Abstract
Objectives
Histone methyltransferase G9a, also known as Euchromatic Histone Lysine Methyltransferase 2 (EHMT2), mediates H3K9 methylation which is associated with transcriptional repression. It possesses immunomodulatory effects and is overexpressed in multiple types of cancer. In this study, we investigated the role of G9a in the induction of trained immunity, a de facto innate immune memory, and its effects in non‐muscle‐invasive bladder cancer (NMIBC) patients treated with intravesical Bacillus Calmette‐Guérin (BCG).
Methods
EHMT2 expression was assessed upon induction of trained immunity by RNA sequencing and Western blotting. G9a inhibitor BIX‐01294 was used to investigate the effect on trained immunity responses in vitro. Subsequent cytokine production was measured by ELISA, epigenetic modifications were measured by ChIP‐qPCR, Seahorse technology was used to measure metabolic changes, and a luminescence assay was used to measure ROS release. RNA sequencing was performed on BIX‐01294‐treated monocytes ex vivo.
Results
The expression of EHMT2 mRNA and protein decreased in monocytes during induction of trained immunity. G9a inhibition by BIX‐01294 induced trained immunity and amplified trained immunity responses evoked by various microbial ligands in vitro. This was accompanied by decreased H3K9me2 at the promoters of pro‐inflammatory genes. G9a inhibition was also associated with amplified ex vivo trained immunity responses in circulating monocytes of NMIBC patients. Additionally, altered RNA expression of inflammatory genes in monocytes of NMIBC patients was observed upon ex vivo G9a inhibition. Furthermore, intravesical BCG therapy decreased H3K9me2 at the promoter of pro‐inflammatory genes.
Conclusion
Inhibition of G9a is important in the induction of trained immunity, and G9a may represent a novel therapeutic target in NMIBC patients.
Keywords: BCG, EHMT2, G9a, inflammation, non‐muscle‐invasive bladder cancer, trained immunity
In this study, we show that G9a inhibits induction of trained immunity in human monocytes. Pharmacological G9a inhibition enhances trained immunity responses, accompanied by decreased H3K9me2 marks at pro‐inflammatory genes. Additionally, ex vivo G9a inhibition was associated with amplified trained immunity responses in monocytes derived from non‐muscle‐invasive bladder cancer patients and altered RNA expression of inflammatory genes.

Introduction
Innate immune cells retain a high plasticity in response to environmental stimuli in order to be able to react rapidly and appropriately during infections. In the last decade, studies have shown that innate immune cells, such as macrophages and natural killer cells, are able to adapt after a previously encountered infection or vaccination and build up a de facto non‐specific innate immune memory through epigenetic and metabolic rewiring. This phenomenon has been termed trained immunity and is described as long‐term functional reprogramming of cells upon stimulation, resulting in an improved response towards a secondary non‐specific challenge. 1 , 2 , 3 Exogenous stimuli, such as β‐glucan (a major cell wall component of Candida albicans) and the Bacillus Calmette‐Guérin (BCG) vaccine, and endogenous stimuli, such as oxidised low‐density lipoprotein (oxLDL) and uric acid, are able to induce different trained immunity programs. 2 , 4 β‐glucan induces trained immunity by binding to the pattern recognition receptor Dectin‐1, and subsequent signalling via the Akt‐mTor‐HIF‐1α pathway, 5 , 6 while BCG induces trained immunity by binding to the NOD2 receptor. 7 Both β‐glucan and BCG have been shown to lead to an enhanced innate immune response upon re‐stimulation or re‐infection. 5 , 6 , 7 In contrast, lipopolysaccharide (LPS) induces a tolerant state in monocytes and macrophages by binding to Toll‐like receptor (TLR) 4, resulting in a long‐term attenuated immune response. 8
Epigenetic rewiring is essential to regulate gene expression for induction of trained immunity. 5 , 6 , 7 , 9 In vitro experiments have shown that broad‐spectrum inhibition of histone methyltransferases abrogated β‐glucan‐induced trained immunity in monocytes. 5 In β‐glucan‐trained monocytes, increased histone 3 lysine 27 (H3K27) acetylation, as well as increased H3K4 mono‐ and trimethylation, is observed at promoters of genes involved in trained immunity (IL6, TNFA, IL1B). 8 , 9 , 10 These epigenetic modifications are generally known to result in transcriptionally active chromatin. Additionally, BCG‐induced trained immunity is associated with decreased H3K9 trimethylation of trained immunity‐related genes, which is associated with gene repression. 11 Recently, lysine methyltransferase Set7 was identified as an important epigenetic regulator of β‐glucan‐induced trained immunity. 10 However, the involvement of other epigenetic modifiers, especially those regulating repressive histone marks, remains elusive.
Multiple studies have implicated a role for G9a in immunity (reviewed by Scheer et al. 12 ). G9a/Euchromatic Histone Lysine Methyltransferase 2 (EHMT2) is a member of the SET domain‐containing family that enables lysine methyltransferase activity. G9a activity is dependent on heterodimerisation with the related G9a‐like protein (GLP). 13 G9a performs mainly H3K9 dimethylation (H3K9me2), but is also able to monomethylate, and to a lesser extent trimethylate the H3K9 residue. 12 , 14 , 15 These epigenetic modifications are generally associated with gene repression. Notably, G9a is also capable of methylation of non‐histone proteins and comprises methyltransferase‐independent activities. 12 Interestingly, G9a‐dependent H3K9me2 is associated with gene repression during endotoxin tolerance. 16 , 17 Since the effect of G9a on trained immunity is unknown, we investigated its role in inducing innate immune memory. Furthermore, G9a is overexpressed in multiple types of cancer, 18 including non‐muscle‐invasive bladder cancer (NMIBC) where high expression is associated with a poor prognosis. 19 As G9a inhibition has been shown to partially trigger immune‐mediated bladder cancer regression, 19 and trained immunity might affect the efficacy of BCG bladder instillations 20 (the gold standard therapy in high‐risk NMIBC), we also investigated the role of G9a in BCG‐treated NMIBC patients.
In this study, we show that G9a inhibits induction of trained immunity in human monocytes. Pharmacological G9a inhibition enhances trained immunity responses, accompanied by decreased H3K9me2 marks at pro‐inflammatory genes. Additionally, ex vivo G9a inhibition was associated with amplified trained immunity responses in monocytes derived from NMIBC patients, and altered RNA expression of inflammatory genes. Furthermore, intravesical BCG immunotherapy decreased H3K9me2 at the promoter of pro‐inflammatory genes.
Results
EHMT2 expression is decreased upon induction of trained immunity
An in vitro model was used to study G9a/EHMT2 in the context of trained immunity. Isolated monocytes from peripheral blood of healthy individuals were stimulated for 24 h, washed, rested for 5 days, and restimulated for 24 h with non‐specific stimuli (Figure 1a). Subsequently, cytokine production capacity (such as IL‐6 and TNF‐α) was measured to determine the trained immunity response. 21 First, we assessed G9a/EHMT2 expression during induction of trained immunity. As depicted in Figure 1b, EHMT2 mRNA expression upon stimulation with LPS and β‐glucan is decreased compared to the control condition. Moreover, EHMT2 protein expression at day 6 (before restimulation) is decreased upon β‐glucan‐induced trained immunity (Figure 1c). Intravenous administration of endotoxin to healthy volunteers (Figure 1d) also led to decreased EHMT2 mRNA expression after 4 h (Figure 1e), concomitantly with short‐term increased cytokine release by monocytes. 22 To investigate the role of EHMT2 in trained immunity in vivo, we assessed the RNA expression in BCG‐vaccinated healthy individuals (Figure 1d). As depicted in Figure 1f, four of the six individuals vaccinated with BCG exhibited decreased EHMT2 mRNA expression in monocytes after 1 month, which is in line with the known interindividual variability in responsiveness to BCG vaccination. RT‐qPCR EHMT2 measurements of in vitro trained monocytes were performed to validate these findings (Supplementary figure 1).
Figure 1.
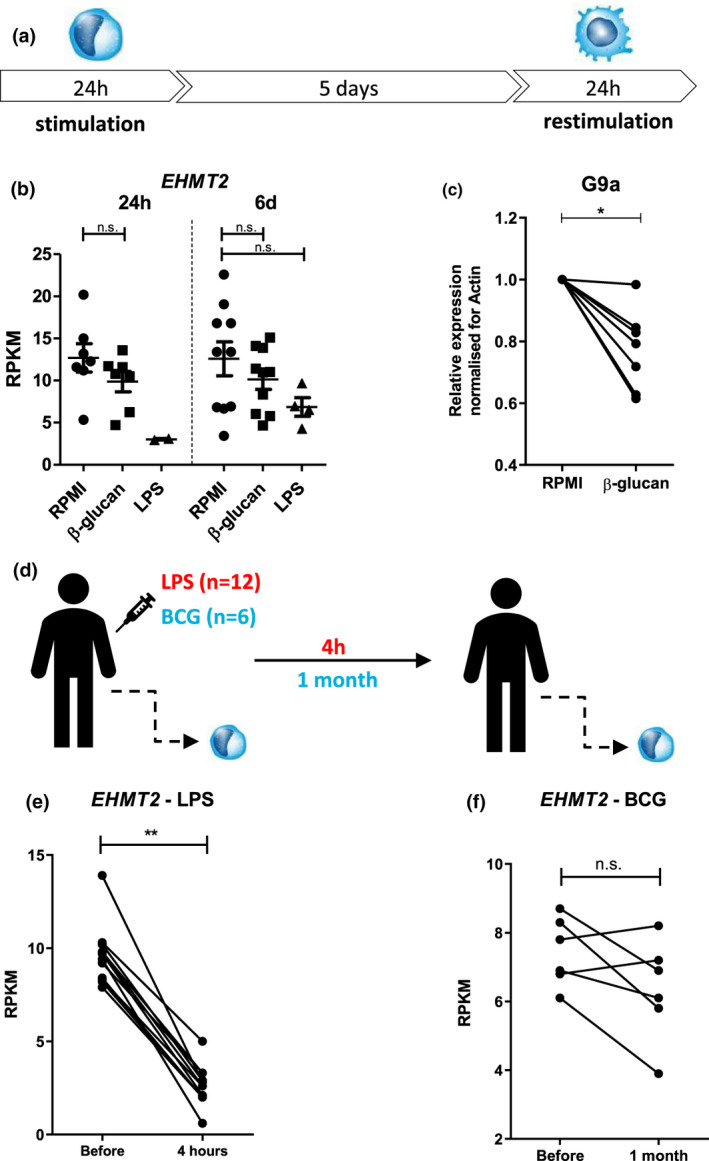
Euchromatic Histone Lysine Methyltransferase 2 (EHMT2) expression is decreased upon induction of trained immunity. (a) Outline of in vitro training method: isolated monocytes were stimulated or left in culture medium alone (RPMI) for 24 h, rested for 5 days, and restimulated with non‐specific stimuli for 24 h. (b) Percoll‐isolated monocytes were stimulated for 24 h with 1 μg mL−1 β‐glucan or 10 ng mL−1 LPS. Cells were analysed by performing RNA sequencing at baseline (prior to stimulation), after 24 h (upon stimulation, n = 2 for LPS and n = 7 for RPMI and β‐glucan) and 6 days (before restimulation, n = 10 for RPMI and β‐glucan). EHMT2 mRNA expression is shown as average reads per kilobase million (RPKM; 2 independent experiments, mean ± SEM, n.s., Wilcoxon signed‐rank test). (c) Relative G9a expression by Western blot of day 6 β‐glucan (2 μg mL−1)‐trained monocytes (n = 7, each line represents one individual) normalised for actin loading control (3 independent experiments, mean ± SEM, *P < 0.05, Wilcoxon signed‐rank test). (d) Healthy individuals received endotoxin, other healthy individuals were administered with BCG, and EHMT2 mRNA expression before, and respectively 4 h, and 1 month later, in monocytes was assessed. (e) EHMT2 mRNA expression in RPKM in monocytes by performing RNA‐seq before and 4 h after 2 ng kg−1 endotoxin administration (n = 12, each line represents one individual, 1 experiment, **P < 0.01, Wilcoxon signed‐rank test). (f) EHMT2 mRNA expression in RPKM in monocytes by performing RNA‐seq, before and 1 month after BCG vaccination in healthy individuals (n = 6, 1 experiment, each line represents one individual, Wilcoxon signed‐rank test, n.s.).
G9a inhibition results in decreased H3K9me2 at gene promoters involved in trained immunity
The synthetic molecule BIX‐01294 23 , 24 was used to inhibit G9a in monocytes in vitro. For this, BIX‐01294 was supplemented during the first 24 h of the trained immunity protocol together with the cells and trained immunity stimuli, and subsequently washed away. We observed that BIX‐01294 did not affect the viability of the cells as assessed by induction of apoptosis (Supplementary figure 2a), and lactate dehydrogenase (LDH) release (Supplementary figure 2b). The addition of BIX‐01294 did not significantly affect EHMT2 mRNA expression in the in vitro training experiment (Figure 2a). Additionally, G9a protein expression was not significantly affected during in vitro training by BIX‐01294 at day 6 (Figure 2b, for Western blots see Supplementary figure 2c). Therefore, we next studied the effect of BIX‐01294 on G9a function. Since G9a mainly performs H3K9 dimethylation, we studied whether this repressive epigenetic modification was affected upon BIX‐01294 exposure. As depicted in Figure 2c, G9a inhibition by BIX‐01294 during β‐glucan‐induced trained immunity was accompanied by decreased H3K9me2 at promoters of genes important for the trained immunity response (TNFA, IL1B). H3K9me2 at the MTOR promoter was decreased in G9a inhibited β‐glucan‐trained cells compared to the control condition (Figure 2c), while glycolytic genes hexokinase 2 (HK2) and phosphofructokinase (PFKP) were not affected (Supplementary figure 2d).
Figure 2.
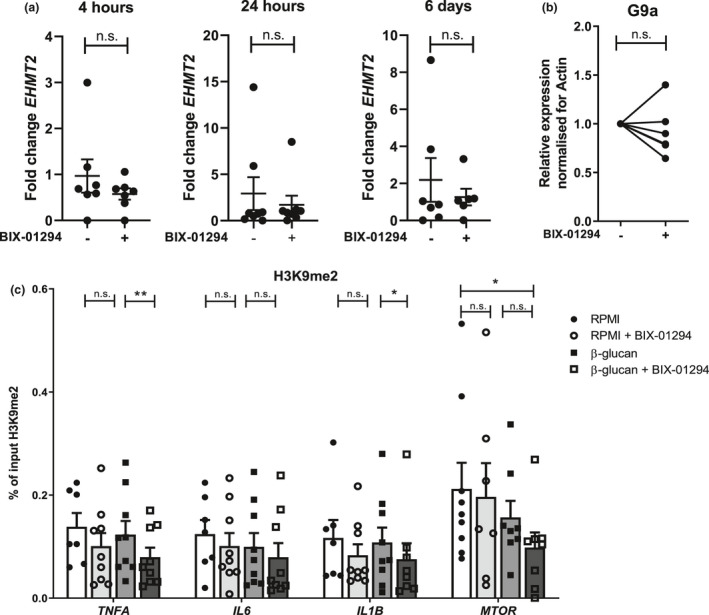
G9a inhibition is accompanied by decreased H3K9me2 at gene promoters involved in trained immunity (a) qRT‐PCR Euchromatic Histone Lysine Methyltransferase 2 (EHMT2) mRNA expression of monocytes after 4h, 1 day, 6 days, upon 24‐h stimulation ± 1 µm BIX‐01294 adjusted for housekeeping gene. Fold change compared to t = 0 (n = 7, mean ± SEM, three independent experiments, Mann–Whitney U‐test, n.s.). (b) G9a protein expression by Western blot of day 6 monocytes incubated for 24 h ± 1 µm BIX‐01294, normalised for actin loading control (n = 7, each line represents one individual, three independent experiments, Wilcoxon signed‐rank test, n.s.). (c) ChIP‐qPCR H3K9me2 levels of day 6 2 μg mL−1 β‐glucan‐trained monocytes ± 1 µm BIX‐01294, at promoters of TNFA, IL1B, IL6 and MTOR (n = 7 RPMI, n = 9 β‐glucan and β‐glucan/RPMI + BIX‐01294, three independent experiments, *P < 0.05, **P < 0.01, Wilcoxon signed‐rank test, mean ± SEM).
G9a inhibition increases trained immunity responses in vitro
To assess the effect of G9a inhibition on trained immunity responses, we measured changes in IL‐6 and TNF‐α cytokine production after restimulation at day 7. Addition of BIX‐01294 for 24 h to monocytes induced an increased pro‐inflammatory response in macrophages after restimulation 1 week later, as shown by increased IL‐6 and TNF‐α production, indicating a trained immunity response (Figure 3a). BCG‐ and oxLDL‐induced trained immunity was amplified by G9a inhibition through BIX‐01294 (Figure 3a), while the impact of the G9a inhibitor on β‐glucan‐induced trained immunity was not significant. LPS‐induced tolerance resulted in decreased IL‐6 and TNF‐α production by macrophages, and this effect could be partially reversed by BIX‐01294 (Figure 3a). The effect on cytokine responses was less pronounced with synthetic G9a inhibitors UNC0638 and UNC0642 (Supplementary figure 3a and b) and was associated with a small but significant increase in the number of cells on day 6 (Supplementary figure 3c). In addition to increased production of pro‐inflammatory cytokines, trained immunity is characterised by a higher metabolic rate. 2 Therefore, basal rates of oxidative phosphorylation and glycolysis [measured by respectively oxidative consumption rate (OCR) and extracellular acidification rate (ECAR)] were assessed at day 6 upon β‐glucan‐induced trained immunity. G9a inhibition by BIX‐01294 increased ECAR and OCR, and increased OCR in β‐glucan‐trained monocytes (Figure 3b and Supplementary figure 3d), which is in agreement with increased trained immunity responses.
Figure 3.
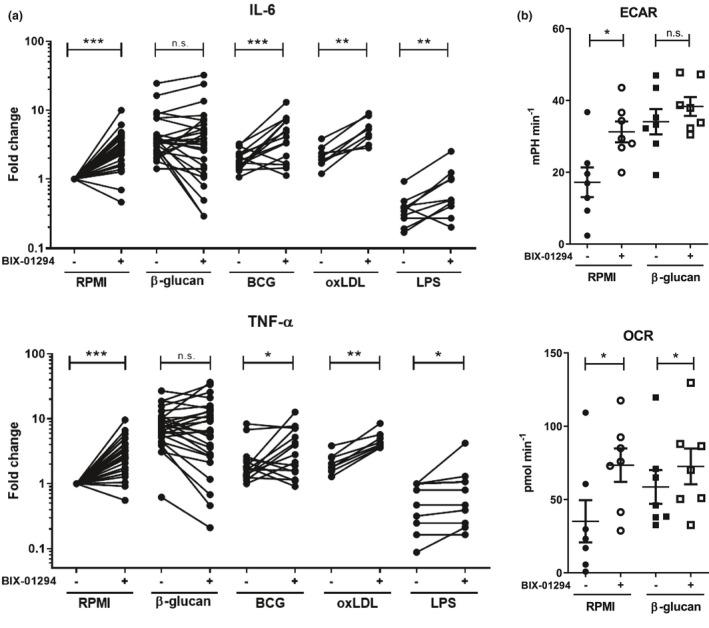
G9a inhibition by BIX‐01294 results in increased trained immunity responses in vitro.(a) Cytokine assessment in supernatant of monocytes incubated for 24 h with 2 μg mL−1 β‐glucan, 5 μg mL−1 BCG, 10 μg mL−1 oxLDL, 1 ng mL−1 LPS, and non‐stimulated (RPMI), ±1 µm BIX‐01294, and restimulated with 10 ng mL−1 LPS for 24 h at day 6 (RPMI n = 27, β‐glucan n = 27, LPS n = 11, BCG n = 15, oxLDL n = 8, each line represents one individual, total of 10 independent experiments). (b) Basal extracellular acidification rate (ECAR) and oxidative consumption rate (OCR) of day 6 2 μg mL−1 β‐glucan‐trained monocytes ± 1 µm BIX‐01294, measured using Seahorse (n = 7, three independent experiments). *P < 0.05, **P < 0.01, ***P < 0.001, Wilcoxon signed‐rank test, mean ± SEM.
G9a inhibition results in increased ROS during induction of trained immunity, but does not depend on NADPH oxidase‐derived ROS
BCG‐induced trained immunity is accompanied by increased reactive oxygen species (ROS) production, 21 which is also observed during G9a inhibition with BIX‐01294. 25 , 26 To explore this phenomenon, we assessed ROS production in BCG‐trained cells with and without G9a inhibition, at day 6 upon restimulation with Zymosan A. As shown in Figure 4a, BCG induces increased ROS release, and G9a inhibition by BIX‐01294 amplified this effect. Short‐term incubation of neutrophils with BIX‐01294 and subsequent stimulation with zymosan did not affect ROS levels (Supplementary figure 3e). To understand whether the enhanced trained immunity response by G9a inhibition is dependent on ROS production, ROS inhibitor N‐acetylcysteine (NAC) was added during the first 24 h of the trained immunity in vitro protocol. Although TNF‐α production is significantly decreased when NAC is added in trained monocytes supplemented with BIX‐01294, the untrained condition was not affected when NAC was added to BIX‐treated cells, suggesting that the effect of G9a inhibition on cytokine production is not dependent on ROS in macrophages (Figure 4b). Furthermore, we assessed trained immunity responses in five chronic granulomatous disease (CGD) patients who have genetic defects in NADPH oxidase leading to impaired ROS production. Induction of trained immunity by β‐glucan and G9a inhibition showed increased IL‐6 and TNF‐α production both in healthy controls and CGD patients (except for one patient for IL‐6 production), further suggesting that the stimulating effects of G9a inhibition on trained immunity are not dependent on NADPH oxidase‐derived ROS (Figure 4c).
Figure 4.
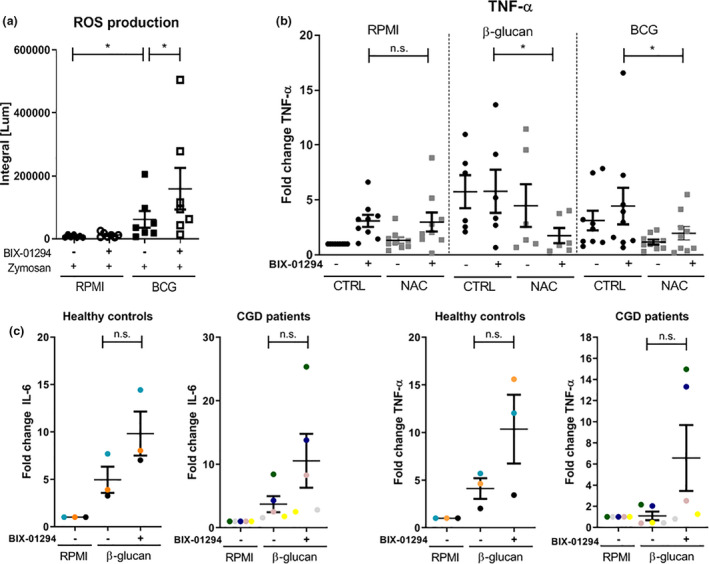
G9a inhibition in BCG‐induced trained immunity increases, but is not dependent on, ROS production. (a) ROS luminescence production after Zymosan (1 mg mL−1) stimulation of day 6 BCG‐trained monocytes ±1 µm BIX‐01294 (n = 7, mean ± SEM, three independent experiments, *P < 0.05, Wilcoxon signed‐rank test). (b) IL‐6 and TNF‐α fold changes of monocytes incubated for 24 h with 2 μg mL−1 β‐glucan, 5 μg mL−1 BCG, ± 1 µm BIX‐01294, ±1 mm NAC, which were restimulated after 6 days with 10 ng mL−1 LPS (n = 9 for RPMI and BCG in three independent experiments, n = 6 for β‐glucan in two independent experiments, mean ± SEM, *P < 0.05, Wilcoxon signed‐rank test). (c) IL‐6 and TNF‐α fold changes of monocytes from chronic granulomatous disease (CGD) patients and healthy individuals incubated for 24 h with 2 μg mL−1 β‐glucan ± 1 µm BIX‐01294 which were restimulated after 6 days with 10 ng mL−1 LPS (n = 3 HC, n = 5 CGD, mean ± SEM, 1 experiment, each colour represents one individual, Mann–Whitney U‐test, n.s.).
G9a inhibition amplifies trained immunity responses, and BCG immunotherapy decreases H3K9me2 in NMIBC patients
We studied the effect of G9a in NMIBC patients who received a standard BCG induction treatment consisting of 6 weekly bladder instillations. First, we performed an ex vivo training experiment using β‐glucan and BCG with peripheral blood monocytes that were obtained prior to the first BCG instillation from five bladder cancer patients. As seen by increased IL‐6 and TNF‐α production, β‐glucan and BCG potentiated the cytokine response of circulating monocytes of all bladder cancer patients, suggestive of induction of trained immunity (Figure 5a). Additionally, G9a inhibition increased trained immunity responses, as shown by augmented IL‐6 and TNF‐α production in the majority of patients in both the control condition and in BCG‐ and β‐glucan‐trained cells (Figure 5a), which is in line with the observations in healthy individuals. Ex vivo G9a inhibition with monocytes obtained after one and six BCG bladder instillations also showed increased IL‐6 and TNF‐α levels in most patients, again suggesting a stimulating effect of G9a inhibition on BCG‐induced trained immunity (see Supplementary figure 4a).
Figure 5.
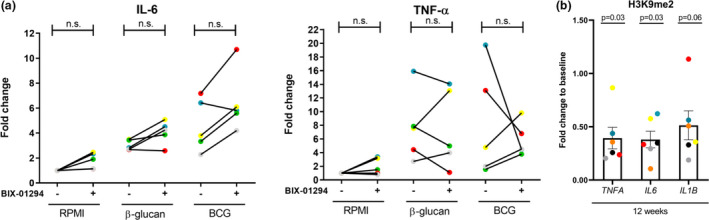
G9a inhibition is associated with amplified trained immunity responses in NMIBC patients, and BCG immunotherapy results in decreased H3K9me2 marks. (a) Fold changes of cytokine production compared to baseline (RPMI without BIX‐01294) in supernatant of circulating monocytes from NMIBC patients incubated for 24 h with 2 μg mL−1 β‐glucan, 5 μg mL−1 BCG, and non‐stimulated (RPMI), ± 1 µm BIX‐01294, washed and restimulated with 10 ng mL−1 LPS for 24 h at day 6 (n = 5, for each donor and visit an independent experiment). Each colour represents one patient. (b) ChIP‐qPCR H3K9me2 levels of isolated circulating monocytes from NMIBC patients after BCG treatment (i.e. approximately 6 weeks after the 6th weekly BCG instillation) compared to baseline, at promoters of TNFA, IL1B, IL6 (n = 6 (one outlier was excluded, Grubb's test, P < 0.05), three independent experiments, Wilcoxon signed‐rank test). Each colour represents one patient.
Next, we assessed whether the main function of G9a, H3K9 dimethylation, was affected in circulating monocytes that were obtained from NMIBC patients 6 weeks after receiving BCG induction treatment (consisting of 6 BCG bladder instillations). As depicted in Figure 5b, statistically significant decreased H3K9me2 levels were observed after BCG therapy at the promoter of IL6, as well as decreased levels for at least 5 out of 7 patients at the promoters of IL1B and TNFA, indicating a potential role for H3K9me2 modifications in modulating cytokine production of circulating monocytes. Additionally, in another group of 5 NMIBC bladder cancer patients (previously treated with intravesical chemotherapy instillation), the levels of H3K9me2 in peripheral blood monocytes were compared with those from healthy controls, as global altered H3K9me2 has been observed in the urothelium of bladder cancer patients compared to healthy controls. 27 We did not observe differences in H3K9me2 levels at the promoters of pro‐inflammatory genes between these healthy individuals and patients (Supplementary figure 4b). The bladder cancer patients showed increased IL‐1β production after 24‐h stimulation of peripheral blood mononuclear cells (PBMCs) with LPS ex vivo, compared to healthy individuals (Supplementary figure 4c).
G9a inhibition in monocytes of bladder cancer patients results in differentially expressed inflammatory genes
RNA sequencing was performed to identify genes playing a role in G9a inhibition in bladder cancer patients. Therefore, RNA expression in monocytes of four bladder cancer patients, previously treated with intravesical chemotherapy instillation, and healthy individuals was compared with and without 24 h of ex vivo exposure to BIX‐01294 (Figure 6a). G9a inhibition in monocytes of bladder cancer patients significantly upregulated 48 genes (Figure 6b and Supplementary table 1) and downregulated six genes (Figure 6c and Supplementary table 2) compared to the cells without G9a inhibition (see Supplementary figure 5a and b for significantly altered gene expression in healthy controls). In Supplementary table 3, pathway analysis of upregulated genes in monocytes of bladder cancer patients is shown. Interestingly, expression of DNA methyltransferase 3 alpha (DNMT3A) and the inhibitor of IL‐18, IL‐18 binding protein (IL18BP), were upregulated in bladder cancer patients derived monocytes exposed ex vivo to BIX‐01294 (Figure 6d). In contrast, inflammation‐related genes TNFRSF11A [TNF receptor superfamily member 11a/receptor activator of NFκB (RANK)] and IL1B were downregulated upon G9a inhibition in bladder cancer patients (Figure 6d), although for IL1B this was not significant. Downregulation of IL‐1β is also observed on protein level in 24 h LPS‐stimulated PBMCs of healthy individuals incubated with BIX‐01294 (Supplementary figure 5c).
Figure 6.
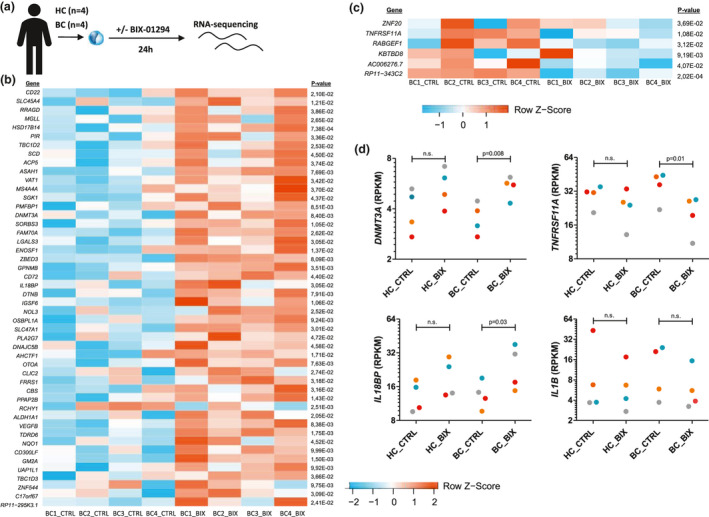
G9a inhibition in bladder cancer patients results in differentially expressed inflammatory‐related genes. (a) CD14+ monocytes were isolated from 4 healthy individuals and 4 bladder cancer patients, and supplemented with 1 µm BIX‐01294 for 24 h, subsequently RNA was isolated for RNA sequencing (four independent experiments). (b, c) Heatmap of significantly upregulated (b) and downregulated (c) genes by RNA sequencing of monocytes from bladder cancer patients ± BIX‐01294 treatment for 24 h [n = 4 control (CTRL), n = 4 BIX‐01294 treatment (BIX)]. (d) Individual plots of differentially expressed genes TNFRSF11A, DNMT3A, IL18BP, IL1B, expressed in log2 RPKM (n = 4 for each group, each colour represents one donor). BC, bladder cancer patient; HC, healthy control. Unadjusted P‐values derived from DEseq are displayed.
Discussion
In the last decade, the concept of trained immunity has been extensively investigated. 2 Although epigenetic rewiring is essential for induction of trained immunity, the modulators involved in this process have remained largely uncharacterised. Here, we identified an inhibitory role for lysine methyltransferase G9a/EHMT2 on induction of trained immunity. Moreover, we recognise an inhibitory role for G9a in BCG immunotherapy for bladder cancer, and we therefore argue that it may represent a therapeutic target.
Previously, it has been shown that EHMT2 transcription is significantly altered during differentiation of monocytes into various lineages. 28 Here, we show that upon induction of trained immunity in vitro, EHMT2 mRNA as well as protein expression is decreased. EHMT2 mRNA expression was decreased in four out of six healthy individuals upon BCG vaccination, a trained immunity inducer in humans, 7 and a strong inhibition of EHMT2 was apparent shortly after in vivo induction of endotoxemia in healthy volunteers. These data argue that downregulation of EHMT2 expression is a constant component of the immune cells during activation, and pharmacological inhibition of this protein may amplify immune responses.
By using G9a inhibitor BIX‐01294 in an in vitro trained immunity model, an important inhibitory role for G9a on trained immunity has been identified. This was shown by (1) increased pro‐inflammatory cytokine production upon non‐specific restimulation of monocytes after training in the presence of G9a inhibition, and (2) changes in the metabolic rates as measured by increases in ECAR and OCR suggestive of increased glycolysis and oxidative phosphorylation, another hallmark of trained immunity. 2 Multiple pharmacological G9a inhibitors exist and are currently of much interest. 29 BIX‐01294 was the first identified G9a inhibitor and specifically inhibits G9a and GLP by competitive binding to the SET domain of G9a directly, 23 , 24 which might explain why we did not observe a direct effect of G9a inhibition on EHMT2 mRNA levels. When using the later developed inhibitors UNC0638 and UNC0642, 30 , 31 which also inhibit G9a by competitive binding to the SET domain, less pronounced differences in trained immunity responses or tolerance were observed, which might be explained by slight changes in effectivity and/or selectivity of the inhibitors. Additionally, we show that G9a inhibition is able to partially reduce LPS tolerance in vitro. This is in line with a previous study showing that G9a interacts directly with RelB to generate epigenetic silencing in endotoxin tolerance in a human cell line. 16 Moreover, Yoshida et al. showed that treatment of mice with LPS abrogated G9a recruitment via ATF7 release, resulting in decreased repressive H3K9me2, which was maintained for long periods and led to enhanced resistance to pathogens. 17
BCG‐trained monocytes show increased ROS production upon restimulation. 21 Previously, it has been shown in cancer cells that treatment with EHMT2 siRNA and BIX‐01294 accumulated intracellular ROS and that treatment with ROS inhibitor NAC or diphenyleneiodonium chloride decreased BIX‐induced autophagy and cell death. 25 , 26 In this study, BIX‐01294 itself did not induce ROS production, but increased ROS production in BCG‐trained macrophages upon restimulation with zymosan. Inhibition of ROS by using NAC did not affect the immune response upon restimulation in the untrained condition. The possible dependence on NADPH oxidase 2‐produced ROS by G9a was excluded by the observed increased trained immunity responses of monocytes isolated from CGD patients upon G9a inhibition (which are unable to produce NADPH oxidase 2‐derived ROS).
Furthermore, we showed that G9a inhibition in β‐glucan‐trained cells is accompanied by decreased H3K9me2 at promoters of genes involved in trained immunity, indicating increased transcription. It has been previously shown that, next to enrichment of active marks during induction of trained immunity, a decrease in the repressive mark H3K9me3 is observed at pro‐inflammatory genes and genes involved in glycolysis. 6 , 11 Here, no significant effect of β‐glucan training on H3K9me2 of important glycolytic genes HK2 and PFKP was observed, also not in combination with G9a inhibition. However, β‐glucan‐induced training together with G9a inhibition led to decreased H3K9me2 at the mTOR promoter compared to the control condition, suggesting increased activity of mTOR, which is known to be essential in β‐glucan‐induced trained immunity. 6
An increasing number of studies have shown a role for G9a in multiple types of cancer. 18 In bladder cancer, EHMT2 expression is associated with poor clinical outcome. 19 By using a novel dual G9a/DNA methyltransferase (DNMT) inhibitor, H3K9me2 is decreased, and apoptosis and autophagy are induced. A combination of the G9a/DNMT inhibitor together with cisplatin treatment in mice increased regression of tumors and metastases. 19 Furthermore, inhibition of G9a by UNC0642 was shown to induce cell apoptosis in bladder cancer cells in vitro, accompanied by decreased levels of H3K9me2. In mice, administration of UNC0642 resulted in suppressive effects on tumor growth. 32 For high‐risk NMIBC patients, repeated bladder instillations with the trained immunity inducer BCG are standard treatment; however, the exact working mechanism is still not completely clear. Recently, it has been proposed that BCG‐induced trained immunity might play a role in the working mechanism. 20 Here, we show that circulating monocytes of bladder cancer patients respond to ex vivo induction of trained immunity by increased pro‐inflammatory cytokine production, which can be amplified by G9a inhibition. The increased production of pro‐inflammatory cytokines is also likely to increase immunological activity at the tumor site. Furthermore, we showed decreased H3K9me2 at the promoters of pro‐inflammatory genes in circulating monocytes 6 weeks after the sixth and final BCG induction instillation (6 weeks' treatment and 6 weeks' rest), which indicates a sustained systemic increase in cytokine production. Several studies have shown an association with increased cytokine production in BCG‐treated NMIBC patients. 20 Altogether, these data suggest that BCG instillation and G9a inhibition might be used synergistically for the treatment of NMIBC. Currently, no G9a inhibitor is yet available for therapeutic purposes.
Previously, altered H3K9 mono‐, di‐ and trimethylation have been observed in urothelium tissue of bladder cancer patients compared to healthy individuals. 27 We could not identify this difference systemically in H3K9me2 levels at promoters of inflammatory genes in peripheral blood monocytes of the relatively low number of bladder cancer patients versus healthy individuals. RNA sequencing of peripheral monocytes derived from bladder cancer patients treated with BIX‐01294 revealed significant upregulation of 48 genes, whereas only 6 genes were significantly downregulated, compared to monocytes from patients without G9a inhibition. Interestingly, methyltransferase DNMT3A RNA expression is upregulated upon G9a inhibition. Previously, DNMT3A/3B have been shown to be crucial for methylation in embryonic stem cells by interacting with G9a, 33 but not for maintenance in adult somatic cells. 34 In cancer, DNMT3A is frequently mutated, and G9a knockdown cancer cells are more sensitive to DNA demethylation treatment by showing increased DNA hypomethylation and cell growth inhibition. 34 Additionally, it has been shown that DNMT3A is methylated in mice by overexpressing GLP or G9a. 35 Remarkably, here we observe an increase in DNMT3A expression after 24‐h G9a inhibition, although the duration of this effect remains to be established. Additionally, IL1B gene expression shows an insignificant decrease in all donors upon G9a inhibition, which we also observed on protein level after 24‐h treatment of PBMCs with BIX‐01294. This could be of interest for cancer treatment, since IL‐1β inhibition has been implicated to reduce tumorigenesis. 36 Additionally, TNFRSF11A, also called RANK, is downregulated upon G9a inhibition. This gene encodes the receptor which is able to induce PI3K and NFκB activation and plays an important role in osteoclast development, but might also be a therapeutic target in breast cancer. 37 These results suggest that the effect of G9a inhibition depends on timing and setting: inhibiting inflammation upon direct G9a inhibition, while promoting trained immunity.
In conclusion, G9a downregulation is important for induction of trained immunity. Pharmacological inhibition of G9a amplifies trained immunity responses, as shown by increased pro‐inflammatory cytokine production and increased metabolic rate, and is accompanied by decreased H3K9me2 at gene promoters of pro‐inflammatory genes. Monocytes of NMIBC patients amplify trained immunity responses upon G9a inhibition and, when treated with trained immunity inducer BCG, show decreased H3K9me2 marks at the promoters of pro‐inflammatory genes. G9a inhibition in monocytes derived from bladder cancer patients previously treated with chemotherapy results in differentially expressed inflammatory genes. Further studies are necessary to define the function of G9a in trained immunity comprehensively, taking into account the numerous SET‐independent and SET‐dependent functions of G9a. Additionally, further elucidation of the role of G9a in BCG immunotherapy for bladder cancer will provide important mechanistic insights and could possibly lead to novel therapeutic strategies for these patients.
Methods
Reagents
RPMI 1640 (Dutch modified; Gibco, Life Technologies, Waltham, MA, USA) was used as culture medium supplemented with 5 μg mL−1 gentamicin (Centraform, Etten‐Leur, the Netherlands), 2 mm l‐glutamin (Gibco) and 1 mm pyruvate (Gibco). Cells were stimulated with synthetic Pam3SK4 (Pam3Cys; EMC Microcollections, Tübingen, Germany), Escherichia coli LPS (serotype 055:B5, Sigma‐Aldrich, St Louis, MO, USA), β‐1,3‐(d)‐glucan (kindly provided by Professor David Williams of East Tennessee State University, USA), BCG vaccine (InterVax, Canada, or for bladder cancer patients, BCG‐Medac, Medac, Wedel, Germany) and oxLDL, which was isolated from pooled human serum by ultracentrifugation and oxidation by incubating with 20 µmol CuSO4 L−1 for 15 h at 37°C followed by dialysis, as previously described. 38 G9a inhibitors BIX‐01294 (1 µM, Sigma) and UNC0638 (1 µM; Sigma) were used. N‐acetyl cysteine (NAC) was used in a concentration of 1 mM (Sigma).
Blood samples
Peripheral blood mononuclear cells were isolated from buffy coats of healthy blood donors (Sanquin, Nijmegen, The Netherlands). Additionally, venous blood was drawn of healthy volunteers after given informed consent (Radboudumc, Nijmegen, The Netherlands). CGD patients with NCF1 or NCF2 mutations (p47 and p67‐phox, respectively) in which severely impaired ROS production has been demonstrated were also included after given informed consent (Radboudumc, n = 3 HC (n = 2 men, mean age 27, SD 5) and n = 5 CGD patients (n = 4 men, mean age 25, SD 9)). Bladder cancer patients treated with BCG instillations were included as part of the Tribute study (n = 7 men, mean age of 65 years, SD 7; Radboudumc and Erasmus MC Rotterdam, The Netherlands, NL60341.091.17). Other bladder cancer patients were included by given informed consent (Radboudumc, CWOM9803‐0060 (n = 5 (mean age of 68, SD 17, four men)), previously treated with mitomycin). Four of these bladder cancer patients (three men, one women) were included for RNA sequencing analysis, in addition to four healthy controls [Radboudumc, NL32357.091.10 (n = 4 (mean age of 38, SD16, two men))]. Cells isolated from individuals which are part of the BCG‐Yellow Fever study 39 (Radboudumc, Nijmegen, The Netherlands, NL50160.092.24) were assessed for RNA expression before and after BCG (SSI, Denmark) vaccination. EHMT2 RNA expression in in vivo E. coli‐induced endotoxin tolerance was examined in 12 healthy non‐smoking volunteers who participated in the endotoxemia study 8 (NCT02602977; Radboudumc Nijmegen, The Netherlands, NL53584.091.15), conducted as described previously. 22 RNA sequencing results of in vitro training experiments from cells of healthy individuals who participated in previous studies 8 , 40 (NCT02602977 and 4899‐07/16) are also included. Inclusion of volunteers and experiments were conducted according to the principles expressed in the Declaration of Helsinki. All volunteers gave written informed consent before any material was taken. In Supplementary table 4, an overview is shown of different blood samples used for this study.
PBMC isolation and stimulation
Cells were isolated by density centrifugation on Ficoll‐Paque (GE Healthcare, Chalfont St Giles, UK), washed three times in PBS and resuspended in culture medium. After isolation, 5 × 105 PBMCs were added to a round bottom 96‐well plate (Greiner, Kremsmünster, Austria). Pam3Cys (10 μg mL−1) or LPS (10 ng mL−1) was added for 24 h at 37°C and 5% CO2.
Monocyte isolation and trained immunity assay
Human primary monocytes were isolated by layering hyper‐osmotic Percoll solution (48.5% Percoll (Sigma‐Aldrich), 41.5% sterile H2O, 0.16 m filter‐sterilised NaCl) on PBMCs. After 15 min of centrifugation at 580 g, the interphase layer was isolated, and cells were washed with PBS and resuspended in culture medium. To increase the purity of Percoll‐isolated monocytes, the monocytes were adhered to polystyrene flat bottom plates (Corning, Sigma‐Aldrich, New York, USA) or Petri dishes (Falcon, Merck, Darmstadt, Germany) for 1 h at 37°C followed by washing with warm PBS. Next, cells were pre‐incubated with culture medium supplemented with 10% human pooled serum as control, or together with 1 μm BIX‐01294, UNC0638 or NAC. Next, culture medium supplemented with 10% human pooled serum was added as a control, or together with either 2 μg mL−1 β‐glucan, 5 μg mL−1 BCG Intervax, 37.500 microorganisms mL−1 BCG‐Medac, oxLDL (10 μg mL−1), or LPS (1 ng mL−1). After 24 h, cells were washed with warm PBS and culture medium was added. Culture medium was refreshed after 3 days of incubation. On day 6, cells were restimulated with RPMI, LPS (10 ng mL−1), or Pam3Cys (10 μg mL−1). After 24 h, supernatants were collected and stored at −20°C until further use. For RNA sequencing experiments, and trained immunity experiments with CGD patients and healthy controls, monocytes were isolated from PBMCs by negative selection (Pan monocyte isolation kit; Miltenyi Biotech, Bergisch Gladbach, Germany) using a MACS system (Miltenyi Biotech) according to the manufacturer's protocol, to gain a high purity of monocytes.
Cytokine measurements
Cytokine production was measured in supernatants using commercial sandwich ELISA kits for human IL‐6, TNF‐α and IL‐1β in accordance with the manufacturer's instructions (R&D systems, Minneapolis, MN, USA).
RNA sequencing
RNA‐seq of in vitro β‐glucan‐induced trained immunity and LPS tolerance has been described previously. 8 , 40 For RNA‐seq experiments with BIX‐01294, 1 × 106 MACS‐isolated monocytes from bladder cancer patients and healthy controls were treated for 24 h with BIX‐01294 or control, RLT buffer (Qiagen, Hilden, Germany) was added after washing, and cells were stored at −80°C. RNA was isolated using RNeasy Mini Kit (Qiagen) according to manufacturer's protocol and included DNAse treatment. RNA sequencing was performed by BGI Genomics (Copenhagen, Denmark). RNA‐seq analysis was performed as described previously 8 ; briefly, fastq files were mapped with bowtie, and expression value (RPKM) tables were generated using mmseq (PMID: 21310039). 41 Differential genes were identified as those with a P < 0.05, FC > 1.5 and RPKM > 1 using DEseq. 41 P‐values displayed in Figure 6b are unadjusted P‐values from DEseq, which are generated using a model that assumes negative binomial (NB) distribution of data. 42 Functional enrichment analysis was performed using HOMER, 43 which scans several libraries (ontologies) for enrichment, including KEGG, MSigDB and ‘Gene Ontocology’. Top terms are highlighted in the manuscript (Supplementary table 3), based on P‐values.
qRT‐PCR
Quantitative RT‐PCR of in vitro trained monocytes was performed by isolating RNA using TRIzol reagent according to the manufacturer's instructions. cDNA was synthesised with the SuperScript III First‐Strand Synthesis System (Thermo Fisher Scientific, Waltham, MA, USA) according to manufacturer's protocol. Quantitative RT‐PCR was performed using StepOne PLUS machine (Thermo Fisher Scientific) with SYBR Green (Invitrogen, Waltham, MA, USA). The values are expressed as log2 fold increase in mRNA levels relative to those in non‐trained cells. 18S was used as a housekeeping gene. The primer sequences are listed in Supplementary table 5.
ChIP‐qPCR
Chromatin immunoprecipitation of healthy individuals was performed of Percoll‐isolated monocytes at day 6 of the in vitro training protocol. Percoll‐isolated monocytes of bladder cancer patients were immediately processed at the day of blood drawing. Cells were cross‐linked with methanol‐free 1% formaldehyde, sonicated and immunoprecipitated using 1 µg of H3K9me2 antibody (Diagenode, Liège, Belgium). Immunoprecipitated chromatin was processed further for qRT‐PCR analysis using the MiniElute DNA purification kit (Qiagen). Primers used in the reaction are listed in Supplementary table 6. Samples were analysed with a comparative Ct method on the StepOne PLUS qPCR machine (Thermo Fisher Scientific) using SYBR green (Invitrogen) in accordance with the manufacturer's instructions.
Western blots
Trained monocytes at day 6 were detached using Versene (Gibco), and subsequently washed and counted. Cells were lysed on ice using standard lysis buffer with phosSTOP (Roche, Basel, Switerzland) and cOmplete (Roche), and vortexed every 10 min for half an hour for total lysis, and subsequently stored at −20°C until further use. The frozen homogenate was subsequently centrifuged, and the supernatant was boiled in Laemmli buffer under denaturing and reducing conditions. Trans Turbo Blot System (Bio‐Rad, Hercules, CA, USA) was used according to the manufacturer's instructions. Mini‐PROTEAN Precast Gels (Bio‐Rad) were used, and samples were loaded onto gels together with Precision Plus Protein Dual Color Standard (Bio‐Rad). After running of the gels, proteins were transferred onto nitrocellulose membranes using the semi‐dry method (Bio‐Rad). Next, blots were blocked with 5% milk powder (ELK) in Tris‐buffered saline (TBS) with 0.1% Tween‐20 (TBS‐T; Invitrogen) for 1 h at room temperature. Thereafter, blots were washed in TBS‐T and incubated overnight with an anti‐G9a antibody (Abcam, Cambridge, UK) and anti‐actin (Sigma) on a roller bank at room temperature. Blots were subsequently washed with TBS‐T and incubated for 1h with secondary antibody swine anti‐rabbit‐HRP (Agilent Technologies, Santa Clara, CA, USA), at room temperature. Last, cells were washed and incubated with ECL (GE Health Care, Chicago, IL, USA) before imaging of the blots (ChemiDoc; Bio‐Rad) and analysing (Image Lab version 5.2.1).
Viability assays
Cytotoxicity was analysed by detecting LDH directly in fresh supernatant using the CytoTox 96® Non‐Radioactive Cytotoxicity Assay (Promega, Leiden, the Netherlands), in accordance with the manufacturer's instructions. Additionally, cell viability was assessed using Annexin V‐FITC (Biovision, San Francisco, CA, USA) and propidium iodide (PI) ECD (Sigma) staining. Cells were stained on ice for 15 min using Annexin V‐FITC and subsequently for 5 min with 10 μg mL−1 PI. Samples were measured on the CytoFLEX (Beckman Coulter, Brea, CA, USA) and analysed with Kaluza 2.1 (Beckman Coulter).
Metabolic measurements
Percoll‐isolated monocytes (10 × 106) were seeded into Petri dishes (Greiner) and stimulated in 10 mL culture medium for 24 h, washed with warm PBS and incubated in normal culture medium at 37°C, 5% CO2. At day 6, cells were detached with Versene solution (Thermo Fisher Scientific) and 1 × 105 cells were plated in quintuplicate to overnight‐calibrated cartridges in assay medium [RPMI with 2 mmol L−1 glutamine, 11 mmol L−1 glucose and 1 mmol L−1 pyruvate (pH adjusted to 7.4)] and incubated for 1 h in a non‐CO2 incubator at 37°C. OCR and ECAR were measured using a Cell Mito Stress Kit in an XFe96 Extracellular Flux Analyzer (Seahorse Bioscience, Agilent Technologies, Santa Clara, CA, USA), with final concentrations of 1 μmol L−1 oligomycin A (Sigma), 1 μmol L−1 carbonyl cyanide‐4‐(trifluoromethoxy) phenylhydrazone (FCCP, Sigma), and 0.5 μmol L−1 rotenone (Sigma)/antimycin A (from Streptomyces sp., Sigma). The protocol was followed using the manufacturer's descriptions and compounds.
Reactive oxygen species assay
Reactive oxygen species production was determined by using a luminol‐enhanced luminescence assay. Neutrophils were isolated by removing PBMCs after density centrifugation on Ficoll‐Paque, and addition of hypotonic lysis buffer (H2O with 155 mm NH4Cl and 10 mm KHCO3) for 15′ on ice, twice. Thereafter, cells were washed with PBS twice and cells were resuspended in culture medium and counted. 5 × 105 neutrophils were added per well in quadruplicate in a white 96‐well plate (Corning, Sigma‐Aldrich). Subsequently, BIX‐01294 or control was added for 30 min. at 37°C, 5% CO2. Thereafter, the neutrophils, or the BCG‐trained macrophages at day 6, were unstimulated or stimulated with 1 mg mL−1 serum‐opsonised Zymosan (from Saccharomyces cerevisiae, Sigma). Next, luminol was added, and chemiluminescence was measured for 1 h every 142 s. Luminescence was expressed as relative light units (RLU) per second.
Statistics
Data were analysed using a Wilcoxon signed‐rank test for paired samples, or a Mann–Whitney U‐test for unpaired samples, using GraphPad Prism software (GraphPad Inc. version 8). Data are expressed as mean ± SEM, and values of *P < 0.05, **P < 0.01 and ***P < 0.001 were considered statistically significant. RNA‐seq data were analysed using a negative binomial linear model in DEseq. A P‐value of < 0.05 was considered significant.
Author Contributions
Vera Mourits: Conceptualization; Data curation; Formal analysis; Investigation; Methodology; Project administration; Writing‐original draft; Writing‐review & editing. Jelmer van Puffelen: Data curation; Investigation. Boris Novakovic: Formal analysis; Investigation; Methodology; Project administration; Visualization. Mariolina Bruno: Investigation; Methodology. Anaisa Valido Ferreira: Investigation. Laszlo Groh: Investigation. Rob Arts: Investigation. Tania O Crisan: Resources. Jelle Zwaag: Investigation. Elisa Jentho: Investigation. Matthijs Kox: Investigation. Peter Pickkers: Supervision. Frank van de Veerdonk: Supervision. Sebastian Weis: Supervision. Egbert Oosterwijk: Resources. Sita Vermeulen: Supervision. Mihai G Netea: Conceptualization; Supervision; Writing‐review & editing. Leo AB Joosten: Conceptualization; Supervision; Writing‐review & editing.
Conflict of Interest
MGN and LABJ are scientific founders of Trained Therapeutics Discovery.
Supporting information
Acknowledgments
We thank all the healthy volunteers and patients for blood donation. MGN was supported by a Spinoza grant of the Netherlands Organization for Scientific Research and an ERC Advanced Grant (TRAIN‐OLD nr. 833247). LABJ and TOC were supported by a Competitiveness Operational Programme grant of the Romanian Ministry of European Funds (HINT, P_37_762, MySMIS 103587). BN was supported by an NHMRC (Australia) Investigator Grant (no. 1173314) and Project Grant (no. 1157556). AVF was supported by an FCT (Portugal) PhD grant (PD/BD/135449/2017). SHV is supported by a grant of the Netherlands Organization for Scientific Research (NWO Vidi 91717334). EJ was supported by DFG grant GRK 1715/2.
References
- 1. Netea MG, Quintin J, van der Meer JW. Trained immunity: a memory for innate host defense. Cell Host Microbe 2011; 9: 355–361. [DOI] [PubMed] [Google Scholar]
- 2. Netea MG, Dominguez‐Andres J, Barreiro LB et al Defining trained immunity and its role in health and disease. Nat Rev Immunol 2020; 20: 375–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Divangahi M, Aaby P, Khader SA et al Trained immunity, tolerance, priming and differentiation: distinct immunological processes. Nat Immunol 2021; 22: 2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Joosten LAB, Crisan TO, Bjornstad P, Johnson RJ. Asymptomatic hyperuricaemia: a silent activator of the innate immune system. Nat Rev Rheumatol 2020; 16: 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Quintin J, Saeed S, Martens JH et al Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe 2012; 12: 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheng SC, Quintin J, Cramer RA et al mTOR‐ and HIF‐1alpha‐mediated aerobic glycolysis as metabolic basis for trained immunity. Science 2014; 345: 1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kleinnijenhuis J, Quintin J, Preijers F et al Bacille Calmette‐Guerin induces NOD2‐dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc Natl Acad Sci USA 2012; 109: 17537–17542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Novakovic B, Habibi E, Wang SY et al Beta‐glucan reverses the epigenetic state of LPS‐induced immunological tolerance. Cell 2016; 167: 1354–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Saeed S, Quintin J, Kerstens HH et al Epigenetic programming of monocyte‐to‐macrophage differentiation and trained innate immunity. Science 2014; 345: 1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Keating ST, Groh L, van der Heijden CDCC et al The Set7 lysine methyltransferase regulates plasticity in oxidative phosphorylation necessary for trained immunity induced by β‐glucan. Cell Rep 2020; 31: 107548–107548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arts RJ, Carvalho A, La Rocca C et al Immunometabolic pathways in BCG‐induced trained immunity. Cell Rep 2016; 17: 2562–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Scheer S, Zaph C. The lysine methyltransferase G9a in immune cell differentiation and function. Front Immunol 2017; 8: 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tachibana M, Ueda J, Fukuda M et al Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3–K9. Genes Dev 2005; 19: 815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rice JC, Briggs SD, Ueberheide B et al Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell 2003; 12: 1591–1598. [DOI] [PubMed] [Google Scholar]
- 15. Patnaik D, Chin HG, Esteve PO, Benner J, Jacobsen SE, Pradhan S. Substrate specificity and kinetic mechanism of mammalian G9a histone H3 methyltransferase. J Biol Chem 2004; 279: 53248–53258. [DOI] [PubMed] [Google Scholar]
- 16. Chen X, El Gazzar M, Yoza BK, McCall CE. The NF‐kappaB factor RelB and histone H3 lysine methyltransferase G9a directly interact to generate epigenetic silencing in endotoxin tolerance. J Biol Chem 2009; 284: 27857–27865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yoshida K, Maekawa T, Zhu Y et al The transcription factor ATF7 mediates lipopolysaccharide‐induced epigenetic changes in macrophages involved in innate immunological memory. Nat Immunol 2015; 16: 1034–1043. [DOI] [PubMed] [Google Scholar]
- 18. Casciello F, Windloch K, Gannon F, Lee JS. Functional role of G9a histone methyltransferase in cancer. Front Immunol 2015; 6: 487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Segovia C, San Jose‐Eneriz E, Munera‐Maravilla E et al Inhibition of a G9a/DNMT network triggers immune‐mediated bladder cancer regression. Nat Med 2019; 25: 1073–1081. [DOI] [PubMed] [Google Scholar]
- 20. van Puffelen JH, Keating ST, Oosterwijk E et al Trained immunity as a molecular mechanism for BCG immunotherapy in bladder cancer. Nat Rev Urol 2020; 17: 513–525. [DOI] [PubMed] [Google Scholar]
- 21. Bekkering S, Blok BA, Joosten LA, Riksen NP, van Crevel R, Netea MG. In vitro experimental model of trained innate immunity in human primary monocytes. Clin Vaccine Immunol 2016; 23: 926–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kox M, van Eijk LT, Zwaag J et al Voluntary activation of the sympathetic nervous system and attenuation of the innate immune response in humans. Proc Natl Acad Sci USA 2014; 111: 7379–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chang Y, Zhang X, Horton JR et al Structural basis for G9a‐like protein lysine methyltransferase inhibition by BIX‐01294. Nat Struct Mol Biol 2009; 16: 312–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kubicek S, O'Sullivan RJ, August EM et al Reversal of H3K9me2 by a small‐molecule inhibitor for the G9a histone methyltransferase. Mol Cell 2007; 25: 473–481. [DOI] [PubMed] [Google Scholar]
- 25. Kim Y, Kim Y‐S, Kim DE et al BIX‐01294 induces autophagy‐associated cell death via EHMT2/G9a dysfunction and intracellular reactive oxygen species production. Autophagy 2014; 9: 2126–2139. [DOI] [PubMed] [Google Scholar]
- 26. Park SE, Yi HJ, Suh N et al Inhibition of EHMT2/G9a epigenetically increases the transcription of Beclin‐1 via an increase in ROS and activation of NF‐kappaB. Oncotarget 2016; 7: 39796–39808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ellinger J, Bachmann A, Goke F et al Alterations of global histone H3K9 and H3K27 methylation levels in bladder cancer. Urol Int 2014; 93: 113–118. [DOI] [PubMed] [Google Scholar]
- 28. Wierda RJ, Goedhart M, van Eggermond MCJA et al A role for KMT1c in monocyte to dendritic cell differentiation. Human Immunol 2015; 76: 431–437. [DOI] [PubMed] [Google Scholar]
- 29. Cao H, Li L, Yang D et al Recent progress in histone methyltransferase (G9a) inhibitors as anticancer agents. Eur J Med Chem 2019; 179: 537–546. [DOI] [PubMed] [Google Scholar]
- 30. Vedadi M, Barsyte‐Lovejoy D, Liu F et al A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat Chem Biol 2011; 7: 566–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu F, Barsyte‐Lovejoy D, Li F et al Discovery of an in vivo chemical probe of the lysine methyltransferases G9a and GLP. J Med Chem 2013; 56: 8931–8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cao YP, Sun JY, Li MQ et al Inhibition of G9a by a small molecule inhibitor, UNC0642, induces apoptosis of human bladder cancer cells. Acta Pharmacol Sin 2019; 40: 1076–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang T, Termanis A, Ozkan B et al G9a/GLP complex maintains imprinted DNA methylation in embryonic stem cells. Cell Rep 2016; 15: 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sharma S, Gerke DS, Han HF et al Lysine methyltransferase G9a is not required for DNMT3A/3B anchoring to methylated nucleosomes and maintenance of DNA methylation in somatic cells. Epigenetics Chromatin 2012; 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang Y, Sun L, Kokura K et al MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a. Nat Commun 2011; 2: 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ridker PM, MacFadyen JG, Thuren T et al Effect of interleukin‐1β inhibition with canakinumab on incident lung cancer in patients with atherosclerosis: exploratory results from a randomised, double‐blind, placebo‐controlled trial. Lancet 2017; 390: 1833–1842. [DOI] [PubMed] [Google Scholar]
- 37. González‐Suárez E, Sanz‐Moreno A. RANK as a therapeutic target in cancer. FEBS J 2016; 283: 2018–2033. [DOI] [PubMed] [Google Scholar]
- 38. Bekkering S, Quintin J, Joosten LA, van der Meer JW, Netea MG, Riksen NP. Oxidized low‐density lipoprotein induces long‐term proinflammatory cytokine production and foam cell formation via epigenetic reprogramming of monocytes. Arterioscler Thromb Vasc Biol 2014; 34: 1731–1738. [DOI] [PubMed] [Google Scholar]
- 39. Arts RJW, Moorlag S, Novakovic B et al BCG vaccination protects against experimental viral infection in humans through the induction of cytokines associated with trained immunity. Cell Host Microbe 2018; 23: 89–100.e105. [DOI] [PubMed] [Google Scholar]
- 40. Jentho E, Novakovic B, Ruiz‐Moreno C et al Heme induces innate immune memory. BioRxiv 2019. 10.1101/2019.12.12.874578 [DOI] [Google Scholar]
- 41. Turro E, Su SY, Gonçalves Â, Coin LJ, Richardson S, Lewin A. Haplotype and isoform specific expression estimation using multi‐mapping RNA‐seq reads. Genome biology 2011; 12(2), 1–15. 10.1186/gb-2011-12-2-r13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010; 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Heinz S, Benner C, Spann N et al Simple combinations of lineage‐determining transcription factors prime cis‐regulatory elements required for macrophage and B cell identities. Mol Cell 2010; 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


