], and lower than those reported in poultry and bovine meat products (0% and 2.7%, respectively) [24], chiken meat (3.66%) [36], dairy products (4.1%) [34] and raw milk (8.33%) [37]. However, our findings are comparable to those reported in salads (0%) [24] and traditional whey (5.20%) [37]. The difference in the occurrence of L. monocytogenes in food products may be due to the foods, sampling strategy, geographical differences and hygienic conditions of preparation and storage. Indeed, food samples collected from street vendors, restaurants and butchers do not meet food safety standards. In some countries, street foods represent a significant proportion of the food consumed by the urban population, and their distribution is relatively related to socio-economic and cultural factors [38,39]. In addition, many studies have reported that foodstuffs promote the growth of L. monocytogenes through their nutrient values and physicochemical properties [40,41].
The present study indicated the highest occurrence of serogroup (1/2a, 1/2c, 3a, and 3c), followed by serogroup (1/2b, 3b, 4b, and 4d). In Ireland, a study performed by Leong et al. showed that L. monocytogenes strains isolated from dairy, meat, seafood and vegetable are of serogroup (1/2a, 3a), (1/2b, 3b, 7), (1/2c, 3c), (4b, 4d, 4e), (1/2a), (1/2b), (1/2c) and (4b/4e) [42]. A study carried out in Iran reported that L. monocytogenes isolated from seafood products, market and processing environments belonged to serotype 1/2a (45.7%), followed by 4b (40.3%), 1/2c (5.39%), 1/2b(4.68%), and 4c (3.96%) [15]. Another study performed in Poland by Skowron and their colleagues, showed that L. monocytogenes strains isolated from fish processing plant belonged to the serogroups 1/2a-3a (38.6%) and 1/2b, 3b (32.8%) [16]. In China, Su et al. reported the distribution of serogroups 1/2c, 3c (39.1%), 1/2a, 3a (36.7%) and 1/2b, 3b, 7 (24.2%) in L. monocytogenes isolated from foods and humans samples [43]. However, in Spain, the serogroups distributed in L. monocytogenes isolated from the environment of dairy processing were 1/2a, 3a (72.73%) followed by 1/2b, 3b, 7 (11.36%), 4b, 4d, 4e (11.36%) and 1/2c, 3c (4.55%) [44]. Previous studies reported that the serotypes 1/2a, 1/2b, 1/2c, and 4b are involved for approximately 95% of all described cases of human listeriosis in the world [45,46,47]. The presence of these serotypes in food products, especially ready-to-eat foods, is a potential risk to public health and can cause severe cases of human listeriosis.
The presence of virulence genes in L. monocytogenes strains may have had a significative effect on their degree of pathogenicity. In fact, the results obtained in this study were similar to those decribed previously for actA gene in L. monocytogenes isolated from fish and fish processing plant [16], retail raw foods [48] and fresh seafoods [49,50]. Furthermore, hlyA, inlA, inlC and inlJ genes were reported in all L. monocytogenes strains isolated from ready-to-eat food in Malaysia and China [51,52]. A study performed in Italy showed the presence of prfA, hlyA, actA, inlA, plcB genes only in one strain of L. monocytogenes isolated from Ricotta Salata cheese [53]. Another study performed by Jamali and his group in open-air fish market environments, showed that 100% of isolated L. monocytogenes were positive for hlyA, inlA and inlC, and 97.7% were positive for inlJ and prfA genes [54]. However, hlyA, prfA and inlA genes were detected in 60.8% of L. monocytogenes strains isolated from raw milk in Egypt [9]. On the other hand, inlA, inlC and inlJ genes were detected in L. monocytogenes isolated from human, animals and vegetables [43,47,55,56]. Therefore, the isolation of L. monocytogenes showing a high rate of virulence genes is very harmful to public health, and the consumption of ready to eat food contaminated by theses strains is considered a major risk for humans and can cause severe cases of morbidity and fatality.
L. monocytogenes acquired natural resistance to many β-lactams and cephalosporins [20,57]. Moreover, penicillin, ampicillin, amoxicillin with or without gentamicin or trimethoprim-sulfamethoxazole were commonly used as the first solution for the therapeutic treatment of listeriosis, while ciprofloxacin, tetracycline and chloramphenicol were used as a second solution [4,8,24]. In the present study, two strains of L. monocytogenes isolated from traditional whey (W500) and smen (S220) showed a high resistance to many antibiotics, including ampicillin, trimethoprim-sulfamethoxazole, gentamicin and chloramphenicol for strain W500 and ampicillin, trimethoprim-sulfamethoxazol and ciprofloxacin for strain S220. Thus, resistance to tetracycline was observed in L. monocytogenes isolated from raw minced meat (M80 and M140), traditional whey (W5) and raw poultry meat (P2). Also, L. monocytogenes strains from traditional whey (W33), raw sausage (Sg310) and raw fish (F300) were found to be resistant to trimethoprim-sulfamethoxazole. The resistance to ampicillin is also detected in two strains from raw milk (Mk9 and Mk100) and one strain from raw bovine meat (B510).
Resistance of L. monocytogenes to these antibiotics may be a result of their overuses in livestock to promote the growth and for the treatment of bacterial infections. A study carried out by Maćkiw and his group in Poland showed that the isolated strains of L. monocytogenes were sensitive to chloramphenicol, gentamicin, ciprofloxacin, while 9.5% of them were resistant to ampicillin [8]. Another study performed in Iran, in seafoods showed a high resistance of penicillin (57%) and ampicillin (100%) [58]. Moreover, our study revealed that all the isolated strains were sensitive to penicillin, which is in agreement with that of Gómez et al. [57]. However, a study performed in Malaysia in chicken carcasses showed that the islolated strains were resistant to penicillin (17.2%), ampicillin (6.9%) and erythromycin (6.9%) [59]. In Egypte, Tahoun et al. showed that L. monocytogenes present a high resistance for tetracycline (81%) and ciprofloxacin (66.7%), and a susceptibility for ampicillin, erythromycin and trimethoprim-sulfamethoxazole [9].
The results of this study showed that MAR index varies between 0.00 and 0.73, with the highest value detected in strains W500 (0.73) and S220 (0.40) isolated from traditional whey and smen, respectively. It should be noted that traditional whey and smen were consumed without any treatment to eliminate the pathogenic bacteria. In other studies, MAR values of 0.38–0.63 and 0.5 were recorded in chicken, meat products and raw milk, respectively [9,60]. Morover, 66.7% of isolated L. monocytogenes strains were resistant to three or more than three class of antibiotics which is a serious risk for public health.
5. Conclusion
The present study provided the data about occurrence, antimicrobial resistance, serotype distribution and virulence genes of L. monocytogenes isolated from foods in Meknes city of Morocco. This study highlighted that the rate of presence of L. monocytogenes strains is 2.9% from 520 food samples. These isolates belonged to serogroups (1/2a, 1/2c, 3a and 3c) and (1/2b, 3b, 4b and 4d), and harbored several virulence genes (inlA, inlC, inlJ, prfA, plcB, hlyA and actA) in addition to their high resistance to antimicrobial agents. However, the presence of these strains in food products presents a major risk for consumers and public health. This study provides baseline information to Moroccan regulatory authorities to allow the application of guidance for controlling L.monocytogenes and to improve the microbiological safety of foods.
Declarations
Author contribution statement
Aziz Bouymajane, Fouzia Rhazi Filali, Aboulkacem Amal, Bouchra Ouhmidou, Mohieddine Moumni: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Said Oulghazi, Nada Lafkih, Abdelaziz Ed-Dra, Abdallah El Allaoui: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data will be made available on request.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We thank the Moulay Ismail University for the financial assistance, which has been allocated to us, to cover the costs of the analyzes carried out at the National Scientific and Technical Research Center (CNRST) of Morocco.
References
- 1.Kramarenko T., Roasto M., Meremäe K., Kuningas M., Põltsama P., Elias T. Listeria monocytogenes prevalence and serotype diversity in various foods. Food Contr. 2013;30:24–29. [Google Scholar]
- 2.Garrido-Maestu A., Azinheiro S., Carvalho J., Prado M. Rapid and sensitive detection of viable Listeria monocytogenes in food products by a filtration-based protocol and qPCR. Food Microbiol. 2018;73:254–263. doi: 10.1016/j.fm.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 3.Akrami-Mohajeri F., Derakhshan Z., Ferrante M., Hamidiyan N., Soleymani M., Conti G.O., Tafti R.D. The prevalence and antimicrobial resistance of Listeria spp in raw milk and traditional dairy products delivered in Yazd, central Iran (2016) Food Chem. Toxicol. 2018;114:141–144. doi: 10.1016/j.fct.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 4.European The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016 Acknowledgements: EFSA and the ECDC wish to thank the members of the Scientific Network for Zoonoses Monitoring Data and the Food and Wat. EFSA J. 2017;15:5077. doi: 10.2903/j.efsa.2017.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lam G.Y., Czuczman M.A., Higgins D.E., Brumell J.H. first ed. Elsevier Inc.; 2012. Interactions of listeria Monocytogenes with the Autophagy System of Host Cells. [DOI] [PubMed] [Google Scholar]
- 6.Goldfine H., Shen H. Springer; 2007. Listeria Monocytogenes: Pathogenesis and Host Response. [Google Scholar]
- 7.Dongyou L. CRC Press; 2008. Handbook of listeria Monocytogenes. [Google Scholar]
- 8.Maćkiw E., Modzelewska M., Maka Ł., Ściezyńska H., Pawłowska K., Postupolski J., Korsak D. Antimicrobial resistance profiles of Listeria monocytogenes isolated from ready-to-eat products in Poland in 2007-2011. Food Contr. 2015;59:7–11. [Google Scholar]
- 9.Tahoun A.B.M.B., Abou Elez R.M.M., Abdelfatah E.N., Elsohaby I., El-Gedawy A.A., Elmoslemany A.M. Listeria monocytogenes in raw milk, milking equipment and dairy workers: molecular characterization and antimicrobial resistance patterns. J. Glob. Antimicrob. Resist. 2017;10:264–270. doi: 10.1016/j.jgar.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 10.Kim S.W., Haendiges J., Keller E.N., Myers R., Kim A., Lombard J.E., Karns J.S., Van Kessel J.A.S., Haley B.J. Genetic diversity and virulence profiles of Listeria monocytogenes recovered from bulk tank milk, milk filters, and milking equipment from dairies in the United States (2002 to 2014) PloS One. 2018;13:1–17. doi: 10.1371/journal.pone.0197053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WHO Antimicrobial resistance. Curr. Opin. Infect. Dis. 2020;4 [Google Scholar]
- 12.Ed-dra A., Filali F.R., Karraouan B., El Allaoui A., Aboulkacem A., Bouchrif B. Prevalence, molecular and antimicrobial resistance of Salmonella isolated from sausages in Meknes, Morocco. Microb. Pathog. 2017;105:340–345. doi: 10.1016/j.micpath.2017.02.042. [DOI] [PubMed] [Google Scholar]
- 13.NCEZID Information for health professionals. Rev. Lit. Arts Am. 2009:2008–2010. https://www.cdc.gov/listeria/technical.html [Google Scholar]
- 14.Benabdejlil Y. Listeriosis in pregnant women in Morocco: a case report. J. Gynecol. Obstet. 2015;3:18. [Google Scholar]
- 15.Fallah A.A., Saei-Dehkordi S.S., Mahzounieh M. Occurrence and antibiotic resistance profiles of Listeria monocytogenes isolated from seafood products and market and processing environments in Iran. Food Contr. 2013;34:630–636. [Google Scholar]
- 16.Skowron K., Kwiecińska-Piróg J., Grudlewska K., Świeca A., Paluszak Z., Bauza-Kaszewska J., Wałecka-Zacharska E., Gospodarek-Komkowska E. The occurrence, transmission, virulence and antibiotic resistance of Listeria monocytogenes in fish processing plant. Int. J. Food Microbiol. 2018;282:71–83. doi: 10.1016/j.ijfoodmicro.2018.06.011. [DOI] [PubMed] [Google Scholar]
- 17.Norme marocaine (NM.08.0.110) 2004. Microbiologie des aliments : Recherche de Listéria monocytogenes – Méthode de routine. [Google Scholar]
- 18.Doumith M., Buchrieser C., Glaser P., Jacquet C., Martin P. Differentiation of the major listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 2004;42:3819–3822. doi: 10.1128/JCM.42.8.3819-3822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.jun Du X., Zhang X., yi Wang X., lan Su Y., Li P., Wang S. Isolation and characterization of Listeria monocytogenes in Chinese food obtained from the central area of China. Food Contr. 2017;74:9–16. [Google Scholar]
- 20.Eucast European society of clinical microbiology and infectious diseases (EUCAST) Société Française Microbiol. 2019:1–181. [Google Scholar]
- 21.Bouymajane A., Filali F.R., Oulghazi S., Ed-Dra A., Benhallam F., El Allaoui A., Anissi J., Sendide K., Ouhmidou B., Moumni M. Occurrence, molecular and antimicrobial resistance of enterococcus spp. Isolated from raw cow’s milk trade by street trading in Meknes city, Morocco. Germs. 2018;8:77–84. doi: 10.18683/germs.2018.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ed-Dra A., Filali F.R., Bouymajane A., Benhallam F., El Allaoui A., Chaiba A., Giarratana F. Antibiotic susceptibility profile of Staphylococcus aureus isolated from sausages in Meknes, Morocco. Vet. World. 2018;11:1459–1465. doi: 10.14202/vetworld.2018.1459-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouayad L., Hamdi T.M. Prevalence of Listeria spp. in ready to eat foods (RTE) from Algiers (Algeria) Food Contr. 2012;23:397–399. [Google Scholar]
- 24.Amajoud N., Leclercq A., Soriano J.M., Bracq-Dieye H., El Maadoudi M., Senhaji N.S., Kounnoun A., Moura A., Lecuit M., Abrini J. Prevalence of Listeria spp. and characterization of Listeria monocytogenes isolated from food products in Tetouan, Morocco. Food Contr. 2018;84:436–441. [Google Scholar]
- 25.Shimojima Y., Ida M., Nakama A., Nishino Y., Fukui R., Kuroda S., Hirai A., Kai A., Sadamasu K. Prevalence and contamination levels of Listeria monocytogenes in ready-to-eat foods in Tokyo, Japan. J. Vet. Med. Sci. 2016;78:1183–1187. doi: 10.1292/jvms.15-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nayak D.N., Savalia C.V., Kalyani I.H., Kumar R., Kshirsagar D.P. Isolation, identification, and characterization of Listeria spp. from various animal origin foods. Vet. World. 2015;8:695–701. doi: 10.14202/vetworld.2015.695-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen N. Characterization and antibiotic susceptibility of Listeria monocytogenes isolated from poultry and red meat in Morocco. Infect. Drug Resist. 2008:45. doi: 10.2147/idr.s3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madden R.H., Hutchison M., Jordan K., Pennone V., Gundogdu O., Corcionivoschi N. Prevalence and persistence of Listeria monocytogenes in premises and products of small food business operators in Northern Ireland. Food Contr. 2018;87:70–78. [Google Scholar]
- 29.Braga V., Vázquez S., Vico V., Pastorino V., Mota M.I., Legnani M., Schelotto F., Lancibidad G., Varela G. Prevalence and serotype distribution of Listeria monocytogenes isolated from foods in Montevideo-Uruguay. Braz. J. Microbiol. 2017;48:689–694. doi: 10.1016/j.bjm.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J.-Q., Regan P., Laksanalamai P., Healey S., Hu Z. Prevalence and methodologies for detection, characterization and subtyping of Listeria monocytogenes and L. ivanovii in foods and environmental sources. Food Sci. Hum. Wellness. 2017;6:97–120. [Google Scholar]
- 31.Montero D., Bodero M., Riveros G., Lapierre L., Gaggero A., Vidal R.M., Vidal M. Molecular epidemiology and genetic diversity of Listeria monocytogenes isolates from a wide variety of ready-to-eat foods and their relationship to clinical strains from listeriosis outbreaks in Chile. Front. Microbiol. 2015;6:1–8. doi: 10.3389/fmicb.2015.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Filiousis G., Johansson A., Frey J., Perreten V. Prevalence, genetic diversity and antimicrobial susceptibility of Listeria monocytogenes isolated from open-air food markets in Greece. Food Contr. 2009;20:314–317. [Google Scholar]
- 33.Garrido V., Vitas A.I., García-Jalón I. Survey of Listeria monocytogenes in ready-to-eat products: prevalence by brands and retail establishments for exposure assessment of listeriosis in Northern Spain. Food Contr. 2009;20:986–991. [Google Scholar]
- 34.El Habib F., Ennaji M.M., El Ouardi A., Senouci S. Listeria monocytogenes in food matrix: frequency and effect of antagonist microbial. Pakistan J. Nutr. 2014;13:141–145. [Google Scholar]
- 35.Fatine Hadrya S.S., Elouardi Abdelmoula, Benali Doha, Hami Hide, Soulaymani Abdelmajid. Bacterial quality of informally marketed raw milk in Kenitra city, Morocco. Pakistan J. Nutr. 2012;11:760–767. [Google Scholar]
- 36.Mohamed K., Control D., Najia A., Ennaji M.M. 2016. Prevalence of Listeria monocytogenes isolated from chicken meat marketed Prévalence de Listeria monocytogenes isolée à partir de la viande de poulet commercialisée à Rabat , Maroc [ Prevalence of Listeria monocytogenes isolated from chicken meat marketed. [Google Scholar]
- 37.Marnissi E. Presence of Listeria monocytogenes in raw milk and traditional dairy products marketed in the north-central region of Morocco. Afr. J. Food Sci. 2013;7:87–91. [Google Scholar]
- 38.Bouymajane A., Filali F.R., Benhallam F., Ed-dra A., El Allaoui A., Chaiba A., Aboulkacem A., Hou M.A. Quantitative and qualitative microbial diversity of the raw cow’s milk sold by street trading in Meknes, Morocco. Malays. J. Microbiol. 2019;15:425–431. [Google Scholar]
- 39.Who . International Food Safety Authorities Network.; 2010. Basic Steps to Improve Safety of Street-Vended Food. [Google Scholar]
- 40.Jacob M.E., Bai J., Renter D.G., Rogers A.T., Shi X., Nagaraja2 T.G. Effect of pH and water activity on the growth limits of Listeria monocytogenes in a cheese matrix at two contamination levels. J. o f Food Prot. 2014;77:314–319. [Google Scholar]
- 41.Poimenidou S.V., Chrysadakou M., Tzakoniati A., Bikouli V.C., Nychas G.J., Skandamis P.N. Variability of Listeria monocytogenes strains in biofilm formation on stainless steel and polystyrene materials and resistance to peracetic acid and quaternary ammonium compounds. Int. J. Food Microbiol. 2016;237:164–171. doi: 10.1016/j.ijfoodmicro.2016.08.029. [DOI] [PubMed] [Google Scholar]
- 42.Leong D., NicAogáin K., Luque-Sastre L., McManamon O., Hunt K., Alvarez-Ordóñez A., Scollard J., Schmalenberger A., Fanning S., O’Byrne C., Jordan K. A 3-year multi-food study of the presence and persistence of Listeria monocytogenes in 54 small food businesses in Ireland. Int. J. Food Microbiol. 2017;249:18–26. doi: 10.1016/j.ijfoodmicro.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 43.Su X., Zhang J., Shi W., Yang X., Li Y., Pan H., Kuang D., Xu X., Shi X., Meng J. Molecular characterization and antimicrobial susceptibility of Listeria monocytogenes isolated from foods and humans. Food Contr. 2016;70:96–102. [Google Scholar]
- 44.Melero B., Stessl B., Manso B., Wagner M., Esteban-Carbonero Ó.J., Hernández M., Rovira J., Rodriguez-Lázaro D. Listeria monocytogenes colonization in a newly established dairy processing facility. Int. J. Food Microbiol. 2019;289:64–71. doi: 10.1016/j.ijfoodmicro.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 45.Vines A., Swaminathan B. Identification and characterization of nucleotide sequence differences in three virulence-associated genes of Listeria monocytogenes strains representing clinically important serotypes. Curr. Microbiol. 1998;36:309–318. doi: 10.1007/s002849900315. [DOI] [PubMed] [Google Scholar]
- 46.Pontello M., Guaita A., Sala G., Cipolla M., Gattuso A., Sonnessa M., Gianfranceschi M.V. Listeria monocytogenes serotypes in human infections (Italy, 2000-2010) Ann. Ist. Super Sanita. 2012;48:146–150. doi: 10.4415/ANN_12_02_07. [DOI] [PubMed] [Google Scholar]
- 47.Mammina C., Aleo A., Romani C., Pellissier N., Nicoletti P., Pecile P., Nastasi A., Pontello M.M. Characterization of Listeria monocytogenes isolates from human listeriosis cases in Italy. J. Clin. Microbiol. 2009;47:2925–2930. doi: 10.1128/JCM.00102-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu S., Wu Q., Zhang J., Chen M., Yan Z., Hu H. Listeria monocytogenes prevalence and characteristics in retail raw foods in China. PloS One. 2015;10:1–16. doi: 10.1371/journal.pone.0136682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Momtaz H., Yadollahi S. Molecular characterization of Listeria monocytogenes isolated from fresh seafood samples in Iran. Diagn. Pathol. 2013;8:1–6. doi: 10.1186/1746-1596-8-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Das S., Lalitha K.V., Thampuran N., Surendran P.K. Isolation and characterization of Listeria monocytogenes from tropical seafood of Kerala, India. Ann. Microbiol. 2013;63:1093–1098. [Google Scholar]
- 51.Jamali H., Thong K.L. Genotypic characterization and antimicrobial resistance of Listeria monocytogenes from ready-to-eat foods. Food Contr. 2014;44:1–6. [Google Scholar]
- 52.Wu S., Wu Q., Zhang J., Chen M., Guo W. Analysis of multilocus sequence typing and virulence characterization of listeria monocytogenes isolates from Chinese retail ready-to-eat food. Front. Microbiol. 2016;7:1–11. doi: 10.3389/fmicb.2016.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coroneo V., Carraro V., Aissani N., Sanna A., Ruggeri A., Succa S., Meloni B., Pinna A., Sanna C. Detection of virulence genes and growth potential in Listeria monocytogenes strains isolated from Ricotta Salata cheese. J. Food Sci. 2016;81:M114–M120. doi: 10.1111/1750-3841.13173. [DOI] [PubMed] [Google Scholar]
- 54.Jamali H., Paydar M., Ismail S., Looi C.Y., Wong W.F., Radmehr B., Abedini A. Prevalence, antimicrobial susceptibility and virulotyping of Listeria species and Listeria monocytogenes isolated from open-air fish markets. BMC Microbiol. 2015;15:1–7. doi: 10.1186/s12866-015-0476-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jamali H., Radmehr B. Frequency, virulence genes and antimicrobial resistance of Listeria spp. isolated from bovine clinical mastitis. Vet. J. 2013;198:541–542. doi: 10.1016/j.tvjl.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 56.Sant’Ana A.S., Igarashi M.C., Landgraf M., Destro M.T., Franco B.D.G.M. Prevalence, populations and pheno- and genotypic characteristics of Listeria monocytogenes isolated from ready-to-eat vegetables marketed in São Paulo, Brazil. Int. J. Food Microbiol. 2012;155:1–9. doi: 10.1016/j.ijfoodmicro.2011.12.036. [DOI] [PubMed] [Google Scholar]
- 57.Gómez D., Azón E., Marco N., Carramiñana J.J., Rota C., Ariño A., Yangüela J. Antimicrobial resistance of Listeria monocytogenes and Listeria innocua from meat products and meat-processing environment. Food Microbiol. 2014;42:61–65. doi: 10.1016/j.fm.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 58.Abdollahzadeh E., Ojagh S.M., Hosseini H., Ghaemi E.A., Irajian G., Naghizadeh Heidarlo M. Antimicrobial resistance of Listeria monocytogenes isolated from seafood and humans in Iran. Microb. Pathog. 2016;100:70–74. doi: 10.1016/j.micpath.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 59.Nunes Barbosa L., Cristina Bergamo Alves F., Fernanda Murbach Teles Andrade B., Albano M., Giovannetti Castilho I., Lucia Mores Rall V., Bortoleto Athayde N., Laiane Casagrande Delbem N., Oliveira Roqa R., Fernandes N Io R A.J. Effects of ocimum basilicum linn essential oil and sodium hexametaphosphate on the shelf life of fresh chicken sausage. J. Food Protect. 2014;77:981–986. doi: 10.4315/0362-028X.JFP-13-498. [DOI] [PubMed] [Google Scholar]
- 60.Marian M.N., Sharifah Aminah S.M., Zuraini M.I., Son R., Maimunah M., Lee H.Y., Wong W.C., Elexson N. MPN-PCR detection and antimicrobial resistance of Listeria monocytogenes isolated from raw and ready-to-eat foods in Malaysia. Food Contr. 2012;28:309–314. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.