Abstract
Background
Apoptosis-related genes(Args)play an essential role in the occurrence and progression of hepatocellular carcinoma(HCC). However, few studies have focused on the prognostic significance of Args in HCC. In the study, we aim to explore an efficient prognostic model of Asian HCC patients based on the Args.
Methods
We downloaded mRNA expression profiles and corresponding clinical data of Asian HCC patients from The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC) databases. The Args were collected from Deathbase, a database related to cell death, combined with the research results of GeneCards、National Center for Biotechnology Information (NCBI) databases and a lot of literature. We used Wilcoxon-test and univariate Cox analysis to screen the differential expressed genes (DEGs) and the prognostic related genes (PRGs) of HCC. The intersection genes of DEGs and PGGs were seen as crucial Args of HCC. The prognostic model of Asian HCC patients was constructed by least absolute shrinkage and selection operator (lasso)- proportional hazards model (Cox) regression analysis. Kaplan-Meier curve, Principal Component Analysis (PCA) analysis, t-distributed Stochastic Neighbor Embedding (t-SNE) analysis, risk score curve, receiver operating characteristic (ROC) curve, and the HCC data of ICGC database and the data of Asian HCC patients of Kaplan-Meier plotter database were used to verify the model.
Results
A total of 20 of 56 Args were differentially expressed between HCC and adjacent normal tissues (p < 0.05). Univariate Cox regression analysis showed that 10 of 56 Args were associated with survival time and survival status of HCC patients (p < 0.05). There are seven overlapping genes of these 20 and 10 genes, including BAK1, BAX, BNIP3, CRADD, CSE1L, FAS, and SH3GLB1. Through Lasso-Cox analysis, an HCC prognostic model composed of BAK1, BNIP3, CSE1L, and FAS was constructed. Kaplan-Meier curve, PCA, t-SNE analysis, risk score curve, ROC curve, and secondary verification of ICGC database and Kaplan-Meier plotter database all support the reliability of the model.
Conclusions
Lasso-Cox regression analysis identified a 4-gene prognostic model, which integrates clinical and gene expression and has a good effect. The expression of Args is related to the prognosis of HCC patients, but the specific mechanism remains to be further verified.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-021-07886-6.
Keywords: Asia, Hepatocellular carcinoma, Apoptosis-related genes(Args), Prognostic model, Risk score
Background
At present, liver cancer has become one of the common causes of malignancy-related death globally [1], and its incidence is increasing year by year [2]. The World Health Organization (WHO) predicts that more than 1 million people will die of liver cancer by 2030 [3]. The incidence of liver cancer has remained high in Asia, as well [4]. HCC, as the main histological subtype of liver cancer, accounts for 90% of primary liver cancer. HCC is usually caused by various risk factors, including hepatitis B virus, hepatitis C virus, fatty liver, alcohol-related cirrhosis, smoking, obesity, different dietary exposures, etc. [5, 6]. Because HCC is a highly heterogeneous disease [7, 8], it is difficult to predict the prognosis. HCC is highly prevalent in Asia, and it is challenging to predict prognosis. Therefore, it is necessary to develop a useful prognostic model for Asian HCC patients.
Apoptosis is a way of programmed cell death related to the changes in cell morphology and structure [9]. Apoptosis mainly includes exogenous pathways caused by death receptors on the cell surface and endogenous apoptotic pathways caused by drugs, chemicals, endoplasmic reticulum stress, perforin, and granzyme [10]. Effector caspases activated by apoptosis signals will destroy the inhibitor of apoptosis, start the activity of caspase-activated deoxyribonuclease (CAD), and then destroy the structure of DNA, inhibit the activity of proteins regulating cell structure, destroy cell structure, transform cells into apoptotic bodies [11]. Apoptosis is closely related to HCC, the defect of apoptosis-inducing pathways will lead to the abnormal proliferation of tumor cells, and the resistance of cells to apoptosis will also increase the ability of tumor cells to evade immune system surveillance [12]. Args are closely related to the progression of HCC. Therefore, we intended to use the data of Asian HCC patients in TCGA database to construct a poly-genes prognostic model of Args and verify it by the data of ICGC database and Kaplan-Meier plotter database.
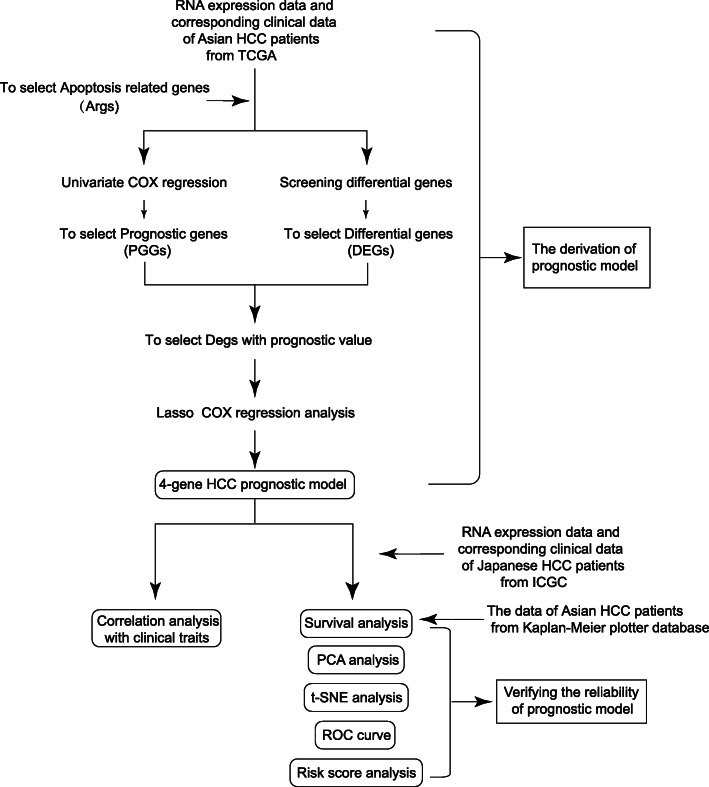
In the study, we downloaded the mRNA expression profiles and corresponding clinical data of Asian HCC patients from TCGA database. Japanese HCC patients’ data from ICGC and Asian HCC patients’ data from Kaplan-Meier plotter database were used for validation. Through Wilcoxon-test, we found the Args which were differentially expressed between HCC and adjacent normal tissues. Univariate Cox regression analysis showed some Args were associated with the survival time and survival status of HCC patients. By taking the intersection, we found that some Args were not only differentially expressed but also correlated with the prognosis of HCC. On this basis, we used Lasso-Cox analysis to mine a HCC prognostic model. Next, we used Kaplan-Meier curve, PCA, t-SNE analysis, risk score curve, ROC curve, and the HCC data from ICGC database and Kaplan-Meier plotter database to verify the prognosis effect of the model. The overall analysis flow was shown in Fig. 1.
Fig. 1.
Flow chart of data collection and analysis
Methods
Data preparation and processing
As of August 31, 2020, we downloaded the RNA-seq data and corresponding clinical characteristics of 158 Asian HCC patients from TCGA database (https://portal.gdc.cancer.gov/), including 6 normal paracancerous samples and 160 tumor samples, and the RNA-seq data and corresponding clinical information of 231 HCC patients from ICGC database (https://dcc.icgc/). The data of the 231 HCC patients downloaded from ICGC database are mainly from Japan [13]. The data of 155 Asian HCC patients from Kaplan-Meier plotter database (http://kmplot.com/analysis/) was selected as well [14]. To obtain the gene expression and clinical traits, we used R package limma [15] to process and normalize the data. We also downloaded Args from Deathbase dataset (http://deathbase.org/) and referred to the research results of GeneCards database (https://www.genecards.org), NCBI database (https://www.ncbi.nlm.nih.gov/), literature.
Identification of prognostic apoptosis-related DEGs in the TCGA dataset
The expression of Args was screened from the TCGA expression matrix. The average value of repetitive genes was taken; the abnormal values were deleted. Through Wilcoxon-test, the genes with p < 0.05 were selected as DEGs. We used Univariate Cox analysis to combine the expression of Args with survival time and survival status of HCC patients to screen out PRGs. The screening condition was p < 0.05. The DEGs and PRGs were intersected to select the Args, which were not only differentially expressed between HCC tissues and adjacent normal tissues but also related to the prognosis of HCC. The protein-protein interaction network was drawn by STRING database (version11.0) (https://string-db.org/) to clarify the interaction of the proteins, and the expression correlation coefficient diagram was drawn by R package igraph to show the expression relationship of the genes. R package pheatmap was used to illustrate the expression heatmap to show the expression of the intersect ARGs. The correlation filtering threshold is 0.2. R package survival was used to clarify the relationship between the genes and the prognosis of HCC patients. The screening condition was p < 0.05, as well.
Construction an Args related prognostic model of Asian HCC patients
The Lasso-penalized Cox regression analysis was applied to construct a prognostic model [16]. We used R package glmnet and lasso algorithm to contract variables, screen the genes that are positively related to the prognosis of HCC, and delete the genes with high correlation, to effectively avoid over-fitting of the prognostic model. The risk score of patients was calculated according to the expression of each gene and its corresponding regression coefficient. The formula was: Risk score = esum (expression of each gene * corresponding coefficient). Next, the HCC patients of TCGA and ICGC were divided into the high-risk and the low-risk groups according to the median cut-off risk score of the TCGA dataset.
Validation of the prognostic model
According to the gene expression of the prognostic model, R package Stats and Rtsne were used for PCA analysis and t-SNE analysis to reduce the dimensionality of data, to explore the distribution of the high-risk and low-risk samples. R package survival and survminer were used to draw the Kaplan-Meier curve to show the overall survival (OS) of different groups. We also illustrated the risk curves to show the relationship between OS and risk scores of patients in two groups. Univariate and multivariate prognostic analysis, ROC curve drawn by R package timeROC were used to determine whether the model can be regarded as a useful biomarker relating to the prognosis of Asian HCC patients. To rule out accidental error, we used the expression data of Japanese HCC patients downloaded from ICGC database for secondary detection. We also used the data of 155 Asian HCC patients of Kaplan-Meier plotter database to draw the Kaplan-Meier curve to show the correlation between the expression of BAK1, BNIP3, CSE1L, FAS and overall survival (OS) to verify the reality of this prognostic model further.
Correlation analysis with clinical traits
At first, we classified the clinical characteristics of HCC patients. Next, the model gene expression and risk score were compared with the clinical traits. T-test was used to determine whether the gene expression and risk score were associated with the clinical characteristics of HCC patients. The screening condition was p < 0.05. And the relationship between OS and clinical traits (Stage, Gender, Age) were displayed by the Kaplan-Meier curves.
Statistical analysis
Wilcoxon-test was used to screen DEGs. Kaplan-Meier analysis was used to compare the differences in OS among different groups. Cox regression analysis of univariate and multivariate variables was performed to determine the independent prognostic factors. All statistical analyses were carried out with R software (3.6.2). P < 0.05 is considered to be statistically significant. All the data were calculated to the nearest ten-thousandth.
Results
The results of data preparation and processing
A total of 158 Asian HCC patients from the TCGA-LIHC dataset, 231 Japanese HCC patients from the ICGC (LIRI-JP) dataset, and 155 Asian HCC patients from Kaplan-Meier plotter database were selected. We synthesized the data of Deathbase dataset, GeneCards dataset, NCBI database, and related literature. A total of 56 Args were selected (Table 1).
Table 1.
The Information of ARGs
| Symbol | Description | Reference |
|---|---|---|
| ANXA1 | Annexin A1 | [11, 17] |
| APAF1 | Apoptotic protease-activating factor 1 | [18] |
| AVEN | Cell death regulator Aven | [19] |
| BAD | Bcl2 antagonist of cell death | [20] |
| BAK1 | Bcl-2 homologous antagonist/killer | [21] |
| BAX | Apoptosis regulator BAX | [22] |
| BCL2 | Apoptosis regulator Bcl-2 | [23, 24] |
| BCL2A1 | Bcl-2-related protein A1 | [25, 26] |
| BCL2L1 | Apoptosis regulator Bcl-X | [27] |
| BCL2L10 | Apoptosis regulator Bcl-B | [28] |
| BCL2L11 | Bcl-2-like protein 11 | [29] |
| BCL2L12 | Bcl-2-like protein 12 | [30] |
| BCL2L14 | Bcl-2-like protein 14 | [31] |
| BCL2L2 | Bcl-2-Like Protein 2 | [32] |
| BID | BH3-interacting domain death agonist | [33, 34] |
| BIK | Bcl-2-interacting killer | [35, 36] |
| BIRC2 | Baculoviral IAP repeat-containing protein 2 | [37, 38] |
| BIRC3 | Baculoviral IAP repeat-containing protein 3 | [38, 39] |
| BMF | Bcl-2-modifying factor | [40] |
| BNIP3 | BCL2/adenovirus E1B 19 kDa protein-interacting protein 3 | [41] |
| BNIP3L | BCL2/adenovirus E1B 19 kDa protein-interacting protein 3-like | [42, 43] |
| CALR | Calreticulin Precursor | [44] |
| CASP10 | Caspase-10 Precursor | [45] |
| CASP3 | Caspase-3 Precursor | [46] |
| CASP6 | Caspase-6 Precursor | [47] |
| CASP7 | Caspase-7 Precursor | [48] |
| CASP8 | Caspase-8 Precursor | [49] |
| CASP9 | Caspase-9 Precursor | [50] |
| CFLAR | CASP8 and FADD-like apoptosis regulator Precursor | [51] |
| CRADD | Death domain-containing protein CRADD | [52, 53] |
| CSE1L | Exportin-2 | [54] |
| CYCS | Cytochrome c | [55] |
| DIABLO | Diablo homolog, mitochondrial Precursor | [56] |
| FADD | Protein FADD | [57, 58] |
| FAS | Tumor necrosis factor receptor superfamily member 6 Precursor | [59] |
| FASL | Tumor necrosis factor ligand superfamily member 6 | [60, 61] |
| HRK | Activator of apoptosis harakiri | [62] |
| LRP1 | Prolow-density lipoprotein receptor-related protein 1 Precursor | [63] |
| MCL1 | Induced myeloid leukemia cell differentiation protein Mcl-1 | [64] |
| MOAP1 | Modulator of apoptosis 1 | [65] |
| NOXA | Phorbol-12-myristate-13-acetate-induced protein 1 | [66] |
| PHAP | Acidic leucine-rich nuclear phosphoprotein 32 family member A | [67] |
| PUMA | Bcl-2-binding component 3 | [68] |
| RIPK1 | Receptor-interacting serine/threonine-protein kinase 1 | [69] |
| SH3GLB1 | Endophilin-B1 | [70] |
| TNF | Tumor necrosis factor Precursor | [71] |
| TNFRSF1A | Tumor necrosis factor receptor superfamily member 1A Precursor | [72] |
| TNFRSF1B | Tumor necrosis factor receptor superfamily member 1B Precursor | [73] |
| TNFSF10 | Tumor necrosis factor ligand superfamily member 10 | [74] |
| TP53 | Cellular tumor antigen p53 | [75] |
| TRADD | Tumor necrosis factor receptor type 1-associated DEATH domain protein | [76] |
| TNFRSF10A | Tumor necrosis factor receptor superfamily member 10A Precursor | [77] |
| TNFRSF10B | Tumor necrosis factor receptor superfamily member 10B Precursor | [77] |
| TNFRSF10C | Tumor necrosis factor receptor superfamily member 10C Precursor | [78] |
| TNFRSF10D | Tumor necrosis factor receptor superfamily member 10D Precursor | [78] |
| XIAP | Baculoviral IAP repeat-containing protein 4 | [79] |
The results of the screening of prognostic apoptosis-related DEGs
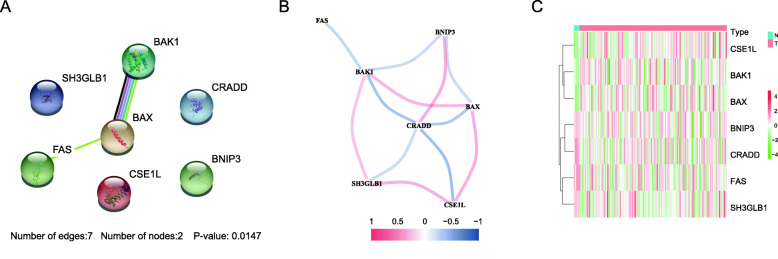
We found that a total of 20 Args were differentially expressed between HCC tissues and adjacent normal tissues (p < 0.05) (Table 2). Through univariate Cox regression analysis, we found that a total of 10 Args genes were related to the prognosis of HCC patients (p < 0.05) (Table 3). The intersection of the DEGs and PGRs was taken, seven apoptosis-related prognostic DEGs were screened out (Fig. 2). There are BAK1, BAX, BNIP3, CRADD, CSE1L, FAS, and SH3GLB1. Using the confirmed protein interaction relationship of String database [80], a PPI protein interaction network was constructed (Fig. 3a). The results showed that BAK1, BAX have the most edges in the network. The R package igraph was used to draw the correlation network according to the coefficient of the expression of the above gene. Figure 3b showed that according to the expression, BAK1, BAX, and CRADD might be the crucial genes of the network. Figure 3c showed that CSE1L, BAX, BAK1 were up-expressed in the HCC tissues and BNIP3, CRADD, FAS, SH3GLB1 down-expressed. Hazard ratio (HR) > 1 indicated that the gene was a high-risk gene related to the prognosis of HCC. Figure 4 showed among the 7 intersect Args, BAK1, BAX, CSE1L, and SH3GLB1 were high-risk prognosis-related genes of HCC, BNIP3, CRADD, FAS were low-risk prognosis-related genes.
Table 2.
Differentially expressed ARGs
| Gene | conMean | treatMean | logFC | p.value |
|---|---|---|---|---|
| BAK1 | 3.6840 | 7.3115 | 0.9889 | 0.0049 |
| BAX | 11.4039 | 22.1529 | 0.9580 | 0.0029 |
| BCL2 | 0.7565 | 0.6726 | −0.1696 | 0.0416 |
| BCL2L12 | 4.5640 | 9.7040 | 1.0883 | 0.0013 |
| BNIP3 | 32.5056 | 24.7871 | −0.3911 | 0.0283 |
| BNIP3L | 11.6700 | 7.6242 | −0.6141 | 0.0033 |
| CALR | 253.8644 | 564.4069 | 1.1527 | 0.0002 |
| CASP7 | 7.0616 | 4.9815 | −0.5034 | 0.0037 |
| CASP8 | 1.9536 | 2.8375 | 0.5385 | 0.0471 |
| CASP9 | 4.3837 | 2.3624 | −0.8919 | 0.0160 |
| CFLAR | 4.0543 | 3.2914 | −0.3008 | 0.0178 |
| CRADD | 6.0676 | 3.7604 | −0.6902 | 0.0036 |
| CSE1L | 11.4809 | 20.6950 | 0.8500 | 0.0010 |
| FAS | 6.2966 | 3.0988 | −1.0229 | 0.0035 |
| SH3GLB1 | 8.8173 | 7.2783 | −0.2767 | 0.0135 |
| TNFRSF1B | 24.5535 | 12.4104 | −0.9844 | 0.0049 |
| PHAP | 10.3958 | 13.9188 | 0.4210 | 0.0129 |
| PUMA | 1.6533 | 3.6201 | 1.1307 | 0.0034 |
| TRAIL-R4 | 5.2293 | 3.1865 | −0.7147 | 0.0156 |
| FASL | 0.4797 | 0.3434 | −0.4824 | 0.0096 |
Table 3.
HCC prognostic ARGs
| Gene | HR | HR.95 L | HR.95H | p.value |
|---|---|---|---|---|
| AVEN | 0.5495 | 0.3382 | 0.8928 | 0.0156 |
| BAK1 | 2.4836 | 1.5969 | 3.8626 | 0.0001 |
| BAX | 1.7013 | 1.0807 | 2.6781 | 0.0217 |
| BMF | 1.4375 | 1.0879 | 1.8995 | 0.0107 |
| BNIP3 | 0.6521 | 0.4665 | 0.9113 | 0.0123 |
| CRADD | 0.3687 | 0.2097 | 0.6483 | 0.0005 |
| CSE1L | 2.9130 | 1.8551 | 4.5741 | <0.0001 |
| FAS | 0.5185 | 0.3474 | 0.7737 | 0.0013 |
| SH3GLB1 | 2.3639 | 1.2372 | 4.5170 | 0.0092 |
| NOXA | 2.0811 | 1.3305 | 3.2553 | 0.0013 |
Fig. 2.
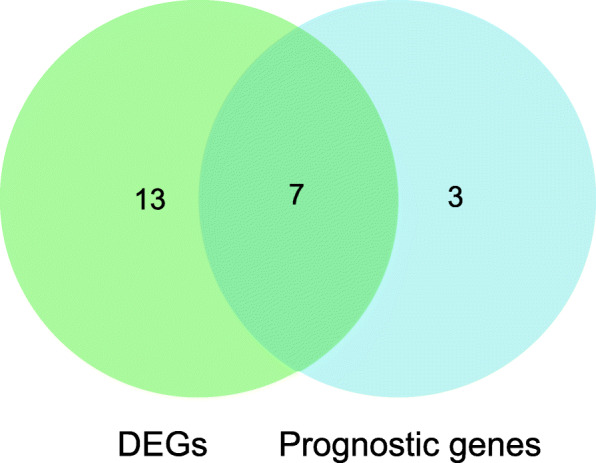
Venn diagram showing the intersect genes of differential genes (DEGs) and prognostic gens (PGGs)
Fig. 3.
The analysis of 7 intersect ARGs. a The PPI network. b The expression correlation network. c The expression heatmap
Fig. 4.
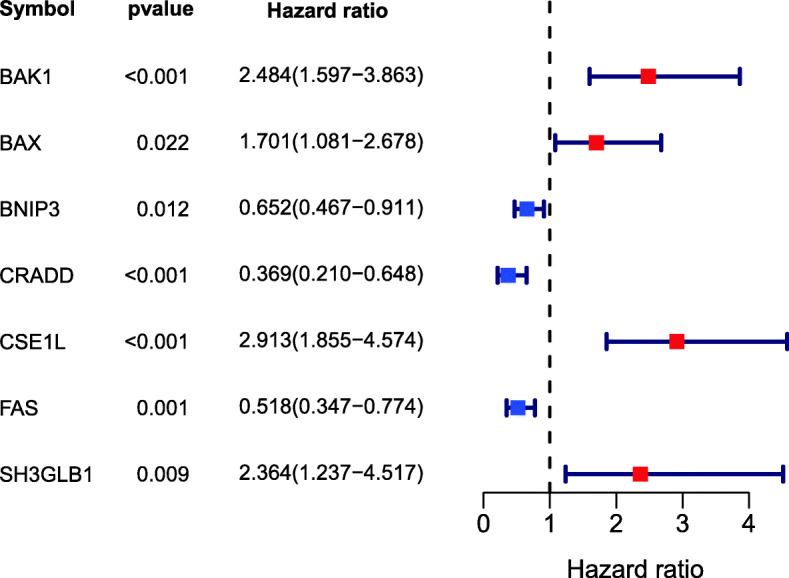
Forest plot showing the results of the univariate Cox regression analysis between gene expression and OS (overall survival)
The results of the construction of the prognostic signature
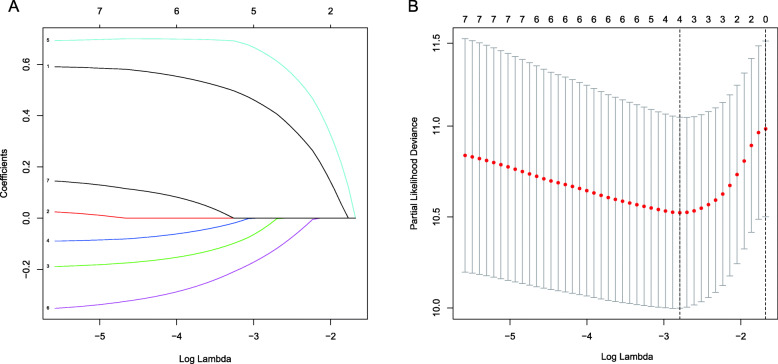
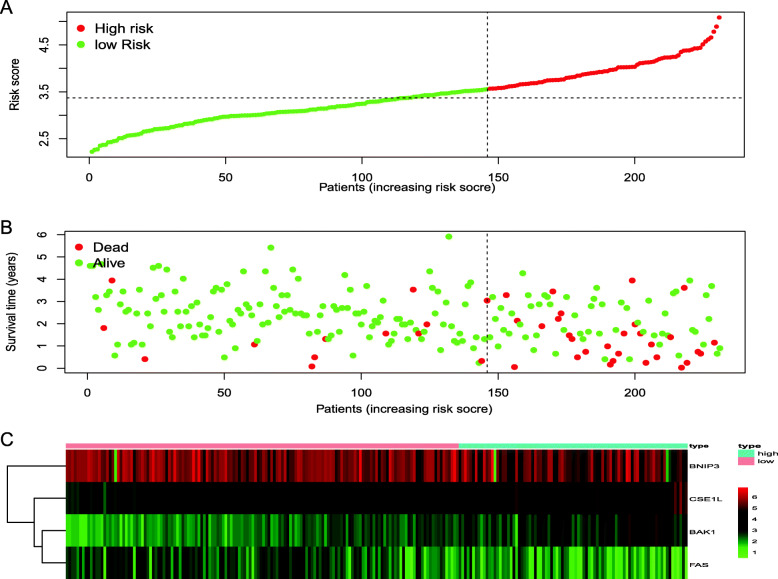
Lasso-Cox regression analysis was applied to establish a prognostic model based on the expression of the 7 genes. According to the penalty parameter (Lambda) in the model, we constructed a prognostic model of HCC patients consisting of 4 genes (Fig. 5). These genes are BAK1, BNIP3, CSE1L, and FAS (Table 4). We then calculated the risk score based on the expression of the 4 genes in the TCGA dataset and corresponding coefficient (coef) (Risk score = e (the expression of BAK1*0.4252 + the expression of BNIP3*-0.0237 + the expression of CSE1L*0.6321 + the expression of FAS*-0.1360)) and divided the patients into high- and low-risk groups according to the median cutoff value of TCGA (risk score < = 3.5614 was low-risk, > 3.5614 was high-risk). The HCC samples of ICGC were also divided into the high- and low-risk groups according to the same median cut-off value.
Fig. 5.
The Construction of a 4-gene prognostic model in the TCGA dataset. a Lasso coefficient profiles of the expression of 7 genes. b Selection of the penalty parameter (Lambda) in the Lasso model via 10-fold cross-validation
Table 4.
The information of the 4 genes used to construct the prognostic signature
| Symbol | Description | Coef |
|---|---|---|
| BAK1 | Bcl-2 homologous antagonist/killer | 0.4252 |
| BNIP3 | BCL2/adenovirus E1B 19 kDa protein-interacting protein 3 | −0.0237 |
| CSE1L | Exportin-2 | 0.6321 |
| FAS | Tumor necrosis factor receptor superfamily member 6 Precursor | −0.1360 |
The results of the validation of the 4-gene prognostic model
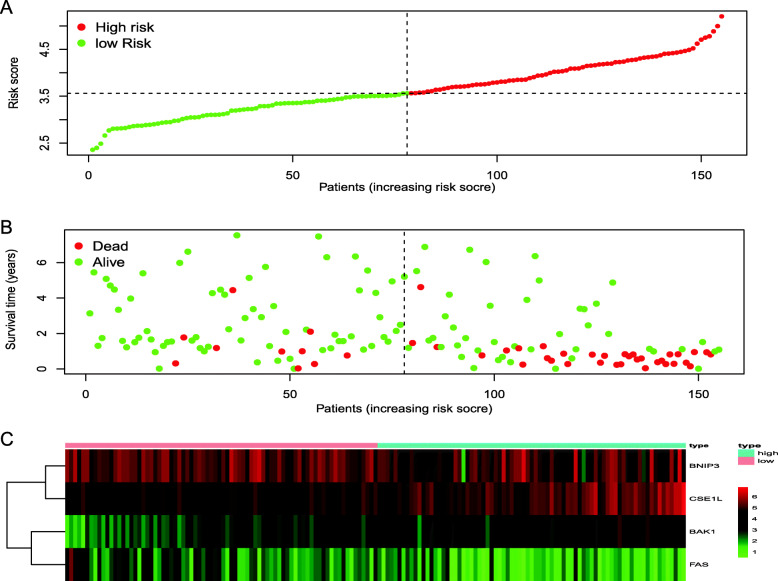
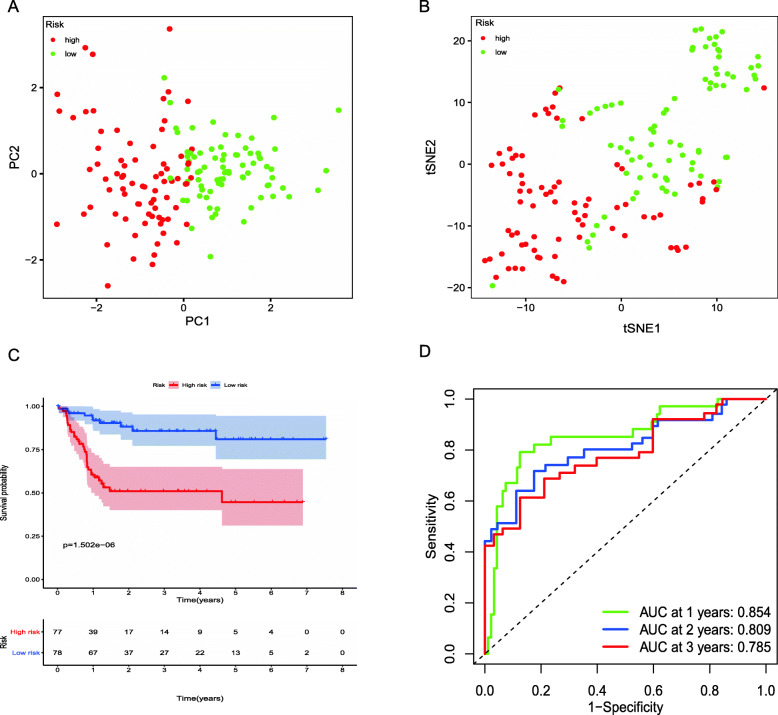
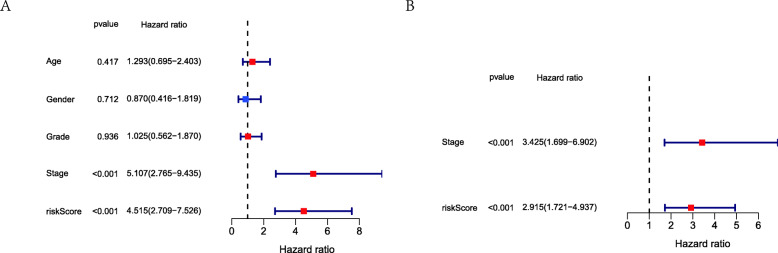
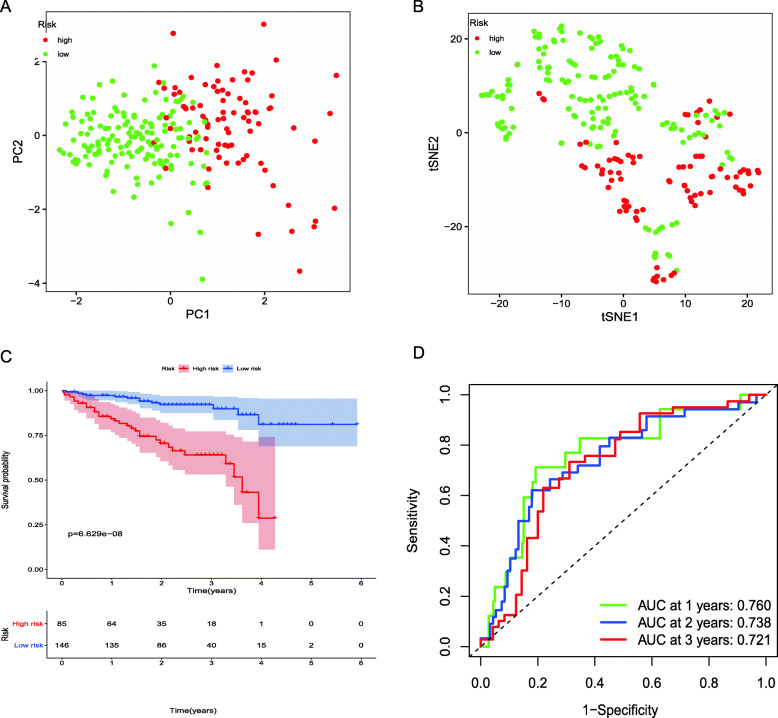
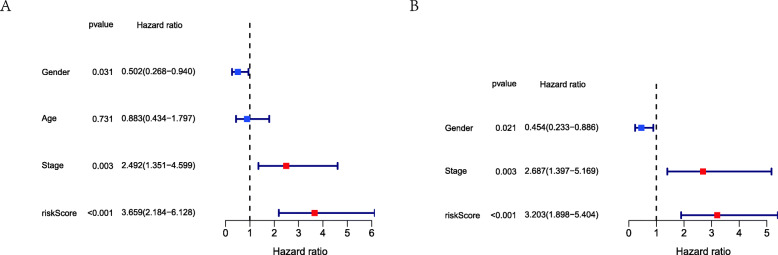
We had classified the Asian HCC patients of TCGA database into the high- and low-risk groups according to the median cut-off value (Fig. 6a). Figure 6b showed that in HCC patients of TCGA, the OS of high-risk patients was significantly lower than that of low-risk patients, suggesting the probability of premature death in high-risk patients was higher than that in low-risk patients. What is more, there are significant differences in gene expression. The expression of BAK1, CSE1L in the high-risk group is higher than that in the low-risk group, but BNIP3, FAS was down-regulated in the high-risk group (Fig. 6c). We used PCA analysis and t-SNE analysis for data dimensionality reduction to observe a significant difference between the high- and low-risk groups. The results (Fig. 7a, b) showed that high-risk and low-risk groups of TCGA were a two-way distribution. Kaplan-Meier curve (Fig. 7c) showed that the OS of high-risk patients was significantly lower than that of patients with low risk at the same timing (p < 0.001). ROC curves evaluated the predictive performance of the risk score for OS, and the area under the curve (AUC) reached 0.854 at 1 year, 0.809 at 2 years, and 0.785 at 3 years (Fig. 7d). The AUC of the three timings were all higher than 0.700, suggesting that the prognostic model can be regarded as a qualified prognostic biomarker of Asian HCC patients. Then we conducted univariate and multivariate Cox regression analysis to determine whether the risk score model could be used as an independent prognostic factor of OS. In univariate Cox regression analysis, the risk score was significantly associated with the OS of the TCGA HCC patients(p < 0.001)(Fig. 8a). After adjusting for other interfering factors, multivariate Cox regression analysis showed that the risk score was still an independent predictor of OS (p < 0.001) (Fig. 8b). In order to exclude the contingency, we used the Japanese HCC samples from ICGC database for secondary detection. The HCC samples of the ICGC dataset were also categorized into high- and low-risk groups by the same median value (Fig. 9a). The result showed that most patients with ICGC were at low-risk. Figure 9b showed that OS in the high-risk group of ICGC was also lower than that in the low-risk group. The expression levels of BAK1, CSE1L, BNIP3, and FAS were consistent with those in the TCGA patients (Fig. 9c). The results of PCA analysis and t-SNE analysis showed that the patients in the high-risk group and the low-risk group in the ICGC dataset also showed a two-way distribution (Fig. 10a, b). The Kaplan-Meier curve showed that the OS of HCC patients in the ICGC dataset was lower than that in the low-risk group (p < 0.001) (Fig. 10c) at the same timing. Figure 10d displayed the AUC reached 0.760 at 1 year, 0.738 at 2 years, and 0.721 at 3 years. The AUC of the three-time points were all higher than 0.700 as well. We performed univariate and multivariate Cox regression analysis to determine whether the risk score model was also an independent prognostic factor of OS in HCC patients of ICGC. Through the analysis, we found that risk score could be regarded as an independent prognostic factor of the OS of IGCG HCC patients as well (p < 0.001) (Fig. 11a, b). Moreover, to verify the relationship between the prognosis model and different clinical traits, we divided the Japanese HCC samples from ICGC database into a high-risk group and low-risk group according to the risk value and drew the Kaplan-Meier curves based on the gender, age, and HCC stage (Fig. 12). The results showed that the HCC prognostic model could also be used as the prognosis of HCC with different age, gender, and stage. Meanwhile, because only used the Japanese HCC dataset for validation to reduce the regional influence of HCC disease, the data of Kaplan Meier plotter database was used to verify again. We selected HCC as the research disease and 155 Asian patients as the research objective. The expression of 4 genes (BAK1, BNIP3, CSE1L, FAS) used to construct the predictive model were selected, and the Kaplan-Meier curves were drawn, respectively (Fig. 13). The results suggested that the prognostic model can be seen as an effective prognostic factor for Asian HCC patients.
Fig. 6.
The validation of the reliability of the 4-gene signature by analyzing the risk score and the TCGA dataset. a The distribution and the median value of the risk scores. b The distributions of OS status, OS and risk score. c The expression of four prognosis-related ARGs between high-risk and low-risk group
Fig. 7.
The validation of the reliability of the 4-gene signature using the TCGA dataset. a PCA plot. b t-SNE analysis. c Kaplan-Meier curve showing the OS of patients in the high-risk group and low-risk group. d AUC of time-dependent ROC curves verified the prognostic performance of the risk score
Fig. 8.
Results of the univariate and multivariate Cox regression analysis regarding OS in the TCGA dataset. a The result of univariate Cox regression analysis. b The result of multivariate Cox regression analysis
Fig. 9.
The validation of the reliability of the 4-gene signature by analyzing the risk score and the ICGC dataset. a The distribution and the median value of the risk scores. b The distributions of OS status, OS and risk score. c The expression of four prognosis-related ARGs between high-risk and low-risk group
Fig. 10.
The validation of the reliability of the 4-gene signature using the ICGC dataset. a PCA plot. b t-SNE analysis. c Kaplan-Meier curve showing the OS of patients in the high-risk group and low-risk group. d AUC of time-dependent ROC curves verified the prognostic performance of the risk score
Fig. 11.
Results of the univariate and multivariate Cox regression analysis regarding OS in the ICGC dataset. a The result of univariate Cox regression analysis. b The result of multivariate Cox regression analysis
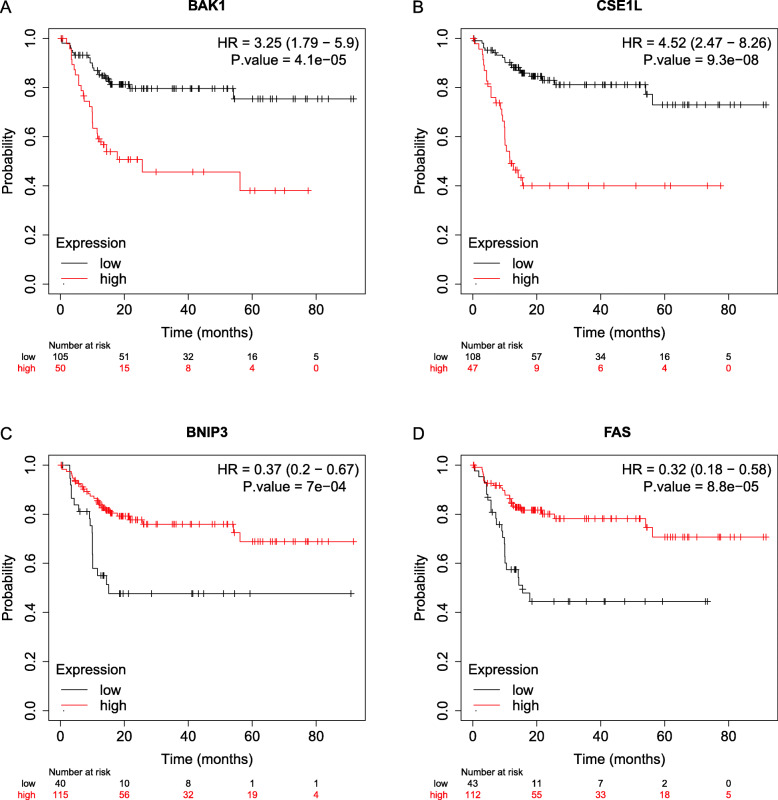
Fig. 12.
The Kaplan-Meier curves of 4 prognostic model genes. a. Kaplan-Meier curve showing the OS of BAK1 in the high-risk group and low-risk group. b. Kaplan-Meier curve showing the OS of CSE1L in the high-risk group and low-risk group. c. Kaplan-Meier curve showing the OS of BNIP3 in the high-risk group and low-risk group. d Kaplan-Meier curve showing the OS of FAS in the high-risk group and low-risk group
Fig. 13.
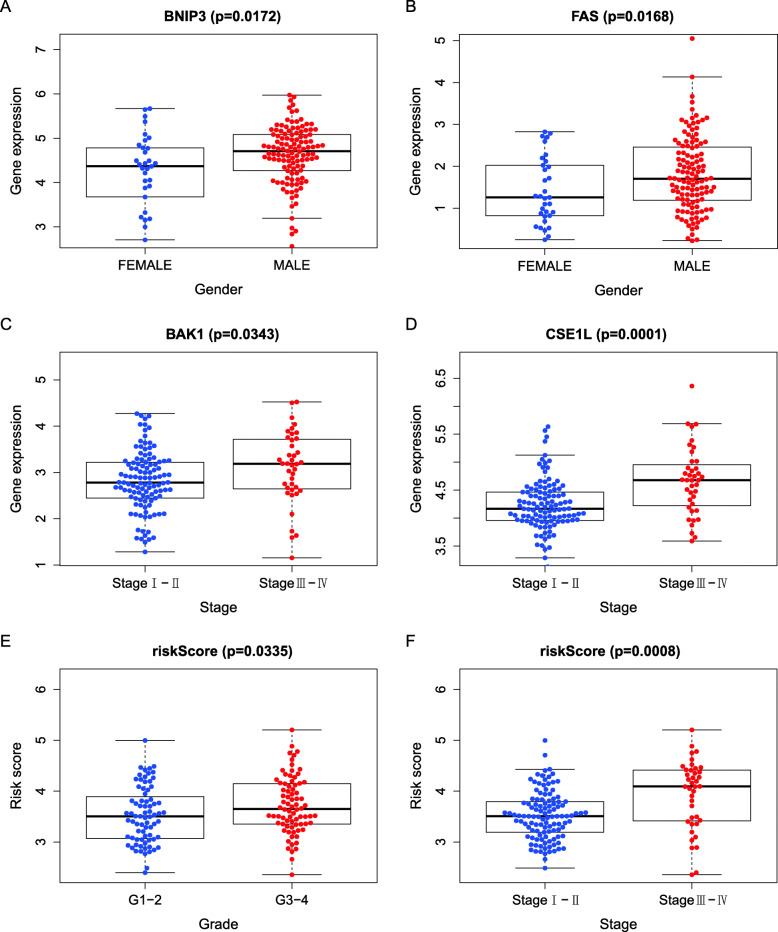
The correlation between gene expression, risk score, and clinical traits. a The correlation between the expression of BNIP3 and gender. b The correlation between the expression of FAS and gender. c The correlation between the expression of BAK1 and HCC stage. d The correlation between the expression of CSE1L and HCC stage. e The correlation between risk score and HCC grade. f The correlation between risk score and HCC stage
The results of correlation analysis with clinical traits
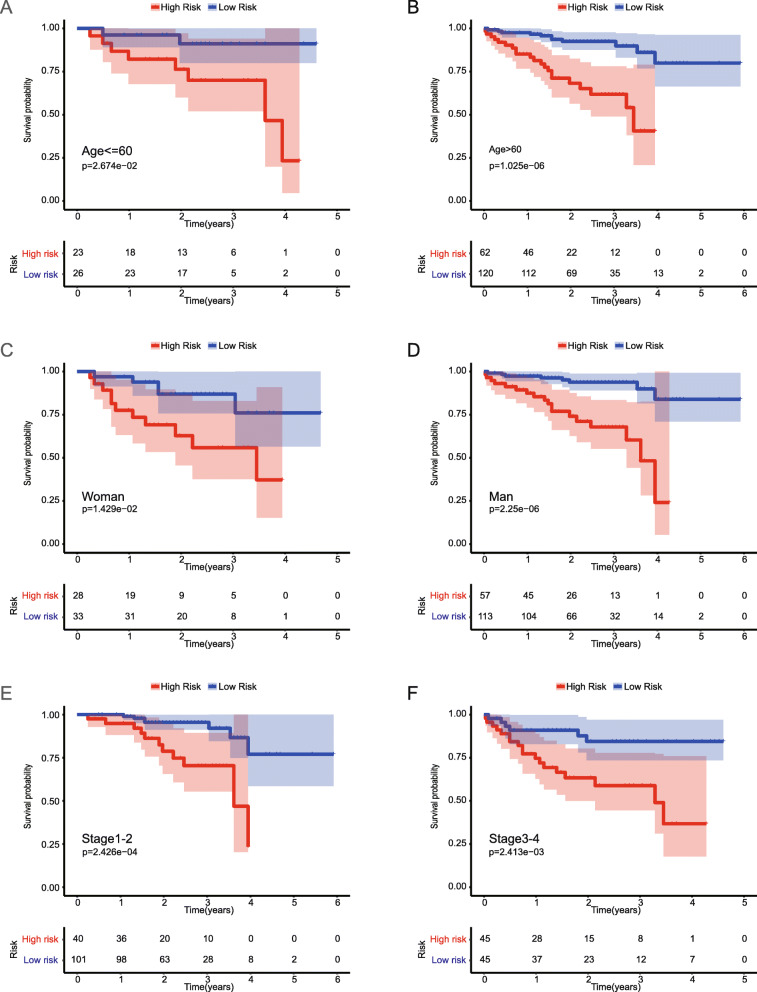
The TCGA-BC-A10W and TCGA-ZP-A9CZ cases without HCC stage information were deleted. T-test was used to determine whether the expression of the genes and the risk score were associated with the clinical characteristics of HCC patients. Some clinical features of HCC, including age, sex, grade, and stage, were assessed for their probable correlation with BAK1, BNIP3, CSE1L, FAS, and risk score, as shown in Table 5. We selected the correlation, which satisfies p < 0.05 to draw the box plots. The results showed that the expression of BNIP3 and FAS were significantly different between male and female (Fig. 13a). BAK1 and CSE1L were all significantly differentially expressed between Stage I-II and Stage III-IV patients (Fig. 13b-d). The values of risk score were significantly different between the Grade 1–2 and Grade 3–4 (p < 0.05) (Fig. 13e). The values of risk score were different between Stage I-II and Stage III-IV, as well. (p<0.001) (Fig. 14f). Correlation analysis of clinical traits showed that risk score was near related to the grade and stage of HCC. We also used the clinical character data of Japanese HCC patients in the ICGC database. The relationship between different clinical traits (Age, Gender, Stage) and OS was showed by Kaplan-Meier curves, respectively. The results (Fig. 14) showed that the prognosis of HCC was Significant different with age, gender, and cancer stage in both high-risk and low-risk groups divided according to the median risk value of the prognostic model (P < 0.05). The result further demonstrated the prognostic model we constructed for Asian HCC patients is reliable.
Table 5.
The clinical correlation analysis
| Gene | Age (<=60/> 60) (p.value) |
Gender (male/female) (p.value) |
Grade (G1–2/ G3–4) (p.value) |
Stage (Stage I-II/Stage III-IV) (p.value) |
|---|---|---|---|---|
| BAK1 | 0.9288(0.3553) | 1.6066(0.1146) | −1.1404(0.2559) | −2.1657(0.0343) |
| BNIP3 | −1.8423(0.0683) | −2.4848(0.0172) | 0.0997(0.9207) | 1.4734(0.1463) |
| CSE1L | 1.3898(0.1675) | 0.4319(0.6674) | −1.9311(0.0554) | −4.1353(0.0001) |
| FAS | −1.7086(0.0904) | −2.4645(0.0168) | 1.7182(0.0879) | 0.1881(0.8515) |
| riskScore | 1.7350(0.0859) | 1.6985(0.0956) | −2.1455(0.0335) | −3.5536(0.0008) |
Fig. 14.
The Kaplan-Meier curves of clinical traits. a Showing the OS of younger (age < =60) in the high-risk group and low-risk group. b Showing the OS of older (age > 60) in the high-risk group and low-risk group. c Showing the OS of women in the high-risk group and low-risk group. d Showing the OS of men in the high-risk group and low-risk group. e Showing the OS of stageI-II in the high-risk group and low-risk group. f Showing the OS of stageIII-IV in the high-risk group and low-risk group
Discussion
Large geographic disparities in incidence and mortality of HCC exist. Asia has the highest incidence of HCC in the world because of the presence of multiple risk factors, such as hepatitis B and the contamination of aflatoxin [81]. Another risk factor for HCC, chronic hepatitis C infection, in Asia is most significant in Japan, the only Asian country with more hepatitis C virus (HCV) than hepatitis B virus (HBV)-related hepatocellular carcinoma. To eliminate the cells infected by viruses, the body will activate apoptosis, but to reduce the death of cancer cells, HCC cells will inhibit apoptosis. The balance of survival and apoptosis are closely related to the progression of HCC [12]. The etiology of HCC in Asia is quite different from that in Europe, the US, and the balance of survival and apoptosis is crucial for the progression of HCC. Therefore, it is necessary to study the cases of Asian HCC patients and find a useful Args related prognostic model based on the genes of Asian populations.
In the study, we systematically studied the expression of 56 Args and the effect of prognosis. And we constructed a new prognostic model based on apoptosis-related DEGs and verified it by the data of TCGA and ICGC databases. The HCC prognostic model consists of four Args (BAK1, BNIP3, CSE1L, and FAS). Pro-Apoptotic Protein BAK (BAK1) is a pro-apoptotic protein [21], which plays a vital role in the process of mitochondrial apoptosis. When receiving an apoptosis signal, BAK1 can change the permeability of mitochondrial outer membrane (MOM), release apoptotic factors, and activate effector caspases to realize apoptosis [82, 83]. BNIP3, a pro-apoptotic member of the Bcl-2 family of apoptotic proteins [84], can overcome the inhabitation of apoptosis caused by BCL2. However, some studies have found that BNIP3 has an inhibitory effect on cancer [85]. BNIP3 can delay the progression of primary breast cancer by preventing the accumulation of dysfunctional mitochondria and reducing the resulting excess reactive oxygen species (ROS) [86]. CSE1L, the cellular apoptosis susceptibility protein, is highly expressed in various cancers [87]. It has been found that CSE1L plays an essential role in regulating apoptosis induced by chemotherapeutic drugs [88]. CSE1L can inhibit paclitaxel-induced apoptosis by affecting G2/M phase cell cycle arrest and microtubule aster formation induced by paclitaxel [88, 89]. The Fas-antigen is a cell surface receptor that transduces apoptotic signals into cells [90]. Fas/FasL signaling pathway will promote cell apoptosis.
In the study, we found that BAK1, CSE1L, BNIP3, and Fas were all related to the prognosis of HCC, but the expression changes were different. BAK1 and CSE1L were up-regulated in HCC tissues, while BNIP3 and Fas were down-regulated. We recognized that it is not clear whether these genes affect the prognosis of HCC patients mainly by affecting cancer cell apoptosis because these genes affect the progress of HCC in many ways, not only apoptosis. Therefore, further studies on these four genes are needed.
Conclusion
We successfully constructed a novel apoptosis gene-related prognostic model of accurately predicting the prognosis of Asian HCC patients, with higher risk scores demonstrating adverse prognosis. Kaplan-Meier curve, PCA analysis, t-SNE analysis, risk score curve, ROC curve, and the data of ICGC were used to verify the reliability of the model. We believed this model could act as a useful independent prognostic predictor for Asian HCC patients.
Supplementary Information
Acknowledgements
None.
Abbreviations
- HCC
Hepatocellular Carcinoma
- Args
Apoptosis-related genes
- DEGs
Differential expressed genes
- PRGs
Prognostic related genes
- TCGA
The Cancer Genome Atlas
- ICGC
International Cancer Genome Consortium
- NCBI
National Center for Biotechnology Information
- Lasso
Least absolute shrinkage and selection operator
- COX
Proportional hazards model
- CAD
Caspase-activated deoxyribonuclease
- PCA
Principal Component Analysis
- t-SNE
t-distributed Stochastic Neighbor Embedding
- ROC
Receiver operating characteristic
- AUC
Area under curve
- OS
Overall survival
- GO
Gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- HR
Hazard ratio
- coef
Corresponding coefficient
- HBV
Hepatitis B virus
- HCV
Hepatitis C virus
- MOM
Mitochondrial outer membrane
- ROS
Reactive oxygen species
Authors’ contributions
Study concept and design: J.Y, J.C, and Z.C; data analysis/interpretation: J.Y; manuscript drafting: J.Y; manuscript revision: J.Y; approval of the final version of submitted manuscript: J.Y and Z.C. All authors have read and approved the manuscript.
Funding
None.
Availability of data and materials
All the data used in the study are obtained from the TCGA database (https://portal.gdc.cancer.gov/), ICGC database (https://dcc.icgc.org/), Deathbase database (http://deathbase.org/), and Kaplan-Meier plotter database (http://kmplot.com/analysis/) which are opening and available to all.
All datasets generated for this study are included in the manuscript and the supplementary files.
Ethics approval and consent to participate
The research didn’t involve animal experiments and human specimens, no ethics related issues.
Consent for publication
Not applicable.
Competing interests
None.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Villanueva A. Hepatocellular Carcinoma. N Engl J Med. 2019;380(15):1450–1462. doi: 10.1056/NEJMra1713263. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57(1):43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 3.Gunnarsson N, Sandin F, Höglund M, Stenke L, Björkholm M, Lambe M, Olsson-Strömberg U, Richter J, Själander A. Population-based assessment of chronic myeloid leukemia in Sweden: striking increase in survival and prevalence. Eur J Haematol. 2016;97(4):387–392. doi: 10.1111/ejh.12743. [DOI] [PubMed] [Google Scholar]
- 4.Center MM, Jemal A. International trends in liver cancer incidence rates. Cancer Epidemiol Biomarkers Prev. 2011;20(11):2362–2368. doi: 10.1158/1055-9965.EPI-11-0643. [DOI] [PubMed] [Google Scholar]
- 5.Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruix J, Han KH, Gores G, Llovet JM, Mazzaferro V. Liver cancer: approaching a personalized care. J Hepatol. 2015;62(1 Suppl):S144–S156. doi: 10.1016/j.jhep.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, Villanueva A, Newell P, Ikeda K, Hashimoto M, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69(18):7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nault JC, Villanueva A. Intratumor molecular and phenotypic diversity in hepatocellular carcinoma. Clin Cancer Res. 2015;21(8):1786–1788. doi: 10.1158/1078-0432.CCR-14-2602. [DOI] [PubMed] [Google Scholar]
- 9.Fleisher TA. Apoptosis. Ann Allergy Asthma Immunol. 1997;78(3):245–249. doi: 10.1016/S1081-1206(10)63176-6. [DOI] [PubMed] [Google Scholar]
- 10.Majtnerová P, Roušar T. An overview of apoptosis assays detecting DNA fragmentation. Mol Biol Rep. 2018;45(5):1469–1478. doi: 10.1007/s11033-018-4258-9. [DOI] [PubMed] [Google Scholar]
- 11.Vaughan AT, Betti CJ, Villalobos MJ. Surviving apoptosis. Apoptosis. 2002;7(2):173–177. doi: 10.1023/A:1014374717773. [DOI] [PubMed] [Google Scholar]
- 12.Igney FH, Krammer PH. Death and anti-death: tumour resistance to apoptosis. Nat Rev Cancer. 2002;2(4):277–288. doi: 10.1038/nrc776. [DOI] [PubMed] [Google Scholar]
- 13.Fujimoto A, Furuta M, Totoki Y, Tsunoda T, Kato M, Shiraishi Y, Tanaka H, Taniguchi H, Kawakami Y, Ueno M, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48(5):500–509. doi: 10.1038/ng.3547. [DOI] [PubMed] [Google Scholar]
- 14.Nagy Á, Lánczky A, Menyhárt O, Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8(1):9227. doi: 10.1038/s41598-018-27521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simon N, Friedman J, Hastie T, Tibshirani R. Regularization paths for Cox's proportional hazards model via coordinate descent. J Stat Softw. 2011;39(5):1–13. doi: 10.18637/jss.v039.i05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia Q, Li X, Zhou H, Zheng L, Shi J. S100A11 protects against neuronal cell apoptosis induced by cerebral ischemia via inhibiting the nuclear translocation of annexin A1. Cell Death Dis. 2018;9(6):657. doi: 10.1038/s41419-018-0686-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gortat A, Sancho M, Mondragón L, Messeguer À, Pérez-Payá E, Orzáez M. Apaf1 inhibition promotes cell recovery from apoptosis. Protein Cell. 2015;6(11):833–843. doi: 10.1007/s13238-015-0200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kutuk O, Temel SG, Tolunay S, Basaga H. Aven blocks DNA damage-induced apoptosis by stabilising Bcl-xL. Eur J Cancer (Oxford, England : 1990) 2010;46(13):2494–2505. doi: 10.1016/j.ejca.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 20.Michurina SV, Kolesnikov SI, Bochkareva AL, Ishchenko IY, Arkhipov SA. Expression of apoptosis regulator proteins Bcl-2 and bad in rat ovarian follicular apparatus during recovery after extreme hypothermia. Bull Exp Biol Med. 2019;168(2):205–209. doi: 10.1007/s10517-019-04675-x. [DOI] [PubMed] [Google Scholar]
- 21.Yang X, Tang S, Li D, Yu X, Wang F, Xiao X. DIDS inhibits overexpression BAK1-induced mitochondrial apoptosis through GSK3β/β-catenin signaling pathway. J Cell Physiol. 2018;233(6):5070–5077. doi: 10.1002/jcp.26396. [DOI] [PubMed] [Google Scholar]
- 22.Lin Y, Kokontis J, Tang F, Godfrey B, Liao S, Lin A, Chen Y, Xiang J. Androgen and its receptor promote Bax-mediated apoptosis. Mol Cell Biol. 2006;26(5):1908–1916. doi: 10.1128/MCB.26.5.1908-1916.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kontos CK, Christodoulou MI, Scorilas A. Apoptosis-related BCL2-family members: key players in chemotherapy. Anticancer Agents Med Chem. 2014;14(3):353–374. doi: 10.2174/18715206113139990091. [DOI] [PubMed] [Google Scholar]
- 24.Warren CFA, Wong-Brown MW, Bowden NA. BCL-2 family isoforms in apoptosis and cancer. Cell Death Dis. 2019;10(3):177. doi: 10.1038/s41419-019-1407-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vier J, Groth M, Sochalska M, Kirschnek S. The anti-apoptotic Bcl-2 family protein A1/Bfl-1 regulates neutrophil survival and homeostasis and is controlled via PI3K and JAK/STAT signaling. Cell Death Dis. 2016;7(2):e2103. doi: 10.1038/cddis.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He CH, Waxman AB, Lee CG, Link H, Rabach ME, Ma B, Chen Q, Zhu Z, Zhong M, Nakayama K, et al. Bcl-2-related protein A1 is an endogenous and cytokine-stimulated mediator of cytoprotection in hyperoxic acute lung injury. J Clin Invest. 2005;115(4):1039–1048. doi: 10.1172/JCI23004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon MK, Kim BY, Lee JY, Ha JH, Kim SA, Lee DH, Lee MS, Lee MK, Choi JS, Cho JH, et al. Cytoplasmic pro-apoptotic function of the tumor suppressor p73 is mediated through a modified mode of recognition of the anti-apoptotic regulator Bcl-X(L) J Biol Chem. 2018;293(51):19546–19558. doi: 10.1074/jbc.RA118.003061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding Q, Xie XL, Wang MM, Yin J, Tian JM, Jiang XY, Zhang D, Han J, Bai Y, Cui ZJ, et al. The role of the apoptosis-related protein BCL-B in the regulation of mitophagy in hepatic stellate cells during the regression of liver fibrosis. Exp Mol Med. 2019;51(1):1–13. doi: 10.1038/s12276-018-0199-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banjara S, Mao J, Ryan TM, Caria S, Kvansakul M. Grouper iridovirus GIV66 is a Bcl-2 protein that inhibits apoptosis by exclusively sequestering Bim. J Biol Chem. 2018;293(15):5464–5477. doi: 10.1074/jbc.RA117.000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu CW, Yang MC, Chou CH, Huang WS, Hsiao BX, Wang YT, Chiou SJ, Loh JK, Hong YR. GSK3β-mediated Ser156 phosphorylation modulates a BH3-like domain in BCL2L12 during TMZ-induced apoptosis and autophagy in glioma cells. Int J Mol Med. 2018;42(2):905–918. doi: 10.3892/ijmm.2018.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woznicki JA, Flood P, Bustamante-Garrido M, Stamou P, Moloney G, Fanning A, Zulquernain SA, McCarthy J, Shanahan F, Melgar S, et al. Human BCL-G regulates secretion of inflammatory chemokines but is dispensable for induction of apoptosis by IFN-γ and TNF-α in intestinal epithelial cells. Cell Death Dis. 2020;11(1):68. doi: 10.1038/s41419-020-2263-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang C, Zhou B, Liu M, Liu Y, Gao R. miR-126-5p restoration promotes cell apoptosis in cervical cancer by targeting Bcl2l2. Oncol Res. 2017;25(4):463–470. doi: 10.3727/096504016X14685034103879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tiwari M, Prasad S, Tripathi A, Pandey AN, Ali I, Singh AK, Shrivastav TG, Chaube SK. Apoptosis in mammalian oocytes: a review. Apoptosis. 2015;20(8):1019–1025. doi: 10.1007/s10495-015-1136-y. [DOI] [PubMed] [Google Scholar]
- 34.Debernardi J, Hollville E, Lipinski M, Wiels J, Robert A. Differential role of FL-BID and t-BID during verotoxin-1-induced apoptosis in Burkitt's lymphoma cells. Oncogene. 2018;37(18):2410–2421. doi: 10.1038/s41388-018-0123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou H, Zhang Y, Fu Y, Chan L, Lee AS. Novel mechanism of anti-apoptotic function of 78-kDa glucose-regulated protein (GRP78): endocrine resistance factor in breast cancer, through release of B-cell lymphoma 2 (BCL-2) from BCL-2-interacting killer (BIK) J Biol Chem. 2011;286(29):25687–25696. doi: 10.1074/jbc.M110.212944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stadler LK, Tomlinson DC, Lee T, Knowles MA, Ko Ferrigno P. The use of a neutral peptide aptamer scaffold to anchor BH3 peptides constitutes a viable approach to studying their function. Cell Death Dis. 2014;5(1):e1037. doi: 10.1038/cddis.2013.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiong Y, Ren YF, Xu J, Yang DY, He XH, Luo JY, Rana JS, Zhang Y, Zheng ZS, Liu DH, et al. Enhanced external counterpulsation inhibits endothelial apoptosis via modulation of BIRC2 and Apaf-1 genes in porcine hypercholesterolemia. Int J Cardiol. 2014;171(2):161–168. doi: 10.1016/j.ijcard.2013.11.033. [DOI] [PubMed] [Google Scholar]
- 38.Yamato A, Soda M, Ueno T, Kojima S, Sonehara K, Kawazu M, Sai E, Yamashita Y, Nagase T, Mano H. Oncogenic activity of BIRC2 and BIRC3 mutants independent of nuclear factor-κB-activating potential. Cancer Sci. 2015;106(9):1137–1142. doi: 10.1111/cas.12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rada M, Nallanthighal S, Cha J, Ryan K, Sage J, Eldred C, Ullo M, Orsulic S, Cheon DJ. Inhibitor of apoptosis proteins (IAPs) mediate collagen type XI alpha 1-driven cisplatin resistance in ovarian cancer. Oncogene. 2018;37(35):4809–4820. doi: 10.1038/s41388-018-0297-x. [DOI] [PubMed] [Google Scholar]
- 40.Lau GJ, Godin N, Maachi H, Lo CS, Wu SJ, Zhu JX, Brezniceanu ML, Chénier I, Fragasso-Marquis J, Lattouf JB, et al. Bcl-2-modifying factor induces renal proximal tubular cell apoptosis in diabetic mice. Diabetes. 2012;61(2):474–484. doi: 10.2337/db11-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen BC, Weng YJ, Shibu MA, Han CK, Chen YS, Shen CY, Lin YM, Viswanadha VP, Liang HY, Huang CY. Estrogen and/or estrogen receptor α inhibits BNIP3-induced apoptosis and autophagy in H9c2 cardiomyoblast cells. Int J Mol Sci. 2018;19(5):1298. [DOI] [PMC free article] [PubMed]
- 42.Du B, Dai XM, Li S, Qi GL, Cao GX, Zhong Y, Yin PD, Yang XS. MiR-30c regulates cisplatin-induced apoptosis of renal tubular epithelial cells by targeting Bnip3L and Hspa5. Cell Death Dis. 2017;8(8):e2987. doi: 10.1038/cddis.2017.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fei P, Wang W, Kim SH, Wang S, Burns TF, Sax JK, Buzzai M, Dicker DT, McKenna WG, Bernhard EJ, et al. Bnip3L is induced by p53 under hypoxia, and its knockdown promotes tumor growth. Cancer Cell. 2004;6(6):597–609. doi: 10.1016/j.ccr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 44.Ceylan Y, Akpınar G, Doger E, Kasap M, Guzel N, Karaosmanoglu K, Kopuk SY, Yucesoy I. Proteomic analysis in endometrial cancer and endometrial hyperplasia tissues by 2D-DIGE technique. J Gynecol Obstet Hum Reprod. 2020;49(2):101652. doi: 10.1016/j.jogoh.2019.101652. [DOI] [PubMed] [Google Scholar]
- 45.Miano M, Cappelli E, Pezzulla A, Venè R, Grossi A, Terranova P, Palmisani E, Maggiore R, Guardo D, Lanza T, et al. FAS-mediated apoptosis impairment in patients with ALPS/ALPS-like phenotype carrying variants on CASP10 gene. Br J Haematol. 2019;187(4):502–508. doi: 10.1111/bjh.16098. [DOI] [PubMed] [Google Scholar]
- 46.Juraver-Geslin HA, Durand BC. Early development of the neural plate: new roles for apoptosis and for one of its main effectors caspase-3. Genesis (New York, NY : 2000) 2015;53(2):203–224. doi: 10.1002/dvg.22844. [DOI] [PubMed] [Google Scholar]
- 47.Vigneswara V, Akpan N, Berry M, Logan A, Troy CM, Ahmed Z. Combined suppression of CASP2 and CASP6 protects retinal ganglion cells from apoptosis and promotes axon regeneration through CNTF-mediated JAK/STAT signalling. Brain. 2014;137(Pt 6):1656–1675. doi: 10.1093/brain/awu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahib MR, Hosojima S, Kushiyama H, Kinoshita T, Shiroishi T, Suda T, Tsuchiya K. Caspase-7 mediates caspase-1-induced apoptosis independently of bid. Microbiol Immunol. 2020;64(2):143–152. doi: 10.1111/1348-0421.12756. [DOI] [PubMed] [Google Scholar]
- 49.Bell BD, Leverrier S, Weist BM, Newton RH, Arechiga AF, Luhrs KA, Morrissette NS, Walsh CM. FADD and caspase-8 control the outcome of autophagic signaling in proliferating T cells. Proc Natl Acad Sci U S A. 2008;105(43):16677–16682. doi: 10.1073/pnas.0808597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu XZ, Zhang Q, Jiang Q, Bai BL, Du XJ, Wang F, Wu LH, Lu XL, Bao YH, Li HL, et al. Genetic screening and functional analysis of CASP9 mutations in a Chinese cohort with neural tube defects. CNS Neurosci Ther. 2018;24(5):394–403. doi: 10.1111/cns.12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y, Zhao YR, Zhang AY, Ma J, Wang ZZ, Zhang X. Targeting of miR-20a against CFLAR to potentiate TRAIL-induced apoptotic sensitivity in HepG2 cells. Eur Rev Med Pharmacol Sci. 2017;21(13):2980. [PubMed] [Google Scholar]
- 52.Ahmad M, Srinivasula SM, Wang L, Talanian RV, Litwack G, Fernandes-Alnemri T, Alnemri ES. CRADD, a novel human apoptotic adaptor molecule for caspase-2, and FasL/tumor necrosis factor receptor-interacting protein RIP. Cancer Res. 1997;57(4):615–619. [PubMed] [Google Scholar]
- 53.Di Donato N, Jean YY, Maga AM, Krewson BD, Shupp AB, Avrutsky MI, Roy A, Collins S, Olds C, Willert RA, et al. Mutations in CRADD result in reduced caspase-2-mediated neuronal apoptosis and cause megalencephaly with a rare lissencephaly variant. Am J Hum Genet. 2016;99(5):1117–1129. doi: 10.1016/j.ajhg.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tai CJ, Hsu CH, Shen SC, Lee WR, Jiang MC. Cellular apoptosis susceptibility (CSE1L/CAS) protein in cancer metastasis and chemotherapeutic drug-induced apoptosis. J Expe Clin Cancer Res: CR. 2010;29(1):110. doi: 10.1186/1756-9966-29-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Allavena G, Cuomo F, Baumgartner G, Bele T, Sellgren AY, Oo KS, Johnson K, Gogvadze V, Zhivotovsky B, Kaminskyy VO. Suppressed translation as a mechanism of initiation of CASP8 (caspase 8)-dependent apoptosis in autophagy-deficient NSCLC cells under nutrient limitation. Autophagy. 2018;14(2):252–268. doi: 10.1080/15548627.2017.1405192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rathore R, McCallum JE, Varghese E, Florea AM, Büsselberg D. Overcoming chemotherapy drug resistance by targeting inhibitors of apoptosis proteins (IAPs) Apoptosis. 2017;22(7):898–919. doi: 10.1007/s10495-017-1375-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang YW, Zhang CM, Huang XJ, Zhang XX, Zhang LK, Li JH, Hua ZC. Tumor-targeted delivery of a C-terminally truncated FADD (N-FADD) significantly suppresses the B16F10 melanoma via enhancing apoptosis. Sci Rep. 2016;6:34178. doi: 10.1038/srep34178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chinnaiyan AM, O'Rourke K, Tewari M, Dixit VM. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell. 1995;81(4):505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 59.Nagata S. Fas-mediated apoptosis. Adv Exp Med Biol. 1996;406:119–124. doi: 10.1007/978-1-4899-0274-0_12. [DOI] [PubMed] [Google Scholar]
- 60.Schneider P, Bodmer JL, Holler N, Mattmann C, Scuderi P, Terskikh A, Peitsch MC, Tschopp J. Characterization of Fas (Apo-1, CD95)-Fas ligand interaction. J Biol Chem. 1997;272(30):18827–18833. doi: 10.1074/jbc.272.30.18827. [DOI] [PubMed] [Google Scholar]
- 61.Liu W, Ramagopal U, Cheng H, Bonanno JB, Toro R, Bhosle R, Zhan C, Almo SC. Crystal Structure of the Complex of Human FasL and Its Decoy Receptor DcR3. Structure (London, England : 1993) 2016;24(11):2016–2023. doi: 10.1016/j.str.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 62.Hao H, Chen C, Rao XM, Gomez-Gutierrez JG, Zhou HS, McMasters KM. E2F-1- and E2Ftr-mediated apoptosis: the role of DREAM and HRK. J Cell Mol Med. 2012;16(3):605–615. doi: 10.1111/j.1582-4934.2011.01338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mueller PA, Zhu L, Tavori H, Huynh K, Giunzioni I, Stafford JM, Linton MF, Fazio S. Deletion of macrophage low-density lipoprotein receptor-related protein 1 (LRP1) accelerates atherosclerosis regression and increases C-C chemokine receptor type 7 (CCR7) expression in plaque macrophages. Circulation. 2018;138(17):1850–1863. doi: 10.1161/CIRCULATIONAHA.117.031702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Su J, Ruan S, Dai S, Mi J, Chen W, Jiang S. NF1 regulates apoptosis in ovarian cancer cells by targeting MCL1 via miR-142-5p. Pharmacogenomics. 2019;20(3):155–165. doi: 10.2217/pgs-2018-0161. [DOI] [PubMed] [Google Scholar]
- 65.Tan KO, Tan KM, Chan SL, Yee KS, Bevort M, Ang KC, Yu VC. MAP-1, a novel proapoptotic protein containing a BH3-like motif that associates with Bax through its Bcl-2 homology domains. J Biol Chem. 2001;276(4):2802–2807. doi: 10.1074/jbc.M008955200. [DOI] [PubMed] [Google Scholar]
- 66.Morsi RZ, Hage-Sleiman R, Kobeissy H, Dbaibo G. Noxa: role in cancer pathogenesis and treatment. Curr Cancer Drug Targets. 2018;18(10):914–928. doi: 10.2174/1568009618666180308105048. [DOI] [PubMed] [Google Scholar]
- 67.Mazroui R, Di Marco S, Clair E, von Roretz C, Tenenbaum SA, Keene JD, Saleh M, Gallouzi IE. Caspase-mediated cleavage of HuR in the cytoplasm contributes to pp32/PHAP-I regulation of apoptosis. J Cell Biol. 2008;180(1):113–127. doi: 10.1083/jcb.200709030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anding AL, Chapman JS, Barnett DW, Curley RW, Jr, Clagett-Dame M. The unhydrolyzable fenretinide analogue 4-hydroxybenzylretinone induces the proapoptotic genes GADD153 (CHOP) and Bcl-2-binding component 3 (PUMA) and apoptosis that is caspase- dependent and independent of the retinoic acid receptor. Cancer Res. 2007;67(13):6270–6277. doi: 10.1158/0008-5472.CAN-07-0727. [DOI] [PubMed] [Google Scholar]
- 69.Xu D, Jin T, Zhu H, Chen H, Ofengeim D, Zou C, Mifflin L, Pan L, Amin P, Li W, et al. TBK1 Suppresses RIPK1-Driven Apoptosis and Inflammation during Development and in Aging. Cell. 2018;174(6):1477–1491.e1419. doi: 10.1016/j.cell.2018.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Touyama K, Khan M, Aoki K, Matsuda M, Hiura F, Takakura N, Matsubara T, Harada Y, Hirohashi Y, Tamura Y, et al. Bif-1/Endophilin B1/SH3GLB1 regulates bone homeostasis. J Cell Biochem. 2019;120(11):18793–18804. doi: 10.1002/jcb.29193. [DOI] [PubMed] [Google Scholar]
- 71.Laubach V, Kaufmann R, Bernd A, Kippenberger S, Zöller N. Extrinsic or intrinsic apoptosis by curcumin and light: still a mystery. Int J Mol Sci. 2019;20(4):905. [DOI] [PMC free article] [PubMed]
- 72.Fu Q, Qin T, Chen L, Liu CJ, Zhang X, Wang YZ, Hu MX, Chu HY, Zhang HW. miR-29a up-regulation in AR42J cells contributes to apoptosis via targeting TNFRSF1A gene. World J Gastroenterol. 2016;22(20):4881–4890. doi: 10.3748/wjg.v22.i20.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Borghi A, Verstrepen L, Beyaert R. TRAF2 multitasking in TNF receptor-induced signaling to NF-κB, MAP kinases and cell death. Biochem Pharmacol. 2016;116:1–10. doi: 10.1016/j.bcp.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 74.He W, Wang Q, Xu J, Xu X, Padilla MT, Ren G, Gou X, Lin Y. Attenuation of TNFSF10/TRAIL-induced apoptosis by an autophagic survival pathway involving TRAF2- and RIPK1/RIP1-mediated MAPK8/JNK activation. Autophagy. 2012;8(12):1811–1821. doi: 10.4161/auto.22145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Robin M, Issa AR, Santos CC, Napoletano F, Petitgas C, Chatelain G, Ruby M, Walter L, Birman S, Domingos PM, et al. Drosophila p53 integrates the antagonism between autophagy and apoptosis in response to stress. Autophagy. 2019;15(5):771–784. doi: 10.1080/15548627.2018.1558001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Anderton H, Bandala-Sanchez E, Simpson DS, Rickard JA, Ng AP, Di Rago L, Hall C, Vince JE, Silke J, Liccardi G, et al. RIPK1 prevents TRADD-driven, but TNFR1 independent, apoptosis during development. Cell Death Differ. 2019;26(5):877–889. doi: 10.1038/s41418-018-0166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li T, Su L, Lei Y, Liu X, Zhang Y, Liu X. DDIT3 and KAT2A proteins regulate TNFRSF10A and TNFRSF10B expression in endoplasmic reticulum stress-mediated apoptosis in human lung cancer cells. J Biol Chem. 2015;290(17):11108–11118. doi: 10.1074/jbc.M115.645333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Świtońska K, Szlachcic WJ, Handschuh L, Wojciechowski P, Marczak Ł, Stelmaszczuk M, Figlerowicz M, Figiel M. Identification of altered developmental pathways in human juvenile HD iPSC with 71Q and 109Q using Transcriptome profiling. Front Cell Neurosci. 2018;12:528. doi: 10.3389/fncel.2018.00528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang WZ, Zhou H, Yan Y. XIAP underlies apoptosis resistance of renal cell carcinoma cells. Mol Med Rep. 2018;17(1):125–130. doi: 10.3892/mmr.2014.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39(Database issue):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goh GB, Chang PE, Tan CK. Changing epidemiology of hepatocellular carcinoma in Asia. Best Pract Res Clin Gastroenterol. 2015;29(6):919–928. doi: 10.1016/j.bpg.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 82.Chittenden T, Flemington C, Houghton AB, Ebb RG, Gallo GJ, Elangovan B, Chinnadurai G, Lutz RJ. A conserved domain in Bak, distinct from BH1 and BH2, mediates cell death and protein binding functions. EMBO J. 1995;14(22):5589–5596. doi: 10.1002/j.1460-2075.1995.tb00246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moldoveanu T, Liu Q, Tocilj A, Watson M, Shore G, Gehring K. The X-ray structure of a BAK homodimer reveals an inhibitory zinc binding site. Mol Cell. 2006;24(5):677–688. doi: 10.1016/j.molcel.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 84.Reed JC. Bcl-2 family proteins. Oncogene. 1998;17(25):3225–3236. doi: 10.1038/sj.onc.1202591. [DOI] [PubMed] [Google Scholar]
- 85.Chourasia AH, Macleod KF. Tumor suppressor functions of BNIP3 and mitophagy. Autophagy. 2015;11(10):1937–1938. doi: 10.1080/15548627.2015.1085136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chourasia AH, Tracy K, Frankenberger C, Boland ML, Sharifi MN, Drake LE, Sachleben JR, Asara JM, Locasale JW, Karczmar GS, et al. Mitophagy defects arising from BNip3 loss promote mammary tumor progression to metastasis. EMBO Rep. 2015;16(9):1145–1163. doi: 10.15252/embr.201540759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wellmann A, Flemming P, Behrens P, Wuppermann K, Lang H, Oldhafer K, Pastan I, Brinkmann U. High expression of the proliferation and apoptosis associated CSE1L/CAS gene in hepatitis and liver neoplasms: correlation with tumor progression. Int J Mol Med. 2001;7(5):489–494. doi: 10.3892/ijmm.7.5.489. [DOI] [PubMed] [Google Scholar]
- 88.Liao CF, Luo SF, Shen TY, Lin CH, Chien JT, Du SY, Jiang MC. CSE1L/CAS, a microtubule-associated protein, inhibits taxol (paclitaxel)-induced apoptosis but enhances cancer cell apoptosis induced by various chemotherapeutic drugs. BMB Rep. 2008;41(3):210–216. doi: 10.5483/BMBRep.2008.41.3.210. [DOI] [PubMed] [Google Scholar]
- 89.Yvon AM, Wadsworth P, Jordan MA. Taxol suppresses dynamics of individual microtubules in living human tumor cells. Mol Biol Cell. 1999;10(4):947–959. doi: 10.1091/mbc.10.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kolben T, Jeschke U, Reimer T, Karsten N, Schmoeckel E, Semmlinger A, Mahner S, Harbeck N, Kolben TM. Induction of apoptosis in breast cancer cells in vitro by Fas ligand reverse signaling. J Cancer Res Clin Oncol. 2018;144(2):249–256. doi: 10.1007/s00432-017-2551-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data used in the study are obtained from the TCGA database (https://portal.gdc.cancer.gov/), ICGC database (https://dcc.icgc.org/), Deathbase database (http://deathbase.org/), and Kaplan-Meier plotter database (http://kmplot.com/analysis/) which are opening and available to all.
All datasets generated for this study are included in the manuscript and the supplementary files.