Abstract
In addition to providing maximal nutritional value for neonatal growth and development, human milk functions as an early defense mechanism against invading pathogens. Human milk oligosaccharides (HMOs), which are abundant in human milk, are a diverse group of heterogeneous carbohydrates with wide ranging protective effects. In addition to promoting the colonization of beneficial intestinal flora, HMOs serve as decoy receptors, effectively blocking the attachment of pathogenic bacteria and viruses. HMOs also function as bacteriostatic agents, inhibiting the growth of gram-positive bacteria. Based on this precedence, an emerging area in the field has focused on characterizing the antiviral properties of HMOs. Indeed, HMOs have been evaluated for their potential as antiviral agents, with many shown to possess activity against life-threatening infections. This targeted review provides insight to the known glycan-binding interactions between select HMOs and influenza, rotavirus, respiratory syncytial virus (RSV), human immunodeficiency virus (HIV), and norovirus. Additionally, we review the role of HMOs in preventing necrotizing enterocolitis (NEC), an intestinal disease linked to viral infections. We close with a discussion of what is known, broadly regarding human milk oligosaccharides and their interactions with coronaviruses.
Keywords: Human Milk Oligosaccharides, HMOs, Anti-viral
Introduction
The past twenty-five years have seen the development of antiviral agents that not only inhibit the viral growth cycle selectively but do so without causing collateral damage to the host. Antivirals function by inhibiting the viral infection cycle through a number of distinct mechanisms, including obstructing entry, uncoating, interfering with receptor recognition, and disrupting viral protein and nucleic acid synthesis.1 Indeed, a myriad of clinically approved drugs exist that treat auto-immune disease syndrome (AIDS),2 human immunodeficiency virus (HIV), herpes simplex virus (HSV), influenza virus, and hepatitis B and C viruses (HBV and HCV, respectively).3 Not surprisingly, these drugs are plagued by the increasing emergence of resistance, limited therapeutic efficacy, and severe side effects.4 For emerging and/or neglected life-threatening viral diseases (e.g., Dengue virus and Zika virus), the lack of approved medications for clinical use represents a largely unmet need.5, 6
In the early stages of an infection, viruses recognize the host’s blood group carbohydrates and other sialylated (substituted variants of neuraminic acid) glycoproteins as receptors. In theory, any carbohydrate that shares structural homology with these cell surface glycans can function as a receptor decoy for viral adhesions, preventing an early stage of infection. Indeed, human milk contains secretary blood group carbohydrates (e.g. fucosylated Lewis antigens and sialylated glycoproteins) that prevent viral infections.7 Human milk contains complex oligosaccharides (HMOs) that possess related functionality. Not surprisingly, HMOs have demonstrated the ability to protect infants against a number of viruses by serving as receptor decoys. In this targeted review, we argue the case for studying the antiviral properties of HMOs, a group of complex carbohydrates that possess both prebiotic and antimicrobial activity. HMOs also possess novel structural features (in comparison to common anti-viral agents), high efficacy, and lack the possibility of collateral damage to the host.
Human Milk Oligosaccharides
Human milk has long been considered the gold standard for infant nutrition, as most health experts, including the American Academy of Pediatrics, recommend exclusive breast feeding for the first six months of life.8 Breast milk not only contains essential nutrients for growth and development, but also is dynamic in its composition—continuously changing to meet the unique needs of the infant. Colostrum, the earliest milk produced by mammals, is a thick, yellow fluid rich in antibodies, proteins, and oligosaccharides.9 The mother’s body starts producing colostrum around mid-pregnancy and continues its secretion up to five days after birth. At this time, the body transitions to producing mature milk over an approximately 14-day window. 95% of the total energy supplied in mature breast milk exists in the form of milk fats and lactose.10, 11 The average macromolecular profile of one liter of human milk contains 9–12 g of proteins, 32–36 g of fats, 67–78 g of lactose, and 5–20 g of human milk oligosaccharides (HMOs).10, 12–14
HMOs are the third largest macromolecule found in human milk.13 While present in the milk of most mammals, complex oligosaccharides such as HMOs are significantly more complex and abundant in primate milk. Indeed, the concentration of HMOs produced by the primate mammary gland are highest in the colostrum (ca. 20 g/L) and averages from 5 to 15 g/L in mature milk.13, 15, 16 To date, over 200 HMOs have been identified, ranging from simple derivatives of lactose containing 3 monosaccharides to complex polymers that incorporate upward of 20 monosaccharides.17–19,20 The isolation of HMOs is a well-established stepwise procedure beginning with the removal of fats via centrifugation, followed by protein precipitation with ethanol. The abundant lactose component in the remaining mixture is hydrolyzed into its glucose and galactose monomers using the enzyme β-galactosidase. The individual HMOs are then purified using size-exclusion chromatography and can be further characterized. These various separation and profiling techniques include: high pH anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD), nuclear magnetic resonance (NMR) spectroscopy, reverse-phase high-performance liquid chromatography (RP-HPLC), hydrophilic interaction chromatography HPLC (HILIC), matrix assisted laser desorption ionization-time-of-flight-mass spectrometry (MALDI-TOF-MS), and permethylation followed by liquid chromatography coupled with high-resolution tandem mass spectrometry (LC-MS/MS).17, 21–24 While these methods have significantly enhanced the availability of HMOs for study, there is still a limited supply of them, therefore chemoenzymatic synthesis and microbial fermentation have been employed to synthesize a library of HMOs for research and supplementation into formula.25, 26
Structurally, HMOs are composed of 5 monosaccharides: L-fucose (Fuc), D-glucose (Glc), D-galactose (Gal), N-acetylglucosamine (GlcNAc), and N-acetylneuraminic acid (Neu5Ac) (Figure 1).13 Lactose (Lac, Gal-β1-4-Glc) forms the reducing end of all HMOs (Figure 1A). To synthesize an HMO, lactose is elongated with lacto-N-biose (LNB; Galβ1-3GlcNAc) or N-acetyllactosamine (LacNAc; Galβ1-4GlcNAc) to generate a type I or type II chain respectively. Generally, LNB is used to terminate a growing HMO chain, while LacNAc is used as an elongation unit to extend the chain at O-3 or O-6 of lactose. Branching occurs when β1-6 linkages are installed (iso-HMOs), while linear chains occur through installation of β1-3 linkages (para-HMOs). While most women generally produce the same “core” oligosaccharides, Lewis blood group and secretor status will determine how Fuc and Neu5Ac are installed. HMOs can be fucosylated via α 1-2, α 1-3 or α 1-4 linkages (Figure 1B) or sialylated via α 2-3 or α 2-6 linkages. These modifications can occur on the lactose core or the elongated chain (Figure 1C). In general, the abundance of fucosylated HMOs is significantly higher (35–50%) than sialylated HMOs (10–15%). Non-fucosylated, neutral HMOs (40–55%) make up the remainder of HMOs (Figure 1A).27, 28 While a neonate consumes several grams of HMOs per day, they are non-nutritive. Indeed, only 3% of HMOs reach systemic circulation. HMOs resist digestion by both enzymatic hydrolysis and intestinal acidity. Thus, ca. 97% of HMOs accumulate in the distal small intestine and colon.29 Previous work in the field has demonstrated that HMOs possess a number of biological functions including modulation of gut microbiota, inhibition of pathogenic adhesion, promotion of brain development, protection of the epithelial barrier, and stimulation of the immune response.30
Figure 1:
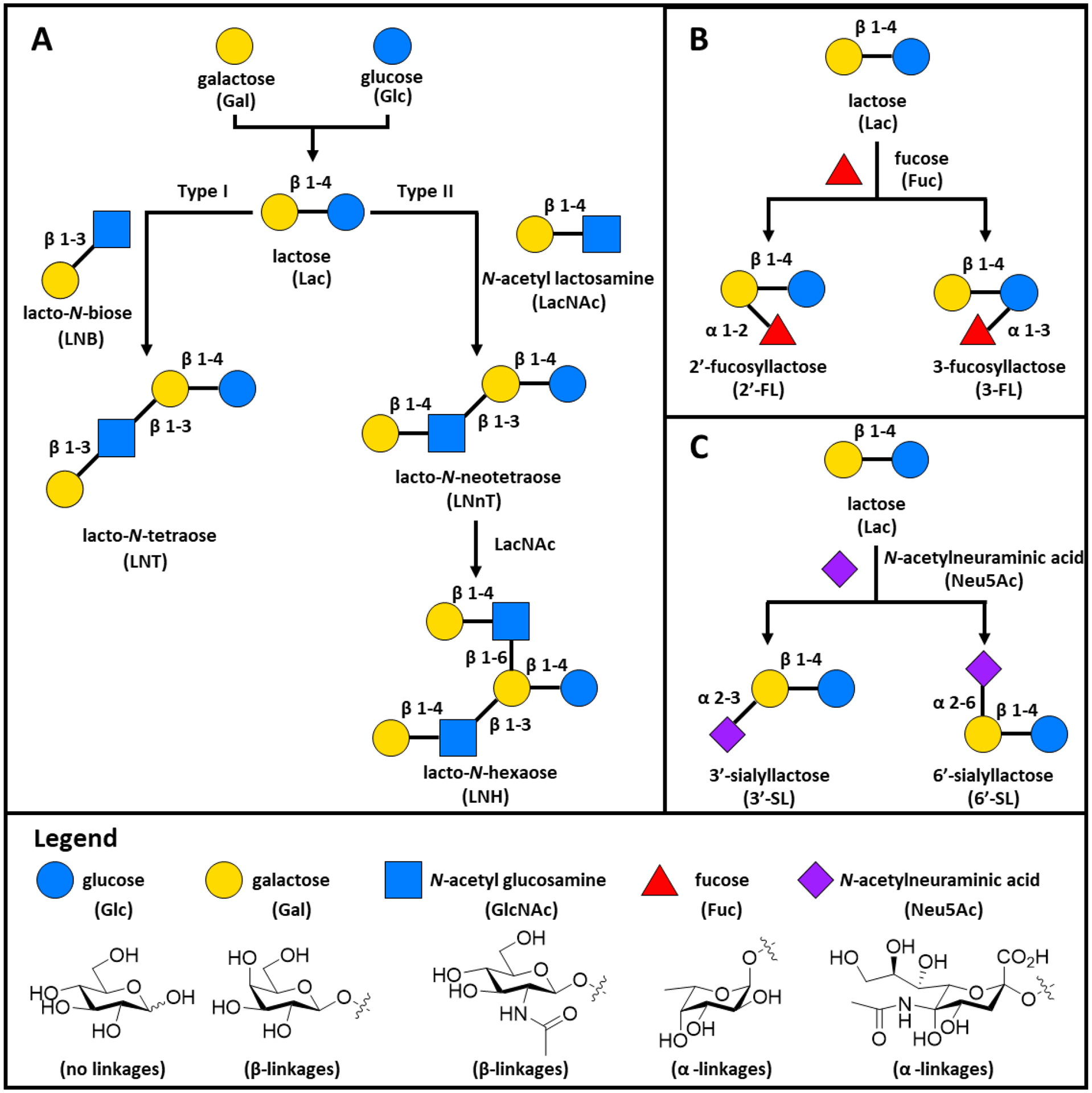
The most common human milk oligosaccharides (HMOs) found in human milk and their biosynthesis. All HMOs are composed from five monosaccharides (Legend): glucose (Glc), galactose (Gal), N-acetyl glucosamine (GlcNAc), fucose (Fuc), and N-acetylneuraminic acid (Neu5Ac). (A) The basic blueprint for HMO biosynthesis with lactose forming the reducing end for all oligosaccharides. Lactose (Lac) can be elongated to with either lacto-N-biose (LNB) to form type I chains, or with N-acetyl lactosamine (LacNAc) to form type II chains. B) Representative fucosylated HMOs synthesized through the addition of fucose. C) Representative sialylated HMOs characterized by the addition of N-acetylneuraminic acid (Neu5Ac).
A primary function of HMOs is to serve as prebiotics, facilitating the establishment of beneficial flora in the infant’s gut microbiome. Accordingly, HMOs select for the growth of a small number of dominant beneficial species which suppress the growth of pathogenic bacteria. As a consequence, the microbial composition of the infant gut varies significantly between breastfed and formula-fed babies. Since infant formula does not replicate the composition of breast milk, and lacks HMOs, the microbiome of formula fed infants possesses greater diversity. In 1954 Kuhn and György reported that a combination of HMOs, which they termed the “bifidus factor”, promotes the growth of Bifidobacterium bifidum (previously classified as Lactobacillus bifidus).31, 32 Bifidobacterium fermentation of HMOs leads to the production of short-chain fatty acids (SCFAs; acetate, propionate, and butyrate). SCFAs protect the commensal flora against invading pathogens and inflammation by regulating gut pH and enhancing the immune system.33–35
In general, the gut flora of a breastfed infant is dominated by Bacteroides, Lactobacillus, and Bifidobacterium spp. as these strains can metabolize HMOs.36, 37 Most Bifidobacterium spp are able to metabolize the principal HMOs present in breast milk, including lacto-N-tetraose (LNT) and 2’-fucosylactose (2’-FL).36, 38 Additional Bifidobacterium and Bacteroides spp., specifically B. bifidum and B. fragilis, use β-galactosidase, α-fucosidase, and sialidase enzymes to hydrolyze HMOs. Next, the released monosaccharides are metabolized.36 Indeed, abundance of the probiotic Bifidobacterium spp. (Bifidobacterium longum subsp. infantis, Bacteroides fragilis, and Bacteroides vulgatus) that function under this mechanism is prominent in breastfed children.39 While Bifidobacterium spp. still dominate the microbiota in formula fed babies, in general there is an overall greater diversity with an increase in Staphylococcus, Streptococcus, Enterococcus, and Clostridium species.40–42 Low microbial in breastfed babies is a result of an abundance of beneficial Bifidobacterium and Lactobacillus species.43
In addition to serving as prebiotics, our group has shown that HMOs possess bacteriostatic properties and antibiofilm.44 Initial studies revealed that both homogeneous and heterogeneous HMOs govern bacterial growth and biofilm assembly.45–49 We discovered that HMO extracts possess antibiofilm and antimicrobial properties against Streptococcus agalactiae (GBS), antimicrobial properties against the Gram-negative aerobe Acinetobacter baumannii, and antibiofilm properties against methicillin-resistant Staphylococcus aureus (MRSA). In a second-generation study, we observed that HMOs potentiate the actions of aminoglycosides, anti-folates, macrolides, lincosamides, and tetracyclines against GBS, S. aureus and A. baumannii.46, 50
Virus Prevention or Treatment with HMOs
HMOs prevent the colonization of viral pathogens.51 The virulence of enteric viruses is dependent, in part, on the pathogen’s ability to adhere to epithelial surfaces (Figure 2A). There are two proposed mechanisms for how HMOs modulate viral pathogenesis. As HMOs share structural homology with epithelial cell surface glycans, they serve as soluble decoy receptors to prevent early cellular attachment (Figure 2B). Additionally, HMOs bind epithelial cell surface receptors to block viral adhesions (Figure 2C).52 The interactions between these two classes of molecules include the well-known carbohydrate - lectin interactions that are critical to the viral infection process as the viral cell surface is decorated with oligosaccharides that recognize lectins on human cells.53
Figure 2:
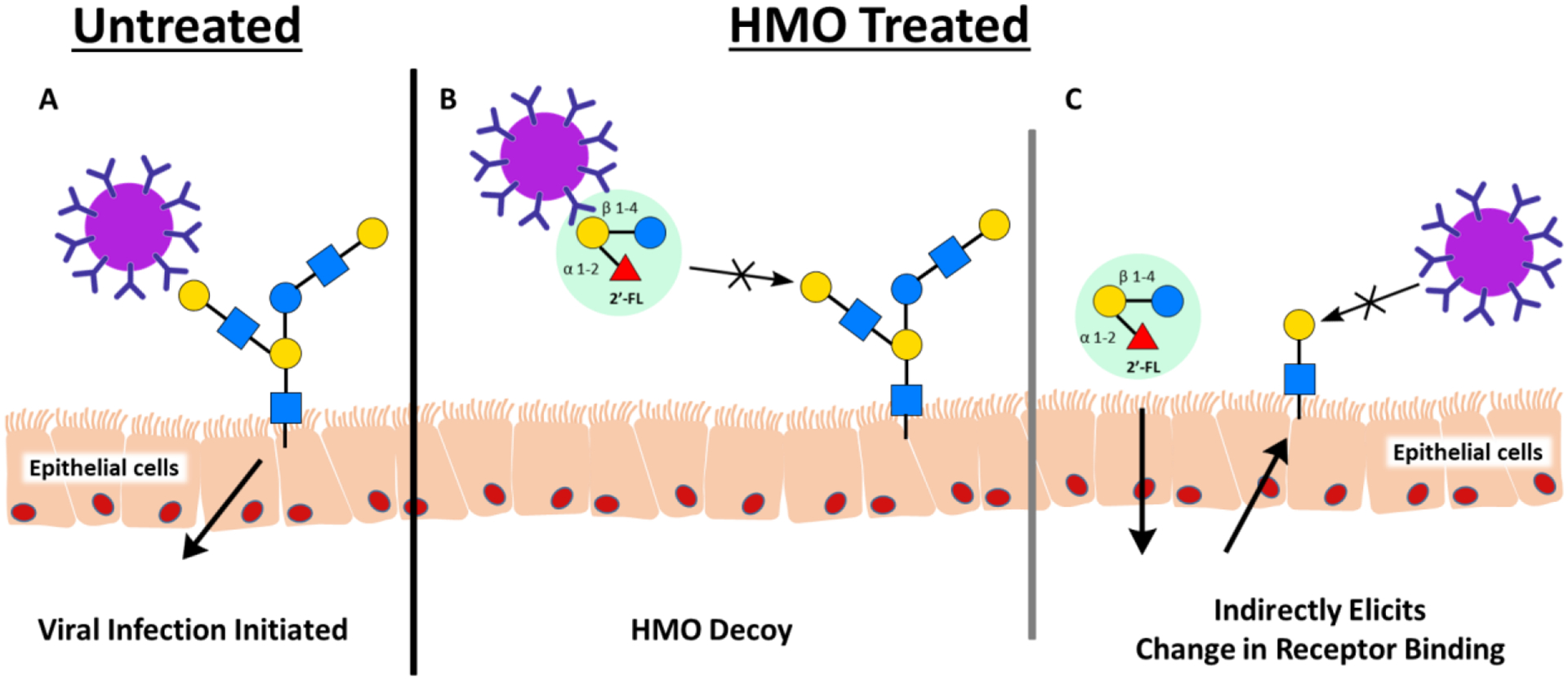
Proposed mechanism of actions of how HMOs can prevent viral adhesion to gut epithelial cells. A) In the absence of HMOs, viruses recognize surface glycans that are necessary for pathogenic adhesion, the first step in establishing infection. B) HMOs resemble surface glycans, acting as soluble decoy receptors and blocking the attaching of viral pathogens to the epithelial cells. C) HMOs additionally can indirectly prevent viral adhesion through binding to the epithelial surface causing a structural change in the receptor.
The innate immune system possesses pattern recognition receptors that identify pathogens.54 In a related process, viral surface lectins recognize human epithelial cell-surface glycans to identify hosts during an infection. HMOs have been explored to tactically protect against virus invasions by mimicking epithelial cell surface glycans. HMOs also possess immunomodulatory activity by reducing viral infection in influenza, rotavirus, respiratory syncytial virus (RSV), human immunodeficiency virus (HIV), norovirus, and the disease necrotizing enterocolitis (NEC), which can accompany viral infection (Figure 3).
Figure 3:
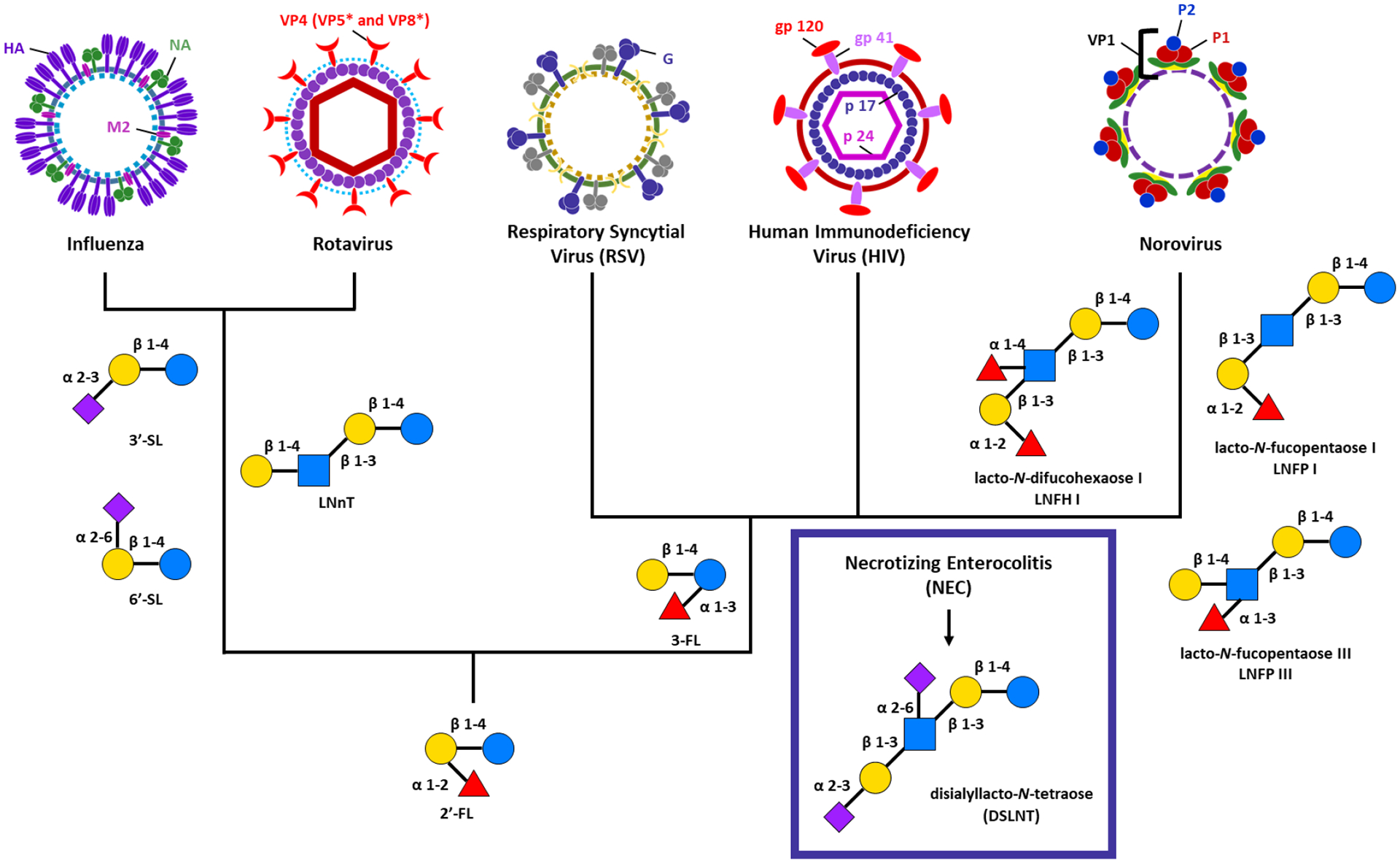
HMOs with known immunomodulatory activity against influenza, rotavirus, respiratory syncytial virus (RSV), human immunodeficiency virus (HIV), norovirus and necrotizing enterocolitis (NEC). With the exception of NEC, all of the viruses discussed in this review have known protection with 2’-fucosyllactose (2’-FL). Influenza and rotavirus additionally have shown reduced infection rates with 3’-sialyllactose (3’-SL), 6’-siallylactose (6’-SL), and Lacto-N-neotetraose (LNnT). 3-fucosyllactose (3-FL) reduces viral load in RSV, HIV, and norovirus. lacto-N-difucohexaose LNFH I, lacto-N-fucopentaose I (LNFP I), and lacto-N-fucopentaose III (LNFP III) all also known to inhibit viral binding. Only one HMO, disialyllacto-N-tetraose (DSLNT) has been shown to reduce NEC infection.
Influenza
Influenza belongs to the Orthomyxoviridae family of negative-sense, single-stranded RNA viruses. Negative-sense RNA must first be converted into positive-sense RNA by an RNA-dependent RNA polymerase before translation.55 The influenza viruses are classified into types A, B, and C. Influenza A and B are principle causes of seasonal epidemics and are responsible for three to five million cases of acute respiratory infections per year.56–58 Influenza C infection is rare and causes mild symptoms.59 Influenza virions are spherical or filamentous in shape with an outer lipid membrane covered in glycoprotein spikes of haemagglutinin (HA) and neuraminidase (NA).60 Both avian and human influenza viruses recognize oligosaccharides containing the sialylated galactose structural motif, where sialic acid is bound to galactose in an α2,6 or α2,3 linkage.61 HA functions by binding sialic acid which enables cell penetrance.60 NA, also known as the receptor-destroying enzyme, cleaves cell-surface sialic acid residues, promoting viral budding and release.60 The complex of HA, NA, and matrix protein M2 is currently the target of a number of antiviral drugs.60 Within the viral core, the nuclear export protein (NEP) and ribonucleoprotein (RNP) complex are required elements of viral transcription and replication.62 Due to the fact that a number of HMOs contain the sialylated galactose sub-structure, it is not surprising that 3’-siallylactose (3’-SL) and 6’-siallyllactose (6’-SL) reduce influenza infection by binding to HA glycoprotein spikes.63–65 Through related binding mechanisms, lacto-N-neotetraose (LNnT), which is non-sialylated, decreases viral load in the epithelium. Interestingly, 2’-FL has been shown to trigger the immune response in an influenza vaccination model.65–67 While the role of 2′-FL-mediated protection is likely due to modulation of the microbiota, the relationship to vaccine efficacy remains unknown.
Rotavirus
Rotaviruses are non-enveloped double-stranded RNA viruses that cause 138 million cases of severe diarrhea, vomiting, and dehydration each year.68–70 The genome of the triple-layered virus is composed of eleven segments that code for six structural viral proteins (VP1-4, 6, 7) and six non-structural proteins (NSP1-6).71 The three major structural proteins include two surface proteins, VP7 and VP4 (middle layer). VP4 is a spike protein responsible for cell attachment and membrane penetrance.72 Additionally, VP4 is proteolytically cleaved by trypsin, producing VP5* and VP8*, which results in higher rates of infection.73
Both fucosylated and sialylated HMOs have been shown to reduce rotavirus infection by acting as decoy receptors to inhibit binding of intestinal histo-blood group antigens (HBGAs) to the viral proteins. 3’-SL and 6’-SL, in particular, reduce viral infectivity; however, the combination of the two HMOs has proven to be more effective in binding to VP8* in a porcine rotavirus model.74 The most abundant HMO found in human milk globally, 2’-FL, causes a significant decrease in infectivity when used as a therapy after appearance of viral symptoms.74 An additional non-fucosylated HMO, LNnT, while failing to reduce viral load in vitro, surprisingly reduced rotavirus infection in an in vivo model by binding to VP8*.75
Respiratory syncytial virus
Respiratory syncytial virus (RSV) is a leading cause of respiratory tract and lung infections in children.76 In 2015, it was estimated that the virus resulted in 33.1 million cases and 59,600 deaths worldwide, with children under six months of age making up 1.4 million cases and 27,300 deaths.77 RSV belongs to the Paramyxoviridae family of single-stranded negative-sense RNA viruses, which includes measles, mumps, Nipah virus, and Hendra virus.78 Structurally, RSV is an enveloped sphere consisting of ten open reading frames and eleven viral proteins of which nine are structural and two are nonstructural.79, 80 The three proteins that are required for replication and to protect the ribonucleoprotein (RNP) core are the nucleoprotein (N), phosphoprotein (P), and the RNA-dependent RNA polymerase large protein (L).80 The fusion protein (F), attachment glycoprotein (G), and short hydrophobic protein (SH) span the membrane, and the F and G proteins are integral for attachment and infection initiation.81 The two membrane-associated proteins (M2-1 and M2-2) are involved in improving the efficiency of transcription and replication.80, 82 Both 2’-FL and 3-FL bind to glycoprotein G, reducing the RSV viral load in airway epithelial cells.83
Human Immunodeficiency Virus
Human Immunodeficiency Virus (HIV), which can lead to acquired immunodeficiency syndrome (AIDS), belongs to the Retroviridae family of pathogens that attack the immune system. The virus is classified as either HIV-1, which is more widespread and aggressive, or HIV-2, which is found mostly in western Africa and is associated with lower pathogenicity.84 This two-stranded RNA virus contains 9 open reading frames and 15 viral proteins.85 The lipid bilayer encloses the RNA core, which is protected by the protein capsid. Two glycoproteins that are embedded in the envelope, gp120 and gp41, are necessary for attachment to the cluster of differentiation 4 (CD4) receptor on the host cell and viral fusion to mediate cell penetration.86 The matrix Gag protein p17, surrounds the capsid, plays a crucial role in replication, and anchors both gp120 and gp41 to the envelope.87, 88 Within the viral core, the major structural protein p24 has recently become a target for HIV vaccines as it is found in high abundance in the blood of infected patients.89 The mechanism in which HIV gains viral entry across the infant’s mucosal barrier is through binding of the receptor, dendritic cell-specific ICAM-3 grabbing non-integrin (DC-SIGN) to the high mannose containing glycans on HIV envelope glycoprotein, gp120. This initiates infection in the CD4+ T lymphocytes of the host.90, 91 There is a correlation between high concentrations of HMOs present in breast milk to lower risk of transmission of HIV to the infant through breastfeeding.92 Carbohydrates in general, but specifically the Lewis blood group antigens found in HMOs, have been shown to inhibit binding of HIV to DC-SIGN.92, 93 DC-SIGN is highly reactive towards the fucose Lewis antigens Lea,b,x,y, including 2’-FL and 3-FL.93
Norovirus
Norovirus is a highly contagious virus that causes gastroenteritis along with some symptoms such as vomiting, diarrhea, and nausea. According to the CDC, norovirus is responsible for 685 million cases worldwide, with 200 million of those cases occurring in children under five.94 Noroviruses belong to the Caliciviridae family which also include additional positive single-stranded RNA viruses associated with respiratory disease, hemorrhages, and gastroenteritis.95 The norovirus genome encodes three open reading frames (ORFs). ORF1 encodes the six nonstructural proteins, while ORF2 and ORF3 encode the two structural proteins, VP1 and VP2, that form the major and minor capsid proteins.96, 97 Structurally, VP1 is composed of 90 dimers which are further separated into a shell domain (S domain) and a protruding domain (P domain). These domains are connected by a flexible hinge linker, 10 to 14 amino acids in length.98, 99 The β-barrel folded P domain is composed of a highly variable P2 subdomain extending out from the P1 subdomain.100 Since P2 is located on the distal surface, it promotes immune recognition as well as contains the receptor binding site.100, 101 Noroviruses, like rotaviruses, recognize histo-blood group antigens (HGBAs) found in the saliva or protective mucosa of the digestive tract - leading to increased infection rates. Based on X-ray crystallography analysis, both 2’-FL and 3-FL bind to a pocket at the top of the P1 domain, acting as a decoy as they structurally mimic HBGAs, effectively inhibit the virus from binding to HGBA.102 In another notable study, it was found that two fucosylated HMOs, lacto-N-fucopentaose III (LNFP III) and 2’-FL bind to one norovirus strain VA387.103 In the same study, another strain of norovirus (Norwalk) bound to lacto-N-fucopentaose I (LNFP I ) and lacto-N-difucohexaose (LNDFH I) to effectively inhibit viral infection.103
Necrotizing enterocolitis (NEC)
The five viruses described above are the most common viral pathogens tested for antiviral activity, however, another disease, necrotizing enterocolitis (NEC) is of importance when discussing HMO treatment options. The exact cause of NEC infections in still unclear but it has been associated with specific bacteria, viruses, and fungi. Rotavirus, norovirus, echovirus, astrovirus, enterovirus, cytomegalovirus, and coronavirus have all been implicated as possible causes of NEC.34 NEC is a devastating intestinal disease that is most common in premature and very low birth weight infants. NEC is associated with a mortality rate between 20 and 40%, with a heightened risk associated with surgical intervention.104–106 The disease is characterized by ulceration, intestinal inflammation, abdominal distention, hemorrhages, and bacterial overgrowth.107, 108 While a low diversity of HMOs in breast milk is associated with NEC development in infants, one specific HMO, disialyllacto-N-tetraose (DSLNT), has been found most effective in preventing NEC.109, 110 It was determined that the removal of one or both sialic acid residues of DSLNT caused those HMOs to have no effect on treating NEC.109 The underlying mechanism of how DSLNT reduces the incidences remains to be elucidated, however, several receptor-mediated hypotheses remain in play. It is unlikely that that DSLNT is selectin-mediated since these transmembrane glycoproteins only bind to fucosylated glycans.65 Other classes of lectins such as siglecs which bind sialylated HMOs, or galectins which bind to sulfated, sialylated, fucosylated, and lactose-containing HMOs, are more likely candidates for DSLNT targeting.65, 111, 112
Coronaviruses.
Coronaviruses are a large family of viruses that are named for the crown-like spikes on their surface. There are four main sub-groupings of coronaviruses, known as alpha, beta, gamma, and delta. Coronaviruses typically cause mild to moderate upper-respiratory tract infections such as the common cold. There are hundreds of coronaviruses, most of which circulate among mammals such as bats and porcine. Occasionally coronaviruses jump to humans, a so-called spillover event, where they cause serious disease. Four of the seven known coronaviruses that sicken people cause only mild to moderate disease. These are 229E (alpha coronavirus), NL63 (alpha coronavirus), OC43 (beta coronavirus), and HKU1 (beta coronavirus).
Three coronaviruses have emerged from animal reservoirs over the past several decades that cause serious and often fatal disease. The first is a virus that causes severe acute respiratory syndrome (SARS) known as SARS coronavirus (SARS-CoV). The virus emerged in 2002 and disappeared by the end of 2004. SARS-CoV is thought to be an animal virus from a thus far, unknown animal reservoir, most likely a bat. The second virus is known as Middle East respiratory syndrome (MERS) and is caused by the MERS coronavirus (MERS-CoV). Transmitted from an animal reservoir in camels, MERS was identified in 2012 and continues to cause irregular, albeit contained outbreaks. The third novel coronavirus to emerge this century is known as SARS-CoV-2. It causes coronavirus disease 2019 (COVID-19), which emerged from China in late 2019 and was declared a global pandemic by the World Health Organization (WHO) in early 2020.
In the context of human milk, it is currently unclear whether severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is shed into breastmilk and transmitted to a baby through breastfeeding. Indeed, recent investigations of appreciable viral load in human milk - but sample sizes are small. 113 It is currently believed that pregnant people are at an increased risk for severe illness from COVID-19 compared to non-pregnant people. Additionally, pregnant people with COVID-19 might be at increased risk for other adverse outcomes, such as preterm birth. However, there is no appreciable evidence that COVID-19 is transmitted through breast milk. Whether or not HMOs are able to serve as receptor decoys or are able to bind the pathogen is currently unknown.
Future Directions
In the last two decades, humankind face six major viral threats – SARS (2002, 2020), swine flu (2009), MERS (2012), avian influenza (2013), and Ebola (2014). While the 2020 SARS pandemic has been catastrophic, it will assuredly not be the final contagion we face. Modern society functions such that disease spillover into humans is far more facile than in the past. Humankind must discover new tools to meet this challenge.
The first step in controlling interspecies transmission of viruses and increasing our ability to treat those that adversely affect human health and wellness, we must characterize the molecular interactions between viruses and their glycoconjugate receptors on host cells. Competitive inhibitors that mimic cell surface receptor glycans may hold the key to preventing viral entry. For example, the earliest stages of norovirus and rotavirus infection feature attachment to HBGAs. As described above, HMOs that share structural homology with HBGAs protect against rotavirus and norovirus infections. Moreover, HMOs target a conserved region of the capsid glycoprotein and prevent viral invasion, which means they are less susceptible to resistance evolution. One would thus expect these molecules to be of interest as an alternative therapeutic.
Before the antimicrobial activity of HMOs can be translated to prophylactic or therapeutic application, the community must elucidate the structural basis of viral inhibition, characterize the potency of HMOs, and define strain specificity. Similar to other carbohydrate-protein interactions, HMOs generally bind to viral capsid glycoproteins with low affinity. By comparison, viral adhesion to host cells is of high affinity due to multivalent interactions with cell surface glycan receptors. Thus, from a structural perspective, synthetic multivalent HMO constructs (similar to the tumor associated carbohydrate antigen vaccine constructs produced by the Danishefsky lab) are of great interest to our group and may serve as highly effective entry inhibitors. In addition to novel multivalent HMO based constructs, we believe that a second subset of HMOs worth exploring are mixed or multi-sialylated / fucosylated HMOs which incorporate several virus-recognizing epitopes in a single molecule.
The greatest roadblock to exploiting HMOs to prevent norovirus, rotavirus, and even influenza infections is their accessibility and general structural characterization. HMOs are a highly diverse group of compounds. Sadly, their benefits (which generally appear to be structure-dependent) cannot be readily attributed to a specific structure as most HMOs have not been characterized. Indeed, fewer than 10% of proposed HMOs have been evaluated for antiviral activity. Thus, advances to the chemical and chemoenzymatic synthesis of HMOs is necessary to achieve broad structure-activity studies.
HMO research has undergone significant advances since Moro and Tissier observed that breastfeeding governs the infant gut microbiome nearly 125 years ago. In the present day, the community’s dedication to characterizing the biological activity of several “small” HMOs has been validated by the first clinical trials in infants using either 2′-FL or 2`-FL/LNnT. These studies have revealed that HMO supplementation of formula is safe and produces a similar growth pattern as breast-fed infants. Thus, a number of exciting opportunities exists for future generations of human milk scientists. Within this context, our view of the future of HMO-based antivirals is optimistic and the first clinical trial of an HMO antimicrobial agent is likely on the horizon.
Acknowledgements
This work is supported by the National Institutes of Health under Grant No R35GM133602. S.D.T. is supported by a Dean’s Faculty Fellowship from the College of Arts & Science at Vanderbilt University and is a Camille Dreyfus Teacher-Scholar.
References
- 1.Menéndez-Arias L; Gago F, Antiviral agents: structural basis of action and rational design. Subcell Biochem 2013, 68, 599–630. [DOI] [PubMed] [Google Scholar]
- 2.HIV Treatment. https://www.fda.gov/drugs/human-immunodeficiency-virus-hiv/hiv-treatment (accessed Dec 18, 2020).
- 3.De Clercq E, Antivirals and antiviral strategies. Nat Rev Microbiol 2004, 2 (9), 704–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wainberg MA, Perspectives on antiviral drug development. Antivir Res 2009, 81 (1), 1–5. [DOI] [PubMed] [Google Scholar]
- 5.Lim SP, Dengue drug discovery: Progress, challenges and outlook. Antivir Res 2019, 163, 156–178. [DOI] [PubMed] [Google Scholar]
- 6.Mehand MS; Al-Shorbaji F; Millett P; Murgue B, The WHO R&D Blueprint: 2018 review of emerging infectious diseases requiring urgent research and development efforts. Antivir Res 2018, 159, 63–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang X; Huang P; Zhong W; Tan M; Farkas T; Morrow AL; Newburg DS; Ruiz-Palacios GM; Pickering LK, Human milk contains elements that block binding of noroviruses to human histo-blood group antigens in saliva. J Infect Dis 2004, 190 (10), 1850–9. [DOI] [PubMed] [Google Scholar]
- 8.Gartner LM; Morton J; Lawrence RA; Naylor AJ; O’Hare D; Schanler RJ; Eidelman AI; American Academy of Pediatrics Section on, B., Breastfeeding and the use of human milk. Pediatrics 2005, 115 (2), 496–506. [DOI] [PubMed] [Google Scholar]
- 9.Kulski JK; Hartmann PE, Changes in human milk composition during the initiation of lactation. Aust J Exp Biol Med Sci 1981, 59 (1), 101–14. [DOI] [PubMed] [Google Scholar]
- 10.Rao PN, Manufacturing technology. Tata McGraw-Hill Education: 2013; Vol. 1. [Google Scholar]
- 11.Koletzko B; Rodriguez-Palmero M; Demmelmair H; Fidler N; Jensen R; Sauerwald T, Physiological aspects of human milk lipids. Early Hum Dev 2001, 65, S3–S18. [DOI] [PubMed] [Google Scholar]
- 12.Ballard O; Morrow AL, Human milk composition: nutrients and bioactive factors. Pediatr Clin North Am 2013, 60 (1), 49–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bode L, Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology 2012, 22 (9), 1147–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newburg DS; Neubauer SH, CHAPTER 4 - Carbohydrates in Milks: Analysis, Quantities, and Significance In Handbook of Milk Composition, Jensen RG, Ed. Academic Press: San Diego, 1995; pp 273–349. [Google Scholar]
- 15.Coppa G; Pierani P; Zampini L; Carloni I; Carlucci A; Gabrielli O, Oligosaccharides in human milk during different phases of lactation. Acta Paediatrica 1999, 88 (s430), 89–94. [DOI] [PubMed] [Google Scholar]
- 16.Gabrielli O; Zampini L; Galeazzi T; Padella L; Santoro L; Peila C; Giuliani F; Bertino E; Fabris C; Coppa GV, Preterm Milk Oligosaccharides During the First Month of Lactation. Pediatrics 2011, 128 (6), e1520–e1531. [DOI] [PubMed] [Google Scholar]
- 17.Ruhaak LR; Lebrilla CB, Advances in Analysis of Human Milk Oligosaccharides. Adv Nutr 2012, 3 (3), 406S–414S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doherty AM; Lodge CJ; Dharmage SC; Dai X; Bode L; Lowe AJ, Human Milk Oligosaccharides and Associations With Immune-Mediated Disease and Infection in Childhood: A Systematic Review. Front Pediatr 2018, 6 (91). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.German JB; Freeman SL; Lebrilla CB; Mills DA, Human milk oligosaccharides: evolution, structures and bioselectivity as substrates for intestinal bacteria. Nestle Nutr Workshop Ser Pediatr Program 2008, 62, 205–18; discussion 218–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varki A; Cummings RD; Aebi M; Packer NH; Seeberger PH; Esko JD; Stanley P; Hart G; Darvill A; Kinoshita T; Prestegard JJ; Schnaar RL; Freeze HH; Marth JD; Bertozzi CR; Etzler ME; Frank M; Vliegenthart JF; Lütteke T; Perez S; Bolton E; Rudd P; Paulson J; Kanehisa M; Toukach P; Aoki-Kinoshita KF; Dell A; Narimatsu H; York W; Taniguchi N; Kornfeld S, Symbol Nomenclature for Graphical Representations of Glycans. Glycobiology 2015, 25 (12), 1323–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porfirio S; Archer-Hartmann S; Moreau GB; Ramakrishnan G; Haque R; Kirkpatrick BD; Petri WA Jr.; Azadi P, New strategies for profiling and characterization of human milk oligosaccharides. Glycobiology 2020, 30 (10), 774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan J; Ding J; Jin G; Yu D; Yu L; Long Z; Guo Z; Chai W; Liang X, Profiling of Sialylated Oligosaccharides in Mammalian Milk Using Online Solid Phase Extraction-Hydrophilic Interaction Chromatography Coupled with Negative-Ion Electrospray Mass Spectrometry. Anal Chem 2018, 90 (5), 3174–3182. [DOI] [PubMed] [Google Scholar]
- 23.Mariño K; Lane JA; Abrahams JL; Struwe WB; Harvey DJ; Marotta M; Hickey RM; Rudd PM, Method for milk oligosaccharide profiling by 2-aminobenzamide labeling and hydrophilic interaction chromatography. Glycobiology 2011, 21 (10), 1317–1330. [DOI] [PubMed] [Google Scholar]
- 24.Stahl B; Thurl S; Zeng J; Karas M; Hillenkamp F; Steup M; Sawatzki G, Oligosaccharides from human milk as revealed by matrix-assisted laser desorption/ionization mass spectrometry. Anal Biochem 1994, 223 (2), 218–26. [DOI] [PubMed] [Google Scholar]
- 25.Bode L; Contractor N; Barile D; Pohl N; Prudden AR; Boons GJ; Jin YS; Jennewein S, Overcoming the limited availability of human milk oligosaccharides: challenges and opportunities for research and application. Nutr Rev 2016, 74 (10), 635–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao Z; Guo Y; Liu Y; Li L; Zhang Q; Wen L; Wang X; Kondengaden SM; Wu Z; Zhou J; Cao X; Li X; Ma C; Wang PG, Chemoenzymatic Synthesis of a Library of Human Milk Oligosaccharides. J Org Chem 2016, 81 (14), 5851–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smilowitz JT; Lebrilla CB; Mills DA; German JB; Freeman SL, Breast Milk Oligosaccharides: Structure-Function Relationships in the Neonate. Annu Rev Nutr 2014, 34 (1), 143–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vandenplas Y; Berger B; Carnielli VP; Ksiazyk J; Lagström H; Sanchez Luna M; Migacheva N; Mosselmans JM; Picaud JC; Possner M; Singhal A; Wabitsch M, Human Milk Oligosaccharides: 2’-Fucosyllactose (2’-FL) and Lacto-N-Neotetraose (LNnT) in Infant Formula. Nutrients 2018, 10 (9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bode L, Human milk oligosaccharides: prebiotics and beyond. Nutr Rev 2009, 67 (s2), S183–S191. [DOI] [PubMed] [Google Scholar]
- 30.Jantscher-Krenn E; Bode L, Human milk oligosaccharides and their potential benefits for the breast-fed neonate. Minerva Pediatr 2012, 64 (1), 83–99. [PubMed] [Google Scholar]
- 31.Gauhe A; György P; Hoover JRE; Kuhn R; Rose CS; Ruelius HW; Zilliken F, Bifidus factor. IV. Preparations obtained from human milk. Arch Biochem Biophys 1954, 48 (1), 214–224. [DOI] [PubMed] [Google Scholar]
- 32.Kunz C, Historical aspects of human milk oligosaccharides. Adv Nutr 2012, 3 (3), 430s–9s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henrick BM; Hutton AA; Palumbo MC; Casaburi G; Mitchell RD; Underwood MA; Smilowitz JT; Frese SA Elevated Fecal pH Indicates a Profound Change in the Breastfed Infant Gut Microbiome Due to Reduction of Bifidobacterium over the Past Century mSphere [Online], 2018. PubMed. 2018 Mar-Apr). [DOI] [PMC free article] [PubMed]
- 34.Coggins SA; Wynn JL; Weitkamp J-H, Infectious causes of necrotizing enterocolitis. Clin Perinatol 2015, 42 (1), 133–ix. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng N; Gao Y; Zhu W; Meng D; Walker WA, Short chain fatty acids produced by colonizing intestinal commensal bacterial interaction with expressed breast milk are anti-inflammatory in human immature enterocytes. PLOS ONE 2020, 15 (2), e0229283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Majta J; Odrzywolek K; Milanovic B; Hubar V; Wrobel S; Strycharz-Angrecka E; Wojciechowski S; Milanowska K, Identification of Differentiating Metabolic Pathways between Infant Gut Microbiome Populations Reveals Depletion of Function-Level Adaptation to Human Milk in the Finnish Population. mSphere 2019, 4 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ayechu-Muruzabal V; van Stigt AH; Mank M; Willemsen LEM; Stahl B; Garssen J; van’t Land B, Diversity of Human Milk Oligosaccharides and Effects on Early Life Immune Development. Front Pediatr 2018, 6 (239). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asakuma S; Hatakeyama E; Urashima T; Yoshida E; Katayama T; Yamamoto K; Kumagai H; Ashida H; Hirose J; Kitaoka M, Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J Biol Chem 2011, 286 (40), 34583–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gopalakrishna KP; Hand TW, Influence of Maternal Milk on the Neonatal Intestinal Microbiome. Nutrients 2020, 12 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balmer SE; Wharton BA, Diet and faecal flora in the newborn: breast milk and infant formula. Arch Dis Child 1989, 64 (12), 1672–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang I; Corwin EJ; Brennan PA; Jordan S; Murphy JR; Dunlop A, The Infant Microbiome: Implications for Infant Health and Neurocognitive Development. Nurs Res 2016, 65 (1), 76–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Timmerman HM; Rutten N; Boekhorst J; Saulnier DM; Kortman GAM; Contractor N; Kullen M; Floris E; Harmsen HJM; Vlieger AM; Kleerebezem M; Rijkers GT, Intestinal colonisation patterns in breastfed and formula-fed infants during the first 12 weeks of life reveal sequential microbiota signatures. Sci Rep 2017, 7 (1), 8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van den Elsen LWJ; Garssen J; Burcelin R; Verhasselt V, Shaping the Gut Microbiota by Breastfeeding: The Gateway to Allergy Prevention? Front Pediatr 2019, 7 (47). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Craft KM; Townsend SD, Mother Knows Best: Deciphering the Antibacterial Properties of Human Milk Oligosaccharides. Acc Chem Res 2019, 52 (3), 760–768. [DOI] [PubMed] [Google Scholar]
- 45.Craft KM; Thomas HC; Townsend SD, Interrogation of Human Milk Oligosaccharide Fucosylation Patterns for Antimicrobial and Antibiofilm Trends in Group B Streptococcus. ACS Infect Dis 2018, 4 (12), 1755–1765. [DOI] [PubMed] [Google Scholar]
- 46.Craft KM; Gaddy JA; Townsend SD, Human Milk Oligosaccharides (HMOs) Sensitize Group B Streptococcus to Clindamycin, Erythromycin, Gentamicin, and Minocycline on a Strain Specific Basis. ACS Chem Biol 2018, 13 (8), 2020–2026. [DOI] [PubMed] [Google Scholar]
- 47.Ackerman DL; Craft KM; Doster RS; Weitkamp JH; Aronoff DM; Gaddy JA; Townsend SD, Antimicrobial and Antibiofilm Activity of Human Milk Oligosaccharides against Streptococcus agalactiae, Staphylococcus aureus, and Acinetobacter baumannii. ACS Infect Dis 2018, 4 (3), 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ackerman DL; Doster RS; Weitkamp JH; Aronoff DM; Gaddy JA; Townsend SD, Human Milk Oligosaccharides Exhibit Antimicrobial and Antibiofilm Properties against Group B Streptococcus. ACS Infect Dis 2017, 3 (8), 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Craft KM; Thomas HC; Townsend SD, Sialylated variants of lacto-N-tetraose exhibit antimicrobial activity against Group B Streptococcus. Org Biomol Chem 2019, 17 (7), 1893–1900. [DOI] [PubMed] [Google Scholar]
- 50.Chambers SA; Moore RE; Craft KM; Thomas HC; Das R; Manning SD; Codreanu SG; Sherrod SD; Aronoff DM; McLean JA; Gaddy JA; Townsend SD, A Solution to Antifolate Resistance in Group B Streptococcus: Untargeted Metabolomics Identifies Human Milk Oligosaccharide-Induced Perturbations That Result in Potentiation of Trimethoprim. mBio 2020, 11 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newburg DS; Ruiz-Palacios GM; Morrow AL, Human milk glycans protect infants against enteric pathogens. Annu Rev Nutr 2005, 25 (1), 37–58. [DOI] [PubMed] [Google Scholar]
- 52.Le Doare K; Holder B; Bassett A; Pannaraj P, Mother’s Milk: A Purposeful Contribution to the Development of the Infant Microbiota and Immunity. Front Immunol 2018, 9, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rogers KM; Heise M, Modulation of cellular tropism and innate antiviral response by viral glycans. J Innate Immun 2009, 1 (5), 405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar H; Kawai T; Akira S, Pathogen recognition by the innate immune system. Int Rev Immunol 2011, 30 (1), 16–34. [DOI] [PubMed] [Google Scholar]
- 55.Palese P; Zheng H; Engelhardt OG; Pleschka S; García-Sastre A, Negative-strand RNA viruses: genetic engineering and applications. Proc Natl Acad Sci U S A 1996, 93 (21), 11354–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Southgate JA; Bull MJ; Brown CM; Watkins J; Corden S; Southgate B; Moore C; Connor TR, Influenza classification from short reads with VAPOR facilitates robust mapping pipelines and zoonotic strain detection for routine surveillance applications. J Bioinform 2019, 36 (6), 1681–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tafalla M; Buijssen M; Geets R; Vonk Noordegraaf-Schouten M, A comprehensive review of the epidemiology and disease burden of Influenza B in 9 European countries. Hum Vaccines Immunother 2016, 12 (4), 993–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iuliano AD; Roguski KM; Chang HH; Muscatello DJ; Palekar R; Tempia S; Cohen C; Gran JM; Schanzer D; Cowling BJ; Wu P; Kyncl J; Ang LW; Park M; Redlberger-Fritz M; Yu H; Espenhain L; Krishnan A; Emukule G; van Asten L; Pereira da Silva S; Aungkulanon S; Buchholz U; Widdowson MA; Bresee JS, Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 2018, 391 (10127), 1285–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dioxirane Epoxidation of Alkenes - Adam - - Major Reference Works - Wiley Online Library; 2019. [Google Scholar]
- 60.Bouvier NM; Palese P, The biology of influenza viruses. Vaccine 2008, 26 Suppl 4 (Suppl 4), D49–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suzuki Y, The highly pathogenic avian influenza H5N1 - initial molecular signals for the next influenza pandemic. Chang Gung Med J 2009, 32 (3), 258–263. [PubMed] [Google Scholar]
- 62.Noda T; Kawaoka Y, Structure of influenza virus ribonucleoprotein complexes and their packaging into virions. Rev Med Virol 2010, 20 (6), 380–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu Y; Mishra S; Song X; Lasanajak Y; Bradley KC; Tappert MM; Air GM; Steinhauer DA; Halder S; Cotmore S; Tattersall P; Agbandje-McKenna M; Cummings RD; Smith DF, Functional glycomic analysis of human milk glycans reveals the presence of virus receptors and embryonic stem cell biomarkers. J Biol Chem 2012, 287 (53), 44784–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zevgiti S; Zabala JG; Darji A; Dietrich U; Panou-Pomonis E; Sakarellos-Daitsiotis M, Sialic acid and sialyl-lactose glyco-conjugates: design, synthesis and binding assays to lectins and swine influenza H1N1 virus. J Pept Sci 2012, 18 (1), 52–58. [DOI] [PubMed] [Google Scholar]
- 65.Triantis V; Bode L; van Neerven RJJ, Immunological Effects of Human Milk Oligosaccharides. Front Pediatr 2018, 6, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xiao L; Leusink-Muis T; Kettelarij N; van Ark I; Blijenberg B; Hesen NA; Stahl B; Overbeek SA; Garssen J; Folkerts G; Van’t Land B, Human Milk Oligosaccharide 2’-Fucosyllactose Improves Innate and Adaptive Immunity in an Influenza-Specific Murine Vaccination Model. Front Immunol 2018, 9, 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wiciński M; Sawicka E; Gębalski J; Kubiak K; Malinowski B, Human Milk Oligosaccharides: Health Benefits, Potential Applications in Infant Formulas, and Pharmacology. Nutrients 2020, 12 (1), 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Woyessa AB; Abebe A; Beyene B; Tefera M; Assefa E; Ketema H; Teshome B; Bekele A; Dugasa Y; Habebe S; Assefa Z; Sufa D; Alemu D; Tilahun H; Biru M; Shume G, Rotavirus-associated acute diarrhea outbreak in West Shewa Zone of Oromia Regional State, Ethiopia, 2017. Pan Afr Med J 2019, 32, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tate JE; Burton AH; Boschi-Pinto C; Parashar UD; Network, f. t. W. H. O. C. G. R. S.; Agocs M; Serhan F; de Oliveira L; Mwenda JM; Mihigo R; Ranjan Wijesinghe P; Abeysinghe N; Fox K; Paladin F; Network, f. t. W. H. O. C. G. R. S., Global, Regional, and National Estimates of Rotavirus Mortality in Children <5 Years of Age, 2000–2013. Clin Infect Dis 2016, 62 (suppl_2), S96–S105. [DOI] [PubMed] [Google Scholar]
- 70.Freed G; Turbitt E, The global imperative to address vaccine-preventable diseases. Aust Fam Physician 2016, 45, 14–16. [PubMed] [Google Scholar]
- 71.Matthijnssens J; Ciarlet M; McDonald SM; Attoui H; Bányai K; Brister JR; Buesa J; Esona MD; Estes MK; Gentsch JR; Iturriza-Gómara M; Johne R; Kirkwood CD; Martella V; Mertens PPC; Nakagomi O; Parreño V; Rahman M; Ruggeri FM; Saif LJ; Santos N; Steyer A; Taniguchi K; Patton JT; Desselberger U; Van Ranst M, Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch Virol 2011, 156 (8), 1397–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ludert JE; Ruiz MC; Hidalgo C; Liprandi F, Antibodies to Rotavirus Outer Capsid Glycoprotein VP7 Neutralize Infectivity by Inhibiting Virion Decapsidation. J Virol 2002, 76 (13), 6643–6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Feng N; Hu L; Ding S; Sanyal M; Zhao B; Sankaran B; Ramani S; McNeal M; Yasukawa LL; Song Y; Prasad BVV; Greenberg HB, Human VP8* mAbs neutralize rotavirus selectively in human intestinal epithelial cells. J Clin Investig 2019, 129 (9), 3839–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Laucirica DR; Triantis V; Schoemaker R; Estes MK; Ramani S, Milk Oligosaccharides Inhibit Human Rotavirus Infectivity in MA104 Cells. J Nutr 2017, 147 (9), 1709–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hester SN; Chen X; Li M; Monaco MH; Comstock SS; Kuhlenschmidt TB; Kuhlenschmidt MS; Donovan SM, Human milk oligosaccharides inhibit rotavirus infectivity in vitro and in acutely infected piglets. Br J Nutr 2013, 110 (7), 1233–1242. [DOI] [PubMed] [Google Scholar]
- 76.Glezen WP; Taber LH; Frank AL; Kasel JA, Risk of Primary Infection and Reinfection With Respiratory Syncytial Virus. Am J Dis Child 1986, 140 (6), 543–546. [DOI] [PubMed] [Google Scholar]
- 77.Shi T; McAllister DA; O’Brien KL; Simoes EAF; Madhi SA; Gessner BD; Polack FP; Balsells E; Acacio S; Aguayo C; Alassani I; Ali A; Antonio M; Awasthi S; Awori JO; Azziz-Baumgartner E; Baggett HC; Baillie VL; Balmaseda A; Barahona A; Basnet S; Bassat Q; Basualdo W; Bigogo G; Bont L; Breiman RF; Brooks WA; Broor S; Bruce N; Bruden D; Buchy P; Campbell S; Carosone-Link P; Chadha M; Chipeta J; Chou M; Clara W; Cohen C; de Cuellar E; Dang DA; Dash-Yandag B; Deloria-Knoll M; Dherani M; Eap T; Ebruke BE; Echavarria M; de Freitas Lázaro Emediato CC; Fasce RA; Feikin DR; Feng L; Gentile A; Gordon A; Goswami D; Goyet S; Groome M; Halasa N; Hirve S; Homaira N; Howie SRC; Jara J; Jroundi I; Kartasasmita CB; Khuri-Bulos N; Kotloff KL; Krishnan A; Libster R; Lopez O; Lucero MG; Lucion F; Lupisan SP; Marcone DN; McCracken JP; Mejia M; Moisi JC; Montgomery JM; Moore DP; Moraleda C; Moyes J; Munywoki P; Mutyara K; Nicol MP; Nokes DJ; Nymadawa P; da Costa Oliveira MT; Oshitani H; Pandey N; Paranhos-Baccalà G; Phillips LN; Picot VS; Rahman M; Rakoto-Andrianarivelo M; Rasmussen ZA; Rath BA; Robinson A; Romero C; Russomando G; Salimi V; Sawatwong P; Scheltema N; Schweiger B; Scott JAG; Seidenberg P; Shen K; Singleton R; Sotomayor V; Strand TA; Sutanto A; Sylla M; Tapia MD; Thamthitiwat S; Thomas ED; Tokarz R; Turner C; Venter M; Waicharoen S; Wang J; Watthanaworawit W; Yoshida LM; Yu H; Zar HJ; Campbell H; Nair H, Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet 2017, 390 (10098), 946–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rima B; Balkema-Buschmann A; Dundon WG; Duprex P; Easton A; Fouchier R; Kurath G; Lamb R; Lee B; Rota P; Wang L; Consortium IR, ICTV Virus Taxonomy Profile: Paramyxoviridae. J Gen Virol 2019, 100 (12), 1593–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thornhill EM; Verhoeven D, Respiratory Syncytial Virus’s Non-structural Proteins: Masters of Interference. Front Cell Infect Microbiol 2020, 10 (225). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kiss G; Holl JM; Williams GM; Alonas E; Vanover D; Lifland AW; Gudheti M; Guerrero-Ferreira RC; Nair V; Yi H; Graham BS; Santangelo PJ; Wright ER, Structural analysis of respiratory syncytial virus reveals the position of M2–1 between the matrix protein and the ribonucleoprotein complex. J Virol 2014, 88 (13), 7602–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McLellan JS; Yang Y; Graham BS; Kwong PD, Structure of Respiratory Syncytial Virus Fusion Glycoprotein in the Postfusion Conformation Reveals Preservation of Neutralizing Epitopes. J Virol 2011, 85 (15), 7788–7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Collins PL; Hill MG; Cristina J; Grosfeld H, Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. PNAS 1996, 93 (1), 81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Duska-McEwen G; Senft AP; Ruetschilling TL; Barrett EG; Buck RH, Human Milk Oligosaccharides Enhance Innate Immunity to Respiratory Syncytial Virus and Influenza Food Nutr Sci 2014, Vol.05No.14, 13. [Google Scholar]
- 84.Esbjörnsson J; Jansson M; Jespersen S; Månsson F; Hønge BL; Lindman J; Medina C; da Silva ZJ; Norrgren H; Medstrand P; Rowland-Jones SL; Wejse C, HIV-2 as a model to identify a functional HIV cure. AIDS Res Ther 2019, 16 (1), 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Frankel AD; Young JAT, HIV-1: Fifteen Proteins and an RNA. Annu Rev Biochem 1998, 67 (1), 1–25. [DOI] [PubMed] [Google Scholar]
- 86.Pancera M; Majeed S; Ban YE; Chen L; Huang CC; Kong L; Kwon YD; Stuckey J; Zhou T; Robinson JE; Schief WR; Sodroski J; Wyatt R; Kwong PD, Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci U S A 2010, 107 (3), 1166–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cannon PM; Matthews S; Clark N; Byles ED; Iourin O; Hockley DJ; Kingsman SM; Kingsman AJ, Structure-function studies of the human immunodeficiency virus type 1 matrix protein, p17. J Virol 1997, 71 (5), 3474–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bugatti A; Giagulli C; Urbinati C; Caccuri F; Chiodelli P; Oreste P; Fiorentini S; Orro A; Milanesi L; D’Ursi P; Caruso A; Rusnati M, Molecular Interaction Studies of HIV-1 Matrix Protein p17 and Heparin: Identification of the heparain-binding motif of p17 as a target for the development of multitarget antagonists J Biol Chem 2013, 288 (2), 1150–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tang S; Zhao J; Wang A; Viswanath R; Harma H; Little RF; Yarchoan R; Stramer SL; Nyambi PN; Lee S; Wood O; Wong EY; Wang X; Hewlett IK, Characterization of immune responses to capsid protein p24 of human immunodeficiency virus type 1 and implications for detection. Clin Vaccine Immunol 2010, 17 (8), 1244–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Geijtenbeek TBH; Kwon DS; Torensma R; van Vliet SJ; van Duijnhoven GCF; Middel J; Cornelissen ILMHA; Nottet HSLM; KewalRamani VN; Littman DR; Figdor CG; van Kooyk Y, DC-SIGN, a Dendritic Cell–Specific HIV-1-Binding Protein that Enhances trans-Infection of T Cells. Cell 2000, 100 (5), 587–597. [DOI] [PubMed] [Google Scholar]
- 91.María Colmenares‡ AP-K, Oscar Muñiz Pello§, Angel L. Corbí¶ and Luis Rivas, Dendritic Cell (DC)-specific Intercellular Adhesion Molecule 3 (ICAM-3)-grabbing Nonintegrin (DC-SIGN, CD209), a C-type Surface Lectin in Human DCs, Is a Receptor for LeishmaniaAmastigotes. J Biol Chem 2002, 277, 36766–36769. [DOI] [PubMed] [Google Scholar]
- 92.Bode L; Kuhn L; Kim HY; Hsiao L; Nissan C; Sinkala M; Kankasa C; Mwiya M; Thea DM; Aldrovandi GM, Human milk oligosaccharide concentration and risk of postnatal transmission of HIV through breastfeeding. Am J Clin Nutr 2012, 96 (4), 831–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.van Liempt E; Bank CM; Mehta P; Garciá-Vallejo JJ; Kawar ZS; Geyer R; Alvarez RA; Cummings RD; Kooyk Y; van Die I, Specificity of DC-SIGN for mannose- and fucose-containing glycans. FEBS Lett 2006, 580 (26), 6123–31. [DOI] [PubMed] [Google Scholar]
- 94.Seviour T; Derlon N; Dueholm MS; Flemming H-C; Girbal-Neuhauser E; Horn H; Kjelleberg S; van Loosdrecht MCM; Lotti T; Malpei MF; Nerenberg R; Neu TR; Paul E; Yu H; Lin Y, Extracellular polymeric substances of biofilms: Suffering from an identity crisis. Water Res 2019, 151, 1–7. [DOI] [PubMed] [Google Scholar]
- 95.Desselberger U, Caliciviridae Other Than Noroviruses. Viruses 2019, 11 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hansman GS; Natori K; Shirato-Horikoshi H; Ogawa S; Oka T; Katayama K; Tanaka T; Miyoshi T; Sakae K; Kobayashi S; Shinohara M; Uchida K; Sakurai N; Shinozaki K; Okada M; Seto Y; Kamata K; Nagata N; Tanaka K; Miyamura T; Takeda N, Genetic and antigenic diversity among noroviruses. J Gen Virol 2006, 87 (4), 909–919. [DOI] [PubMed] [Google Scholar]
- 97.Xi J; Graham D; Wang K; Estes M, Norwalk virus genome cloning and characterization. Science 1990, 250 (4987), 1580–1583. [DOI] [PubMed] [Google Scholar]
- 98.Prasad BV; Rothnagel R; Jiang X; Estes MK, Three-dimensional structure of baculovirus-expressed Norwalk virus capsids. J Virol 1994, 68 (8), 5117–5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hardy ME, Norovirus protein structure and function. FEMS Microbiol Lett 2005, 253 (1), 1–8. [DOI] [PubMed] [Google Scholar]
- 100.Graziano VR; Wei J; Wilen CB, Norovirus Attachment and Entry. Viruses 2019, 11 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tan M; Hegde RS; Jiang X, The P domain of norovirus capsid protein forms dimer and binds to histo-blood group antigen receptors. J Virol 2004, 78 (12), 6233–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Weichert S; Koromyslova A; Singh BK; Hansman S; Jennewein S; Schroten H; Hansman GS, Structural Basis for Norovirus Inhibition by Human Milk Oligosaccharides. J Virol 2016, 90 (9), 4843–4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shang J; Piskarev VE; Xia M; Huang P; Jiang X; Likhosherstov LM; Novikova OS; Newburg DS; Ratner DM, Identifying human milk glycans that inhibit norovirus binding using surface plasmon resonance. Glycobiology 2013, 23 (12), 1491–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hong CR; Han SM; Jaksic T, Surgical considerations for neonates with necrotizing enterocolitis. Semin Fetal Neonat M 2018, 23 (6), 420–425. [DOI] [PubMed] [Google Scholar]
- 105.Neu J; Walker WA, Necrotizing enterocolitis. N Engl J Med 2011, 364 (3), 255–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Claud EC; Keegan KP; Brulc JM; Lu L; Bartels D; Glass E; Chang EB; Meyer F; Antonopoulos DA, Bacterial community structure and functional contributions to emergence of health or necrotizing enterocolitis in preterm infants. Microbiome 2013, 1 (1), 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tanner SM; Berryhill TF; Ellenburg JL; Jilling T; Cleveland DS; Lorenz RG; Martin CA, Pathogenesis of necrotizing enterocolitis: modeling the innate immune response. Am J Pathol 2015, 185 (1), 4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ballance WA; Dahms BB; Shenker N; Kliegman RM, Pathology of neonatal necrotizing enterocolitis: A ten-year experience. J Pediatr 1990, 117 (1, Part 2), S6–S13. [DOI] [PubMed] [Google Scholar]
- 109.Bode L, Human Milk Oligosaccharides in the Prevention of Necrotizing Enterocolitis: A Journey From in vitro and in vivo Models to Mother-Infant Cohort Studies. Front Pediatr 2018, 6, 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Autran CA; Kellman BP; Kim JH; Asztalos E; Blood AB; Spence ECH; Patel AL; Hou J; Lewis NE; Bode L, Human milk oligosaccharide composition predicts risk of necrotising enterocolitis in preterm infants. Gut 2018, 67 (6), 1064–1070. [DOI] [PubMed] [Google Scholar]
- 111.Rapoport EM; Kurmyshkina OV; Bovin NV, Mammalian galectins: structure, carbohydrate specificity, and functions. Biochemistry (Mosc) 2008, 73 (4), 393–405. [DOI] [PubMed] [Google Scholar]
- 112.Koliwer-Brandl H; Siegert N; Umnus K; Kelm A; Tolkach A; Kulozik U; Kuballa J; Cartellieri S; Kelm S, Lectin inhibition assays for the analysis of bioactive milk sialoglycoconjugates. Int Dairy J 2011, 21 (6), 413–420. [Google Scholar]
- 113.Gross R; Conzelmann C; Muller JA; Stenger S; Steinhart K; Kirchhoff F; Munch J, Detection of SARS-CoV-2 in human breastmilk. Lancet 2020, 395 (10239), 1757–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]


