Summary
Neuropeptides are essential signaling molecules secreted by dense-core vesicles (DCVs). They contribute to information processing in the brain, controlling a variety of physiological conditions. Defective neuropeptide signaling is implicated in several psychiatric disorders. Here, we provide a protocol for the quantitative analysis of DCV fusion events in rodent neurons using pH-sensitive DCV fusion probes and custom-written analysis algorithms. This method can be used to study DCV fusion mechanisms and is easily adapted to investigate fusion principles of other secretory organelles.
For complete details on the use and execution of this protocol, please refer to Persoon et al. (2019).
Subject areas: Cell culture, Cell membrane, Microscopy, Neuroscience, Molecular/chemical probes
Graphical Abstract

Highlights
-
•
Protocol for quantitative analysis of neuropeptide exocytosis in rodent CNS neurons
-
•
Generation and lentiviral infection of neurons on prepatterned astrocyte microdot islands
-
•
Visualization of neuronal DCV fusion events using pH-sensitive fusion probes
-
•
Quantitative analysis of fusion events using custom-written algorithms
Neuropeptides are essential signaling molecules secreted by dense-core vesicles (DCVs). They contribute to information processing in the brain, controlling a variety of physiological conditions. Defective neuropeptide signaling is implicated in several psychiatric disorders. Here, we provide a protocol for the quantitative analysis of DCV fusion events in rodent neurons using pH-sensitive DCV fusion probes and custom-written analysis algorithms. This method can be used to study DCV fusion mechanisms and is easily adapted to investigate fusion principles of other secretory organelles.
Before you begin
This protocol describes the experimental steps to quantify DCV fusion events in single cultured neurons grown on microdot islands of astrocytes using pH-sensitive DCV fusion probes. The protocol covers the production of lentiviral particles, preparation of single neuron cultures on astrocyte microdots, image acquisition, and data analysis using custom-written algorithms.
Lentivirus particle production
Timing: 3 days
For efficient infection of cultured neurons use lentiviral infection to express your gene of interest. In this protocol we use the DCV fusion probes NPY-pHluorin and BDNF-pHluorin, and synapsin-mCherry as live synapse marker. All constructs are driven by a human synapsin promoter for neuron specific expression.
These steps describe lentiviral particle production according to Naldini et al. (1996). We use a 2nd generation lenti viral production system (see https://www.addgene.org/guides/lentivirus/ for info on the use of 2nd or 3rd generation systems).
-
1.
Plate 293T cells from an 80% confluent T175 flask in 245 mm square tissue culture (TC) treated-dishes in DMEM complete (please refer to “Materials and equipment” for media recipes). Incubate in a 37°C, 5% CO2 incubator for 1 day, or until 50% confluence.
CRITICAL: 293T cell passage number should be < 25.
-
2.
For each lenti viral batch (lenti-hSyn-NPY-pHluorin, lenti-hSyn-BDNF-pHluorin, or lenti-hSyn-synapsin-mCherry), transfect cells with a plasmid mix in PBS containing a VSV-G envelope plasmid (e.g., p.MD2.G, 11.4 μg), packaging plasmid (e.g., pCMVΔR8.2, 22.4 μg) and a transfer plasmid containing your gene of interest (e.g., pRRL, 45.8 μg) using polyethyleneimine (PEI; 240 μg). Make up a total volume of 5 mL PBS per plate with a DNA to PEI ratio of 1:3.
-
3.
Replace media with 100 mL Opti-MEM without fetal calf serum (FCS) after 1 day.
-
4.
Harvest conditioned medium 40 h after transfection.
-
5.
Centrifuge supernatant using an Amicon spin filter (100 kDa cutoff) at 4,000 × g for 20–30 min.
-
6.
Transfer the ± 150 μL of concentrated virus to a tube, dilute in 1 mL PBS and filter sterilize through a 0.2 μm syringe filter.
-
7.
Typical viral titers obtained with this method range from 5 × 108 IFU/mL to 1 × 109 IFU/mL.
CRITICAL: Viral particles should be aliquoted in single use 0.5 mL tubes, 25–50 μL per tube, and stored at −80°C. Do not re-freeze. Thawed aliquots can be stored at 4°C for up to 4 weeks without loss of efficiency.
Coverslip preparation for neuronal culture on astrocyte microdots
Timing: 2 days
-
8.
Etch 18 mm glass coverslips in 1 N HCl for 12–16 h, wash with H2O followed by 1 h in 1 N NaOH.
-
9.
Rinse extensively with H2O and briefly in 70% EtOH. Place coverslips, once air dried, in 12-well plates.
-
10.
Apply 0.5 mL of agarose type II-A (1.5 gr/l in H2O) to cover the entire surface of each coverslip. Remove agarose with a 1 mL pipet within 30 s and let the coverslip air dry.
-
11.
Prepare the Poly-D-lysine (PDL)-collagen-acetic acid-mix (1:1:3). PDL (0.5 mg/mL; stored at −20°C), collagen (∼3.5 mg/mL; stored at 4°C), acetic acid (17 mM; stored at 4°C).
-
12.
Put 500 μL of mix onto a filter paper (3 mm cellulose chromatography paper, Whatman) and suck off the extra fluid, the paper should be wet, but not soaked.
-
13.
Put a rubber stamp containing a grid of 250 μm diameter dots (Burgalossi et al., 2012) on the paper (do not push) and then on the agarose-treated coverslip (give a little pressure).
-
14.
UV-sterilize the plates for 30 min. Plates can be stored up to 2 weeks at 19°C–22°C in a sterile environment.
Culture single primary hippocampal neurons on astrocyte microdots
Timing: 3 weeks
Astrocyte microdot and primary hippocampal culture preparations have been extensively described (Burgalossi et al., 2012). This is a brief overview highlighting the most important steps.
-
15.
Astrocyte cell preparation: Under a dissection microscope, isolate cortices of two P1 rat pups in Hanks+ medium.
-
16.
Incubate for 45 min in 5 mL of papain containing ES medium at 37°C with gentle agitation.
-
17.
Remove ES and add 5 mL of IS medium and incubate for another 15 min. Remove IS and add 2 mL DMEM+ medium. Triturate cortices with a fire-polished Pasteur pipette.
-
18.
Plate the single cell suspension in T175 flasks in 35 mL DMEM+ and incubate at 37°C, 5% CO2.
-
19.
Refresh medium after 1 day.
-
20.
When astrocytes are confluent (± 7 days), dissociate the cells from the flask with trypsin-EDTA, count the cells and plate at a density of 6,000 cells/well of a 12 well plate in DMEM+.
-
21.
Incubate the astrocyte culture in a 37°C, 5% CO2 incubator for 3–4 days, or until confluent on the microdots (Figure 1A). Replace DMEM+ with NB+ medium.
Note: DMEM+ should be replaced with NB+ medium on the day of neuronal culture, or 1 day before.
-
22.
Under a dissection microscope, isolate E18-P1 mouse hippocampi in Hanks+ medium. Add 0.025% Trypsin to the Hanks+ solution and incubate hippocampi for 20 min at 37°C, triturate the hippocampi, count the cells and plate neurons at a density of 1,000 cells/well of a 12 well plate in NB+ medium.
-
23.
Incubate astrocyte-neuron co-cultures at 37°C, 5% CO2 for up to 21 days.
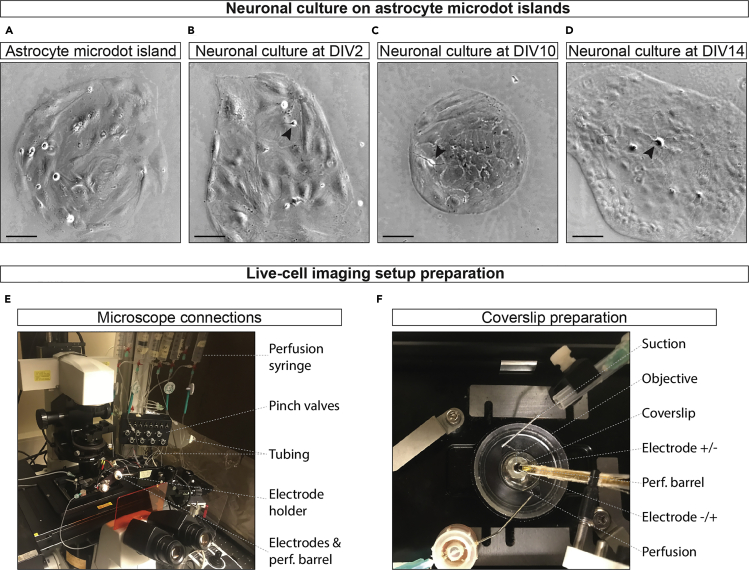
Figure 1.
Neuronal culture and microscope setup
(A) Example of astrocyte microdot island. Scale bar, 100 μm.
(B) Example of hippocampal neuron, indicated by the arrowhead, 2 days after plating (DIV2) on astrocyte microdot. Scale bar, 100 μm.
(C) Example of hippocampal neuron (arrowhead) at DIV10 on astrocyte microdot. Scale bar, 100 μm
(D) Example of hippocampal neuron (arrowhead) at DIV14 on astrocyte microdot. Scale bar, 100 μm.
(E) Live-cell imaging setup with pinch-valve controlled perfusion system, electrodes, and perfusion barrel.
(F) Example of a coverslip mounted on the microscope stage with perfusion and suction needles, electrodes, and barrel system to deliver Tyrode’s NH4Cl, or other solutions.
Infect neurons with DCV fusion reporter constructs
Timing: 5 min
This step labels neuronal DCVs with super-ecliptic pHluorin to monitor DCV fusion.
-
24.
At day-in-vitro (DIV) 9–10, check the quality of island cultures under a bright-field microscope. The majority of the astrocyte islands should be intact and confluent and contain no more than one hippocampal neuron (Figure 1C).
-
25.
Infect neurons by adding 0.5–3 μL of lentivirus particles encoding for BDNF- or NPY-pHluorin to label DCVs and synapsin-mCherry to label synapses into each well.
CRITICAL: Titrate new lentiviral batches by infecting DIV9–10 neurons with a limiting dilution series. In our hands, 0.5 to 3 μL of a virus solution (∼5 × 108 IFU/mL) prepared according to our lentivirus particle production protocol will result in infection of >90% of neurons imaged using a fluorescence microscope at DIV14. For each lentiviral batch the optimal concentration needs to be assessed. Batches will remain stable for at least 12 months at −80°C and for up to 4 weeks when stored at 4°C. For lenti-hSyn-NPY-pHluorin and lenti-hSyn-BDNF-pHluorin, many moving and stationary puncta should be visible upon Tyrode's NH4Cl perfusion. For lenti-hSyn-synapsin-mCherry, immobile puncta (presynaptic terminals) should be visible. Select the optimal viral titer for use in live imaging experiments.
-
26.
Leave 1 or 2 coverslips per condition uninfected. These will be used for Fluo5 Ca2+ imaging at DIV14.
Optional: Infection of additional lentiviral constructs (Cre, rescue constructs) can be performed at earlier or later timepoints. However, the total volume of viral solution should not exceed 20 μL.
Prepare solutions for live-cell imaging
Timing: 30 min
-
27.Prepare Tyrode's solution and Tyrode's solution containing NH4Cl (refer to “Materials and equipment” for recipes).
-
a.Dissolve all salts in dH2O at 19°C–22°C.
-
b.Adjust the pH to 7.4 with 1 N NaOH.
-
c.Check the osmolarity and adjust to 280 mOsmol with glucose.
-
d.Filter sterilize and store at 4°C.
-
e.Bring to 19°C–22°C before live-cell imaging.
-
a.
-
28.
Prepare 2 mM Fluo5-AM aliquots of 5 μL by dissolving Fluo5-AM in DMSO.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Agarose type II-A | Sigma | Cat# A9918 |
| Laminin | Sigma | Cat# L2020 |
| Poly-D-lysine | Sigma | Cat# P6407 |
| Rat tail collagen | BD Biosciences | Cat# 354236 |
| Acetic acid | VWR | Cat# 20104.334 |
| B-27 | Gibco | Cat# 17504-044 |
| PBS | Gibco | Cat# 14190-094 |
| Polyethyleneimine | Polysciences | Cat# 23966-1 |
| Opti-MEM | Fisher | Cat# 12087549 |
| Hanks | Sigma | Cat# H9394 |
| HEPES 1 M | Gibco | Cat# 15630-056 |
| DMEM + UltraGlutamine I | Lonza | Cat# LO BE12-604F/U1/12 |
| FCS | Gibco | Cat# 10270 |
| NEAA | Life Technologies | Cat# 11140-035 |
| Penicillin/streptomycin | Gibco | Cat# 15630-056 |
| L-Cysteine | Sigma | Cat# C7352 |
| EDTA | AppliChem | Cat# A2937 0500 |
| Neurobasal medium | Gibco | Cat# 21103-049 |
| Glutamax | Gibco | Cat# 35050-038 |
| Papain | Worthington | Cat# 3127 |
| Trypsin-EDTA | Gibco | Cat# 25300-054 |
| Trypsin | Gibco | Cat# 1509-046 |
| Albumin | AppliChem | Cat# A1391 0100 |
| Trypsin inhibitor | Sigma | Cat# T9523 |
| NaOH | Sigma | Cat# S5881 |
| HCl | Sigma | Cat# 38283 |
| EtOH | VWR | Cat# 83804.360 |
| NaCl | Fisher | Cat# 15673410 |
| KCl | SERVA | Cat# 7447-40-7 |
| CaCl2·2H2O | Sigma | Cat# C7902 |
| MgCl2·6H2O | Sigma | Cat# M9272 |
| HEPES (1 M) | Fisher | Cat# 15630-056 |
| Glucose-H2O | VWR | Cat# 1040741000 |
| NH4Cl | Sigma | Cat# 12125-02-9 |
| Fluo-5F-AM | Molecular Probes | Cat# F14222 |
| Dimethyl sulfoxide (DMSO) | Sigma | Cat# D2438 |
| Experimental models: cell lines | ||
| Mouse: primary neurons from hippocampus, cortex, or striatum | N/A | N/A |
| Rat: primary astrocytes | N/A | N/A |
| 293T cells | ATCC | CRL-11268 |
| Experimental models: organisms/strains | ||
| Mouse: Rab3ABCD null embryonic day 18 (E18) pups | Schlüter et al., 2004 | N/A |
| Mouse: Rim1/2 conditional null mutant E18 pups | Kaeser et al., 2011; Kaeser et al., 2008 | N/A |
| Rat: Wistar (Crl:WI) P1 pups | Charles River | Strain code: 003 |
| Recombinant DNA | ||
| hSyn(pr):NPY-pHluorin | Van de Bospoort et al., 2012 | N/A |
| hSyn(pr):rBDNF-pHluorin | de Wit et al., 2009 | N/A |
| hSyn(pr):Synapsin-mCherry | (Persoon et al., 2019) | N/A |
| p.MD2.G (encoding VSV-G envelope) | Naldini et al., 1996 | Addgene: #12259 |
| pCMVΔR8.2 (2nd gen. packaging encoding HIV-1 Gag, Pol, Tat, and Rev proteins) | Naldini et al., 1996 | Addgene: #12263 |
| pRRL (2nd/3rd gen. transfer) | Naldini et al., 1996 | Addgene: #12252 |
| Software and algorithms | ||
| MATLAB | MathWorks | https://mathworks.com |
| ImageJ | NIH | https://imagej.net/ RRID:SCR_003070 |
| DCV_pHluorin | N/A | https://github.com/alemoro/ |
| Microsoft Excel | Microsoft | https://www.microsoft.com |
| NIS Elements | Nikon Instruments |
https://www.nikoninstruments.com/Products/Software RRID:SCR_014329 |
| Other | ||
| 0.2 μm syringe filters | Fisher | Cat# 10331075 |
| T175 flask | Greiner | Cat# 690175 |
| TC-treated culture dishes | Fisher | Cat# 10350602 |
| Amicon spin filter | Millipore | Cat# UFC910096 |
| 3 mm cellulose chromatography paper, Whatman | VWR | Cat# 514-8013 |
| 18 mm glass coverslip | VWR | Cat# 631-1342 |
| 12 well plates | Greiner | Cat# 665165 |
| NikonTi-E Eclipse | Nikon | https://nikon.com |
| EMCCD DU-897 camera | Andor | https://andor.com |
| Master-8 | AMPI | https://ampi.co.il |
| Stimulus isolator A-395 | WPI | https://wpiinc.com |
| Pinch valve | N/A | N/A |
| 50 mL perfusion syringes | VWR | Cat# 613-5398 |
| Exadrop | B. Braun | Cat# 4061306 |
| Tubing | Sigma | Cat# 58703 |
| Luer Lock male | Brunschwig | Cat# CT58.1 |
| Luer Lock female | Brunschwig | Cat# CT62.1 |
| Glass capillary | GmbH | Cat# GB150F-8P |
| Platinum electrodes | N/A | N/A |
| Dissection microscope | Nikon | SMZ745 |
| Deposited data | ||
| DCV_pHluorin code | Persoon et al., 2019 | https://github.com/alemoro |
Materials and equipment
DMEM complete medium (DMEM+)
| Reagent | Final concentration (mM or μM) | Volume (mL) |
|---|---|---|
| DMEM + UltraGlutamine I | 1× | 440 |
| FCS | 10% | 50 |
| NEAA | 1% | 5 |
| Penicillin/streptomycin | 50,000 U/mL | 5 |
| Total | n/a | 500 |
Store at 4°C for up to 1 month. Prepare 50 mL aliquots to reduce rounds of heating (37°C) and cooling, and risk of contamination.
Hanks+
| Reagent | Final concentration (mM or μM) | Volume (mL) |
|---|---|---|
| Hanks | 1× | 990 |
| HEPES 1 M | 10 mM | 10 |
| Total | n/a | 1,000 |
Store at 19°C–22°C in the dark for up to 24 months.
Enzyme solution (ES)
| Reagent | Final concentration | Volume (mL) |
|---|---|---|
| DMEM + UltraGlutamine I | 1× | 245 |
| L-Cysteine | 50 mg | n/a |
| CaCl2 | 100 mM | 2.5 |
| EDTA | 50 mM | 2.5 |
| Total | n/a | 250 |
Store at −20°C in 5 mL aliquots for up to 12 months.
Preparation:
-
-
Defrost ES (5 mL) and add 20–25 U/mL papain.
-
-
Incubate at 37°C until papain is dissolved (solution should be clear).
-
-
Bubble 15 min with carbogen (solution should go from purple to more pinkish) and sterilize with 0.2 μm filter before use.
Inhibitor solution (IS)
| Reagent | Final concentration | Volume (mL) |
|---|---|---|
| DMEM + UltraGlutamine 1 | 1× | 225 |
| FCS | 10% | 25 |
| Albumin | 625 mg | n/a |
| Trypsin inhibitor | 625 mg | n/a |
| Total | n/a | 250 |
Store at −20°C for up to 12 months.
Complete neurobasal medium (NB+)
| Reagent | Final concentration | Volume (mL) |
|---|---|---|
| Neurobasal medium | 1× | 479.25 |
| B-27 Supplement | 2% | 10 |
| HEPES 1 M | 17 mM | 9 |
| Glutamax | 1% | 1.25 |
| Penicillin/streptomycin | 50,000 U/mL | 0.5 |
| Total | n/a | 500 |
Store at 4°C for up to 1 month. Prepare 50 mL aliquots to reduce rounds of heating (37°C) and cooling, and risk of contamination.
Tyrode’s imaging medium
| Reagent | Final concentration (mM) | Volume (mL) |
|---|---|---|
| NaCl | 119 | n/a |
| KCl | 2.5 | n/a |
| CaCl2 | 2 | n/a |
| MgCl2 | 2 | n/a |
| HEPES | 25 | n/a |
| Glucose | 30 | n/a |
| Total | n/a | n/a |
Store at 4°C for up to 6 months.
Tyrode’s NH4Cl medium
| Reagent | Final concentration (mM) | Volume (mL) |
|---|---|---|
| NaCl | 69 | n/a |
| KCl | 2.5 | n/a |
| CaCl2 | 2 | n/a |
| MgCl2 | 2 | n/a |
| NH4Cl | 50 | n/a |
| HEPES | 25 | n/a |
| Glucose | 30 | n/a |
| Total | n/a | n/a |
Store at 4°C for up to 6 months.
Step-by-step method details
Microscope preparation
Timing: 10 min
Here, directions are provided to setup a Nikon Ti-E Eclipse inverted microscope system for live-cell imaging experiments. These steps are similar for other microscopes and acquisition software such as Metamorph, AxioVision, and Olympus Zen. Please refer to the manufacturer's instructions how to send a TTL signal to the Master-8.
Live imaging experiments are performed on a Nikon Ti-E Eclipse inverted microscope system fitted with a Confocal A1R (LU4A Laser) unit and an EMCCD (Andor DU-897), a 40× oil objective (NA 1.3) and appropriate filter sets. NIS Elements software (version 4.30) controls the microscope and image acquisition. Electrical field stimulation is applied via two parallel platinum electrodes controlled by a Master-8 (AMPI) connected to a stimulus isolator (A-395, World Precision Instruments). Superfusion of NH4Cl containing Tyrode’s through a glass capillary placed just above the neuron is controlled with a pinch valve (Figure 1E).
-
1.
In the image acquisition software NIS Elements open ND Acquisition and select time. Set the interval as time between frames to 0.5 s for 2 Hz image acquisition, and the duration of the image acquisition.
-
2.
Set TTL out to trigger the Master-8 at the start of the image acquisition.
-
3.Set up a Master-8 paradigm (Figure 2):
-
a.Start with a delay of 30 s to record baseline fluorescence.
-
b.Send 50, 1 ms pulses at 50 Hz to the stimulus isolator for 16 times with 0.5 s between bursts.
-
c.Add a delay of 26 s to record post-stimulus fusion events.
-
d.Send a 10 s pulse to the pinch valve controller to start superfusion of NH4Cl containing Tyrode’s 26 s after stimulation.
-
a.
-
4.
Fill Tyrode’s and Tyrode’s NH4Cl syringes for gravity flow delivery via software controlled pinch valves (Figure 1E).
Figure 2.
Master 8 setup
Graphical representation of the Master 8 interface and stimulation paradigm used in this protocol. Channel 1 is connected to the computer (PC), channel 3 is connected to the stimulus isolator, and channel 4 is connected to the pinch valve system for Tyrode’s NH4Cl application. Channels 2, 5, 6, 7, 8 are not connected to an external device. Channel 1 receives the TTL signal from the PC at the start of the time-lapse recording. After a 30 s (L) delay, channel 1 delivers a 1 ms (D) trigger to channel 2 and channel 4. Channel 2 responds with 16 (M) 1 ms (D) pulses every 1.5 s (I) that triggers channel 3 with 50 (M) 1 ms (D) pulses every 20 ms (I). Thus, the communication between channel 2 and 3 generates the 16 trains of 50 action potentials at 50 Hz. Channel 4 responds 50 s (L) after input from channel 1 by delivering a 5 s (D) trigger to the pinch valve to start Tyrode's NH4Cl perfusion.
Live-cell imaging, Ca2+ imaging
Timing: 30 min to 1 h
This step visualizes intracellular Ca2+ elevations upon stimulation and subsequent buffering from the cytosol. It provides a read out of the culture’s physiological state and is typically performed at the start of each DCV fusion imaging session.
-
5.
Incubate one coverslip containing uninfected DIV14 neurons with Fluo5-AM (2 μΜ final concentration) for 10 min at 37°C, 5% CO2.
-
6.
Place the coverslip into an imaging chamber and add 500 μL of Tyrode’s solution.
-
7.Prepare neurons for live-cell imaging at the microscope setup (Figure 1F).
-
a.Place the imaging chamber in the stage holder and bring the neurons in focus using a 40× oil objective in DIC or fluorescence mode (Figure 1D).
-
b.Establish Tyrode's perfusion of 0.5 mL/min by placing one perfusion needle at one end and a suction needle at the opposite end of the imaging chamber.
-
c.Place two platinum electrodes connected to the stimulus isolator at opposite sides of an astrocyte microdot containing a single neuron.
 CRITICAL: Make sure that the distance between the electrodes spans one astrocyte microdot (± 0.5mm) and are placed in such a way that the center of your field of view lies between the electrodes. Lower the electrodes as far as possible, just above the glass coverslip but do not touch the coverslip.
CRITICAL: Make sure that the distance between the electrodes spans one astrocyte microdot (± 0.5mm) and are placed in such a way that the center of your field of view lies between the electrodes. Lower the electrodes as far as possible, just above the glass coverslip but do not touch the coverslip. -
d.Adjust the 488nm laser power and camera exposure time for optimal dynamic range.
 CRITICAL: Make sure that the camera pixels are far from saturation since stimulation will typically induce a 100-fold increase in fluorescence.
CRITICAL: Make sure that the camera pixels are far from saturation since stimulation will typically induce a 100-fold increase in fluorescence.
-
a.
-
8.
Start time-lapse recording (image acquisition at 2Hz, or higher) and trigger the Master-8 via TTL pulse.
-
9.
Repeat steps 7c–8 for a maximum of 5 times, or for no longer than 1 h.
CRITICAL: Make sure to leave at least one astrocyte microdot between a previously stimulated neuron and the next to guarantee that each neuron is naive at the start of each recording.
-
10.
Assess Ca2+ influx and efflux of each neuron by plotting intensity over time of multiple regions of interests (ROIs) drawn on the neurites (Figure 3A).
Optional: If you also use synapsin-mCherry you can draw ROIs on synapsin-mCherry positive puncta to image Ca2+ signals in synapses specifically.
Figure 3.
Neuronal live-cell imaging
(A) Top, typical neuron after incubation with the Ca2+ reporter Fluo5-AM. Insets show zoom of a neurite before field stimulation (B), during stimulation (S) and 2 s after stimulation (R). Bottom, average Fluo5-AM signal during a live-cell imaging experiment consisting of 30 s baseline recording (B), 16 trains of 50 AP at 50 Hz stimulation (S, light blue bars), and 50 s recovery (R). Note the sharp rise and fall of the calcium signal. Scale bar, 20 μm in all.
(B) Top, typical neuron with NPY-pHluorin fusion events, calculated using the Standard Deviation Z-projection option in Fiji, with a zoom of a neurite before field stimulation (B), during stimulation (S), after stimulation (R), and during Tyrode’s NH4Cl superfusion (N). Bottom, five example intensity traces showing typical NPY-pHluorin fusion events (start indicated with green arrow head). The intensity was calculated in 3 × 3 pixel ROIs. Scale bar, 20 μm in all.
Live-cell imaging, DCV fusion
Timing: 1 day
This step visualizes single fusion events of pHluorin-labeled DCVs upon stimulation.
-
11.
Place one coverslip containing DIV14 neurons infected with NPY-pHluorin (or BDNF-pHluorin) into an imaging chamber and add 500 μL Tyrode's solution.
-
12.
Similar to step 7, establish perfusion, position the platinum electrodes and the glass capillary for Tyrode’s NH4Cl superfusion close to the neuron in focus.
-
13.
Start a time-lapse recording with Tyrode’s NH4Cl superfusion at the end of the acquisition to dequench NPY-pHluorin in all DCVs (Figure 3B).
DCV fusion events detection
Timing: 30 min/cell
This step provides information on semi-automatic analysis of DCV fusion events in time-lapse recordings. DCV fusion events are detected based on the standard deviation of the signal intensity throughout the stimulation period. After the automatic detection, it is possible to manually inspect the ROIs for false positives, or to manually add new ROIs.
-
14.Install the DCV_pHluorin toolset by extracting the DCVpHluorin.zip file from Github in the Fiji.app/macros/toolsets folder. Start Fiji and run the toolset via selecting DCV_pHluorin in the toolset dropdown menu (Figure 4A)
-
a.After the first installation, the toolset needs to be initialized by opening the “Option menu” (Figures 4B and 4C).
-
b.As a final step, install the “Check ROIs” plugin via Plugin > Install Plugin… in Fiji interface. Locate the “DCVpHluorin_CheckROIs.groovy” in the Fiji.app/macros/toolsets/DCVpHluorin folder.
-
a.
-
15.Open a time-lapse file in Fiji and manually inspect it for focus drift, bleaching, and stage movements. Open the ROI Manager and start the analysis of DCV fusion events via selecting “Fusion detection” under the “Start Analysis Menu,” and adjust the parameters as follows (Figure 3D):
-
a.Movie normalization method. “B&W Opening” is set as default, use “Baseline subtraction” for noisy images.
-
b.Set the “Baseline frames” and “Start of NH4 (frame)” according to your time-lapse acquisition settings. In this case 30 and 160, respectively.
-
c.Select the “Number of stimulations” and the “Stimulation time (in frames)” accordingly. For more accurate measurements, it is recommended to round the duration of the stimulation period to the closest multiple of 10. For example, the 16 times 50 APs at 50 Hz last for 24 s (48 frames), so the “Stimulation time (frame)” is set to 50 frames. The program will then ask to insert the “Start of the stimulation (frame)” at a later stage.Note: in this case, the 16 times 50 APs at 50 Hz is considered as 1 stimulation. However, it is possible to split one stimulation in multiple blocks. Dividing the stimulation in sub-stimulations improves the detection but is more computationally intensive, thus slower and prone to memory errors.
-
d.Adjust the “Smoothing radius” to improve the signal difference between background and foreground, and to reduce scatter noise.
-
e.Enter the value for the “SNR” (signal to noise evaluation) as a number between 0 and 255, indicating the difference in signal between foreground and background.Note: it is possible to manually determine the value of the SNR by selecting “Evaluate SNR.” This will prompt the image used to detect the potential location of the DCV fusion events. Run the Fiji “Find Maxima” function with “Preview point selection” and adjust the value of the “prominence” until the majority of puncta are selected without selecting background noise. Close the “Find Maxima” and report the number on the “Evaluate SNR” dialog. The program will then restart.
-
f.If the “Baseline subtraction” method was selected, insert a desired “Detection threshold” as number of standard deviations above the mean intensity signal. Only pixels with higher intensity values will be considered for ROI placement.
-
g.Enter a desired “Cleaning threshold” as number of standard deviations above the mean intensity signal. This threshold is calculated as a rolling standard deviation with the value of “Baseline frames” as window size.
-
h.Adjust the “ROI size” to match the observed size of the DCV fusion events.
-
i.To avoid multiple detection of the same fusion event, set the “Maximum pxs overlap between ROIs” to be at most 1/3 of the total pixel area.
-
a.
-
16.Evaluate the outcome of the detection by playing the time-lapse video. To visualize all detected regions, make sure that the “Show All” option in the ROI Manager is selected.
-
a.Add any missing regions with the “Place ROIs tool;”Note: After the initial fusion detection, the program will name the ROI in the format of ROIID-EventStart, where ROIID is a 4 digits identifier which starts at “0000,” and the EventStart is the frame number where the signal rose above the detection threshold. For manually placed events the number corresponds to the frame selected by the user. EventStart can be repeated multiple times.
-
b.Regions can be deleted at this step with “Alt + click” inside a ROI.Optional: The user can evaluate each ROI intensity and detection via the “Check ROIs” in the “ROIs Interaction Menu” (Figure 5A). The program will report the Intensity over Time (in frames), with the start of the DCV fusion event indicated with a red circle. Here the user can adjust the detected frame, remove, or add DCV fusion events.
-
a.
Figure 4.
DCV_pHluorin installation and workflow
(A) Fiji main interface, locate the DCV_pHluorin toolset on the toolset dropdown menu, “≫”.
(B) Fiji main interface after loading the DCV_pHluorin toolset.
(C) Visualization of the DCV_pHluorin toolset submenu.
(D) DCV_pHluorin main workflow. Trapezoid blocks represent steps where the user is required to perform an action.
Figure 5.
DCV_pHluorin example and result tables
(A) Example Fiji interface after running the detection of DCV fusion events. In red, Fiji main interface. In yellow, the time-lapse window, the arrowhead indicates the currently selected ROI. In light-green the ROI Manager, each ROI is saved as a four digits identifier, an “-” and the fusion start frame number. In dark-green the dialog to modify the current event, from the ROIs Interaction menu > Check ROIs. In light blue the intensity over time plot of the selected ROI, the red circle indicates the start of the fusion event.
(B) Explanation of the “Vesicle Parameters” result table.
(C) Explanation of the “Pool Values” result table.
-
17.
Save the ROI Manager file via the “Save ROIs” in the “ROIs Interaction Menu.” Alternatively, the intensity of the individual ROIs can be calculated via the “Measure ROIs” function, which will report a comma separated file with the different ROIs as columns and the time-lapse frames as rows. This file can be stored for further use.
-
18.Once the DCV fusion events are detected and the ROI Manager file is saved, calculate the total number of pHluorin-labeled DCVs by doing the following:
-
a.Make sure that the ROI Manager is open and empty, without any selections on the image;
-
b.On the “Start Analysis Menu” select “Get NH4 pool.” An option dialog will appear with the same options as in step 15.
-
c.The important parameters are the “Start of NH4 (frame)” and “SNR.” If the value of the SNR is set to 0, the program will calculate the SNR value based on the intensity during NH4Cl superfusion. Alternatively, it is possible to manually determine the SNR value selecting “Evaluate SNR” as in step 15e.
-
a.
-
19.
Evaluate the outcome of the detection by observing both the pHluorin response in the time-lapse, as well as on the “MAX” figure, which is the normalized maximum intensity projection used to detect the pHluorin positive DCVs. The program will automatically save the ROI Manager file and the list of ROI intensities in the same folder as the image file.
-
20.If synapsin-mCherry was used to identify synapses it is possible to create a synapse mask with the following steps:
-
a.Duplicate the image and invert the gray values, in order to have a black object on white background;
-
b.Perform a morphological opening by applying a minimum filter, a maximum filter, and a Gaussian blur filter of the same radius, preferably of roughly the same size as the synapses in pixels;
-
c.Subtract the morphological opened image from the original and threshold the result using the “Triangle” method from Fiji “Auto threshold;”
-
d.Manually inspect the synapse mask and delete any extra objects.
-
a.
-
21.
Once the detection for all recordings is completed, collect all the data via the “ROIs Frame readers Menu” and select “From Folder.” A dialog will appear asking the “baseline frames” used to calculate the baseline intensity level to estimate the DCV fusion duration, the start of the NH4Cl superfusion, and whether to calculate the “Synaptic location” of the fusion events, and the estimation of the total amount of pHluorin-labeled DCVs (“Pool Estimation”). This will result in a table with one entry per single DCV fusion event, and one table with three estimations of the total DCV pool (Figure 5B).
Note: During data collection the program will look for the ROI Manager files of the DCV fusion events, then it will automatically calculate the DCV fusion duration and the DCV fusion intensity. If “Synaptic location” was selected, the program will open the synapse mask associated to the time-lapse and calculate the distance between the center of the DCV fusion event to the closest synaptic area. For better performance it is convenient to organize the data so that all time-lapse files and their ROI Manager files are in one single folder. Another folder will contain the NH4Cl response csv files and the associated ROI Manager files, and a third folder with the images of the synapse mask.
Expected outcomes
A single hippocampus will yield between 1.5–3.0 × 105 neurons/mL, so dissecting half a brain is enough for one experiment. After 2 days in culture (DIV2), neurons attached to the astrocyte microdots forming neurites can be observed (Figure 1B). At DIV9, neurons should have a visible, complex dendritic arborization populating roughly 2/3 of the astrocyte microdot. The astrocyte microdot islands should be round with smooth edges and fully populated with astrocytes. Some microdot islands will have no neurons or harbor multiple neurons. For quantitative DCV fusion analysis select the island harboring one single neuron.
During a typical Fluo-5 based live-cell imaging experiment, neurons should have a diffuse, low fluorescence signal, just above the signal from the underlying astrocytes. During stimulation, the signal should uniformly increase in the neuron, with the soma as the brightest spot, indicating Ca2+ influx into the cell. In response to neurotransmitter release from the neuron, it is possible to observe a wave of increase in fluorescence from the underlying astrocytes. Ca2+ efflux from the neurons should be fast, and visible for all of the 16 trains of APs, reaching a 70%–80% fluorescence intensity decay at the end of the stimulation.
For NPY-pHluorin based live-cell imaging experiments, the neurons will have a faint diffuse fluorescence signal that distinguishes the neurites from the underlying astrocytes. The soma will have a higher signal intensity due to the NPY-pHluorin molecules transiting through the trans-Golgi network, which is not acidic. Some non-acidic, pHluorin-labeled vesicles can be observed, which are trafficking along the neurites, as well as some bright puncta that could reflect pHluorin accumulation at the plasma membrane, the extracellular matrix, or in the astrocytes. During stimulation, DCV fusion events should be visible during the recordings, however depending on the intensity scaling of the recording preview, this signal might not directly be visible by eye. During Tyrode’s NH4Cl superfusion, the entire DCV population should be visible within 1 s from the opening of the pinch valve with vesicles moving along the neurites.
Using the 16 times 50 APs at 50 Hz stimulation paradigm, the expected number of DCV fusion events per cell is between 30 and 200, however it is possible to observe numbers up to 1,000 and below 10. The intensity of the individual fusion events, a measure of the amount of NPY-pHluorin inside the vesicle, ranges between 1.5 and 3 ΔF/F0 units above the baseline.
Quantification and statistical analysis
Data exclusion criteria
-
1.
Neurons should respond to electrical stimulation with a sharp rise in intracellular calcium levels that should decrease rapidly after stimulation back to baseline levels. Based on our experience, if more than 30% of the tested neurons do not show Ca2+ influx or efflux upon electrical stimulation, it is recommended to discard the culture. Ca2+ is the essential trigger for DCV fusion (Hartmann et al., 2001; de Wit et al., 2009). Therefore, insufficient Ca2+ influx or efflux contributes to variation in DCV fusion numbers. In addition, maintaining a proper Ca2+ concentration gradient is essential for neuronal health (Brini et al., 2014).
-
2.Individual time-lapse recordings are excluded from quantification If:
-
a.There are no detectable puncta after Tyrode’s NH4Cl superfusion. Neurons typically contain between a 1,400 and 18,000 NPY-pHluorin-labeled DCVs (Persoon et al., 2018) that appear as bright, ± 0.6 μm (3 px) puncta during Tyrode’s NH4Cl superfusion.
-
b.There is noticeable focus drift.
-
c.Cell debris, or other fluorescence objects are hovering on top of the neuron.
-
d.There is a large number of non-acidic, already bright, pHluorin-labeled vesicles in the neuron.
-
e.DCVs are not visibly trafficking upon Tyrode’s NH4Cl superfusion.
-
a.
-
3.
For correct total DCV pool estimates it is important that pixels (outside the soma) are not saturated upon NH4Cl superfusion. Time-lapse recordings are excluded if this is the case.
DCV fusion events data extraction
-
4.
Import the data generated with DCV_pHluorin into a computational software, such as MATLAB, R, or Python.
-
5.
The “Fusion Start” value corresponds to the frame where a DCV fusion event was detected. To get the time in s of the start of the fusion event use where F ≡ Imaging frequency. Then, generate a histogram of the DCV fusion events per cell.
Note: Use an adequate bin size to represent the data. The ultimate goal is to avoid having too many 0 data points per bin. For 2 Hz data, a binning of 0.5 s or 1.5 s is recommended. This is because with 0.5 s the binning is per frame, while with 1.5 s the binning spans from one train of APs to the next one.
-
6.
Calculate the cumulative count based on the histogram.
-
7.
Extract the total number of DCV fusion events per cell and normalize it to the total DCV pool to obtain the DCV Release Fraction. DCV_pHluorin offers 3 different pool calculations, “Raw Pool,” “Fused Corrected,” and “Puncta Corrected.” It is important to carefully choose which one to use based on the overall quality of the signal during Tyrode’s NH4Cl superfusion. If the intensity per individual DCV fusion event is comparable to the intensity of single DCV puncta during Tyrode’s NH4Cl superfusion, then the “Fused Corrected” offers the best approximation of the number of pHluorin-labeled DCVs. If the intensity per individual DCV fusion event is lower than single DCV puncta during Tyrode’s NH4Cl superfusion, then it is suggested to use the “Puncta Corrected.”
-
8.
Each individual neuron is considered as n = 1 for statistical evaluation. It is recommended to analyze at least 30 neurons derived from at least 3 independent cultures.
-
9.
The different conditions are evaluated according to statistical standards. If the data is normally distributed, the difference in mean is evaluated using a Student t-test for experiments evaluating 2 conditions, or analysis of variance (ANOVA) followed by Tukey’s post-hoc correction for evaluating 3 or more conditions. If the data is not normally distributed the equivalent nonparametric tests, Kruskal-Wallis test and Mann Whitney U test, are used.
Optional: It is possible to normalize to the total number of fusion events to observe differences in fusion kinetics between different conditions. DCV_pHluorin also generates several parameters which might be of interest, such as DCV fusion event duration, DCV fusion event location (synaptic or extra-synaptic), and the distance of the DCV fusion event from the closest synaptic location and the closest other DCV fusion location.
Limitations
This protocol is optimized for the quantification of the number of DCV fusion events, their duration, and localization, in isolated neurons. Because of the heterogeneity of the neuronal cell types present in the culture, detecting small (<20%) effects might be challenging. Therefore, it is recommended to perform a power analysis to calculate the number of independent experiments required to obtain sufficient power. In our experience, a significant difference of 30% or more requires 30 neurons from 3 independent cultures. Selection of a specific neuronal cell type by expressing DCV fusion reporters under a specific promoter, such as somatostatin, parvalbumin (for GABAergic neurons), or CaMKII (for mature glutamatergic neurons) may reduce variability.
Troubleshooting
Problem 1
Neurons do not respond to electrical stimulation (step 8 in Step-by-step method details).
Potential solution
First, make sure that the electrodes are ∼ 1 mm from the surface of the coverslip and that they are surrounding the neuron in the field of view. If neurons are still not responding to electrical stimulation, calibrate the stimulus strength required to depolarize the membrane. To do so, label neurons with the calcium indicator Fluo5-AM and stimulate with 1 ms pulses starting at 100 mA, and decrease the current until the minimum current is reached that reliably elicits Ca2+ influx during 1 ms pulses.
Problem 2
No DCV fusion upon stimulation (step 13 in Step-by-step method details).
Potential solution
DCV fusion is variable between neurons. Therefore image at least 10 neurons before concluding that there is no DCV fusion upon stimulation. It is also important to exclude low expression of the reporter, by carefully inspecting the response to Tyrode’s NH4Cl application. The response should be fast, reaching a maximum intensity within 1 s after the application, and the entire neurite arbor should contain labeled vesicles. It is also important to notice that high expression levels of the reporter could cause more DCVs to be less acidic. This will also affect the number of DCV fusion events upon stimulation. Therefore, it is suggested to titer the number of lentivirus particles to add to the culture as the minimum amount that reliably infects ∼90% of the neurons on a coverslip.
Problem 3
DCV_pHluorin detects the same DCV fusion event in multiple ROIs (step 16 in Step-by-step method details).
Potential solution
It is possible to observe multiple DCV fusion events less than 1 px apart in regions where the synapse density is higher, or with multiple neurite crossings. To avoid detection of the same DCV fusion event in multiple ROIs, it is recommended to set the “Maximum pxs overlap between ROIs” to be at most 1/3 of the total area of the ROIs. In case of 3 × 3 px ROIs, the “Maximum pxs overlap between ROIs” would then be of 3 pxs. By doing so, even if one DCV fusion event might span multiple ROIs, only the ROIs that contains all the pixels associated with the DCV fusion event will have a mean intensity value high enough to be detected. It is recommended to manually inspect potential overlap of ROIs via the “Check ROIs” function, especially in fusion dense areas.
Problem 4
Non-homogeneous fluorescence signal intensity in different regions of the neurite arbor (steps 10, 13, and 15 in Step-by-step method details).
Potential solution
Different signal intensities across different regions of the neurite arbor could be caused by a miss-aligned illumination path. To test if this is the case, it is recommended to visualize a uniformly distributed protein, such as cytosolic GFP. The problem should be solved by aligning the light path.
If the uneven illumination, or signal intensity, leads to overrepresentation of the higher signal areas, it is recommended to adjust the “Movie normalization” settings. By using the “Baseline subtraction” Fiji will calculate the average intensity value of the first X frames (X set by user in “Baseline Frames” option). It will subtract this average intensity value from all frames. This method works well for noisy images, but it is sensitive to uneven signal intensities, thus favoring regions with high signal.
An additional option for "movie normalization" is the "B&W opening" option. This uses the information of the pixels surrounding the pixels of interest by a value “Smoothing radius” to first perform a maximum filter, then a minimum filter, and lastly a Gaussian Blur filter. Since the “B&W Opening” scales pixels based on local information compared to the entire image, it is more suitable for images with uneven illumination or signal intensity. In addition to both normalization methods, the detection is done on the standard deviation of the signal over time. Using the standard deviation ensures that high intensity regions are not overrepresented compared to low intensity regions.
Problem 5
Non-acidic structures are visible before NH4Cl perfusion (step 13 in Step-by-step method details).
Potential solution
As stated in problem 2, too high expression levels of the DCV reporter construct could cause DCVs to be less acidic. In addition, when designing new DCV reporter constructs by tagging other neuropeptides with pHluorin it is possible to observe mistargeting to other organelles. For example, mistargeting to reticular or tubular structures has been observed for NPY tagged with EGFP (Nagai et al., 2002). To test if your DCV reporter of interest targets to DCVs it important to first validate the sequence of your construct. Next, it is recommended to verify efficient targeting to DCVs by co-immunolabeling with endogenous DCV markers (Persoon et al., 2018). If your reporter construct is indeed mistargeting it is recommended to try and reduce the construct length to a minimal functional construct (for example only containing the signal peptide and N-terminal domain of the pre-propeptide) and subsequently validate the correct targeting.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ruud F. Toonen (ruud.toonen@cncr.vu.nl).
Materials availability
This study did not generate new unique reagents. Plasmids generated in this study are available upon request.
Data and code availability
The DCV_pHluorin code is available in GitHub at https://github.com/alemoro
Acknowledgments
The authors thank Robbert Zalm for cloning and producing lentiviral particles, Desiree Schut and Lisa Laan for astrocyte and neuronal cultures, Joke Wortel for animal breeding, Joost Hoetjes for genotyping, and members of the CNCR DCV project team for fruitful discussions. This work is supported by an ERC Advanced Grant (322966) of the European Union (to M.V.)
Author contributions
R.I.H. and A.M. acquired data. R.I.H., A.M., C.M.P., M.V., and R.F.T. designed figures and wrote the protocol.
Declaration of interests
The authors declare no competing interests.
References
- Brini M., Calì T., Ottolini D., Carafoli E. Neuronal calcium signaling: function and dysfunction. Cell. Mol. Life Sci. 2014;71:2787–2814. doi: 10.1007/s00018-013-1550-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgalossi A., Jung S., Man K.M., Nair R., Jockusch W.J., Wojcik S.M., Brose N., Rhee J.S. Analysis of neurotransmitter release mechanisms by photolysis of caged Ca2+ in an autaptic neuron culture system. Nat. Protoc. 2012;7:1351–1365. doi: 10.1038/nprot.2012.074. [DOI] [PubMed] [Google Scholar]
- de Wit J., Toonen R.F., Verhage M. Matrix-dependent local retention of secretory vesicle cargo in cortical neurons. J. Neurosci. 2009;29:23–37. doi: 10.1523/JNEUROSCI.3931-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M., Heumann R., Lessmann V. Synaptic secretion of BDNF after high-frequency stimulation of glutamatergic synapses. EMBO J. 2001;20:5887–5897. doi: 10.1093/emboj/20.21.5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser P.S., Deng L., Wang Y., Dulubova I., Liu X., Rizo J., Südhof T.C. RIM proteins tether Ca2+ channels to presynaptic active zones via a direct PDZ-domain interaction. Cell. 2011;144:282–295. doi: 10.1016/j.cell.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser P.S., Kwon H.B., Chiu C.Q., Deng L., Castillo P.E., Südhof T.C. RIM1α and RIM1β are synthesized from distinct promoters of the RIM1 gene to mediate differential but overlapping synaptic functions. J. Neurosci. 2008;28:13435–13447. doi: 10.1523/JNEUROSCI.3235-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T., Ibata K., Park E.S., Kubota M., Mikoshiba K., Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F.H., Verma I.M., Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- Persoon C.M., Moro A., Nassal J.P., Farina M., Broeke J.H., Arora S., Dominguez N., van Weering J.R., Toonen R.F., Verhage M. Pool size estimations for dense-core vesicles in mammalian CNS neurons. EMBO J. 2018;37:e99672. doi: 10.15252/embj.201899672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persoon C.M., Hoogstraaten R.I., Nassal J.P., van Weering J.R., Kaeser P.S., Toonen R.F., Verhage M. The RAB3-RIM pathway is essential for the release of neuromodulators. Neuron. 2019;104:1065–1080. doi: 10.1016/j.neuron.2019.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter O.M., Schmitz F., Jahn R., Rosenmund C., Südhof T.C. A complete genetic analysis of neuronal Rab3 function. J. Neurosci. 2004;24:6629–6637. doi: 10.1523/JNEUROSCI.1610-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Bospoort R., Farina M., Schmitz S.K., de Jong A., de Wit H., Verhage M., Toonen R.F. Munc13 controls the location and efficiency of dense-core vesicle release in neurons. J. Cell Biol. 2012;199:883–891. doi: 10.1083/jcb.201208024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The DCV_pHluorin code is available in GitHub at https://github.com/alemoro