Abstract
Terpenoids are a large and diverse group of natural products with commercial applications. Microbial production of terpenes is considered as a feasible approach for the stable supply of these complex hydrocarbons. Cyanobacteria, photosynthetic prokaryotes, are attractive hosts for sustainable bioproduction, because these autotrophs require only light and CO2 for growth. Despite cyanobacteria having been engineered to produce a variety of compounds, their productivities of terpenes are generally low. Further research is needed to determine the bottleneck reactions for enhancing terpene production in cyanobacteria. In this study, we engineered the fast-growing cyanobacterium Synechococcus elongatus UTEX 2973 to produce a commercially-used terpenoid, limonene. We identified a beneficial mutation in the gene encoding geranylgeranyl pyrophosphate synthase crtE, leading to a 2.5-fold increase in limonene production. The engineered strain produced 16.4 mg L−1 of limonene at a rate of 8.2 mg L−1 day−1, which is 8-fold higher than limonene productivities previously reported in other cyanobacterial species. Furthermore, we employed a combinatorial metabolic engineering approach to optimize genes involved in the upstream pathway of limonene biosynthesis. By modulating the expression of genes encoding the enzymes in the MEP pathway and the geranyl pyrophosphate synthase, we showed that optimization of the expression level is critical to enhance limonene production in cyanobacteria.
Keywords: Cyanobacteria, Limonene, Geranylgeranyl pyrophosphate synthase
Highlights
-
•
Engineering of the fast growing cyanobacterium Synechococcus elongatus UTEX 2973 for limonene production.
-
•
Identification of a beneficial mutation with 2.5-fold increase in limonene productivity.
-
•
Pathway optimization for limonene biosynthesis.
1. Introduction
Terpenoids (or terpenes) comprise a diverse group of natural products with a variety of applications, including pharmaceuticals, nutraceuticals, agriculture, and flavor and fragrance (Ajikumar et al., 2008). Microbial production of terpenes offers a versatile approach compared to chemical synthesis and extraction from plant species (Niu et al., 2017). In recent years, cyanobacteria are viewed as potential hosts for microbial bioproduction, because these photosynthetic prokaryotes require only light, CO2, and minerals for growth. In spite of many efforts to engineer cyanobacteria for terpenoid production, their titers and productivities remained low (Lin and Pakrasi, 2019). One of the main challenges to improve cyanobacterial terpene production is to redirect the photosynthetically fixed carbon to terpene synthesis instead of cell biomass (Melis, 2013). Moreover, the 2-C-Methyl-d-erythritol-4-phosphate (MEP) pathway for the synthesis of the terpene precursors is highly regulated (Banerjee and Sharkey, 2014), thus increasing the difficulty of engineering this precursor pathway. Although several attempts have been made to overexpress the MEP pathway in cyanobacteria for terpene production, their improvements have been marginal (Englund et al., 2015; Halfmann et al., 2014; Kiyota et al., 2014). Further enhancements in titer and productivity are urgently needed for a practical photosynthetic terpene production.
As the precursor for the synthesis of pigments, including chlorophyll, and carotenoids, the terpene biosynthesis pathway is important to photosynthetic microorganisms. Cyanobacteria use the MEP pathway to produce the 5-carbon precursors, isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP), for terpene synthesis. Using both DMAPP with IPP as substrates, prenyltransferase CrtE catalyzes the condensation reactions to produce prenyl pyrophosphates with longer carbon chains, such as geranyl pyrophosphate (GPP), farnesyl pyrophosphate (FPP), and geranylgeranyl pyrophosphate (GGPP). One of the major challenges in using cyanobacteria for terpene production is competition between natural pigment biosynthesis and the engineered pathway for limited prenyl pyrophosphates. The competition requires balanced carbon fluxes to achieve high terpene productivity while maintain proper cell growth.
In the present work, we engineered the fast-growing cyanobacterium Synechococcus elongatus UTEX 2973 for enhanced production of a commercially valuable monoterpene, limonene. In optimal growth conditions, Synechococcus 2973 can grow as fast as the yeast Saccharomyces cerevisiae while using only light and CO2 (Yu, 2015; Ungerer et al., 2018a). The high CO2 fixation rate of this strain allows it to accumulate cell biomass at a rate of over 1 g L−1 day−1 (Ungerer et al., 2018a). A recent study reported the high productivity of sucrose by an engineered Synechococcus 2973 strain (Lin et al., 2020), demonstrating its potential for enhanced bioproduction. Therefore, we used this fast growing cyanobacterium as a host to see if the production of limonene can be further increased. We discovered a mutant strain with a 2.5-fold improvement in limonene production. Whole-genome sequencing results revealed that this high limonene producer has two single nucleotide polymorphisms (SNPs) compared to the wild type Synechococcus 2973 in crtE and Synechococcus outer membrane protein B (somB) genes. Using the CRISPR/Cpf1 genome editing tool, we conducted a mutational analysis to investigate which SNP leads to enhanced limonene production. Our result showed that the SNP generating CrtE R299Q drastically improved limonene productivity. Furthermore, by expressing and modulating the expression level of a specific GPP synthase (GPPS) from Abies grandis, we demonstrated that the enzyme level of GPPS is critical for controlling the carbon fluxes toward limonene synthesis. Finally, we expressed the rate-limiting genes in the MEP pathway for enhancing the precursor pool (IPP and DMAPP) for limonene synthesis, providing insights in the engineering of the upstream precursor pathways. This integrated approach of using the CrtE R299Q mutant, modulation of GPPS expression, and optimization of the precursor pathway could be applicable for enhancing other monoterpene production in cyanobacteria.
2. Material and methods
2.1. Growth conditions
Synechococcus elongatus UTEX 2973 was cultured in BG11 medium at 38°C, 250 μmol photons m−2 s−1 light, and 1% CO2 in an AlgaeTron growth chamber (Photon Systems Instruments, Czech Republic). For the mutant strains, the antibiotic kanamycin (10 μg mL−1) was used in BG11 agar plates or liquid medium. To test limonene production, mutant strains were grown in BG11 medium for 24 hours, and then diluted to an OD730 of 0.05 (0.06 g L−1 cell dry weight) to initiate the experiments. To compare the limonene production of cyanobacteria from previous studies, a higher initial OD730 of 0.4 (0.5 g L−1 cell dry weight) was used for the experiment. Strains were grown in 10 mL of BG11 medium in 50-mL flasks with 10% (v/v) of isopropyl myristate (Kato et al., 2017) covered as an organic overlay to trap limonene. IPTG (1 mM) was added to induce gene expression.
2.2. Construction of strains
Plasmids and strains used in this study are listed in Table 1. All cloning experiments were performed using the Gibson isothermal DNA assembly method (Gibson et al., 2009). The RSF1010 self-replicating plasmid was used for expression of genes in this study (Huang et al., 2010). The lims and gpps genes were codon optimized for expression in Synechocystis 6803 and E. coli, respectively (Lin et al., 2017). The idi gene was amplified from the plasmid JEBI2999 (Peralta-Yahya et al., 2011). The dxs gene was amplified from genomic DNA of E. coli. To engineer the RBS of gpps, synthetic RBSs with different translation rates were obtained from the RBS calculator (Salis et al., 2009). Bacterial conjugation was used to introduce plasmids into Synechococcus 2973 (Yu et al., 2015).
Table 1.
Plasmids and strains used in this study.
| Plasmids | Description | Reference |
|---|---|---|
| pRL443 | Conjugal plasmid for bacterial conjugation | Elhai et al. (1997) |
| pRL623 | Helper plasmid for bacterial conjugation | Elhai et al. (1997) |
| pSL3383 | CRISPR/Cpf1 plasmid which generates SomB Y380H mutant | This study |
| pSL3384 | CRISPR/Cpf1 plasmid which generates CrtE R299Q mutant | This study |
| pSL3385 | Ptrc1O-lims-RBS (TIR=4743)-gpps, RSF1010 plasmida | This study |
| pSL3386 | Ptrc1O-lims-RBS (TIR=1008)-gpps, RSF1010 plasmida | This study |
| pSL3387 | Ptrc1O-lims-RBS (TIR=3146)-gpps, RSF1010 plasmida | This study |
| pSL3388 | Ptrc1O-lims-RBS (TIR=5635)-gpps, RSF1010 plasmida | This study |
| pSL3389 | Ptrc1O-lims-RBS (TIR=8185)-gpps, RSF1010 plasmida | This study |
| pSL3390 | Derived from pSL3385, gpps-eyfp fusion construct | This study |
| pSL3391 | Derived from pSL3386, gpps-eyfp fusion construct | This study |
| pSL3392 | Derived from pSL3387, gpps-eyfp fusion construct | This study |
| pSL3393 | Derived from pSL3388, gpps-eyfp fusion construct | This study |
| pSL3394 | Derived from pSL3389, gpps-eyfp fusion construct | This study |
| pSL3395 | pSL3385 carrying PlacUV5-dxs | This study |
| pSL3396 | pSL3385 carrying PlacUV5-idi | This study |
| pSL3397 | pSL3385 carrying PlacUV5-dxs-idi | This study |
| Strains | Description | Reference |
| WT | Synechococcus 2973 wild type | Yu et al. (2015) |
| High Lim | Synechococcus 2973 somB Y380H crtE R299Q. High limonene-producing strain | This study |
| Low Lim | Synechococcus 2973 somB L234S. Low limonene-producing strain | This study |
TIR, translation initiation rate.
2.3. CRISPR/Cpf1 genome editing
CRISPR/Cpf1 was used to construct mutant strains with SomB Y380H or CrtE R299Q protein mutants. The Cpf1 plasmid cloning and genome editing procedures were based on methods reported in previous studies (Ungerer and Pakrasi, 2016; Ungerer et al., 2018b). Briefly, the guide RNA sequences which target somB (5′-gtgaaaccagttttggctac-3′) or crtE (5′-cggcacggaaacactgatgt-3′) were cloned into the CRISPR/Cpf1 plasmid. The repair templates which generate the mutations were then cloned into the plasmids by Gibson assembly. The CRISPR genome editing plasmids were introduced into wild type Synechococcus 2973 to generate SomB and CrtE R299Q mutations. Successful gene editing was confirmed by Sanger sequencing. To cure the plasmids, strains were cultured in BG11 without the addition of antibiotics.
2.4. Whole genome sequencing and identification of variants
The high and low limonene-producing strains were sequenced by the Washington University Genome Technology Access Center (GTAC) using the Illumina HiSeq 2500 sequencer, producing between 13-14 million unpaired 50-bp reads for each sample. Adapters and low quality regions were trimmed from the unpaired Illumina reads using the bbduk program in the BBTools suite (https://sourceforge.net/projects/bbmap/). The quality of the Illumina reads before and after trimming were assessed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The reference Synechococcus 2973 genome assembly was obtained from the NCBI Assembly database (Kitts et al., 2016) under the RefSeq assembly accession number GCF_000817325.1. The trimmed Illumina reads from each Synechococcus sample were mapped against this reference genome assembly using the aln subcommand in bwa (Li and Durbin, 2009) with default parameters. The alignment results were converted into the BAM format using SAMtools (Li et al., 2009). The alignments were filtered using the view subcommand of SAMtools so that only alignments with mapping quality of 10 or above are kept. The MarkDuplicates subcommand of Picard Tools (http://broadinstitute.github.io/picard/) was used to identify optical and PCR duplicates in the filtered BAM alignment files. The resulting BAM files for the two Synechococcus samples were analyzed jointly using FreeBayes (https://github.com/ekg/freebayes) with the ploidy parameter set to 2 to identify sequence variants. The putative variants identified by FreeBayes were filtered using the vcffilter tool in the vcflib library (https://github.com/vcflib/vcflib) so that only the variants with quality scores greater than 20 were kept. An UCSC Assembly Hub for the reference Synechococcus 2973 genome assembly was produced using the UCSC Genome Browser utilities developed by the Genome Bioinformatics Group at the UCSC Genomics Institute (Raney et al. 2014). The gene annotations for the reference Synechococcus 2973 assembly was obtained from the NCBI RefSeq FTP site under the accession number GCF_000817325.1. These gene annotations were then integrated into the Synechococcus 2973 Assembly Hub. The VCF file containing the filtered variants identified by FreeBayes and the reference gene annotations in bigGenePred format were analyzed by the UCSC Variant Annotation Integrator tool (Hinrichs et al., 2016) to identify the functional roles of each variant (e.g., non-synonymous mutations, frame shifts).
2.5. Limonene measurement
Limonene dissolved in the isopropyl myristate (IM) overlay was used for quantification. The samples were prepared by diluting 100 μL of the IM overlay in 300 μL of pure IM, and limonene concentration was quantified using a gas chromatography instrument equipped with a flame ionization detector (Hewlett-Packard model 7890 A, Agilent Technologies, CA, USA) and a 30-meter DB5-MS column (J&W Scientific). The oven temperature initiated at 70 °C and held for 3 minutes, followed by an increase at 5 °C/min to 260 °C and held for 3 minutes. Limonene was quantified by comparing peak area to that of a (R)-limonene internal standard.
2.6. Fluorescence measurement
The engineered strains with GPPS-EYFP fusion constructs were cultured at a starting OD730 of 0.05 in BG11 medium with antibiotics for 24 hours. Cultures were diluted to a similar OD730 = 0.5 for fluorescence measurement. The fluorescence and OD730 of cultures were measured on a Synergy Mx plate reader (BioTek, Winooski, VT). The excitation and emission wavelengths were set to 485 and 528 nm, respectively. The fluorescence intensity was normalized by OD730 to compare between strains.
3. Results
3.1. Identification of a high limonene-producing mutant of Synechococcus 2973
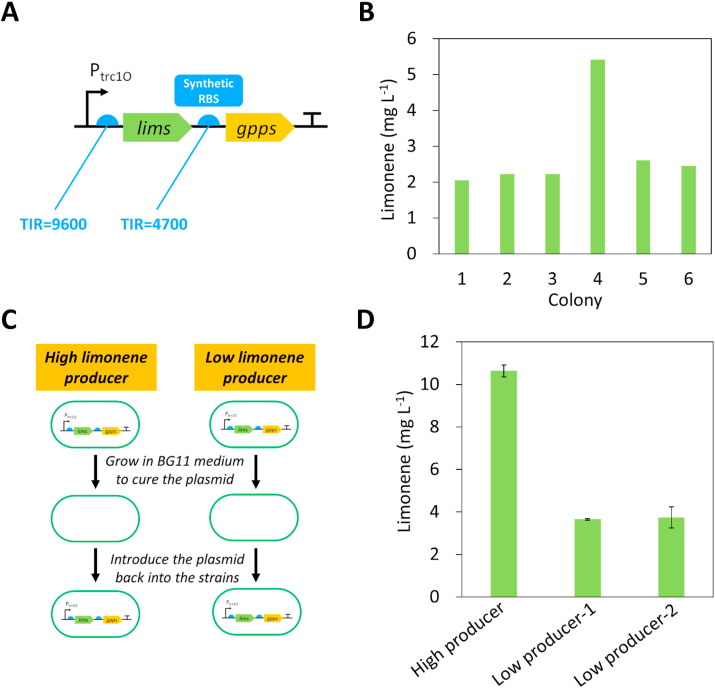
To engineering Synechococcus 2973 for limonene production, the limonene synthase (lims) gene from Mentha spicata (Colby et al., 1993) and the gpps gene from Abies grandis (Burke and Croteau, 2002) were cloned into the RSF1010 self-replicating plasmid under the control of the trc1O promoter (Fig. 1A). Since the GPPS competes with the CrtE for terpene precursors, the expression level of GPPS should not be too strong to ensure a sufficient carbon flux for pigment synthesis. Therefore, we decided to use a synthetic ribosome binding site (RBS) with a lower translation initiation rate (TIR) to drive the expression of GPPS. First, we used the RBS calculator (Salis et al., 2009) to estimate the TIR of the RBS in the trc1O promoter, and obtained a synthetic RBS with half of the TIR compared to that of the RBS in Ptrc1O (Fig. 1A). A Lac repressor (lacI) was included in the plasmid to control expression of lims and gpps. In our previous study, we reported random deleterious mutations in the lims gene in E. coli when cloned into the pCC5.2 neutral-site-targeting plasmid (Lin et al., 2017). However, the lims gene in the RSF1010 plasmid had no mutation after introduction into E. coli, likely because of LacI repression in the RSF1010 plasmid.
Fig. 1.
Identification of a high limonene-producing mutant of Synechococcus 2973. (A) The plasmid pSL3385 containing lims from M. spicata and gpps from A. grandis was introduced in Synechococcus 2973. A synthetic RBS for the gpps gene was obtained from the RBS calculator, with half of the TIR compared to the RBS in trc1O promoter. (B) Limonene production in Synechococcus 2973 expressing the plasmid pSL3385. One mutant produced 2.5-fold higher of limonene compared to the other strains. The experiment was conducted with the initial OD730 of 0.05 in 1% CO2 and 500 μmol photons m−2 s−1 light for 2 days (C) Schematics of testing the difference in genome background in the high and low limonene producers. The plasmid pSL3385 was cured and then re-introduced into the strains. (D) Limonene production of the re-constructed high and low limonene-producing strains. The experiment was conducted with the initial OD730 of 0.05 in 1% CO2 and 250 μmol photons m−2 s−1 light for 2 days. RBS, ribosome biding site; TIR, translation initiation rate.
The plasmid pSL3385 (Table 1) was then introduced into Synechococcus 2973 via bacterial conjugation. We picked 6 colonies to test limonene production. Notably, the limonene titer from one of the mutants was 2.5-fold higher than the other strains (Fig. 1B). Sequencing of the plasmid in the high limonene-producing mutant revealed no mutation in the lims gene or the trc1O promoter. These results suggest that the high limonene producer may have mutations in the genome which benefits limonene synthesis. To test this hypothesis, we cured the plasmid pSL3385 in the high limonene and two low limonene producers by growing the strains in BG11 medium without the antibiotic kanamycin, and re-introduced the plasmid pSL3385 into the strains (Fig. 1C). By testing the limonene production in the re-constructed strains, the high producer remained to produce 2.5-fold higher of limonene than the low producers (Fig. 1D), indicating that the genome background of the high-limonene producer is different from the low producer. Notably, we found that the limonene production by the engineered strain was sensitive to higher light intensity. Decreasing the light intensity from 500 to 250 μmol photons m−2 s−1 resulted in increased limonene production (Fig. 1B & D). Therefore, we used the lower light intensity for further experiments.
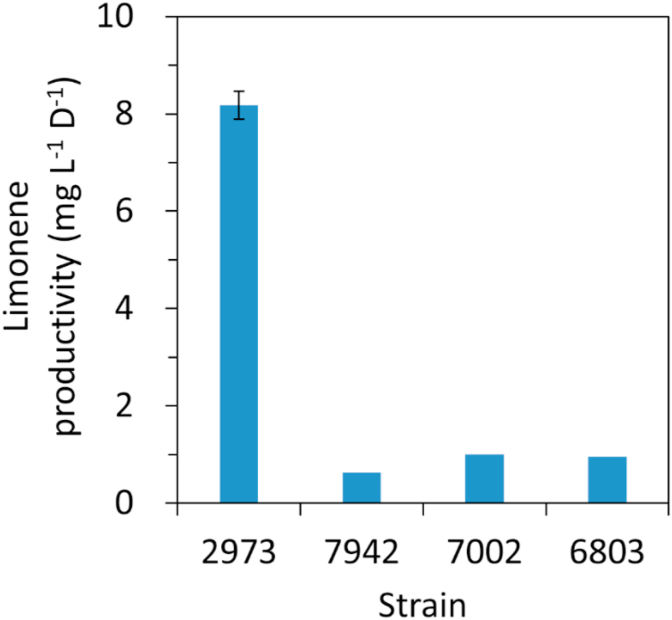
To date, the highest productivity of limonene in cyanobacteria reported in literature is 1 mg L−1 day−1 in Synechococcus sp. PCC 7002 (Davies et al., 2014). To compare limonene productivity between different strains under the same condition, we cultured our high producer Synechococcus 2973 following the same experimental condition reported in the Synechococcus 7002 study (Davies et al., 2014). The experiment was initiated at an OD730 of 0.4 (equivalent to 0.5 g L−1 cell dry weight) under 1% CO2 and 250 μmol photons m−2 s−1 light. The productivity of limonene in Synechococcus 2973 high limonene producer was 8-fold higher than the previously engineered Synechococcus 7002 strain (Fig. 2). The limonene titer was 16.4 mg L−1 after 2 days of growth. This result demonstrates the potential of using Synechococcus 2973 as a photosynthetic microbial host for limonene production.
Fig. 2.
Comparison of limonene productivity in cyanobacteria. The Synechococcus 2973 high limonene producer was cultivated using the same experimental conditions reported in literature. 6803, Synechocystis 6803 (Lin et al., 2017); 7002, Synechococcus 7002 (Davies et al., 2014); Synechococcus 7942 (Wang et al., 2016). The experiment was initiated at an OD730 = 0.4. The limonene titer in Synechococcus 2973 represents mean ± sd of three biological replicates.
3.2. Genome sequencing of high limonene-producing strain
To identify the mutation(s) in the high limonene producer, whole genome sequencing was conducted. A low producer (Fig. 1B, colony 6) was included for genome sequencing to compare with the high producer. Compared to the wild type of Synechococcus 2973, two single nucleotide polymorphisms (SNPs) were found in the high producer, whereas the low producer had one SNP (Table 2). In the high producer, one SNP is on the gene encoding the Synechococcus outer membrane protein B (SomB), whereas the other SNP is on the GGPP synthase (CrtE). Both SNPs lead to amino acid changes (Y380H in SomB and R299Q in CrtE). As for the low producer, the SNP was also observed on the somB gene and caused an amino acid substitution (L234S).
Table 2.
Genome sequencing results of Synechococcus 2973 limonene-producing mutants.
| Nucleotide position | Locus tag | Gene function | 2973 WT | High producer | Low producer | Amino acid change |
|---|---|---|---|---|---|---|
| 1638555 | M744_08675 | Porin (SomB) | T | T | C | L234S |
| 1638992 | M744_08675 | Porin (SomB) | T | C | T | Y380H |
| 2496789 | M744_12865 | GGPPS (CrtE) | G | A | G | R299Q |
The SomB protein was identified as a porin in the outer membrane of Synechococcus PCC 6301 (Hansel et al., 1998). However, its biological function remains unclear. Analysis of a transposon mutant library revealed that it is a non-essential gene in Synechococcus 7942 (Rubin et al., 2015). CrtE, an essential enzyme in cyanobacteria, catalyzes the formation of pyrophosphate intermediates in terpene biosynthesis. This enzyme is critically important in cyanobacteria, because it controls the carbon flux for photosynthetic pigment synthesis.
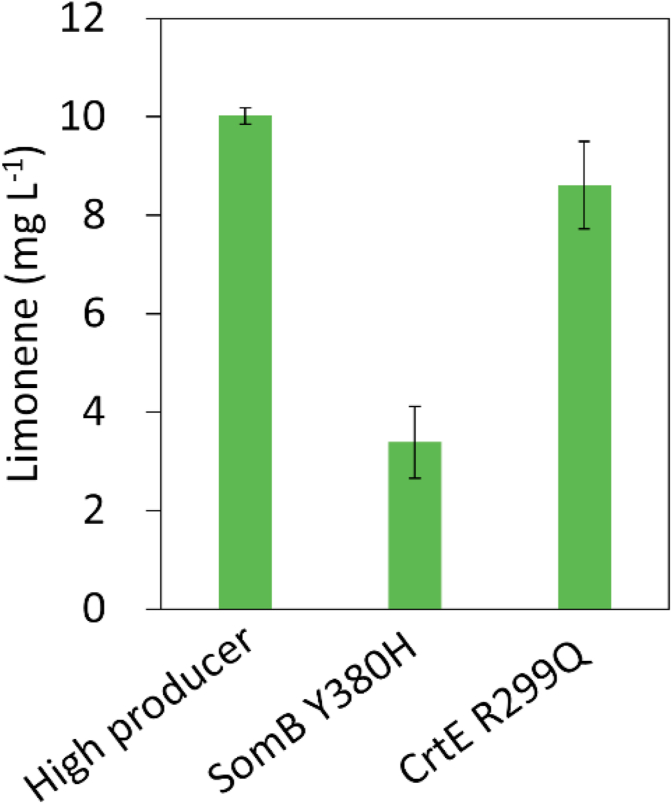
3.3. Identification of the CrtE R299Q mutant that leads to enhanced limonene production
To identify which SNP leads to increased limonene production in the high producer, we used CRISPR/Cpf1 genome editing to create the SNPs in Synechococcus 2973, generating strains with SomB Y380H or CrtE R299Q mutations. Then, the plasmid pSL3385 was further introduced into both strains to test limonene production. Notably, we have repeatedly observed that the efficiency of bacterial conjugation of SomB Y380H mutant was higher than the CrtE R299Q mutant and the WT (data not shown). As shown in Fig. 3, the CrtE R299Q strain enhanced limonene production significantly compared to that in the SomB Y380H mutant. This result determines that the R299Q mutation in the crtE gene confers the high limonene productivity and highlights the importance in balancing the flux for pigment synthesis during limonene production.
Fig. 3.
Limonene production in Synechococcus 2973 SomB Y380H and CrtE R299Q mutants. The experiment was conducted in 1% CO2 and 250 μmol photons m−2 s−1 light for 2 days.
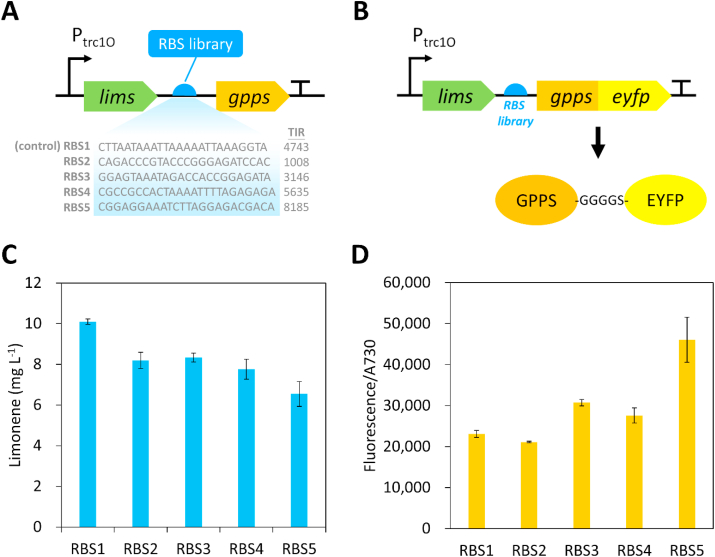
3.4. Modulation of GPPS expression to optimize limonene synthesis pathway
To further increase limonene production in Synechococcus 2973, we used the high limonene producer and decided to further optimize flux from IPP and DMAPP between limonene production and pigment syntheses. To achieve this goal, we modulated the enzyme level of GPPS by using a synthetic RBS library with different translation rates to control GPPS expression (Fig. 4A). RBS1 is the original GPPS RBS sequence on plasmid pSL3385 (Fig. 4A). We cloned 4 additional RBSs with TIRs ranging from 1000 to 8000. Since the TIRs of the RBS library were based on computational calculation, we wanted to examine if these RBS sequences lead to different expression levels of GPPS. Therefore, the C-terminus of GPPS was fused with an enhanced yellow fluorescent protein (EYFP) via a GGGGS peptide linker (Fig. 4B). The strength of RBS can be quantified by measuring the EYFP fluorescence in the limonene-producing mutants. As shown in Fig. 4C, different amounts of limonene were produced in Synechococcus 2973 using the RBS library to drive GPPS expression. Moreover, the result of EYFP fluorescence confirmed that the RBS sequences led to different protein levels of GPPS (Fig. 4D). The control strain, RBS1, produced the highest amount of limonene (10 mg L−1) (Fig. 4C), whereas its GPPS expression level was the second lowest (Fig. 4D). By contrast, the RBS5 strain had the strongest expression of GPPS (Fig. 4D), while the limonene titer was the lowest (Fig. 4C), with 35% decrease compared to the control strain RBS1. Our results indicate that the original RBS1 sequence used in plasmid pSL3385 is the optimized RBS for limonene production. Although the limonene titer could not be further improved by engineering the RBS of the gpps gene, these results demonstrate that optimization of GPPS expression is critical for enhancing limonene production in cyanobacteria.
Fig. 4.
Modulation of gpps expression via RBS engineering to optimize limonene biosynthesis. (A) A synthetic RBS library with varying translation rates was used to modulate the expression of gpps. (B) GPPS was fused with an enhanced yellow fluorescent protein (EYFP) using a peptide linker to verify the strength of synthetic RBSs. (C) Limonene production in Synechococcus 2973 using the RBS library to express gpps. (D) Fluorescence level of GPPS-EYFP fusion protein expressed by the RBS library. The experiment was conducted in 1% CO2 and 250 μmol photons m−2 s−1 light. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
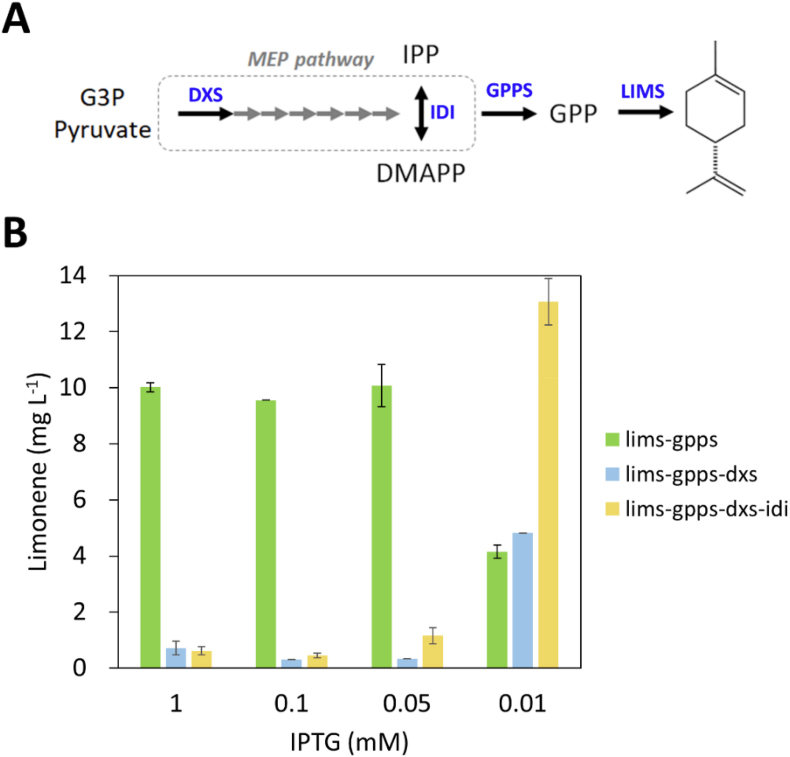
3.5. Engineering of the MEP pathway to increase limonene production
To increase the substrates for limonene synthesis, we engineered the MEP pathway in Synechococcus 2973 high limonene producer. It is known that 1-deoxy-d-xylulose 5-phosphate synthase (DXS) and isopentenyl diphosphate isomerase (IDI) catalyze the rate-limiting reactions in the MEP pathway (Fig. 5A). To overexpress the dxs and idi genes in the high limonene producer, a second operon driven by the lacUV5 promoter was cloned into the plasmid pSL3385. The constructs harboring the dxs and idi genes (Table 1) were then introduced into the high limonene producer. To test limonene production of the mutant strains, we induced the gene expression using different concentrations of IPTG (1 mM, 0.05 mM, and 0.01 mM), because the expression level of enzymes is expected to affect the titer of chemicals in cyanobacteria (Lin et al., 2020). As shown in Fig. 5B, limonene titer varied with IPTG concentrations. The control strain, expressing lims and gpps, produced a similar amount of limonene (10 mg L−1) when IPTG was higher than 0.05 mM, whereas the titer decreased 60% to 0.4 mg L−1 with the addition of 0.01 mM IPTG. The dxs-expressing strains decreased limonene production dramatically and exhibited reduced cell growth, suggesting that overexpression of dxs is toxic to Synechococcus 2973. Surprisingly, the limonene titer in the dxs and idi coexpression strains increased significantly to 13 mg L−1 under 0.01 mM IPTG conditions (Fig. 5B). These results suggest that the expression level of DXS and IDI is critical to improve limonene production in Synechococcus 2973.
Fig. 5.
IPTG titration to modulate the expression of dxs and idi in the MEP pathway in Synechococcus 2973 limonene-producing strains. (A) Limonene biosynthetic pathway. The overexpressed enzymes were highlighted in blue color. (B) Limonene production of strains with IPTG titration. The experiment was conducted in 1% CO2 and 250 μmol photons m−2 s−1 light for 2 days. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
In this study, we engineered the fast-growing cyanobacterium Synechococcus 2973 for limonene production. We identified a high limonene-producing strain with only 2 SNPs in the porin SomB and the GGPP synthase CrtE. By expressing the plasmid pSL3385 in the SomB (Y380H) and CrtE (R299Q) strains, we discovered that the R299Q mutation in CrtE led to a 2.5-fold improvement in limonene production. In cyanobacteria, CrtE is an essential enzyme that catalyzes the formation of GGPP, a precursor of photosynthetic pigments including chlorophyll and carotenoids.
The crystal structure of CrtE from the model cyanobacterium Synechococcus sp. PCC 7002 was solved recently (Feng et al., 2020). It is a homodimeric enzyme with 67% identity compared to the CrtE of Synechococcus 2973. The R299 residue is close to the c-terminal end of the enzyme (302 residues for Synechococcus 2973 CrtE), and is not involved in the elongation reaction from IPP and DMAPP (Feng et al., 2020). Presumably, the R299Q mutation decreases the enzymatic activity of CrtE, thus enabling the GPPS to convert more substrates to limonene production. Further characterization is needed to investigate the activity of CrtE R299Q. Additionally, the CrtE R299Q strain may be useful for increasing production of other terpenes which use GPP or FPP as substrates.
Modulation of GPPS expression varied the limonene production titers in Synechococcus 2973 (Fig. 4), suggesting that the enzyme level of GPPS is important to increase limonene production. In addition, this may indicate that a limited amount of fixed carbon was directed to the MEP pathway. Although the expression of dxs and idi genes in the MEP pathway slightly increased the limonene titer, our results demonstrated that the optimization of their enzyme levels is necessary for enhanced production (Fig. 5). Biosynthesis of limonene requires 9 enzymatic reactions from the central carbon metabolism (Fig. 5A). The entire biosynthesis pathway should be systematically engineered to identify the bottleneck reactions of limonene biosynthesis (Englund et al., 2018).
This study identified that the Synechococcus 2973 CrtE R299Q mutant improves limonene biosynthesis significantly, demonstrating that this fast-growing cyanobacterium is a promising host for photosynthetic terpene production. Importantly, the expression level of upstream enzymes including DXS, IDI, and GPPS are critical for enhanced the production. As a proof of concept, we demonstrated that the fast-growing Synechococcus 2973 with the genetic modification and metabolic engineering led to significant improvement in limonene productivity compared to other cyanobacterial strains. The growth rate of Synechococcus 2973 is 2–4 fold higher than other model cyanobacterial strains (Yu et al., 2015). However, our high producer showed over 8-fold improvement in limonene productivity (Fig. 2), suggesting the CrtE mutation and expression optimization of GPPS are critical for enhancing limonene production. Future work will focus on scaling up the production volume in a photobioreactor and optimizing the growth parameters to see if the productivity can be maintained or further improved. Finally, this work also demonstrates the value of using a fast-growing cyanobacterium for metabolic engineering purposes because the rapid growth property accelerates the design-build-test cycle for strain engineering.
Author contributions
P.C.L. performed experiments and analyzed data. All authors designed research, wrote the manuscript, and read and approved the final manuscript.
Declaration of competing interest
The authors declare no financial or commercial conflict of interest.
Acknowledgement
Funding to support this work was provided by the Office of Science (BER), U.S. Department of Energy (DE-SC0019386) to H.B.P. We thank Wilson Leung from the Department of Biology at Washington University for assisting in the variant calling analysis.
References
- Ajikumar P.K., Tyo K., Carlsen S., Mucha O., Phon T.H., Stephanopoulos G. Terpenoids: opportunities for biosynthesis of natural product drugs using engineered microorganisms. Mol. Pharm. 2008;5:167–190. doi: 10.1021/mp700151b. [DOI] [PubMed] [Google Scholar]
- Banerjee A., Sharkey T. Methylerythritol 4-phosphate (MEP) pathway metabolic regulation. Nat. Prod. Rep. 2014;31:1043–1055. doi: 10.1039/c3np70124g. [DOI] [PubMed] [Google Scholar]
- Burke C., Croteau R. Geranyl diphosphate synthase from Abies grandis: cDNA isolation, functional expression, and characterization. Arch. Biochem. Biophys. 2002;405:130–136. doi: 10.1016/s0003-9861(02)00335-1. [DOI] [PubMed] [Google Scholar]
- Colby S.M., Alonso W.R., Katahira E.J., McGarvey D.J., Croteau R. 4S-limonene synthase from the oil glands of spearmint (Mentha spicata). cDNA isolation, characterization, and bacterial expression of the catalytically active monoterpene cyclase. J. Biol. Chem. 1993;268:23016–23024. [PubMed] [Google Scholar]
- Davies F.K., Work V.H., Beliaev A.S., Posewitz M.C. Engineering limonene and bisabolene production in wild type and a glycogen-deficient mutant of Synechococcus sp. PCC 7002. Front. Bioeng. Biotechnol. 2014;2:21. doi: 10.3389/fbioe.2014.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhai J., Vepritskiy A., Muro-Pastor A.M., Flores E., Wolk C.P. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 1997;179:1998–2005. doi: 10.1128/jb.179.6.1998-2005.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund E., Andersen-Ranberg J., Miao R., Hamberger B., Lindberg P. Metabolic engineering of Synechocystis sp. PCC 6803 for production of the plant diterpenoid manoyl oxide. ACS Synth. Biol. 2015;4:1270–1278. doi: 10.1021/acssynbio.5b00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund E., Shabestary K., Hudson E.P., Lindberg P. Systematic overexpression study to find target enzymes enhancing production of terpenes in Synechocystis PCC 6803, using isoprene as a model compound. Metab. Eng. 2018;49:164–177. doi: 10.1016/j.ymben.2018.07.004. [DOI] [PubMed] [Google Scholar]
- Feng Y., Morgan R.M.L., Fraser P.D., Hellgardt K., Nixon P.J. Crystal structure of geranylgeranyl pyrophosphate synthase (CrtE) involved in cyanobacterial terpenoid biosynthesis. Front. Plant Sci. 2020;11:589. doi: 10.3389/fpls.2020.00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., 3rd, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Halfmann C., Gu L., Zhou R. Engineering cyanobacteria for the production of a cyclic hydrocarbon fuel from CO2 and H2O. Green Chem. 2014;16:3175–3185. [Google Scholar]
- Hansel A., Pattus F., Jürgens U.J., Tadros M.H. Cloning and characterization of the genes coding for two porins in the unicellular cyanobacterium Synechococcus PCC 6301. Biochim. Biophys. Acta Gene Struct. Expr. 1998;1399:31–39. doi: 10.1016/s0167-4781(98)00090-6. [DOI] [PubMed] [Google Scholar]
- Hinrichs A.S., Raney B.J., Speir M.L., Rhead B., Casper J., Karolchik D., Kuhn R.M., Rosenbloom K.R., Zweig A.S., Haussler D., Kent W.J. UCSC data integrator and variant annotation integrator. Bioinformatics. 2016;32:1430–1432. doi: 10.1093/bioinformatics/btv766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.H., Camsund D., Lindblad P., Heidorn T. Design and characterization of molecular tools for a Synthetic Biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res. 2010;38:2577–2593. doi: 10.1093/nar/gkq164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A., Takatani N., Ikeda K., Maeda S.I., Omata T. Removal of the product from the culture medium strongly enhances free fatty acid production by genetically engineered Synechococcus elongatus. Biotechnol. Biofuels. 2017;10:141. doi: 10.1186/s13068-017-0831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitts P.A., Church D.M., Thibaud-Nissen F., Choi J., Hem V., Sapojnikov V., Smith R.G., Tatusova T., Xiang C., Zherikov A., DiCuccio M., Murphy T.D., Pruitt K.D., Kimchi A. Assembly: a resource for assembled genomes at NCBI. Nucleic Acids Res. 2016;44:D73–D80. doi: 10.1093/nar/gkv1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyota H., Okuda Y., Ito M., Hirai M.Y., Ikeuchi M. Engineering of cyanobacteria for the photosynthetic production of limonene from CO2. J. Biotechnol. 2014;185:1–7. doi: 10.1016/j.jbiotec.2014.05.025. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25 doi: 10.1093/bioinformatics/btp324. 1754-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing S. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P.-C., Saha R., Zhang F., Pakrasi H.B. Metabolic engineering of the pentose phosphate pathway for enhanced limonene production in the cyanobacterium Synechocystis sp. PCC 6803. Sci. Rep. 2017;7:17503. doi: 10.1038/s41598-017-17831-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P.C., Pakrasi H.B. Engineering cyanobacteria for production of terpenoids. Planta. 2019;249:145–154. doi: 10.1007/s00425-018-3047-y. [DOI] [PubMed] [Google Scholar]
- Lin P.C., Zhang F., Pakrasi H.B. Enhanced production of sucrose in the fast-growing cyanobacterium Synechococcus elongatus UTEX 2973. Sci. Rep. 2020;10:390. doi: 10.1038/s41598-019-57319-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis A. Carbon partitioning in photosynthesis. Curr. Opin. Chem. Biol. 2013;17:453–456. doi: 10.1016/j.cbpa.2013.03.010. [DOI] [PubMed] [Google Scholar]
- Niu F.X., Lu Q., Bu Y.F., Liu J.Z. Metabolic engineering for the microbial production of isoprenoids: carotenoids and isoprenoid-based biofuels. Synth. Syst. Biotechnol. 2017;2:167–175. doi: 10.1016/j.synbio.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta-Yahya P.P., Ouellet M., Chan R., Mukhopadhyay A., Keasling J.D., Lee T.S. Identification and microbial production of a terpene-based advanced biofuel. Nat. Commun. 2011;2:483. doi: 10.1038/ncomms1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin B.E., Wetmore K.M., Price M.N., Diamond S., Shultzaberger R.K., Lowe L.C., Curtin G., Arkin A.P., Deutschbauer A., Golden S.S. The essential gene set of a photosynthetic organism. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E6634–E6643. doi: 10.1073/pnas.1519220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salis H.M., Mirsky E.A., Voigt C.A. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009;27:946. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer J., Pakrasi H.B. Cpf1 is a versatile tool for CRISPR genome editing across diverse species of cyanobacteria. Sci. Rep. 2016;6:39681. doi: 10.1038/srep39681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer J., Lin P.C., Chen H.Y., Pakrasi H.B. 2018. Adjustments to Photosystem Stoichiometry and Electron Transfer Proteins Are Key to the Remarkably Fast Growth of the Cyanobacterium Synechococcus elongatus UTEX 2973. mBio. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer J., Wendt K.E., Hendry J.I., Maranas C.D., Pakrasi H.B. Comparative genomics reveals the molecular determinants of rapid growth of the cyanobacterium Synechococcus elongatus UTEX 2973. Proc. Natl. Acad. Sci. U.S.A. 2018;115:E11761–E11770. doi: 10.1073/pnas.1814912115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Liu W., Xin C., Zheng Y., Cheng Y., Sun S., Li R., Zhu X.G., Dai S.Y., Rentzepis P.M., Yuan J.S. Enhanced limonene production in cyanobacteria reveals photosynthesis limitations. Proc. Natl. Acad. Sci. U.S.A. 2016;113:14225–14230. doi: 10.1073/pnas.1613340113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Liberton M., Cliften P.F., Head R.D., Jacobs J.M., Smith R.D., Koppenaal D.W., Brand J.J., Pakrasi H.B. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO(2) Sci. Rep. 2015;5:8132. doi: 10.1038/srep08132. [DOI] [PMC free article] [PubMed] [Google Scholar]