Abstract
Summary
In response to the COVID-19 pandemic, we established COVID-KOP, a new knowledgebase integrating the existing Reasoning Over Biomedical Objects linked in Knowledge Oriented Pathways (ROBOKOP) biomedical knowledge graph with information from recent biomedical literature on COVID-19 annotated in the CORD-19 collection. COVID-KOP can be used effectively to generate new hypotheses concerning repurposing of known drugs and clinical drug candidates against COVID-19 by establishing respective confirmatory pathways of drug action.
Availability and implementation
COVID-KOP is freely accessible at https://covidkop.renci.org/. For code and instructions for the original ROBOKOP, see: https://github.com/NCATS-Gamma/robokop.
1 Introduction
In the absence of effective medications for COVID-19, there is an urgent need to identify drugs that can combat this ongoing pandemic. This task can be accomplished most rapidly by repurposing the existing medications. Biomedical knowledge graphs such as Reasoning Over Biomedical Objects linked in Knowledge Oriented Pathways (ROBOKOP) (Bizon et al., 2019) provide an efficient way to identify potential candidate drugs by making inferences on the relationships between knowledge graph nodes. We have merged the ROBOKOP knowledge graph with the new supply of COVID-19-related information from recent publications and other knowledge sources to form COVID-KOP.
2 Materials and methods
We used COVID-19 Open Research Dataset (CORD-19, https://allenai.org/data/cord-19) containing over 60 000 full-text research papers with ontological tagging provided by the SciBiteAI group (https://github.com/SciBiteLabs/CORD19). We parsed CORD-19 data into a format compatible with the ROBOKOP’s knowledge graph by extracting, sentence by sentence, the counts of ontological terms and tag co-occurrences. This resulted in 800 000 new edges in the COVID-KOP knowledge graph. In addition, we used the SciGraph tool (https://github.com/SciGraph/SciGraph), which also allows biomedical ontological term tagging and tag co-occurrence counts at the paper rather than sentence level, leading to 4.5 million new edges.
Gene Ontology Annotation data for all viral proteins, including those of SARS-CoV-2, were downloaded from the EBI FTP site (see https://github.com/TranslatorIIPrototypes/ViralProteome for details). The knowledge graph integration tool KGX (https://github.com/NCATS-Tangerine/kgx) was used to merge the GOA data and create a ROBOKOP-formatted graph. In total, the COVID-KOP database and knowledge graph comprise nodes for 40 000 proteins, 4000 NCBITaxon (Federhen, 2012) terms, 1300 GO annotations (Ashburner et al., 2000) and 232 000 new edges (labeled as ‘related_to’) on top of those in ROBOKOP. We use bidirectional edges for these linkages because the connection directionality is not provided in primary sources.
A set of 26 SARS-CoV-2 symptoms was identified from various resources (https://www.cebm.net/covid-19/covid-19-signs-and-symptoms-tracker/; https://covid.cd2h.org/N3C; https://www.hematology.org/covid-19/covid-19-and-coagulopathy) and a recent commentary (Schett et al., 2020). This information was manually entered into the COVID-KOP database as edges between the COVID-19 and its phenotypes.
Due to multiple identifier systems used by different databases for the same entities, we utilized the Data Translator Node Normalization API (https://github.com/TranslatorIIPrototypes/NodeNormalization) for data integration. COVID-KOP is powered by the knowledge graph database Neo4J (https://neo4j.com/), which uses Cypher to enable complex graph database queries. The fully integrated COVID-KOP KG can be mined in the same way as ROBOKOP KG (Bizon et al., 2019).
3 Case study
We illustrate the utility of COVID-KOP by examining the linagliptin—COVID-19 connection (Fig. 1). Linagliptin is a type 2 diabetes (T2D) drug that is undergoing clinical trials for COVID-19 (https://clinicaltrials.gov/ct2/show/NCT04341935). Linagliptin inhibits dipeptidyl peptidase-4 (DPP-4), which degrades hormones stimulating insulin production (Scott, 2011). It is known to be overexpressed in patients with T2D (Barchetta et al., 2020). COVID-19 patients with T2D have a higher risk of developing more severe symptoms, possibly due to an increased expression of the host receptor ACE2 (Fang et al., 2020).
Fig. 1.
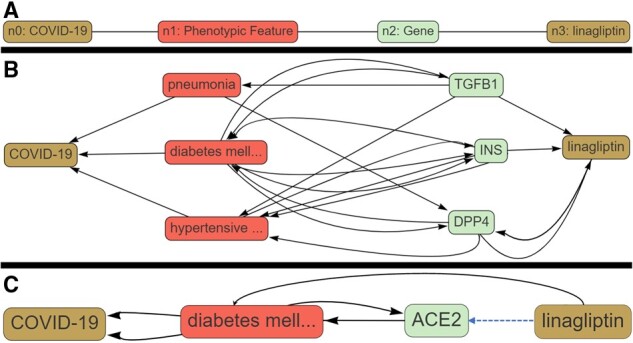
Execution of a COVID-KOP query for linagliptin-COVID-19 pair. (A) Query graph; (B) answer graph for linagliptin-DPP4-T2D-COVID-19 pathway; (C) answer graph for linagliptin-ACE2-T2D-COVID-19 pathway
We applied COVID-KOP to identify possible mechanistic connections between linagliptin and COVID-19 that would support its expected therapeutic effect or identify possible side effects. Using the COVID-KOP user interface, a query is constructed as follows. Individual nodes are placed and linked to specific biomedical objects (such as node n2 being a gene in Fig. 1A). The user can then link these nodes together in any order. In this case study, node n0 is matched to COVID-19; n1 is marked as any phenotypic feature linked to COVID-19; and n2 is any gene related to a phenotypic feature connected to COVID-19. Finally, n2 must also have a connection to the linagliptin.
Executing this query generated 47 subgraphs ranked by an algorithm implemented in ROBOKOP (Morton et al., 2019), with the pathway serving as a rationale for the linagliptin clinical trial against COVID-19 (Linagliptin-T2D-DPP4-COVID-19; Fig. 1B) ranked the highest. Three conditions—pneumonia, T2D and hypertensive disorder—were identified as associated with COVID-19. Genes associated with these conditions and also linked to linagliptin are annotated in Figure 1B. Pneumonia was not directly related to any of these genes except for transforming growth factor beta (TGFβ-1), the inhibition of which results in increased susceptibility to pneumonia in mice with a pneumonia-resistant phenotype (Neill et al., 2012). Relatedly, linagliptin has been reported to ‘significantly decrease’ TGFβ-1 transcript levels (Wang et al., 2015). Our answer graph suggests that linagliptin may inhibit TGFβ-1 transcription and thus possibly increase patients’ risk of developing more severe pneumonia, even though it may alleviate some of the more severe pathologies of COVID-19 seen in T2D patients via DPP-4 inhibition.
We also uncovered an additional inference associating linagliptin and Angiotensin-Converting Enzyme II (ACE2), the host receptor for SARS-CoV-2 entry: Linagliptin-ACE2-T2D-COVID-19 (Fig. 1C). Downregulation of ACE2 expression in lung tissue has been associated with severe COVID-19 clinical outcomes (Nakhleh and Shehadeh, 2020). Expression of ACE2 is increased in patients with T2D; thus, ACE inhibitors and angiotensin-receptor blockers are commonly used to treat individuals with this condition (Pal and Bhansali, 2020). A recent study (Zhang et al., 2015) demonstrated that administration of linagliptin significantly upregulated ACE2 expression and was useful in preventing angiotensin II-induced cardiac fibrosis in animal models. It is yet unclear if upregulating ACE2 would be beneficial or detrimental to patients, especially those with T2D; nevertheless, this inference reveals an additional pathway linking linagliptin and COVID-19. Thus, COVID-KOP could help recovering known biochemical pathways associating a drug with COVID-19 (linagliptin-DPP4-T2D -COVID-19) as well as offer potentially novel inferences (linagliptin-TGFB1-Pneumonia-COVID-19).
4 Conclusions
COVID-KOP is a knowledgebase and web portal that integrates the existing ROBOKOP biomedical knowledge graph with information gathered from recently published biomedical information regarding COVID-19. The presented case study illustrates the utility of COVID-KOP for generating and supporting hypotheses concerning drug repurposing against COVID-19.
Funding
This work was supported by the National Center for Advancing Translational Sciences, National Institutes of Health [OT2R002514] and National Institutes of Health [1U01CA207160].
Conflict of Interest: none declared.
Contributor Information
Daniel Korn, Department of Computer Science, University of North Carolina at Chapel Hill, USA.
Tesia Bobrowski, Division of Chemical Biology and Medicinal Chemistry, UNC Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, USA.
Michael Li, Department of Computer Science, University of North Carolina at Chapel Hill, USA.
Yaphet Kebede, Renaissance Computing Institute, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599-7568, USA.
Patrick Wang, CoVar Applied Technologies, Durham, NC 27701, USA.
Phillips Owen, Renaissance Computing Institute, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599-7568, USA.
Gaurav Vaidya, Renaissance Computing Institute, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599-7568, USA.
Eugene Muratov, Division of Chemical Biology and Medicinal Chemistry, UNC Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, USA.
Rada Chirkova, Department of Computer Science, North Carolina State University, Raleigh, NC 27606-5550, USA.
Chris Bizon, Renaissance Computing Institute, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599-7568, USA.
Alexander Tropsha, Division of Chemical Biology and Medicinal Chemistry, UNC Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, USA.
References
- Ashburner M. et al. (2000) Gene ontology: tool for the unification of biology. Nat. Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchetta I. et al. (2020) COVID-19 and diabetes: is this association driven by the DPP4 receptor? Potential clinical and therapeutic implications. Diabetes Res. Clin. Pract., 163, 108165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizon C. et al. (2019) ROBOKOP KG and KGB: integrated knowledge graphs from federated sources. J. Chem. Inf. Model., 59, 4968–4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L. et al. (2020) Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir. Med., 8, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federhen S. (2012) The NCBI Taxonomy database. Nucleic Acids Res., 40, D136–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton K. et al. (2019) ROBOKOP: an abstraction layer and user interface for knowledge graphs to support question answering. Bioinformatics, 35, 5382–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhleh A., Shehadeh N. (2020) Interactions between antihyperglycemic drugs and the renin-angiotensin system: putative roles in COVID-19. A mini-review. Diabetes Metab. Syndr., 14, 509–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill D.R. et al. (2012) T regulatory cells control susceptibility to invasive pneumococcal pneumonia in mice. PLoS Pathog., 8, e1002660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal R., Bhansali A. (2020) COVID-19, diabetes mellitus and ACE2: the conundrum. Diabetes Res. Clin. Pract., 162, 108132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schett G. et al. (2020) COVID-19: risk for cytokine targeting in chronic inflammatory diseases? Nat. Rev. Immunol., 20, 271–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L.J. (2011) Linagliptin. Drugs, 71, 611–624. [DOI] [PubMed] [Google Scholar]
- Wang X.-Y. et al. (2015) P1206 : anti-inflammatory and direct antifibrotic effect of oral hepatotropic DPP4 inhibitors in models of NASH and biliary fibrosis. J. Hepatol., 62, S809. [Google Scholar]
- Zhang L.H. et al. (2015) Preservation of glucagon-like peptide-1 level attenuates angiotensin II-induced tissue fibrosis by altering AT1/AT2 receptor expression and angiotensin-converting enzyme 2 activity in rat heart. Cardiovasc. Drugs Ther., 29, 243–255. [DOI] [PubMed] [Google Scholar]


