Abstract
Bacteria synthesize and export adhesive macromolecules to enable biofilm formation. These macromolecules, collectively called the biofilm matrix, are structurally varied and often unique to specific bacterial species or subspecies. This heterogeneity in matrix utilization makes it difficult to facilitate direct comparison between biofilm formation mechanisms of different bacterial species. Despite this, some matrix components, in particular the polysaccharides poly-β-1,6-N-acetyl-glucosamine (PNAG) and bacterial cellulose, are utilized by many Gram-negative species for biofilm formation. However, there is a very narrow distribution of these components across Gram-positive organisms, whose biofilm matrix determinants remain largely undiscovered. We found that a genetic locus required for the production of a biofilm matrix component of P. aeruginosa, the Pel polysaccharide, is widespread in Gram-negative bacteria and that there is a variant form of this cluster present in many Gram-positive bacterial species. We demonstrated that this locus is required for biofilm formation by Bacillus cereus ATCC 10987, produces a polysaccharide that is similar to Pel, and is post-translationally regulated by cyclic-3′,5′-dimeric-guanosine monophosphate (c-di-GMP) in a manner identical to P. aeruginosa. However, while the proposed mechanism for Pel production appears remarkably similar between B. cereus and P. aeruginosa, we identified several key differences between Gram-negative and Gram-positive Pel biosynthetic components in other monoderms. In particular, 4 different architectural subtypes of the c-di-GMP-binding component PelD were identified, including 1 found only in Streptococci that has entirely lost the c-di-GMP recognition domain. These observations highlight how existing multi-component bacterial machines can be subtly tweaked to adapt to the unique physiology and regulatory mechanisms of Gram-positive organisms. Collectively, our analyses suggest that the Pel biosynthetic locus is one of the most phylogenetically widespread biofilm matrix determinants in bacteria, and that its mechanism of production and regulation is extraordinarily conserved across the majority of organisms that possess it.
Keywords: Pseudomonas aeruginosa, Bacillus cereus, exopolysaccharide, biofilm formation, biofilm matrix, c-di-GMP
Comment on: Whitfield GB, Marmont LS, Bundalovic-Torma C, Razvi E, Roach EJ, Khursigara CM, Parkinson J, Howell PL. Discovery and characterization of a Gram-positive Pel polysaccharide biosynthetic gene cluster. PLoS Pathog. 2020 Apr;16(4):e1008281. doi: 10.1371/journal.ppat.1008281. eCollection 2020 Apr. PubMed PMID: 32236137; PubMed Central PMCID: PMC7112168.
Introduction
Bacteria benefit immensely from the ability to exist as multicellular communities, or biofilms, wherein metabolic abundances are shared as public goods and the burden of peripheral threats is spread across many individual community members.1 To form biofilms, bacteria self-produce adhesive macromolecules that facilitate attachment to surfaces and maintain community structure through preservation of intercellular connections. These extracellular adhesives, collectively called the biofilm matrix, are enormously diverse in their structure and often extremely limited in their phylogenetic distribution. The complexity is such that the biofilm matrix composition of each bacterial species is almost certainly unique, with major differences noted between different strains of the same species and even the same strain in different niches.2,3 While the development of a consensus model for biofilm matrix production and its role in biofilm formation and structure may seem insurmountable, there are some key biofilm matrix determinants that are widespread and thus make promising targets to decipher this complexity.
Synthase-Derived Polysaccharides Are a Widespread Biofilm Determinant
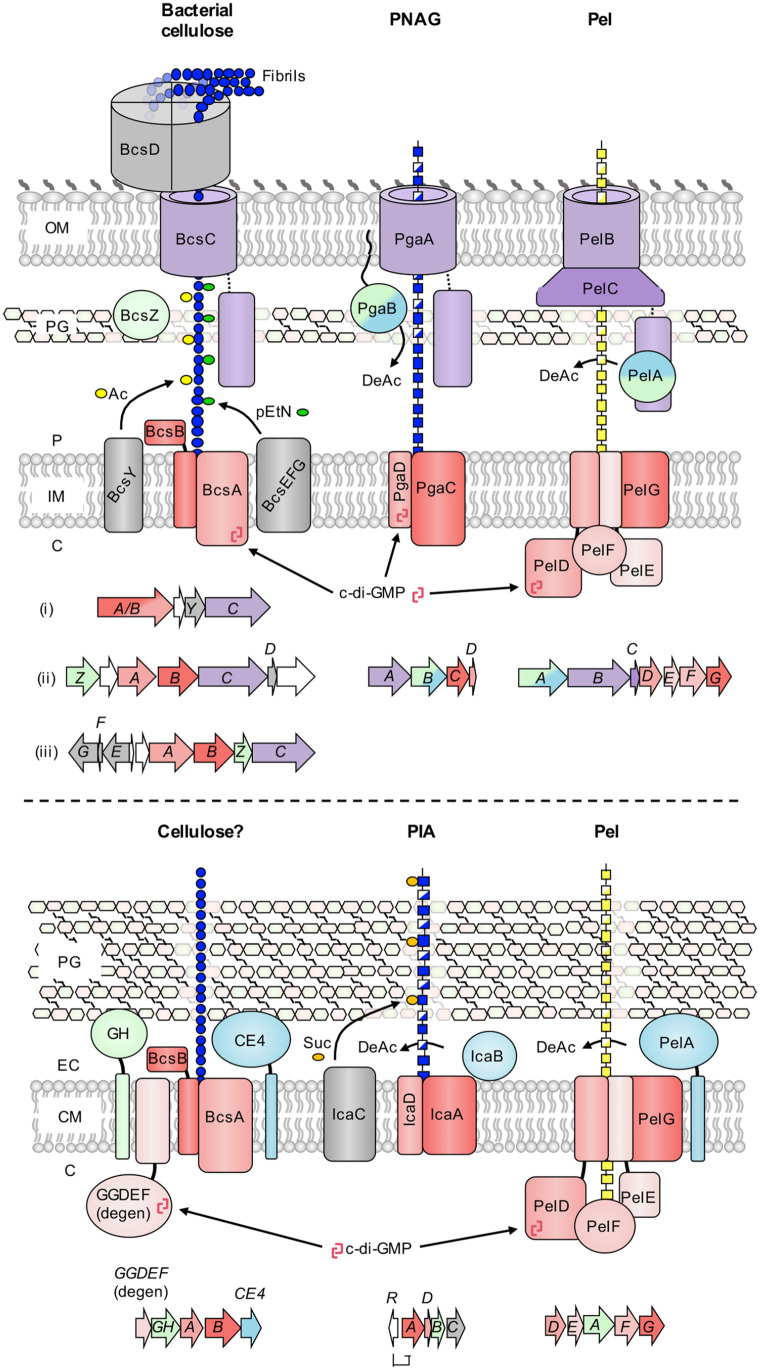
Polysaccharides are a major component of the biofilm matrix of many bacterial species.4 Critically, some of these biofilm polysaccharides, in particular bacterial cellulose and poly-β-1,6-N-acetyl-glucosamine (PNAG), are conserved across a wide variety of bacterial genera and are essential for biofilm formation by many of these organisms. As a result, their mechanisms of biosynthesis and contributions to biofilm physiology have been extensively studied.4 Both polysaccharides are produced using a synthase-dependent mechanism. This system is defined by the presence of a membrane-embedded polysaccharide polymerization and transport enzyme, or synthase (Figure 1).5 Synthase-dependent polysaccharide production does not require a lipid acceptor for polymer construction and the bifunctional activity of the synthase enzyme allows for coupled polymerization and membrane transport, greatly simplifying the process. These systems are often, but not always, regulated post-translationally by direct binding of cyclic-3′,5′-dimeric guanosine monophosphate (c-di-GMP) to the synthase and, in diderm bacteria, periplasmic transit and outer membrane export are facilitated by a β-barrel porin and its coupled tetratricopeptide repeat (TPR) domain.5 Since synthase-dependent systems like bacterial cellulose and PNAG require very few components for complete functionality, are encoded as a single uninterrupted locus (Figure 1), and generate a product that is sufficient to drive biofilm formation, it is no surprise that they are so widely distributed in bacteria. However, these polymers have primarily been restricted to Gram-negatives, with only very limited production of a variant of PNAG called polysaccharide intercellular adhesin (PIA) in Staphylococci,6 and our recent identification of putative bacterial cellulose biosynthetic clusters in some Lactobacilli and Clostridial species (Figure 1).7
Figure 1.
Biosynthetic models for synthase-dependent polysaccharides in Gram-negative and Gram-positive bacteria. (top) Schematics for the biosynthesis of bacterial cellulose, poly-β-1,6-N-acetyl-glucosamine (PNAG), and the Pel polysaccharide in Gram-negative bacteria. Representative genetic loci are shown below each model and are drawn to scale. Three representative gene clusters are shown for bacterial cellulose due to the diversity of these operons: (i) Komagataeibacter xylinus E25 locus one, (ii) Komagataeibacter xylinus E25 locus two and (iii) Salmonella enterica serovar Typhimurium. The PNAG operon is from Escherichia coli K-12 substr. MG1655, and the Pel operon is from Pseudomonas aeruginosa PAO1. (bottom) Schematics for the biosynthesis of bacterial cellulose, polysaccharide intercellular adhesin (PIA), and the Pel polysaccharide in Gram-positive bacteria. Representative genetic loci are shown below each model and are drawn to scale. The bacterial cellulose operon is from Clostridium botulinum ATCC 3502, the PIA operon is from Staphylococcus epidermidis RP62A, and the Pel operon is from Bacillus cereus ATCC 10987. Conserved functionalities across synthase systems are coloured as follows: red, synthase components (polymerization, inner membrane transport, c-di-GMP binding); purple, tetratricopeptide-repeat and β-barrel (outer membrane export), blue, deacetylase; green, glycoside hydrolase. Variable accessory components for each system are depicted in gray. C, cytoplasm; IM, inner membrane; P, periplasm; OM, outer membrane; PG, peptidoglycan; CM, cytoplasmic membrane; EC, extracellular space; Ac, acetyl group; pEtN, phosphoethanolamine; DeAc, deacetylation; c-di-GMP, cyclic-3′,5′-dimeric guanosine monophosphate; Suc, succinyl group, GH, predicted glycoside hydrolase; CE4, predicted carbohydrate esterase family 4 deacetylase; GGDEF (degen), degenerate diguanylate cyclase with intact inhibitory site.
Pel: One of the Most Prevalent Biofilm Polysaccharides?
The Pel polysaccharide has long been recognized as a biofilm determinant of the Gram-negative opportunistic pathogen Pseudomonas aeruginosa,3 although its importance is often overshadowed by the Psl polysaccharide, which in most P. aeruginosa species is the dominant matrix component. Until recently it was unclear how Pel was produced, but work by our group has provided compelling evidence that suggests it is generated via a synthase-dependent mechanism (Figure 1).8 Furthermore, while the pel operon has been identified in a small number of non-Pseudomonad Gram-negative bacterial species,9 some of which utilize Pel for biofilm formation,10 exactly how widespread pel operons are was unclear. To address this, we performed preliminary BLAST searches using the unique outer membrane protein PelC that revealed over 500 Pel biosynthetic loci in diverse Proteobacteria, hinting that Pel may be much more widespread than was originally thought.8 We followed up on this with a broader search using a novel computational method that enabled an unbiased search of fully sequenced bacterial genomes for Pel biosynthetic operons. To our surprise this search identified pel-like clusters in many Gram-positive bacterial species.7 To validate that these Gram-positive pel-like loci were functional, we used Bacillus cereus as a model organism to characterize the contribution of this operon to biofilm formation. Our study revealed not only that this locus was required for B. cereus biofilm formation, but that the polymer produced was similar to Pel from P. aeruginosa.11 Furthermore, polymer production appears to be highly conserved across Gram-negative and -positive organisms, including the mechanism of post-translational regulation by c-di-GMP. The latter observation was striking, given that considerably less is known generally about c-di-GMP-dependent regulation in Gram-positive bacteria, and many studied species appear to utilize this second messenger to a lesser degree than their Gram-negative counterparts.12
Collectively, this data led us to hypothesize that the Pel polysaccharide may be the most mechanistically conserved and phylogenetically diverse synthase-dependent pathway studied to date. Since synthase-produced polysaccharides generally appear to be the most widespread biofilm determinants, it would follow that Pel may be one of the most prevalent biofilm components produced by bacteria. Indeed, while the distribution of bacterial cellulose biosynthesis clusters amongst Proteobacteria is comparably far reaching, their genetic loci exhibit considerable variability and often include a number of accessory genes that modify the polymer in different ways, including the addition of acetyl or phosphoethanolamine substituents, or the formation of cellulose microfibrils (Figure 1).13 Furthermore, there is no compelling documented evidence of cellulose production in Gram-positive bacteria.13 However, the computational analysis that uncovered the presence of pel loci in Gram-positive organisms also identified candidate Gram-positive bacterial cellulose biosynthetic loci in Lactobacilli and Clostridia.7 The ability of these clusters to produce a cellulose-like polymer has not been evaluated. They also appear to differ in several ways from their Gram-negative counterparts. In particular, their BcsA orthologs lack a c-di-GMP binding PilZ domain, and instead encode a separate putative c-di-GMP binding protein with similarity to degenerate diguanylate cyclase receptors, like those associated with P. aeruginosa and B. cereus Pel production (Figure 1). Similarly, while PNAG biosynthetic clusters are found in Staphylococci, where the polymer is referred to as PIA, there are several critical differences between the Gram-negative and Gram-positive variants. First, the Gram-positive synthase is not regulated post-translationally by c-di-GMP as it is in Gram-negative organisms. Instead, the entire operon is regulated at the transcriptional level by the repressor IcaR (Figure 1). Second, a biosynthetic protein specific to Staphylococci, IcaC, is thought to modify the polymer via the addition of succinyl groups.6 Therefore, while bacterial cellulose, PNAG and Pel biosynthetic clusters are all undeniably widespread, Pel production appears to be the most mechanistically conserved of the 3 polymers across Gram-negative and Gram-positive organisms. Further, while PIA is produced by many species of Staphylococci, our latest search has identified >900 syntenically conserved pel operons in a wide variety of Gram-positive bacterial genera, making it significantly more phylogenetically widespread in monoderms than any other biofilm polysaccharide.
Similar But Different: Gram-Positive PelA Is a Mono-Functional Enzyme
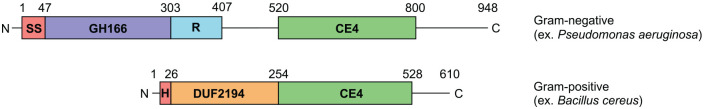
While the proteins involved in Pel production are predicted to be functionally conserved across bacteria, they are not all identical. The most apparent difference between Gram-negative and Gram-positive Pel biosynthetic clusters is in the predicted architecture of the Pel modification enzyme, PelA. In Gram-negatives, PelA is a periplasmically-localized bifunctional carbohydrate esterase family 4 (CE4) deacetylase14 and α-1,4-N-acetyl-galactosamine specific glycoside hydrolase (Figure 2)15 that associates with the Pel biosynthetic machinery by directly interacting with the TPR domain of PelB.16 While PelA deacetylase activity is required for Pel biosynthesis14 and glycoside hydrolase activity is linked to biofilm dispersal,17 it is unclear what role, if any, the glycoside hydrolase domain plays in Pel production. Since PelB is an outer membrane associated protein and thus is not present in monoderms, all Gram-positive PelA orthologs instead have a predicted N-terminal transmembrane helix which presumably functions to keep extracellular PelA tethered to the cell (Figure 2). Furthermore, Gram-positive PelA orthologs do not have a glycoside hydrolase domain. Instead, they invariably contain a domain that is predicted to mediate protein-protein interactions based on similarity to oligomerization domains associated with glycoside hydrolase family 42 enzymes (Figure 2). It is unclear what the function of this domain is in the context of Pel biosynthesis, although we hypothesize that it may mediate interactions with membrane-embedded Pel components to keep it associated with active polymer production, similar to the interaction between PelA and PelB in Gram-negatives.16
Figure 2.
Gram-positive PelA orthologs exhibit a conserved domain architecture that differs from their Gram-negative counterparts. (top) Domain organization of full-length Gram-negative PelA, using the protein from Pseudomonas aeruginosa as an example. (bottom) Domain organization of full-length Gram-positive PelA, using the protein from Bacillus cereus as an example. The approximate boundaries for each domain are indicated, and the size of each domain is drawn to scale. Regions with no confidently predicted structure or function are shown as a line. SS, predicted signal sequence that targets Gram-negative PelA to the periplasmic space; GH166, glycoside hydrolase family 166 domain; R, predicted oxidoreductase domain; CE4, carbohydrate esterase family 4 domain; H, trans-membrane helix; DUF2194, domain of unknown function with predicted similarity to oligomerization domains of glycoside hydrolase family 42 proteins.
Curiously, we noted that while all Gram-positive PelA orthologs lack a glycoside hydrolase domain, many of the associated biosynthetic clusters, including B. cereus,11 contain a separate gene with predicted similarity to the glycoside hydrolase domain found in P. aeruginosa PelA.15 Our analyses have confirmed that this protein is an active α-1,4-N-acetyl-galactosaminidase and revealed that deletion of this gene significantly elevated biofilm formation. This suggests that glycoside hydrolase activity may function to maintain biofilm formation levels within a prescribed range. Furthermore, since not every identified Gram-positive pel cluster contains such a glycoside hydrolase ortholog,11 this putative safeguard against runaway biofilm formation may not be required by every bacterial species.
C-di-GMP Is Not Universal: Adapting Post-Translational Regulation of Pel
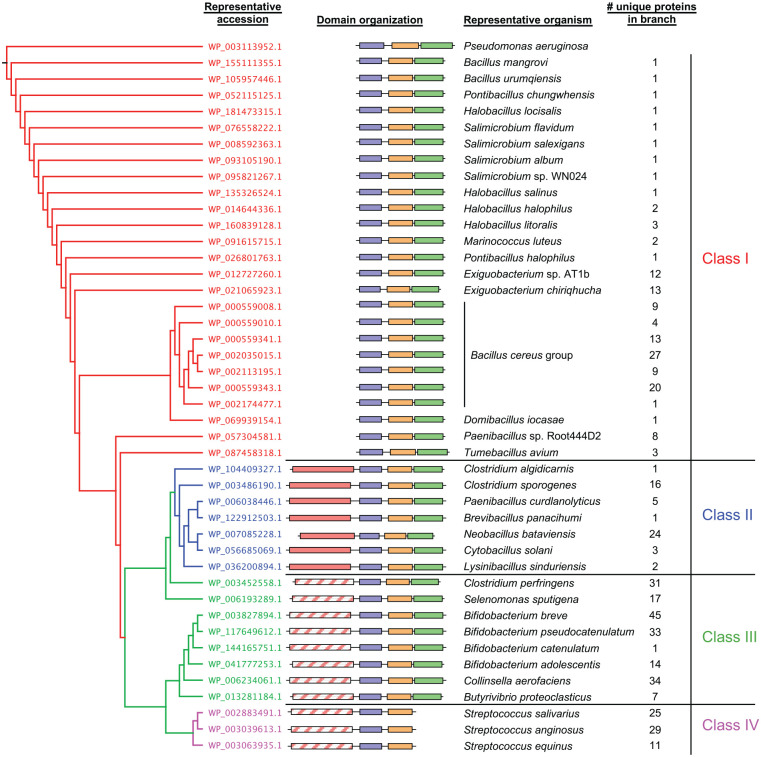
Part of the reason that Staphylococci have likely adapted their PNAG synthase to function independently of post-translational regulation by c-di-GMP is because Staphylococci generally do not signal using c-di-GMP.12 While we did not detect any Pel biosynthetic clusters in Staphylococci, there were many that were found in Streptococci, which are also not known to signal using c-di-GMP.12 This led us to examine in more detail the component of the Pel synthase responsible for c-di-GMP binding, PelD. In all studied Gram-negatives, as well as many Gram-positive organisms including B. cereus, PelD contains 3 domains: a N-terminal domain of unknown function (DUF4118) comprised of 4 predicted transmembrane helices, a central cGMP-specific phosphodiesterases, adenylyl cyclases, and FhlA (GAF) domain and a C-terminal degenerate GGDEF domain (Figure 3).11,18 While this domain is enzymatically inactive, an allosteric product inhibition site retains the ability to bind c-di-GMP, which is required for Pel production in P. aeruginosa and B. cereus.11,18 We found that in Streptococci the GGDEF domain of PelD is universally absent which, combined with the lack of genomically-encoded c-di-GMP metabolic enzymes, suggests that Pel production is unlikely to be regulated post-translationally by c-di-GMP in this genus (Figure 3).11 Instead, Streptococci have several additional genes of unknown function directly downstream of pelG that appear to be part of the same operon, which may provide a means to regulate Pel biosynthesis using an as yet undiscovered mechanism.11
Figure 3.
Gram-positive PelD orthologs cluster into 4 different classes based on their predicted domain architectures. (left) Phylogenetic tree generated from multiple sequence alignment of PelD protein sequences. Terminal branches with many closely clustered leaves were collapsed for ease of presentation, and 1 representative PelD ortholog from each cluster is shown. Each branch is coloured according to the PelD domain architecture class, shown at the far right. (middle) Schematic representations of Gram-positive PelD domain organizations corresponding to the protein accession numbers from the phylogenetic tree. Individual domains are represented as boxes and are drawn to scale. (right) Representative bacterial species from the corresponding branch of the phylogenetic tree whose PelD domain organization is schematically depicted. The number of leaves present in each collapsed branch of the phylogenetic tree is indicated. DUF4118, domain of unknown function containing 4 predicted transmembrane helices; GAF, cGMP-specific phosphodiesterases, adenylyl cyclases and FhlA domain; GGDEF, diguanylate cyclase domain; SDR, short chain dehydrogenase/reductase domain. Protein domains are coloured by predicted function as follows: purple, DUF4118; orange, GAF; green, degenerate GGDEF; red, SDR; hatched red, degenerate SDR.
Our analysis of Streptococcal PelD proteins also revealed a second unexpected feature: the presence of a predicted short chain dehydrogenase/reductase (SDR) domain at the N-terminus, adjacent to the DUF4118 domain (Figure 3). Members of the SDR family are enzymatically diverse, including oxidoreductases, epimerases and lyases, and can metabolize a wide array of substrates, therefore without further characterization the exact function of this domain is difficult to predict. However, this is a moot point, since the conserved SYK catalytic triad of SDR enzymes is absent in this domain of Streptococcal PelD, as is the GxxGxxG motif required for co-factor binding, suggesting that it is enzymatically inactive (Figure 3).11 The discovery of a degenerate SDR domain associated with Streptococcal PelD prompted us to examine all of the Gram-positive PelD orthologs to determine the distribution of this domain. Astonishingly, we identified this domain in nearly 75% of all Gram-positive PelD orthologs, all of which, excluding Streptococci, also have a C-terminal degenerate GGDEF domain (Figure 3). Furthermore, a distinct sub-group of these SDR domains have retained the canonical catalytic and co-factor binding residues, suggesting that they may be enzymatically active. To better facilitate comparison, we categorized all PelD homologs into 4 classes based on their domain architecture: Class I proteins have the original PelD domain architecture identified in P. aeruginosa and B. cereus, Class II proteins have the addition of a potentially enzymatically active SDR domain to the N-terminus, Class III proteins have the addition of a degenerate SDR domain to the N-terminus and Class IV proteins, found exclusively in Streptococci, have an N-terminal degenerate SDR domain and have also lost the C-terminal degenerate GGDEF domain (Figure 3).
While it is difficult to predict with certainty the precise activity of any SDR domain, it is noteworthy that the active domains of Class II orthologs exhibit strong predicted similarity to UDP-hexose-C4-epimerases. This is intriguing, as an important consideration for the biosynthesis of the N-acetyl-galactosamine (GalNAc) rich Pel polymer19 is how the nucleotide-sugar precursor utilized by the synthase is generated. Our recent work in Pseudomonas protegens, a close relative of P. aeruginosa, identified an additional gene in its pel operon that we have shown is a UDP-GlcNAc-C4-epimerase whose activity, in conjunction with a functionally redundant paralog elsewhere in the genome, is required for Pel biosynthesis.20 Our analysis of Gram-positive Pel biosynthetic clusters identified many UDP-hexose-C4-epimerase orthologs within or adjacent to pel operons, which were primarily associated with loci that contain Class I PelD proteins.11 In contrast, pel clusters that contain Class II PelD orthologs do not encode predicted UDP-hexose-C4-epimerases. Therefore, one possibility is that Class II PelD organisms do not have a UDP-GlcNAc-C4-epimerase in their genome, and that this sub-group has evolved to allow for acquisition of a fully operational pel gene cluster in these species. Degeneration of this SDR domain in Class III and IV PelD species may have resulted from functional redundancy. Indeed, many Streptococci encode a UDP-hexose-C4-epimerase ortholog in their pel clusters that could potentially render activity of the SDR domain unnecessary.11 Even so, the universal retention of this domain in clades that contain it, regardless of whether it has retained the capacity for activity, hints at a potential undiscovered secondary function. Since Class II, III and IV PelD orthologs are almost invariantly associated with pel operons harbouring one or more accessory genes of unknown function,11 we hypothesize that this domain may facilitate interactions with new partner proteins.
Future Perspectives
Many of the observations outlined above hint at the possibility that, in monoderm bacteria, the synthase complex may incorporate additional activities. This may include interaction with the unique Gram-positive variant of PelA, a Class II PelD with epimerase activity that can generate substrate precursors, or a Streptococcal Class IV PelD that may recruit new regulatory factors in the absence of c-di-GMP binding. Each of these possibilities may offer opportunities to better understand the widespread, yet architecturally varied, polysaccharide synthase enzymes. Furthermore, although the genetic capacity to produce Pel due to the presence of a pel biosynthetic cluster may be remarkably widespread, we have very little information about which of these species actually utilize their pel operons. We hope our discovery of the broad phylogenetic distribution of Pel will encourage other researchers to explore whether their favourite model organism harbours a pel locus and, if so, whether it is utilized for biofilm formation or perhaps other as yet undiscovered phenotypes.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from the Canadian Institutes of Health Research (CIHR; MOP 43998 and FDN154327 to PLH). PLH is a recipient of a Canada Research Chair. GBW has been supported by graduate scholarships from the Natural Sciences and Engineering Council of Canada (NSERC) and Cystic Fibrosis Canada.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: GBW and PLH prepared the manuscript text and figures.
ORCID iDs: Gregory B Whitfield  https://orcid.org/0000-0003-0274-8402
https://orcid.org/0000-0003-0274-8402
P Lynne Howell  https://orcid.org/0000-0002-2776-062X
https://orcid.org/0000-0002-2776-062X
References
- 1. Flemming H-C, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14:563-575. [DOI] [PubMed] [Google Scholar]
- 2. Wozniak DJ, Wyckoff TJO, Starkey M, et al. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc National Acad Sci. 2003;100:7907-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Colvin KM, Irie Y, Tart CS, et al. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ Microbiol. 2011;14:1913-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Whitfield GB, Marmont LS, Howell PL. Enzymatic modifications of exopolysaccharides enhance bacterial persistence. Front Microbiol. 2015;6:471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Low KE, Howell PL. Gram-negative synthase-dependent exopolysaccharide biosynthetic machines. Curr Opin Struc Biol. 2018;53:32-44. [DOI] [PubMed] [Google Scholar]
- 6. Atkin KE, MacDonald SJ, Brentnall AS, Potts JR, Thomas GH. A different path: revealing the function of staphylococcal proteins in biofilm formation. FEBS Lett. 2014;588:1869-1872. [DOI] [PubMed] [Google Scholar]
- 7. Bundalovic-Torma C, Whitfield GB, Marmont LS, Howell PL, Parkinson J. A systematic pipeline for classifying bacterial operons reveals the evolutionary landscape of biofilm machineries. Plos Comput Biol. 2020;16:e1007721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Whitfield GB, Marmont LS, Ostaszewski A, et al. Pel polysaccharide biosynthesis requires an inner membrane complex comprised of PelD, PelE, PelF, and PelG. J Bacteriol. 2020;202:e00684-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Friedman L, Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol. 2003;51:675-690. [DOI] [PubMed] [Google Scholar]
- 10. Vasseur P, Vallet-Gely I, Soscia C, Genin S, Filloux A. The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology+. 2005;151:985-997. [DOI] [PubMed] [Google Scholar]
- 11. Whitfield GB, Marmont LS, Bundalovic-Torma C, et al. Discovery and characterization of a Gram-positive Pel polysaccharide biosynthetic gene cluster. Plos Pathog. 2020;16:e1008281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Purcell EB, Tamayo R. Cyclic diguanylate signaling in Gram-positive bacteria. Fems Microbiol Rev. 2016;40:753-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Römling U, Galperin MY. Bacterial cellulose biosynthesis: diversity of operons, subunits, products, and functions. Trends Microbiol. 2015;23:545-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Colvin KM, Alnabelseya N, Baker P, Whitney JC, Howell PL, Parsek MR. PelA deacetylase activity is required for Pel polysaccharide synthesis in Pseudomonas aeruginosa. J Bacteriol. 2013;195:2329-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mauff FL, Bamford NC, Alnabelseya N, et al. Molecular mechanism of Aspergillus fumigatus biofilm disruption by fungal and bacterial glycoside hydrolases. J Biol Chem. 2019;294:10760-10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marmont LS, Whitfield GB, Rich JD, et al. PelA and PelB proteins form a modification and secretion complex essential for Pel polysaccharide-dependent biofilm formation in Pseudomonas aeruginosa. J Biol Chem. 2017;292:19411-19422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cherny KE, Sauer K. Untethering and degradation of the polysaccharide matrix are essential steps in the dispersion response of Pseudomonas aeruginosa biofilms. J Bacteriol. 2020;202:e00575-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Whitney JC, Colvin KM, Marmont LS, Robinson H, Parsek MR, Howell PL. Structure of the cytoplasmic region of PelD, a degenerate diguanylate cyclase receptor that regulates exopolysaccharide production in Pseudomonas aeruginosa. J Biol Chem. 2012;287:23582-23593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jennings LK, Storek KM, Ledvina HE, et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc Natl Acad Sci U S A. 2015;112:11353-11358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marmont LS, Whitfield GB, Pfoh R, et al. PelX is a UDP-N-acetylglucosamine C4-epimerase involved in Pel polysaccharide–dependent biofilm formation. J Biol Chem. 2020;295:11949-11962. [DOI] [PMC free article] [PubMed] [Google Scholar]