Bacterial flagellar filaments serve as an initial attachment point for many bacteriophages to bacteria. Some bacteria harbor numerous flagellin genes and are therefore able to generate flagellar filaments with complex compositions, which is thought to be important for evasion from bacteriophages.
KEYWORDS: Caulobacter crescentus, bacteria, bacteriophage, cryo-electron microscopy, cryo-electron tomography, flagellum, helical reconstruction, single particle
ABSTRACT
Caulobacter crescentus is a Gram-negative alphaproteobacterium that commonly lives in oligotrophic fresh- and saltwater environments. C. crescentus is a host to many bacteriophages, including ϕCbK and ϕCbK-like bacteriophages, which require interaction with the bacterial flagellum and pilus complexes during adsorption. It is commonly thought that the six paralogs of the flagellin gene present in C. crescentus are important for bacteriophage evasion. Here, we show that deletion of specific flagellins in C. crescentus can indeed attenuate ϕCbK adsorption efficiency, although no single deletion completely ablates ϕCbK adsorption. Thus, the bacteriophage ϕCbK likely recognizes a common motif among the six known flagellins in C. crescentus with various degrees of efficiency. Interestingly, we observe that most deletion strains still generate flagellar filaments, with the exception of a strain that contains only the most divergent flagellin, FljJ, or a strain that contains only FljN and FljO. To visualize the surface residues that are likely recognized by ϕCbK, we determined two high-resolution structures of the FljK filament, with and without an amino acid substitution that induces straightening of the filament. We observe posttranslational modifications on conserved surface threonine residues of FljK that are likely O-linked glycans. The possibility of interplay between these modifications and ϕCbK adsorption is discussed. We also determined the structure of a filament composed of a heterogeneous mixture of FljK and FljL, the final resolution of which was limited to approximately 4.6 Å. Altogether, this work builds a platform for future investigations of how phage ϕCbK infects C. crescentus at the molecular level.
IMPORTANCE Bacterial flagellar filaments serve as an initial attachment point for many bacteriophages to bacteria. Some bacteria harbor numerous flagellin genes and are therefore able to generate flagellar filaments with complex compositions, which is thought to be important for evasion from bacteriophages. This study characterizes the importance of the six flagellin genes in C. crescentus for infection by bacteriophage ϕCbK. We find that filaments containing the FljK flagellin are the preferred substrate for bacteriophage ϕCbK. We also present a high-resolution structure of a flagellar filament containing only the FljK flagellin, which provides a platform for future studies on determining how bacteriophage ϕCbK attaches to flagellar filaments at the molecular level.
INTRODUCTION
Bacterial motility relies upon signaling cascades that link chemical signals to mechanical responses in various bacterial appendages. One of the most dynamic appendages is the bacterial flagellum, which is assembled from over 60 structural and regulatory genes (1–4). The flagellum is a multiprotein complex with three distinct regions, the motor, hook, and filament (4, 5). The motor, also called the basal body, is located within the cell wall and generates torque via an electrochemical potential gradient, resulting in motor speeds of between 300 and 1,700 Hz (6–8). This mechanical signal is transferred to the filament by the flagellum’s universal joint called the hook, a tubular supercoiled helical structure (9). Motility is achieved by the flagellar filament, which acts as an Archimedes screw to propel the cell body (10).
The flagellar filament is a long cylindrical structure with a diameter of 12 to 15 nm that is composed of thousands of flagellin subunits and, as such, can span approximately 15 µm in length (see Fig. S1 in the supplemental material) (11). Individual flagellin subunits typically have masses between 25 kDa and 65 kDa and consist of multiple domains (12, 13). The two most conserved domains, denoted D0 and D1, are associated with flagellin polymerization and form the lumen of the flagellar filament. Specific regions of D1 are known recognition sites for the innate immune systems of mammals and plants (14–17). However, the two surface-exposed domains, D2 and D3, demonstrate high sequence variation (17, 18) and are known loci for a myriad of posttranslational modifications, such as O- and N-linked glycosylation (19–23). Mass spectroscopy studies of flagellar proteins show that Halobacterium and Clostridium flagellins are among the most heavily glycosylated species, where glycans account for 10% of their molecular weight (24). It was thought that only O-linked glycans were found in bacteria, but studies show that both O-linked and N-linked glycans are present in different bacterial species, similar to the homologous archaeal flagella (25–27). The D2 and D3 regions have also been shown to serve as an initial attachment point between flagellotropic bacteriophages and their hosts (1, 28–32). Thus, the periphery of flagellins serves as a hub for posttranslational modifications and protein-protein interactions that mediate bacteriophage predation.
The process of bacteriophage adsorption, where a bacteriophage attaches to and infects a cell, is dependent upon specific surface interactions and physical diffusion (Fig. 1A) (33). In some cases, phage tail fibers wrap directly around the flagellum as the bacteria swim (34–37). Previously, we characterized an alternative, headfirst, attachment mechanism of the bacteriophage ϕCbK to Caulobacter crescentus flagella and the requirement of pili for phage adsorption (1). In this mechanism, the phage head filament wraps around the flagellum, allowing for the localization of bacteriophages to the cell pole as the flagellar filament rotates (1). Next, the tail of ϕCbK binds to the pilus filament and then is retracted to the pilus portal for irreversible attachment (1). Recent work has underscored the importance of both the flagellar apparatus and pilus for ϕCbK infection, as transposon (Tn) insertions are enriched in related gene loci during TnSeq experiments on infected cultures (38). Interestingly, recent work also suggests that components of the flagellum play a functional role in regulating production of pili (2). Thus, formation of the first attachment site, the flagellum, is coupled to the formation of the ultimate ϕCbK receptor, the pilus filament, and portal. However, much remains unknown regarding the underlying adhesion mechanism to the flagellum and the structural contacts that endow specificity between phage ϕCbK and the C. crescentus flagellum. For example, which residues on the surface of the flagellum and individual flagellins directly interact with the ϕCbK head filament? Answering this question has been impeded by the lack of a high-resolution model of the C. crescentus flagellar filament.
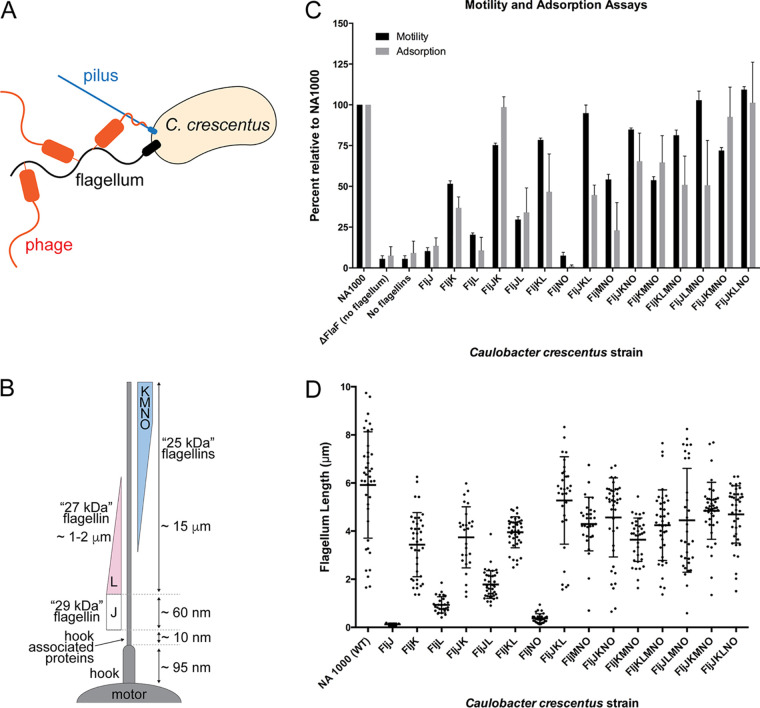
FIG 1.
Contribution of various flagellins to C. crescentus mobility and phage adsorption. (A) Schematic of phage adsorption to C. crescentus, which includes attachment to the flagella, subsequent interaction with the pilus portal, and genome ejection. (B) Schematic of known organization of flagellin proteins in the C. crescentus flagellum. (C) Cell motility and adsorption kinetics of phage ϕCbK to the wild type and flagellin mutants of C. crescentus. Strain motility and adsorption of phage ϕCbK were measured in triplicate and are normalized relative to the wild-type C. crescentus strain (NA1000). (D) Flagellum length measurements relative to wild-type NA1000 C. crescentus, as determined by negative-stain electron microscopy (Fig. 2). The points indicate individual measurements of each flagellum. Error bars indicate standard deviation, and the central cross bars represent the average length measurement for each strain. For clarity, the strains are named by the flagellin(s) they synthesize.
The number of flagellin genes present in bacterial genomes is highly variable. For example, some bacteria have a single flagellin gene, such as Escherichia coli and Bacillus subtilis, while others maintain multiple flagellin genes, including Helicobacter pylori, Rhizobium meliloti, Salmonella enterica serovar Typhimurium, and the Vibrionaceae (39). In the case of S. enterica serovar Typhimurium, phase variation is used to control which flagellin is incorporated into flagellar filaments, and as such, single filaments contain only one type of flagellin protein. However, almost 50% of bacterial species, including C. crescentus, assemble single flagellar filaments that are compositionally heterogeneous (40). The functional purpose of this heterogeneity is unknown, although it may provide a selective advantage for responding to environmental pressures, improved pathogenicity, or resistance to bacteriophage predation. The last has already been demonstrated in bacteria that exhibit phase variation (41–45).
C. crescentus has been a model for understanding the composition and structure of bacterial flagella (46–48). The C. crescentus flagellar filament is composed of six independent flagellin subunits (49), all of which are likely glycosylated through O-linked addition of pseudaminic acid (23). Immuno-electron microscopy (immuno-EM) labeling of C. crescentus flagellar filaments suggests that individual subunits are differentially incorporated along the length of the filament (50–52) (Fig. 1B). The flagellins are expressed from two separate loci on the C. crescentus genome. The alpha locus contains the fljJ (29-kDa flagellin), fljK (25-kDa flagellin), and fljL (27-kDa flagellin) genes as well as the two flagellin translation regulators, flaF and flbT. The beta locus includes the fljM, fljN, and fljO genes, all of which encode 25-kDa flagellins (40, 49). Previously, it was determined that alterations to the six flagellins incorporated into the C. crescentus flagellum could reduce but not enhance motility (40).
The small size of flagellin monomers and high degree of symmetry in assembled filaments provide challenges for high-resolution structure determination by contemporary cryo-electron microscopy and helical reconstruction methods (53–55). Typically, it is necessary to determine an initial estimate of helical symmetry by de novo indexing. However, this requires long and straight filaments, or two-dimensional classes of smaller “boxes” with sufficient signal in their amplitude spectra for layer line indexing. When it is not possible to index a helical assembly de novo, initial estimates of helical symmetry can be obtained from homologous structures (53).
The architecture of flagella is well conserved across bacterial species. Most contain 11-start helices with protofilaments that run nearly parallel to the helical axis (56–58). A few 11-start flagellin structures have been determined, including flagellar filaments from B. subtilis, S. enterica, Pseudomonas aeruginosa, and Kurthia sp. strain 11kri321 (11, 57–59). In the case of the B. subtilis, S. enterica, and P. aeruginosa reconstructions, single residue substitutions were used to straighten the filaments by locking the 11-start protofilament into either right (R-type)- or left (L-type)-handedness conformations, which yielded a uniform filament onto which helical symmetry could be more effectively determined and applied during structure determination (58, 59). In Kurthia, straightening substitutions were not employed to intentionally lock the filament into a particular R-type or L-type handedness (57).
We have previously demonstrated that reduced motility of C. crescentus leads to decreased adsorption of phage ϕCbK (1, 6), as similarly observed with E. coli phage χ (4, 31), B. subtilis phage PBS1 (59, 60), and Agrobacterium tumefaciens phages GS2 and GS6 (31, 61). Here, we characterize the effects of flagellin composition in C. crescentus on phage ϕCbK adsorption. These results were achieved by combining motility and bacteriophage adsorption assays with negative-stain transmission electron microscopy (TEM), cryo-electron microscopy (cryo-EM), and cryo-electron tomography (cryo-ET) of C. crescentus mutants. We show that no one flagellin protein is essential for phage adsorption, although a preference for some flagellins is apparent. As such, bacteriophage ϕCbK likely recognizes a common motif among the six C. crescentus flagellins. We also present the first high-resolution helical reconstructions for the FljK flagellar filament from C. crescentus and a lower-resolution model of a flagellar filament containing more than one flagellin.
RESULTS
Since C. crescentus harbors six distinct flagellins, we sought to deconvolute the effect of motility, filament composition, and structure on ϕCbK adsorption by using a series of C. crescentus mutant strains that lacked one, several, or all but one flagellin (Table 1). For clarity, strains will be referred to by the flagellin(s) that they synthesize.
TABLE 1.
Caulobacter crescentus strains used in this study; all mutant strains are derived from NA1000
| Strain | Genotype | Description | Reference no. or source |
|---|---|---|---|
| NA1000 | Wild type | Synchronizable C. crescentus wild-type strain | 82 |
| LS1218 | ΔflaF | Deletion of the flagellin translation regulator | 83 |
| TPA2357 | ΔfljJ ΔfljL ΔfljM-fljO fljK T103C | Strain carrying a genomic allele of fljK encoding T103C | Unpublished data |
| TPA2357 | ΔfljJ ΔfljK ΔfljL ΔfljM-fljO | Deletion of all C. crescentus flagellins | 40 |
| TPA1299 | ΔfljM-fljNO | Deletion of flagellins M, N, and O | 40 |
| TPA970 | ΔfljJ ΔfljK ΔfljL | Deletion of flagellins J, K, and L | 40 |
| TPA2354 | ΔfljK ΔfljL ΔfljM-fljO | Only flagellin J is present | 40 |
| TPA2353 | ΔfljJ ΔfljL ΔfljM-fljO | Only flagellin K is present | 40 |
| TPA2346 | ΔfljJ ΔfljK ΔfljM-fljO | Only flagellin L is present | 40 |
| TPA2352 | ΔfljK ΔfljM-fljO | Only flagellins J and L are present | 40 |
| TPA2356 | ΔfljL ΔfljM-fljO | Only flagellins J and K are present | 40 |
| TPA2344 | ΔfljJ ΔfljM-fljO | Only flagellins K and L are present | 40 |
| TPA1140 | ΔfljJ ΔfljK ΔfljL ΔfljM | Only flagellins N and O are present | 40 |
| TPA2234 | ΔfljK | Deletion of the 25-kDa flagellin K | 40 |
| TPA916 | ΔfljL | Deletion of the 27-kDa flagellin L | 40 |
| TPA663 | ΔfljJ | Deletion of the 29-kDa flagellin J | 40 |
| TPA541 | ΔfljM | Deletion of the 25-kDa flagellin M | 40 |
| TPA934 | ΔfljJ ΔfljL | Only the 25-kDa flagellins are present | 40 |
| TPA933 | ΔfljJ ΔfljK | Deletion of flagellins J and K | 40 |
| CS3 | ΔfljJ ΔfljK ΔfljL ΔfljM-fljO pCS1 | TPA2357 carrying pCS1 | This study |
| CS12 | ΔfljJ ΔfljK ΔfljL ΔfljM-fljO pCS12 | TPA2357 carrying pCS12 | This study |
Phage adsorption to flagellin mutants of C. crescentus.
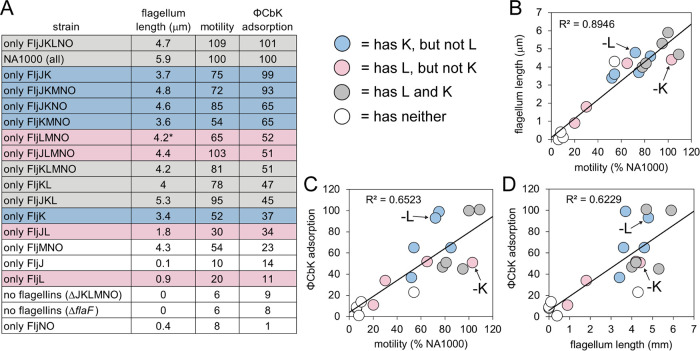
C. crescentus strains lacking flagellar filaments altogether (ΔflaF and ΔfljJKLMNO) exhibit reduced motility and adsorption rates of ϕCbK (Fig. 1C; see Fig. S2 in the supplemental material). We also find that motility and filament length (Fig. 2 and Fig. 3A and B) are strongly correlated. These two results, which agree with previous observations (1, 40, 62), serve as an internal control for the analyses conducted here. Interestingly, we observe weaker correlations between motility and adsorption and also between filament length and adsorption, suggesting that motility or filament length alone does not exclusively drive adsorption (Fig. 1C and D), and as such, phage ϕCbK is likely sensitive to composition of the flagellar filament.
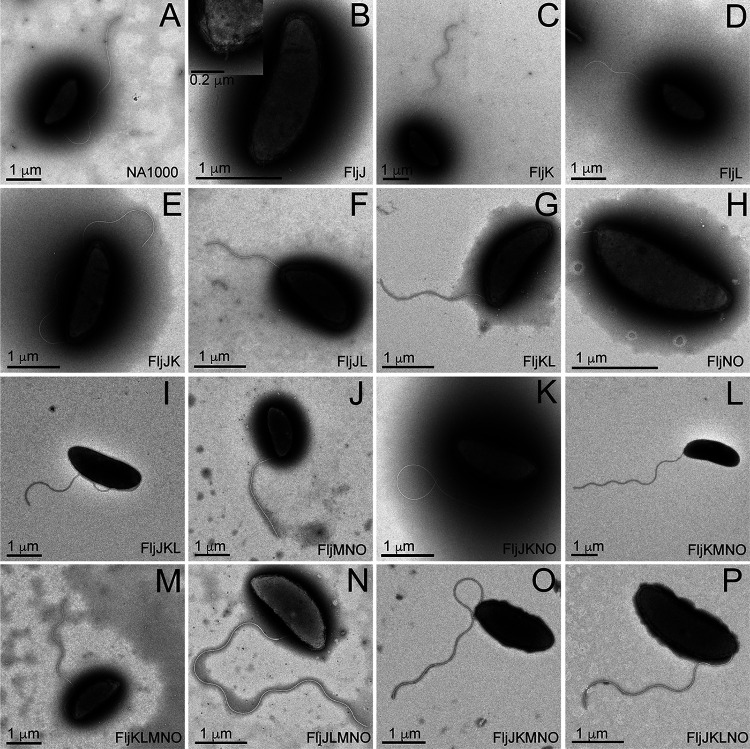
FIG 2.
Transmission electron microscopy images of negatively stained C. crescentus strains from Fig. 1 using 1% phosphotungstic acid, showing that most strains produce flagella. Only FljJ (B) and FljNO (H) presented a severely truncated flagellum that lacked an appreciable flagellar filament. Scale bars are not identical in size among the figure panels.
FIG 3.
Summary of functional assays performed. For clarity, C. crescentus strains are designated by the flagellin(s) they synthesize. (A) Data for motility and adsorption are normalized to that of wild-type C. crescentus strain NA1000. (B to D) Plots showing correlation between observed phenotypes. Strains are colored as in panel A.
In all strains tested, expression of a single flagellin protein reduced ϕCbK adsorption. Adsorption to FljJ and FljL was extremely low at ∼10% of wild type (Fig. 1C and Fig. 3A). This reduction was accompanied by reduced motility, as well as relatively short flagellum length (Fig. 1D and Fig. 2B and D). In the case of FljK, ϕCbK adsorption was reduced to ∼35%, and motility of the strain was ∼50% of wild type, although flagellum length remained similar to that of wild type. Interestingly, the FljJ strain lacked filaments altogether (Fig. 2B).
To further examine the requirements for successful phage adsorption to the C. crescentus flagellum, we measured ϕCbK adsorption to a series of four strains that expressed only two flagellins. The FljJK and FljKL strains exhibited the highest motility, longest filaments, and greatest ϕCbK adsorption. Interestingly, FljJK displayed near-wild-type levels of ϕCbK adsorption that could be attenuated approximately 2-fold by replacement of FljJ with FljL (Fig. 3A). Phage ϕCbK adsorption was significantly reduced for FljJL (34%) and FljNO (1%), which tracked strongly with reduced length and motility.
Increasing in complexity, two strains were examined that expressed three flagellins, FljJKL and FljMNO, which correspond to the alpha or beta locus flagellin genes, respectively (40, 49). The beta locus flagellin strain FljMNO uniformly exhibited decreased motility, filament length, and ϕCbK adsorption relative to wild type. The phenotype of the FljJKL strain, however, yielded two interesting observations. First, deletion of beta locus flagellins confers a minimal effect on speed, motility, and filament length relative to wild type. Second, comparison of FljJKL and FljJK shows that including the FljL flagellin attenuates ϕCbK adsorption 2-fold relative to the FljJK strain, despite other properties nearly matching wild type (Fig. 1C and D and Fig. 3A).
In light of the observations that FljJ and FljK seem important for adsorption, the ϕCbK adsorption rate of the ΔfljJ ΔfljK strain (or FljLMNO) was tested (Fig. S2 and Fig. 3A). The adsorption profile of FljLMNO was reduced relative to wild type but not reduced to the same levels as the nonflagellated strain, thus showing that FljJ and FljK are important but not essential for adsorption.
We then examined ϕCbK adsorption to FljKMNO, which contains only the 25-kDa flagellins (Fig. 1C). Motility of FljKMNO was ∼50% that of wild type, and ϕCbK adsorption was 65% that of wild type (Fig. 1C and Fig. 3A). Simultaneously, we studied the profiles for FljJKNO and determined that even though the strain’s motility was higher, at 85%, the adsorption profile was equivalent to FljKMNO (65%).
To quantitatively determine the relationship between the removal of single flagellins and ϕCbK adsorption, we examined FljKLMNO, FljJLMNO, FljJKMNO, and FljJKLNO. Removal of FljN or FljO as single flagellins was not possible, as previously noted by Faulds-Pain (40). Omission of FljJ reduced motility and adsorption to ∼80% and 50% that of wild type, respectively. Omission of FljK caused a nominal effect on filament length and motility but a significant reduction in phage adsorption (Fig. 1C and D and Fig. 3A). Omission of FljL or FljM exerted only minor effects on function and morphology of flagella.
The impact of FljK to ϕCbK adsorption is especially apparent when we compared the slope values from the ϕCbK adsorption kinetics plots for all four strains expressing only five flagellins and wild-type C. crescentus (Fig. S2). The slope for infected wild-type cells is −0.0142; the slopes for the four strains, in increasing slope steepness, are as follows: FljJLMNO, −0.0053; FljKLMNO, −0.0119; FljJKLNO, −0.0183; and FljJKMNO, −0.0189. These data illustrate that ϕCbK adsorption is generally lower in the presence of FljL and higher in the presence of FljK, with the noted exception of the wild type.
The results of Driks et al. indicate that the intermediate-size (27-kDa) flagellin (FljL) may serve as an adapter between FljJ and the subsequent assembly of the smaller (25-kDa) flagellin subunits (Fig. 1B). However, our imaging results demonstrate that mutants with deletions of either FljJ or FljL produce flagellar filaments of lengths comparable to that of wild type (Fig. 1D). As such, inclusion of FljJ or FljL is not essential for filament formation despite their localization at the base of the assembled wild-type filament.
Tomographic reconstruction of C. crescentus strains during ϕCbK adsorption.
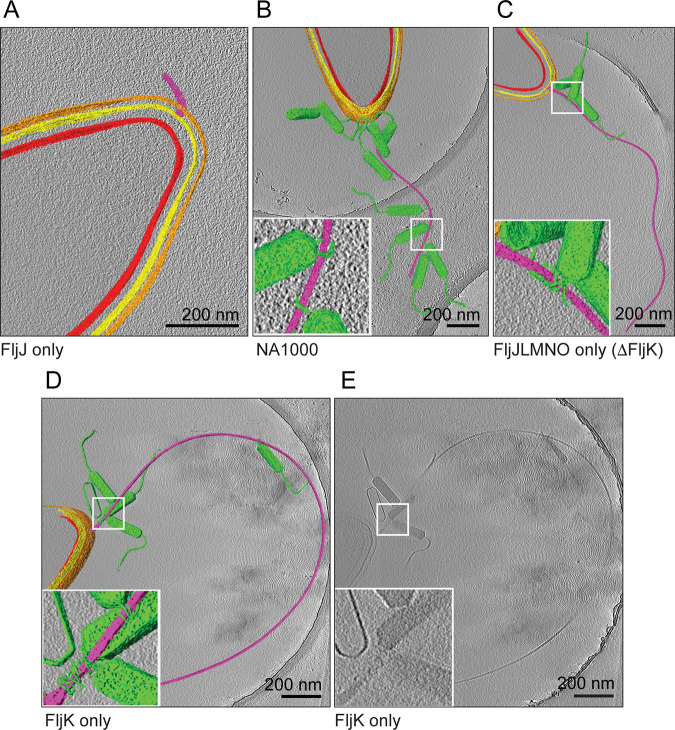
The adsorption of ϕCbK phage along the flagellar filament of the flagellin deletion strains was examined by cryo-ET. Using negative-stain TEM, we observed intact flagella for all strains except FljJ (Fig. 2B) and FljNO (Fig. 2H), which only had flagella stubs that the authors attribute to hooks combined with severely shortened flagellar filaments. Using cryo-ET, we resolved intact flagella for all tested deletion strains except for FljJ (Fig. 4A) and FljNO (data not shown), the former of which had flagella stubs and no ϕCbK phage adsorbed headfirst along the truncated flagellum length. In deletion strains with intact flagella, it was noted that the ϕCbK phage was adsorbed along the length of the flagella and that the overall flagellar structure was comparable to that of flagella of wild-type cells (Fig. 4). It was also determined that a greater number of ϕCbK phages were adsorbed along the flagellum of the FljJK strain (data not shown) than the FljJLMNO mutant, which correlated with the results from the phage adsorption assays. Cryo-ET imaging of ϕCbK-infected strain FljJLMNO (Fig. 4C) revealed that the head filament of ϕCbK could still interact with the flagellum in the absence of FljK, albeit at lower abundance, as expected from our adsorption assays.
FIG 4.
Averaged slices merged with segmentations through three-dimensional tomographic reconstructions of representative ϕCbK-infected C. crescentus cells, selected from all strains examined. Segmentation was performed in Amira, coloring the inner membrane red, outer membrane yellow, S-layer orange, flagellum purple, and phage particles green. Scale bars are 200 nm. (A) C. crescentus FljJ assembles only a shortened flagellum. (B, C, and D) Phages were observed either adsorbed along the length of the flagellum or attached to the cell poles of strains investigated. Insets highlight the ϕCbK head filament wrapped around the C. crescentus flagellum. (E) Unsegmented tomographic slice of FljK for comparison.
Driks et al. (50) proposed that the largest (29-kDa) flagellin subunit (FljJ) is used to initiate the assembly of flagellin subunits in C. crescentus by forming a short flagellar segment to which other flagellins assemble (Fig. 1B). Our tomographic reconstructions of the FljJ-only mutant comport with this model, and we resolved the length of the corresponding flagellar filament and hook to be between 120 nm and 160 nm, respectively (Fig. 4A and Movie S1). Interestingly, the FljJ-only mutant supports ϕCbK adsorption, albeit at a greatly reduced level compared with the other strains (Fig. 1C, Fig. 3A, and Fig. S2). The low-level ϕCbK adsorption is likely due to ϕCbK tail association with the C. crescentus pilus filament and the retraction of the pilus toward the cell pole and the pilus portal, which is the site of irreversible ϕCbK attachment (1, 63, 64).
Helical reconstruction of a C. crescentus filament containing only FljK.
The above data suggest that FljK is an optimal, but not exclusive, substrate for recruitment of ϕCbK. This is supported by the observation that it is possible to recover approximately 100% of adsorption with a minimal complement of flagellins, so long as at least one is FljK (see strain FljJK, Fig. 3). We therefore sought to determine a high-resolution structure of FljK in order to build a platform for subsequent interrogation of the filament-ϕCbK interaction mechanism.
Single-particle helical reconstruction methods are a powerful tool for high-resolution structure determination of filaments, but most successfully determined structures of homologous flagellins to date have employed straightening mutations to facilitate computational aspects of the reconstruction process (11, 58, 59). We therefore utilized error-prone PCR to mutagenize a plasmid (pCS1) encoding a T103C variant of FljK that is amenable to fluorescence and cryo-correlative light and electron microscopy (cryo-CLEM) imaging via maleimide dye labeling of the cysteine residues (data not shown) (65). Several isolates were defective for motility, and both fluorescence and negative-stain TEM imaging were used to determine if such strains indeed harbored straightened filaments (data not shown). One such strain, encoding FljK T103C N130S on plasmid pCS12 (Table 2), exhibited straightened filaments which were used for high-resolution cryo-TEM imaging (Fig. 5A).
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference no. or source |
|---|---|---|
| pMR10 | Broad-host-range cloning vector, Kanr, low copy no.a | 83 |
| pCS1 | Construct with fljK T103C amplified from YB6528 with ∼500 bp of upstream sequence cloned into the BamHI and HindIII sites of pMR10 | This study |
| pCS12 | Construct with fljK T103C N130S; derived from error-prone PCR mutagenesis to pCS1 | This study |
Kanr, resistance to kanamycin.
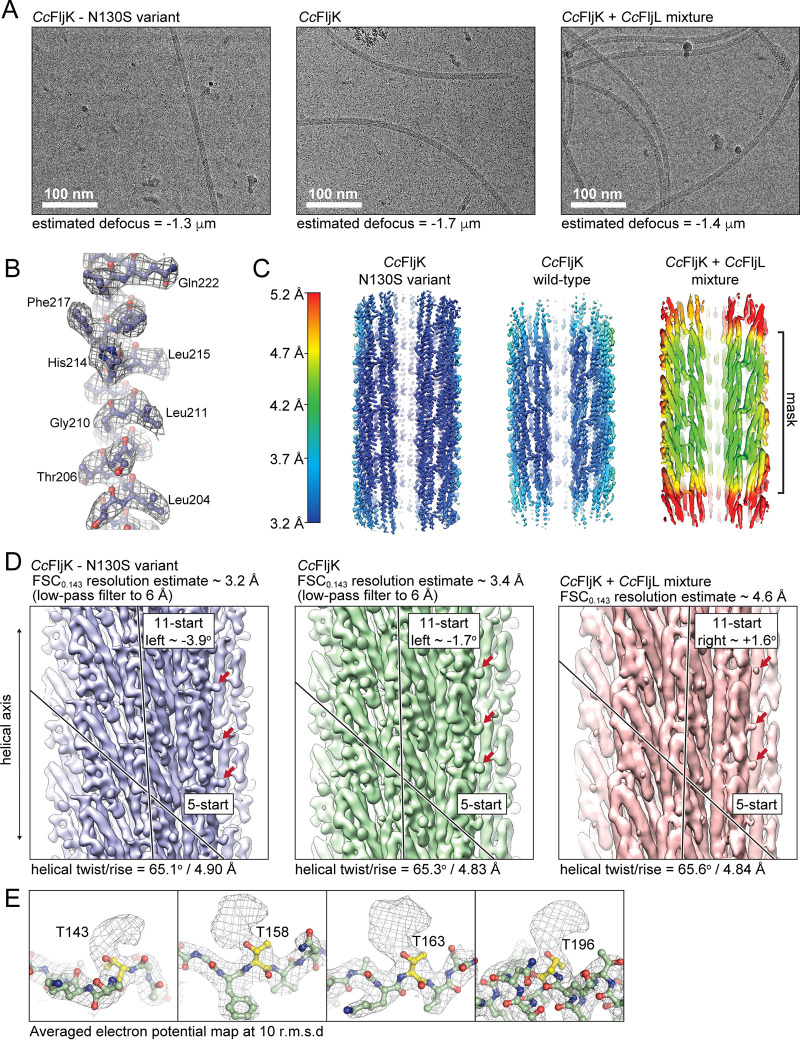
FIG 5.
High-resolution imaging and reconstruction of the FljK flagellar filament. (A) Exemplar micrographs of straightened FljK, wild-type FljK, and a heterogeneous mixture of FljK and FljL. (B) Example of electron potential map in which main-chain connectivity and side chains are clearly visible. (C) Local resolution plots of the three reconstructions generated in Relion 3.1 and Chimera (75, 81). The straightened FljK flagellin exhibits the highest resolution. The heterogeneous mixture of FljK and FljL (FljK/L) does not yield a high-enough resolution map for modeling of amino acid side chains. (D) Comparison of the three reconstructions generated in this work. The single-component FljK reconstructions are low-pass filtered to allow visualization of 5- and 11-start protofilaments that are a hallmark of related flagellins (11, 57–59). Note that the heterogeneous FljK/L reconstruction lacks definition of amino acid side chains even in the absence of low-pass filtering. Interestingly, however, the FljK/L reconstruction shows nodules (denoted by arrows) that correspond to apparent glycosylation loci. (E) Numerous posttranslational modifications are clearly visible on surface-exposed threonine residues that are likely O-linked glycans in the FljK-only reconstructions.
We tested the effect of the “straightened” FljK filament on function by subjecting the FljK T103C N130S strain to motility and ϕCbK adsorption assays. The adsorption profile (Fig. S3) and motility were similar to the strains lacking flagellins altogether. As such, straightening FljK very likely compromises motility and, by extension, adsorption. The straight variant of FljK carries an amino acid change (T103C) that allows maleimide staining of the flagellum. We also tested the effects of this change in isolation to rule out possible confounding effects of T103C when used for labeling. FljK T103C had a similar adsorption profile and motility to FljK alone, and as such, we conclude that the T103C substitution has little effect on motility or adsorption.
In lieu of direct helical indexing, we used initial estimates of helical symmetry obtained from high-resolution structures of homologous flagellins (11, 57–59) and a featureless cylinder as an initial model (Fig. S4). After reconstruction and refinement of helical symmetry parameters in Relion, we were able to recover a map that contained a clear definition of amino acid side chains (Fig. 5B), which was utilized as a cross-validation metric for the inherently biased approach used here for acquisition of the initial helical symmetry parameters. We note that the final helical symmetry obtained here is much closer to that of hydrated homologous flagellins (rise/twist, ∼4.8 Å/65°) than that of dehydrated C. crescentus flagellins in negative-stain TEM (rise/twist, ∼3.8 Å/65°) (48).
The straightened variant of FljK exhibited a “locked” left-handed 11-start helix with an approximate intersubunit rotation of −3.9° (Fig. 5D). Subsequent to reconstruction of this straightened variant of FljK, we attempted helical reconstruction of an unmodified filament that was not “straightened” by an amino acid substitution and therefore not presumably “locked” into either a right- or left-handed 11-start conformer. The resulting reconstruction was of slightly lower resolution and overall map quality relative to the straightened variant of FljK, which is likely due to conformational heterogeneity that is present in the nonstraightened filament. However, it was still possible to clearly delineate amino acid side chains in the majority of the reconstructed map. Interestingly, the wild-type variant of FljK exhibited a left-handed 11-start helix with an approximate rotation of −1.7° (Fig. 5D). The resulting structures of the FljK flagellin monomers and assembled filaments conform to the known architecture of other bacterial flagellins and flagellar filaments (11, 57–59).
We note the appearance of numerous apparent posttranslational modifications that decorate the periphery of the FljK filament (Fig. 5E). These modifications are all localized to conserved threonine residues and are likely O-linked glycans containing pseudaminic acid, consistent with a recent report that the FlmG protein glycosylates all six flagellins in C. crescentus (23, 25–27, 66). As the periphery of the filament has the poorest map quality, we were unable to confirm the exact identity of the glycans nor model them into the atomic models generated here.
Helical reconstruction of a compositionally heterogeneous filament.
From our functional data, we find that adsorption of phage ϕCbK (Fig. 1C and Fig. 3A, C, and D) is sensitive to the presence or absence of specific flagellins. For example, comparison of the FljJKL and FljJK strains shows that inclusion of FljL reduces phage ϕCbK adsorption approximately 2-fold while exerting a small improvement in motility. We therefore sought to understand what differences may exist at the structural level when FljL is expressed with FljK. Using the same processing pipeline as above, we obtained a helical reconstruction of FljKL flagella that is limited to a final resolution of 4.6 Å. The reduced resolution is attributed to the confounding effects of compositional and conformational heterogeneity, and the reconstruction should be interpreted with caution, as it was not indexed de novo nor does the final map exhibit side chain densities to serve as cross-validation for the reconstruction pipeline used here. However, the low-resolution map still provides some interesting observations. First, the flagellin fold, with terminal D0 domains and intervening D1 domains, is apparent in the reconstruction (Fig. S1 and Fig. 5D), noting that the starting model of a featureless cylinder contained no such information. As such, inclusion of FljL does not substantially alter the overall fold or arrangement of monomers in the resulting filament. Second, there are clear nodules on the periphery of the lower-resolution FljK and FljL map that map to the same locations as the apparent posttranslational modifications in the FljK reconstructions (Fig. 5E). Third, the mixed FljK and FljL filament exhibits an 11-start protofilament with a rotation of 1.6°, which is distinct from those of the compositionally homogeneous FljK reconstructions (Fig. 5D).
Interprotein contacts in the FljK flagellum.
Comparison of the straightened and nonstraightened reconstructions of FljK shows that the N130S substitution exerts a minimal effect on the overall structure of FljK, with an all-atom root mean square deviation (RMSD) of 0.7 Å between the two chains (Fig. 6A). However, there are local differences at protein-protein interfaces that alter the overall organization of the filament. Substitution of N130 for serine disrupts a hydrogen-bonding contact between the side chain of N130 and the carbonyl oxygen of K126 (Fig. 6B and C). This, in turn, disrupts a series of hydrogen bonds between adjacent strands in the 5-start protofilament, resulting in a vertical displacement of approximately 2.5 Å along the helical axis. As such, protein-protein contacts along the exterior of the filament can induce structural changes that propagate to the rest of the helical assembly and change the rotation of the 11-start protofilament. This is noteworthy because phage ϕCbK interacts with the periphery of the filament and could potentially alter the structure of the filament after initial association.
FIG 6.
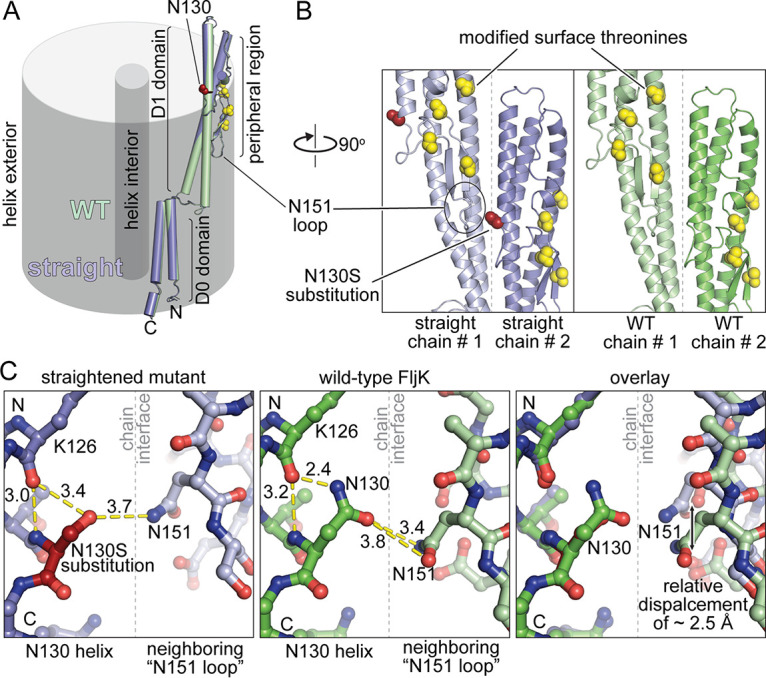
Comparison of local structure in straightened and nonstraightened FljK filaments. (A) The two FljK variants are highly similar with an all-atom RMSD of 0.7 Å. The N130S straightening substitution is depicted by red spheres, and posttranslationally modified surface threonine residues are depicted by yellow spheres. (B) The N130S straightening substitution is located at a protein-protein interface along the surface of the FljK filament. (C) The N130S straightening substitution perturbs a main-chain hydrogen bond with the alpha-helical carbonyl oxygen of K126 and alters the local hydrogen bonding network at a protein-protein interface, resulting in displacement of the neighboring “N151 loop” by approximately 2.5 Å.
DISCUSSION
Assembly of a multiflagellin filament.
We have shown that flagellar filament formation in C. crescentus is remarkably tolerant of deletions to specific flagellins. For example, all strains imaged here exhibited flagella, with the exception of the FljJ and FljNO strains (Fig. 2). As such, it appears that alternate arrangements may be possible for filament assembly. Filament assembly in C. crescentus is therefore likely a dynamic process with respect to flagellin incorporation, with selection criteria for flagellin insertion that are neither inflexible nor well understood.
We also note a trend upon which inclusion of more types of flagellins generally increases “function” of the C. crescentus filament with respect to length and motility (Fig. 3B). The absence of “all or nothing” observations with respect to these properties suggests that the C. crescentus flagellum is an exquisitely complex biophysical apparatus that can exploit compositional heterogeneity to enhance function.
Flagellin complement significantly affects ϕCbK adsorption.
Another possible advantage of preserving multiple flagellin paralogs in a bacterial genome is for evasion of flagellotropic phages. For example, a duplicate flagellin may acquire adaptations that attenuate attachment of phage to flagellins, effectively blocking or inhibiting one step of the adsorption process. The alpha locus flagellin FljL appears to play such a role. This viewpoint is supported by two observations. First, a minimal flagellum containing only FljJ and FljK exhibits wild-type levels of adsorption, and second, addition of FljL to this minimal flagellum attenuates adsorption approximately 2-fold while simultaneously increasing flagellum function (as evinced by length and motility). We have focused our initial high-resolution structure determination efforts on simplified filaments containing FljK and FljL due to their opposing effects on motility and absorption.
Among the minimal flagellin strains tested here, we find that retention of only FljJ and FljK can support near-wild-type levels of motility and adsorption. We therefore tested the cognate deletion strain, which contains all flagellins except FljJ and FljK (or FljLMNO). Interestingly, the FljLMNO strain retains nearly half the adsorption of wild type. Thus, phage ϕCbK exhibits a remarkable degree of flexibility for adsorption to all flagellins, although the importance of FljJ and FljK is evinced by the need to include four other flagellins to achieve half the adsorption of the two-flagellin FljJK strain.
At this point, the role of the beta locus flagellins (FljM, FljN, and FljO) remains unclear, as our data exhibit no clear pattern regarding the function of these flagellins. It is possible that the importance of these flagellins may lie in a function not tested here, for example, functional redundancy, predation by other bacteriophages, or motility in environments that are not captured by our assay conditions.
Interplay between the flagellum and pilus.
Recent work has demonstrated that the expression of the C. crescentus flagellum also coregulates expression of the pilus (2). For example, deletions of the flagellum motor protein MotB, the flagellin protein FljK, and the hook protein FlgE resulted in fewer pili (2). Interestingly, these deletions also decreased ϕCbK adsorption, as expected from the essential nature of the pilus filament and portal for ϕCbK adsorption (1). Thus, the production of pili and flagella is not only central to ϕCbK adsorption but also coupled. Another interesting aspect to the interplay between flagella and pili is that translation of FljK, which, from our work, is important for adsorption and is likely regulated by FlbT interaction with fljK mRNA, and a similar regulatory mechanism is suggested to exist for pilA mRNA (2, 67, 68). It is unclear if other flagellin mRNAs are regulated by FlbT or how the other flagellins may, in turn, affect PilA translation. This work builds a foundation for better understanding the interplay between the flagellum, pilus, and phage ϕCbK by establishing the individual contribution of flagellin subunits on ϕCbK adsorption. It will be interesting to test the effect of different flagellar complements on pilus filament (PilA) expression.
Comparison to other flagellins of known structure.
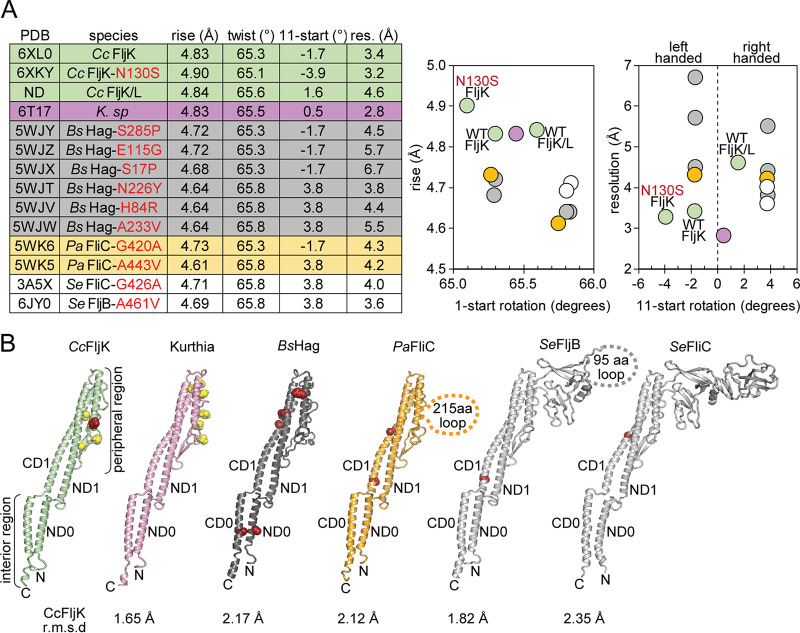
Comparison of our FljK reconstruction to that of other 11-start bacterial flagellins provides several interesting observations. First, helical symmetry is remarkably well conserved, with an average variance in rise and twist below 10% (Fig. 7A). These small differences in helical symmetry, however, can exert a large effect on the handedness of the 11-start protofilament. Until recently, all determined structures of 11-start flagellins fell into two classes, left-handed structures with an 11-start rotation of approximately −1.7° or right-handed structures with an 11-start rotation of approximately +3.8° (11, 57–59). These two dominant classes of 11-start protofilaments strongly influenced the field’s view of filament structure, where toggling between two defined states was thought to play a functional role in bacterial motility (4). Recently, a high-resolution reconstruction of the flagellar filament from Kurthia sp. found the corresponding 11-start protofilament to be nearly parallel to the helical axis with a rotation of approximately +0.5°, which could be considered an outlier from all other successful helical reconstructions to date (11, 57–59). Here, we show that two of our reconstructions also fail to conform to the −1.7° or +3.8° precedent for 11-start rotations, and as such, there is a previously unappreciated degree of large-scale conformations that flagellar filaments can accommodate, perhaps representing untapped structural diversity in flagellar filament organization (Fig. 7A).
FIG 7.
Comparison to other 11-start bacterial flagellins. (A) All 11-start bacterial flagellins of known structure have similar helical symmetry values. Neighboring plots are constructed to highlight differences in symmetry. (B) Structural comparison of C. crescentus FljK to other bacterial flagellins. Known straightening substitutions are depicted by red spheres, and posttranslationally modified residues are depicted by yellow spheres. The N130S straightening substitution in FljK is a relative outlier in that it perturbs a protein-protein contact at the surface of the flagellin, while the majority of other straightening substitutions typically alter protein-protein contacts in the core of the flagellin.
Despite these differences, we find that flagellins are remarkably well conserved at the level of secondary structure within the interior of the filament (Fig. 7B). For example, the much larger D2 and D3 domains that are present in Pseudomonas and Salmonella flagellins do not significantly alter the structure of the D0 and D1 regions, which are quite similar to those in the Caulobacter, Kurthia, and Bacillus flagellins (Fig. 7B). However, the overall architecture of the filament is clearly sensitive to alterations in the peripheral regions, as our “straightening” N130S substitution is located on the surface of the filament (Fig. 6B). Furthermore, helical assemblies can be so exquisitely sensitive to small changes in local structure that it is possible to completely rearrange the entire helical superstructure through substitution of only a single amino acid (69). We therefore propose that the surface threonine-linked modifications play a functional role in altering the overall architecture of the filament, in addition to being critical for effective secretion and assembly (23). The viewpoint is underscored by the lack of visible flagella and loss of motility in strains that are deficient for biosynthesis of pseudaminic acid, which is transferred to all six flagellins in C. crescentus by the FlmG protein (23). The role of threonine-linked modifications with respect to phage adsorption and interaction represents an interesting topic for future research. It would be worthwhile to test whether different O-linked glycosylations, as proposed by Ardissone et al. (23), would allow filament assembly but alter phage adsorption.
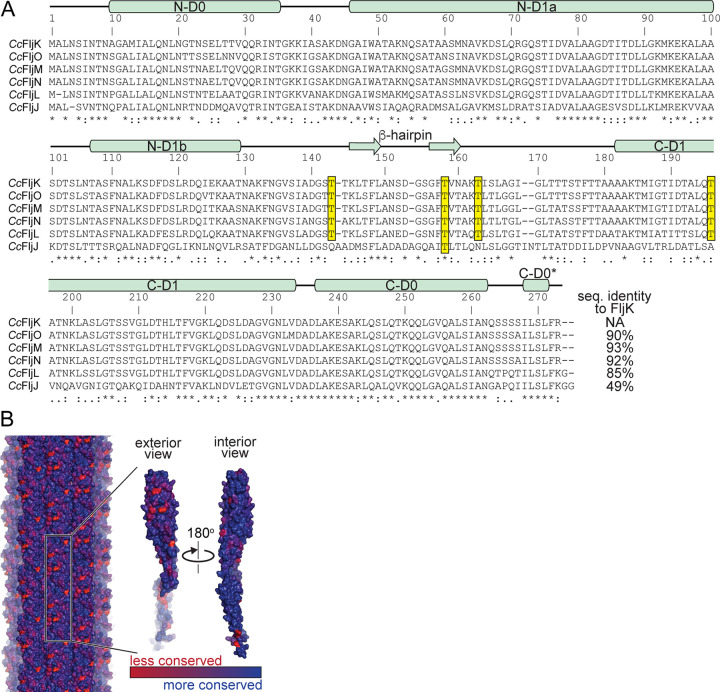
Sequence analysis of Caulobacter flagellins.
Our high-resolution reconstructions of FljK allowed us to perform a more informed sequence analysis of C. crescentus flagellins than was previously possible (Fig. 8A). Interestingly, the surface-exposed threonine residues that harbor posttranslational modifications in our reconstruction of FljK (Fig. 5E) are conserved in the beta locus flagellins (FljM, FljN, and FljO) and also FljL, but are almost entirely absent in the most divergent flagellin, FljJ. Because nearly all flagellins may be modified, this suggests that the mechanism by which FljK enhances phage ϕCbK predation may not occur through selective recognition of this modification. However, it remains to be seen if phage ϕCbK is sensitive to the threonine-linked posttranslational modifications that decorate the periphery of the filament.
FIG 8.
Phylogenetic analysis of flagellin paralogs in C. crescentus. (A) Sequence alignment of the six flagellins present in C. crescentus. The observed secondary structure in FljK is annotated above the sequence alignment. Conserved surface-exposed threonine residues that harbor posttranslational modifications are highlighted yellow. Note that the most divergent flagellin, FljJ, lacks most of such threonine residues. (B) Conservation mapping of C. crescentus flagellins with the program ConSurf (65). The greatest degree of sequence divergence is located along the surface of the flagellin.
We mapped sequence conservation among the six C. crescentus flagellins onto our structure of FljK and found that the periphery of the filament contains several “hot spots” for sequence variation relative to the interior of the filament (Fig. 8B). As such, differential inclusion of flagellins can alter the surface that is presented to phage ϕCbK. These “hot spots” are an attractive location for future work that seeks to determine the preference of some flagellins by ϕCbK. However, ϕCbK is not absolutely dependent upon any one such surface because the vast majority of strains examined here still exhibit some degree of adsorption. We therefore propose that ϕCbK is somewhat tolerant of variations to the flagellins present on the surface of the C. crescentus flagellum.
We also note that FljK and the beta locus flagellins all have N and C termini that are conserved in length. This is not unexpected, as both termini of these proteins are buried in the lumen of the flagellum. However, comparison of these flagellins with those expected to be located at the base of the filament, FljJ and FljL (Fig. 1B), shows that their amino termini are shorter and carboxy termini longer (Fig. 8A). We speculate that these differences exist for the purpose of altering the local curvature and flexibility of the filament close to the hook and membrane-embedded motor and are therefore likely play a role in the transmission of force to the body of the filament.
Concluding remarks.
This work provides numerous insights into determinants of ϕCbK infection of C. crescentus but also raises a host of new questions. For example, what drives ϕCbK preference for FljK and the negative effects of FljL? What is the exact identity of the posttranslational modification, and is it uniformly or differentially installed along the body of the flagellar filament? What are the functional consequences of these modifications? It remains to be seen if addition of these posttranslational modifications can have an effect on motility as observed in Clostridium difficile (70) or if they are required for phage association like in Campylobacter jejuni (71, 72).
We have established a tractable platform for the isolation of various flagellar filaments from C. crescentus and rapid structure determination via cryo-electron microscopy. We found that both conformational and compositional heterogeneity are potential roadblocks to overcome when determining helical structures. This platform will allow future biochemical studies to interrogate the functional importance of flagellar posttranslational modifications in C. crescentus. It will also enable the study of interactions between phages and the flagellar filament in addition to allowing for the determination of other single-flagellin filament structures that do not promote phage ϕCbK adsorption to the same degree as FljK. This information will be valuable to help discern the nuanced, yet functionally relevant, determinants for flagellotropic phage predation in bacteria with compositionally heterogeneous flagella.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All C. crescentus strains were grown in peptone-yeast extract (PYE; 0.2% peptone, 0.1% yeast extract, 1 mM MgSO4, and 0.5 mM CaCl2) modified from Schmidt and Stanier (24, 58). E. coli strains were grown on LB medium (BD, Franklin Lakes, NJ). Selection with kanamycin, 5 µg/ml for C. crescentus and 50 µg/ml for E. coli, was applied where indicated. Phage adsorption assays were performed in PYE containing 4 mM instead of 1 mM MgSO4. Plating medium contained 1.5% (wt/vol) Bacto agar.
Motility assays.
To test the motility phenotypes of flagellin mutants, we carried out motility assays as previously described (1, 6). Briefly, PYE soft-agar plates containing 0.3% (wt/vol) agar were stab inoculated with isolated colonies from plates that had been incubated overnight. Motility zone diameters were measured on three separate plates after 72 h of incubation at 30°C and are presented as percentages of the motility of wild-type strain NA1000.
Evaluation of adsorption kinetics.
Phage adsorption to the C. crescentus flagellum was measured as previously described (1, 34, 73) after the systematic removal of one or more flagellins. Briefly, overnight cultures of C. crescentus were infected with ϕCbK at a multiplicity of infection (MOI) of 0.002 in PYE adsorption medium (4 mM MgSO4). A parallel control flask contained only phages. Adsorption proceeded with slow shaking (60 rpm) at 30°C for 45 min. At 5-min intervals, aliquots of 100 μl were taken and mixed in ice-cold tubes containing 900 μl of PYE medium and 30 μl of chloroform to disrupt bacterial cells; after 45 min, samples were centrifuged (5 min at 12,000 × g, 4°C) to precipitate bacterial debris. The supernatant containing unabsorbed phages was used to infect an indicator strain (C. crescentus bNY30A; optical density at 600 nm [OD600], 0.3) and overlaid on PYE agar. Plates were incubated at 30°C overnight for enumeration of PFU. The adsorption rate constant (k) was calculated by plotting relative plaque-forming units over time and dividing the slope of the regression line by the viable bacterial count.
Negative-stain electron microscopy.
Samples were prepared for negative-stain TEM by first glow discharging 400-mesh copper, carbon-coated, Formvar grids (EM Sciences, Hatfield, PA) for 40 s. Five microliters of overnight cultures of C. crescentus was applied to the grids and allowed to adsorb for 1 min before being washed three times with fresh PYE and stained with 1% phosphotungstic acid (PTA; pH 6.8) for 1 min. Negatively stained C. crescentus cells were imaged on a Jeol JEM-1400 transmission electron microscope (TEM; Jeol, Ltd., Japan) equipped with a LaB6 filament and operated at an accelerating voltage of 120 kV. To capture the total length of the bacterial flagellum, images at a low magnification of ×2,000 were digitally captured on a Gatan Ultrascan US1000 (2k by 2k) charge-coupled-device (CCD) camera (Gatan, Pleasanton, CA). Images were imported into ImageJ, and flagellar length was quantified using the measure analysis option (26). The average flagellum length for each C. crescentus strain was determined from at least 25 (except strain FljJ) flagella that were intact and attached to the cell.
Cryo-electron tomography.
To characterize ϕCbK adsorption to C. crescentus strains by cryo-ET, bacterial overnight cultures were grown to an OD600 of 0.6, infected at an MOI of 10, and incubated without shaking for 15 min at 30°C. Four-microliter aliquots were plunge frozen onto glow-discharged, 200-mesh copper Quantifoil grids (Quantifoil, Germany) in liquid ethane using a Vitrobot Mark III (FEI, Hillsboro, Oregon). Bovine serum albumin (BSA)-treated 10 nm colloidal gold was either previously applied onto the grids and air-dried or mixed with the sample for homogeneous gold distribution in the ice. Data collection was carried out using a Jeol JEM-2200FS 200-kV field emission gun (FEG) TEM with an in-column Omega energy filter (slit width, 20 eV). Images were collected with a Direct Electron LP DE-20 direct electron detector (Direct Electron, San Diego, CA). Images were acquired at magnifications with an effective pixel size equal to or less than 7.6 Å on the specimen. A cumulative electron dose between 120 e−/Å2 and 140 e−/Å2 was used for tilt series acquisition from −62° to +62°, and images were acquired at −6.0 μm defocus. Tilt series images were automatically collected with 2° angular increments using SerialEM (27). Tomograms were reconstructed from the aligned images using the IMOD tomography package (74). Three-dimensional volumes were segmented manually using Amira (FEI Visualization Sciences Group).
Recovery of straight flagellum isolates by a genetic screen.
C. crescentus isolates with straightened FljK flagella were recovered in a screen for loss of motility following mutagenesis. Mutagenesis of fljK was performed by error-prone PCR on a plasmid (pCS1) that contained the gene encoding the FljK T103C variant, a functional amino acid change that enables flagella to be dyed with a cysteine-reactive maleimide dye (65). Mutagenized pCS1 was transformed into C. crescentus ΔfljJKLMNO, and transformants were selected by growth on PYE supplemented with 5 µg/ml kanamycin. Individual isolates were transferred to low-percent agar plates as described above for a motility assay. Nonmotile isolates of C. crescentus were recovered. These isolates were further characterized for straight flagellar filaments by negative-stain TEM as described above. The plasmids from nonmotile isolates with straightened flagella were purified using the NEB monarch DNA miniprep kit (NEB, Ipswich, MA) and sequenced by Sanger sequencing using the primers 5′-GGCACGAATTCCGAGCTGACGACCGTTCAG-3′ and 5′-CAGGCTCAGGATCGAGGACGAAGACTGGTTGGC-3′. One of the nonmotile isolates carried a single mutation coding for N130S in fljK. This isolate was selected for further testing. The plasmid from this isolate (pCS12) was regenerated in the pCS1 background using quick change site-directed mutagenesis with the primers 5′-TTGAACTTGGCGCTCGTCGCGGCCTTTTCG-3′ and 5′-CGAAAAGGCCGCGACGAGCGCCAAGTTCAA-3′. The plasmid pCS12 was transformed into C. crescentus ΔfljJKLMNO, and the resulting phenotype was confirmed to be similar to the original nonmotile isolate.
Isolation of flagellins for high-resolution structural analysis.
Three liters of C. crescentus cells were grown to an OD600 of 0.6 in PYE. Cells were pelleted at 10,000 × g for 15 min, and the supernatant was collected. Then, the supernatant was centrifuged at 48,000 × g for 45 min to pellet flagellar filaments. The resulting pellet was gently resuspended by overlaying with 3 ml of phosphate-buffered saline and incubating overnight at 4°C; harsh mechanical resuspension was avoided to prevent filament damage. The 10,000 × g and 48,000 × g centrifugation steps were repeated to remove additional debris. Then, the flagellum solution was centrifuged at 17,500 × g for 15 min, and the supernatant was collected, followed by a final pelleting spin at 21,000 × g for 2 h and gentle resuspension in 100 μl of phosphate-buffered saline (PBS) as above.
Grid preparation.
Four-microliter aliquots of the purified flagella were plunge frozen onto glow-discharged, 200-mesh copper Quantifoil grids (Quantifoil Micro Tools GmbH, Germany) or C-flat grids (Protochips, Morrisville, NC) in liquid ethane using a Vitrobot Mark IV (Thermo Fisher, Hillsboro, Oregon). In some cases, BSA-treated 10 nm colloidal gold was mixed with the sample for homogeneous gold distribution in the ice.
Data collection and processing.
Imaging was performed on a Thermo Fisher Titan Krios FEG-TEM operated at 300 kV and a Gatan K3 camera. Dose-fractionated micrographs were collected in counting mode over a nominal defocus range of −0.3 to −2.5 µm at a pixel size of 0.87 Å with a total dose of ∼60 e−/Å2 (or 1.33 e−/Å2/frame) on the specimen and ∼1 e−/pixel/frame on the camera (Table 1). Data processing was conducted within the Relion 3.1 framework (53, 74, 75). Gain-normalized micrographs were motion corrected with MotionCor2 (76) and initial estimates for whole-micrograph defocus performed in gCTF (77). Segments were manually picked in Relion 3.1 and extracted with a box size of 320 pixels and an interbox distance of 25 Å. two-dimensional classification was performed on 2-fold down-sampled particles (box size of 160 pixels, 1.74 Å pixel size) using 50 classes with an in-plane angular sampling of 0.25° and a mask of 250 Å (see Fig. S4 in the supplemental material). Segments contributing to the best classes were then reextracted with recentering.
Amplitude spectra of the best two-dimensional classes were of insufficient quality for de novo indexing (Fig. S4B and Fig. S5A in the supplemental material). Therefore, initial estimates for helical symmetry were taken from homologous flagellar filaments of known structure (11, 57–59). An initial model was obtained using three-dimensional classification in Relion 3.1, starting from a featureless cylinder and a fixed helical rise of 4.8 Å and helical twist of 65.6° (Fig. S4C and D). The resulting volumes exhibited clear secondary structure features that are consistent with previously determined structures of flagellins. One such volume was then low pass filtered to 10 Å and used as an input volume for another iteration of three-dimensional classification in which the helical symmetry was not fixed. A consensus from the best classes was combined and subjected to helical autorefinement with optimization of helical symmetry parameters, after which point the resulting maps for the FljK-only reconstructions were of sufficient quality to unambiguously trace the main chain and model the side chains of most residues. Subsequent to autorefinement and per-particle defocus refinement and motion correction, another round of three-dimensional classification without alignment was used to further curate the data set, prior to reextraction to the native pixel size of 0.87 Å and a final round of masked refinement and postprocessing.
Subsequent to reconstruction, layer lines from projections of the reconstructed maps were compared to those obtained from naive two-dimensional classification to ensure that the amplitude spectra were unaltered by application of helical symmetry during reconstruction (11, 57–59) (Fig. S5). As further validation, refined out-of-plane tilt values obtained when using our estimated symmetry were compared to the same out-of-plane tilt values that were obtained when imposing likely incorrect symmetry (Fig. S6A), showing that all reconstructions presented here exhibit a near-Gaussian distribution of out-of-plane tilt (Fig. S6B, C, and D), unlike the clearly bimodal distribution observed upon application of likely incorrect symmetry (Fig. S6A). Masked Fourier shell correlation (FSC) curves are shown in Fig. S5C.
Model building.
Manual model building was first conducted with the straightened variant of FljK as the corresponding map and exhibited the best resolution of side-chain densities throughout the asymmetric unit. Model building was performed de novo for a monomer in Coot (78), and Phenix (79) was then used for real space refinement with Ramachandran (80) and secondary structure restraints. After refinement to convergence, the refined monomer was manually placed in all positions throughout the helical reconstruction and subjected to a final round of refinement in Phenix. A model of the wild-type FljK was built de novo as above, but automated refinement in Phenix additionally employed reference model restraints (79) from a polyalanine variant of the above straightened monomer. The resulting models exhibit excellent stereochemistry and map-model correlation (Table 3). Model building was not performed for the heterogeneous FljK and FljL reconstruction, as the corresponding map lacked sufficient definition to assign the position of amino acid side chains and, as such, lacked an essential cross-validation metric for the initial helical symmetry estimates that were used during reconstruction.
TABLE 3.
Data collection, reconstruction, and model statistics
| Characteristic | Data for strain: |
||
|---|---|---|---|
| FljK T103C N130S | FljK wild type | CcFljK and FljL wild typea | |
| PDB ID | 6XKY | 6XL0 | NA |
| EMDB ID | EMD-22229 | EMD-22231 | EMD-22232 |
| Data collection and processing | |||
| Voltage (kV) | 300 | 300 | 300 |
| Electron exposure (e−/Å2) | 60 | 60 | 60 |
| No. of frames | 45 | 45 | 45 |
| Nominal defocus range (µm) | 0.3–2.5 | 0.3–2.5 | 0.3–2.5 |
| Pixel size (Å) | 0.87 | 0.87 | 0.87 |
| Symmetry (beyond helical) | C1 | C1 | C1 |
| No. of micrographs | 630 | 1,016 | 494 |
| No. of initial particles | 82,716 | 177,826 | 160,871 |
| No. of final particles | 59,868 | 77,146 | 54,743 |
| Box size (Å) | 278 | 278 | 278 |
| Box size (pixels) | 320 | 320 | 320 |
| Helical symmetry | |||
| Rise (Å) | 4.90 | 4.83 | 4.88 |
| Twist (degrees) | 65.2 | 65.3 | 65.6 |
| 11-start filament hand | Left | Left | Right |
| Helical Z parameter (%) | |||
| Masking | 70 | 70 | 70 |
| Refinement | 50 | 50 | 50 |
| Map resolution (Å) | 3.2 | 3.4 | 4.6 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Sharpening B factor (Å2) | −64 | −66 | −177 |
| Structure refinement | |||
| Total atoms | 38,760 | 38,820 | NA |
| Ligands | 0 | 0 | NA |
| Water | 0 | 0 | NA |
| RMS, bonds (Å) | 0.007 | 0.008 | NA |
| RMS, angles (o) | 1.0 | 0.8 | NA |
| Ramachandran favored (%) | 98.5 | 97.0 | NA |
| Ramachandran outliers (%) | 0 | 0 | NA |
| Rotamer outliers (%) | 0.5 | 0.0 | NA |
| Map-model correlation (masked) | 0.86 | 0.80 | NA |
| Model B factors | |||
| Minimum | 19.3 | 27.8 | NA |
| Mean | 30.6 | 50.4 | NA |
| Maximum | 60.1 | 159 | NA |
| MolProbity score | 1.2 | 1.7 | NA |
| Clashscore | 4.7 | 9.2 | NA |
NA, not applicable.
Data availability.
Maps for the mutant FljK, wild-type FljK, and FljK and FljL reconstructions have been deposited into the Electron Microscopy Data Bank under accession no. EMD-22229, EMD-22231, and EMD-22232, respectively. Coordinates for the mutant FljK and wild-type FljK models have been deposited in the Protein Data Bank under accession no. 6XKY and 6XL0, respectively. Other data supporting the findings of the manuscript are available from the corresponding author upon request. Requests for other data supporting the findings of the study and materials should be addressed to the corresponding author.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to J. Kollman and E. Lynch (University of Washington), E. Bullitt (Boston University), and E. Egelman (University of Virginia) for valuable advice and feedback on numerous aspects of helical reconstruction methods. We thank Y. Brun (University of Montreal) and P. Aldridge (Newcastle University) for Caulobacter crescentus strains and valuable discussions about bacterial flagella and cell motility. We acknowledge the Robert P. Apkarian Integrated Electron Microscopy Core of Emory University for microscopy services and support.
This work was supported in part by the University of Wisconsin—Madison, the Morgridge Institute for Research, Emory University, Children’s Healthcare of Atlanta, and the Georgia Research Alliance to E.R.W.; public health service grants R01GM104540 and R01GM104540-03S1 to E.R.W. from the NIH; and NSF grant 0923395 to E.R.W. J.C.S. was supported in part by the Biotechnology Training Program, T32GM135066. Data collection at Florida State University was made possible by NIH grants S10 OD018142-01, S10 RR025080-01, and U24 GM116788 to Kenneth A. Taylor.
R.S.D., C.W.S., Z.K., R.C.G-F., D.P., J.C.S., and E.R.W. prepared strains and conducted motility and functional assays and negative stain imaging. R.S.D., Z.K., R.C.G-F., and E.R.W. prepared samples for cryo-ET and calculated reconstructions. N.T.P., D.P., and J.C.S. prepared samples for helical reconstruction. E.J.M. and N.T.P. collected data for helical reconstruction. E.J.M performed helical reconstruction. All authors contributed to data interpretation and manuscript preparation. E.R.W conceived of and supervised the work.
We declare no competing financial interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Guerrero-Ferreira RC, Viollier PH, Ely B, Poindexter JS, Georgieva M, Jensen GJ, Wright ER. 2011. Alternative mechanism for bacteriophage adsorption to the motile bacterium Caulobacter crescentus. Proc Natl Acad Sci U S A 108:9963–9968. doi: 10.1073/pnas.1012388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellison CK, Rusch DB, Brun YV. 2019. Flagellar mutants have reduced pilus synthesis in Caulobacter crescentus. J Bacteriol 201:e00031-19. doi: 10.1128/JB.00031-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Minamino T, Imada K. 2015. The bacterial flagellar motor and its structural diversity. Trends Microbiol 23:267–274. doi: 10.1016/j.tim.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 4.Nakamura S, Minamino T. 2019. Flagella-driven motility of bacteria. Biomolecules 9:279. doi: 10.3390/biom9070279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao X, Norris SJ, Liu J. 2014. Molecular architecture of the bacterial flagellar motor in cells. Biochemistry 53:4323–4333. doi: 10.1021/bi500059y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Callahan CT, Wilson KM, Ely B. 2016. Characterization of the proteins associated with Caulobacter crescentus bacteriophage CbK particles. Curr Microbiol 72:75–80. doi: 10.1007/s00284-015-0922-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X, Berg HC. 2000. Torque-speed relationship of the flagellar rotary motor of Escherichia coli. Biophysical J 78:1036–1041. doi: 10.1016/S0006-3495(00)76662-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magariyama Y, Sugiyama S, Muramoto K, Maekawa Y, Kawagishi I, Imae Y, Kudo S. 1994. Very fast flagellar rotation. Nature 371:752–752. doi: 10.1038/371752b0. [DOI] [PubMed] [Google Scholar]
- 9.Matsunami H, Barker CS, Yoon Y-H, Wolf M, Samatey FA. 2016. Complete structure of the bacterial flagellar hook reveals extensive set of stabilizing interactions. Nat Commun 7:13425. doi: 10.1038/ncomms13425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macnab RM 2003. How bacteria assemble flagella. Annu Rev Microbiol 57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 11.Maki-Yonekura S, Yonekura K, Namba K. 2010. Conformational change of flagellin for polymorphic supercoiling of the flagellar filament. Nat Struct Mol Biol 17:417–422. doi: 10.1038/nsmb.1774. [DOI] [PubMed] [Google Scholar]
- 12.Song WS, Yoon S-I. 2014. Crystal structure of FliC flagellin from Pseudomonas aeruginosa and its implication in TLR5 binding and formation of the flagellar filament. Biochem Biophys Res Commun 444:109–115. doi: 10.1016/j.bbrc.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Beatson SA, Minamino T, Pallen MJ. 2006. Variation in bacterial flagellins: from sequence to structure. Trends Microbiol 14:151–155. doi: 10.1016/j.tim.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Smith KD, Andersen-Nissen E, Hayashi F, Strobe K, Bergman MA, Barrett SLR, Cookson BT, Aderem A. 2003. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat Immunol 4:1247–1253. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- 15.Gómez-Gómez L, Bauer Z, Boller T. 2001. Both the extracellular leucine-rich repeat domain and the kinase activity of FSL2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell 13:1155–1163. doi: 10.2307/3871370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gómez-Gómez L, Boller T. 2000. FLS2: an LRR receptor–like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 17.Yonekura K, Maki-Yonekura S, Namba K. 2003. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- 18.Samatey FA, Imada K, Nagashima S, Vonderviszt F, Kumasaka T, Yamamoto M, Namba K. 2001. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature 410:331–337. doi: 10.1038/35066504. [DOI] [PubMed] [Google Scholar]
- 19.De Maayer P, Cowan DA. 2016. Flashy flagella: flagellin modification is relatively common and highly versatile among the Enterobacteriaceae. BMC Genomics 17:377. doi: 10.1186/s12864-016-2735-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Messner P 2009. Prokaryotic protein glycosylation is rapidly expanding from “curiosity” to “ubiquity”. ChemBioChem 10:2151–2154. doi: 10.1002/cbic.200900388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Logan SM 2006. Flagellar glycosylation—a new component of the motility repertoire? Microbiology (Reading) 152:1249–1262. doi: 10.1099/mic.0.28735-0. [DOI] [PubMed] [Google Scholar]
- 22.Merino S, Tomas JM. 2014. Gram-negative flagella glycosylation. Int J Mol Sci 15:2840–2857. doi: 10.3390/ijms15022840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ardissone S, Kint N, Viollier PH. 2020. Specificity in glycosylation of multiple flagellins by the modular and cell cycle regulated glycosyltransferase FlmG. Elife 9:e60488. doi: 10.7554/eLife.60488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schirm M, Schoenhofen IC, Logan SM, Waldron KC, Thibault P. 2005. Identification of unusual bacterial glycosylation by tandem mass spectrometry analyses of intact proteins. Anal Chem 77:7774–7782. doi: 10.1021/ac051316y. [DOI] [PubMed] [Google Scholar]
- 25.Poweleit N, Ge P, Nguyen HH, Loo RR, Gunsalus RP, Zhou ZH. 2016. CryoEM structure of the Methanospirillum hungatei archaellum reveals structural features distinct from the bacterial flagellum and type IV pilus. Nat Microbiol 2:16222. doi: 10.1038/nmicrobiol.2016.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salah Ud-Din AIM, Roujeinikova A. 2018. Flagellin glycosylation with pseudaminic acid in Campylobacter and Helicobacter: prospects for development of novel therapeutics. Cell Mol Life Sci 75:1163–1178. doi: 10.1007/s00018-01702696-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nothaft H, Szymanski CM. 2010. Protein glycosylation in bacteria: sweeter than ever. Nat Rev Microbiol 8:765–778. doi: 10.1038/nrmicro2383. [DOI] [PubMed] [Google Scholar]
- 28.Shea TB, Seaman E. 1984. SP3: a flagellotropic bacteriophage of Bacillus subtilis. J Gen Virol 65:2073–2076. doi: 10.1099/0022-1317-65-11-2073. [DOI] [PubMed] [Google Scholar]
- 29.Phothaworn P, Dunne M, Supokaivanich R, Ong C, Lim J, Taharnklaew R, Vesaratchavest M, Khumthong R, Pringsulaka O, Ajawatanawong P, Klumpp J, Brown N, Imam M, Clokie MRJ, Galyov EE, Korbsrisate S. 2019. Characterization of flagellotropic, Chi-like Salmonella phages isolated from Thai poultry farms. Viruses 11:520. doi: 10.3390/v11060520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunstan RA, Pickard D, Dougan S, Goulding D, Cormie C, Hardy J, Li F, Grinter R, Harcourt K, Yu L, Song J, Schreiber F, Choudhary J, Clare S, Coulibaly F, Strugnell RA, Dougan G, Lithgow T. 2019. The flagellotropic bacteriophage YSD1 targets Salmonella Typhi with a Chi-like protein tail fibre. Mol Microbiol 112:1831–1846. doi: 10.1111/mmi.14396. [DOI] [PubMed] [Google Scholar]
- 31.Schade SZ, Adler J, Ris H. 1967. How bacteriophage chi attacks motile bacteria. J Virol 1:599–609. doi: 10.1128/JVI.1.3.599-609.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baldvinsson SB, Sorensen MC, Vegge CS, Clokie MR, Brondsted L. 2014. Campylobacter jejuni motility is required for infection of the flagellotropic bacteriophage F341. Appl Environ Microbiol 80:7096–7106. doi: 10.1128/AEM.02057-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Storms ZJ, Sauvageau D. 2015. Modeling tailed bacteriophage adsorption: insight into mechanisms. Virology 485:355–362. doi: 10.1016/j.virol.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt JM, Stanier RY. 1965. Isolation and characterization of bacteriophages active against stalked bacteria. J Gen Microbiol 39:95–107. doi: 10.1099/00221287-39-1-95. [DOI] [PubMed] [Google Scholar]
- 35.Bendis I, Shapiro L. 1970. Properties of Caulobacter ribonucleic acid bacteriophage phi Cb5. J Virol 6:847–854. doi: 10.1128/JVI.6.6.847-854.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.West D, Lagenaur C, Agabian N. 1976. Isolation and characterization of Caulobacter crecentus bacteriophage phi Cd1. J Virol 17:568–575. doi: 10.1128/JVI.17.2.568-575.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt JM 1966. Observations on the adsorption of Caulobacter bacteriophages containing ribonucleic acid. J Gen Microbiol 45:347–353. doi: 10.1099/00221287-45-2-347. [DOI] [PubMed] [Google Scholar]
- 38.Christen M, Beusch C, Bosch Y, Cerletti D, Flores-Tinoco CE, Del Medico L, Tschan F, Christen B. 2016. Quantitative selection analysis of bacteriophage phiCbK susceptibility in Caulobacter crescentus. J Mol Biol 428:419–430. doi: 10.1016/j.jmb.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 39.Wilson DR, Beveridge TJ. 1993. Bacterial flagellar filaments and their component flagellins. Can J Microbiol 39:451–472. doi: 10.1139/m93-066. [DOI] [PubMed] [Google Scholar]
- 40.Faulds-Pain A, Birchall C, Aldridge C, Smith WD, Grimaldi G, Nakamura S, Miyata T, Gray J, Li G, Tang JX, Namba K, Minamino T, Aldridge PD. 2011. Flagellin redundancy in Caulobacter crescentus and its implications for flagellar filament assembly. J Bacteriol 193:2695–2707. doi: 10.1128/JB.01172-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coward C, Grant AJ, Swift C, Philp J, Towler R, Heydarian M, Frost JA, Maskell DJ. 2006. Phase-variable surface structures are required for infection of Campylobacter jejuni by bacteriophages. Appl Environ Microbiol 72:4638–4647. doi: 10.1128/AEM.00184-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frost JA, Kramer JM, Gillanders SA. 1999. Phage typing of Campylobacter jejuni and Campylobacter coli and its use as an adjunct to serotyping. Epidemiol Infect 123:47–55. doi: 10.1017/S095026889900254X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Namba K 2001. Roles of partly unfolded conformations in macromolecular self-assembly. Genes Cells 6:1–12. doi: 10.1046/j.1365-2443.2001.00384.x. [DOI] [PubMed] [Google Scholar]
- 44.Gillen KL, Hughes KT. 1991. Molecular characterization of flgM, a gene encoding a negative regulator of flagellin synthesis in Salmonella typhimurium. J Bacteriol 173:6453–6459. doi: 10.1128/jb.173.20.6453-6459.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shin H, Lee J-H, Kim H, Choi Y, Heu S, Ryu S. 2012. Receptor diversity and host interaction of bacteriophages infecting Salmonella enterica serovar Typhimurium. PLoS One 7:e43392. doi: 10.1371/journal.pone.0043392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koyasu S, Asada M, Fukuda A, Okada Y. 1981. Sequential polymerization of flagellin A and flagellin B into Caulobacter flagella. J Mol Biol 153:471–475. doi: 10.1016/0022-2836(81)90292-8. [DOI] [PubMed] [Google Scholar]
- 47.Koyasu S, Shirakihara Y. 1984. Caulobacter crescentus flagellar filament has a right-handed helical form. J Mol Biol 173:125–130. doi: 10.1016/0022-2836(84)90407-8. [DOI] [PubMed] [Google Scholar]
- 48.Trachtenberg S, DeRosier DJ. 1988. Three-dimensional reconstruction of the flagellar filament of Caulobacter crescentus. A flagellin lacking the outer domain and its amino acid sequence lacking an internal segment. J Mol Biol 202:787–808. doi: 10.1016/0022-2836(88)90559-1. [DOI] [PubMed] [Google Scholar]
- 49.Ely B, Ely TW, Crymes WB, Jr, Minnich SA. 2000. A family of six flagellin genes contributes to the Caulobacter crescentus flagellar filament. J Bacteriol 182:5001–5004. doi: 10.1128/JB.182.17.5001-5004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Driks A, Bryan R, Shapiro L, DeRosier DJ. 1989. The organization of the Caulobacter crescentus flagellar filament. J Mol Biol 206:627–636. doi: 10.1016/0022-2836(89)90571-8. [DOI] [PubMed] [Google Scholar]
- 51.Lagenaur C, Agabian N. 1978. Caulobacter flagellar organelle: synthesis, compartmentation, and assembly. J Bacteriol 135:1062–1069. doi: 10.1128/JB.135.3.1062-1069.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weissborn A, Steinmann HM, Shapiro L. 1982. Characterization of the proteins of the Caulobacter crescentus flagellar filament. Peptide analysis and filament organization. J Biol Chem 257:2066–2074. [PubMed] [Google Scholar]
- 53.He S, Scheres SHW. 2017. Helical reconstruction in RELION. J Struct Biol 198:163–176. doi: 10.1016/j.jsb.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Egelman EH 2010. Reconstruction of helical filaments and tubes. Methods Enzymol 482:167–183. doi: 10.1016/S0076-6879(10)82006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Desfosses A, Ciuffa R, Gutsche I, Sachse C. 2014. SPRING - an image processing package for single-particle based helical reconstruction from electron cryomicrographs. J Struct Biol 185:15–26. doi: 10.1016/j.jsb.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 56.Galkin VE, Yu X, Bielnicki J, Heuser J, Ewing CP, Guerry P, Egelman EH. 2008. Divergence of quaternary structures among bacterial flagellar filaments. Science 320:382–385. doi: 10.1126/science.1155307. [DOI] [PubMed] [Google Scholar]
- 57.Blum TB, Filippidou S, Fatton M, Junier P, Abrahams JP. 2019. The wild-type flagellar filament of the Firmicute Kurthia at 2.8 Å resolution in vivo. Sci Rep 9:14948. doi: 10.1038/s41598-01951440-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang F, Burrage AM, Postel S, Clark RE, Orlova A, Sundberg EJ, Kearns DB, Egelman EH. 2017. A structural model of flagellar filament switching across multiple bacterial species. Nat Commun 8:960. doi: 10.1038/s41467-017-01075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamaguchi T, Toma S, Terahara N, Miyata T, Ashihara M, Minamino T, Namba K, Kato T. 2020. Structural and functional comparison of Salmonella flagellar filaments composed of FljB and FliC. Biomolecules 10:246. doi: 10.3390/biom10020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilson JJ, Takahashi I. 1978. Adsorption of Bacillus subtilis bacteriophage PBS 1. Can J Microbiol 24:1–8. doi: 10.1139/m78-001. [DOI] [PubMed] [Google Scholar]
- 61.Bradley DE, Douglas CJ, Peschon J. 1984. Flagella-specific bacteriophages of Agrobacterium tumefaciens: demonstration of virulence of nonmotile mutants. Can J Microbiol 30:676–681. doi: 10.1139/m84-101. [DOI] [PubMed] [Google Scholar]
- 62.Llewellyn M, Dutton RJ, Easter J, O'Donnol D, Gober JW. 2005. The conserved flaF gene has a critical role in coupling flagellin translation and assembly in Caulobacter crescentus. Mol Microbiol 57:1127–1142. doi: 10.1111/j.1365-2958.2005.04745.x. [DOI] [PubMed] [Google Scholar]
- 63.Skerker JM, Shapiro L. 2000. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J 19:3223–3234. doi: 10.1093/emboj/19.13.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ellison CK, Kan J, Dillard RS, Kysela DT, Ducret A, Berne C, Hampton CM, Ke Z, Wright ER, Biais N, Dalia AB, Brun YV. 2017. Obstruction of pilus retraction stimulates bacterial surface sensing. Science 358:535–538. doi: 10.1126/science.aan5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N. 2016. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44:W344–50. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leclerc G, Wang SP, Ely B. 1998. A new class of Caulobacter crescentus flagellar genes. J Bacteriol 180:5010–5019. doi: 10.1128/JB.180.19.5010-5019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mangan EK, Malakooti J, Caballero A, Anderson P, Ely B, Gober JW. 1999. FlbT couples flagellum assembly to gene expression in Caulobacter crescentus. J Bacteriol 181:6160–6170. doi: 10.1128/JB.181.19.6160-6170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anderson PE, Gober JW. 2000. FlbT, the post-transcriptional regulator of flagellin synthesis in Caulobacter crescentus, interacts with the 5' untranslated region of flagellin mRNA. Mol Microbiol 38:41–52. doi: 10.1046/j.1365-2958.2000.02108.x. [DOI] [PubMed] [Google Scholar]
- 69.Egelman EH, Xu C, DiMaio F, Magnotti E, Modlin C, Yu X, Wright E, Baker D, Conticello VP. 2015. Structural plasticity of helical nanotubes based on coiled-coil assemblies. Structure 23:280–289. doi: 10.1016/j.str.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Faulds-Pain A, Twine SM, Vinogradov E, Strong PC, Dell A, Buckley AM, Douce GR, Valiente E, Logan SM, Wren BW. 2014. The post-translational modification of the Clostridium difficile flagellin affects motility, cell surface properties and virulence. Mol Microbiol 94:272–289. doi: 10.1111/mmi.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sacher JC, Shajahan A, Butcher J, Patry RT, Flint A, Hendrixson DR, Stintzi A, Azadi P, Szymanski CM. 2020. Binding of phage-encoded FlaGrab to motile Campylobacter jejuni flagella inhibits growth, downregulates energy metabolism, and requires specific flagellar glycans. Front Microbiol 11:397. doi: 10.3389/fmicb.2020.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ewing CP, Andreishcheva E, Guerry P. 2009. Functional characterization of flagellin glycosylation in Campylobacter jejuni 81–176. J Bacteriol 191:7086–7093. doi: 10.1128/JB.00378-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bender RA, Refson CM, O'Neill EA. 1989. Role of the flagellum in cell-cycle-dependent expression of bacteriophage receptor activity in Caulobacter crescentus. J Bacteriol 171:1035–1040. doi: 10.1128/JB.171.2.1035-1040.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, Scheres SH. 2018. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zivanov J, Nakane T, Scheres SHW. 2020. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7:253–267. doi: 10.1107/S2052252520000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. 2017. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang K 2016. Gctf: real-time CTF determination and correction. J Struct Biol 193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, Moriarty NW, Oeffner RD, Poon BK, Prisant MG, Read RJ, Richardson JS, Richardson DC, Sammito MD, Sobolev OV, Stockwell DH, Terwilliger TC, Urzhumtsev AG, Videau LL, Williams CJ, Adams PD. 2019. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ramachandran GN, Ramakrishnan C, Sasisekharan V. 1963. Stereochemistry of polypeptide chain configurations. J Mol Biol 7:95–99. doi: 10.1016/S0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 81.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 82.Evinger M, Agabian N. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol 132:294–301. doi: 10.1128/JB.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jenal U, Shapiro L. 1996. Cell cycle-controlled proteolysis of a flagellar motor protein that is asymmetrically distributed in the Caulobacter predivisional cell. EMBO J 15:2393–2406. doi: 10.1002/j.1460-2075.1996.tb00597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Maps for the mutant FljK, wild-type FljK, and FljK and FljL reconstructions have been deposited into the Electron Microscopy Data Bank under accession no. EMD-22229, EMD-22231, and EMD-22232, respectively. Coordinates for the mutant FljK and wild-type FljK models have been deposited in the Protein Data Bank under accession no. 6XKY and 6XL0, respectively. Other data supporting the findings of the manuscript are available from the corresponding author upon request. Requests for other data supporting the findings of the study and materials should be addressed to the corresponding author.