Abstract
Detection of SARS-CoV-2 RNA in nasopharyngeal samples using the real-time reverse transcription polymerase chain reaction (rRT-PCR) is the gold standard for diagnosing COVID-19. Determination of SARS-CoV-2 RNA by rRT-PCR sometimes results in an inconclusive test result due to a high cycle threshold-value.
We retrospectively analyzed 30,851 SARS-CoV-2 rRT-PCR test results. Borderline positivity was considered as the presence of ≤25 viral copies per milliliter, while no amplification was considered as a negative test result. Of all test results, 204 were answered as borderline, of which 107 were accompanied by a follow-up test within 96 hours.
Of the 107 follow-up samples, 10 (9.35%) were found positive for SARS-CoV-2. COVID-19 symptoms were not predictive for testing positive in the follow-up test. The positive SARS-CoV-2 samples in the follow-up group represented 0.92% of all positive test results, highlighting the need for retesting and increased hygienic measures for borderline SARS-CoV-2 patients [NCT04636294].
Keywords: COVID-19, SARS-CoV-2, rRT-PCR, Borderline, Molecular diagnostics
1. Introduction
The coronavirus disease (COVID)-19 pandemic tremendously impacts health, social and economic sectors (Chu et al., 2020)(Boeckmans et al., 2020). Early detection of severe acute respiratory syndrome coronavirus (SARS-CoV)-2-infected persons is crucial to limit viral transmission among the population. Recent data show an asymptomatic infection rate of about 46% (95% confidence interval [CI] 18.48; 73.60) (He et al., 2020), which hampers quick interventions to prevent disease spread by using disinfection, protective equipment, quarantine or isolation measures (Lotfi et al., 2020).
SARS-CoV-2 is an enveloped, positive-sense single-stranded RNA beta-coronavirus (Romano et al., 2020). Detection of viral RNA in nasopharyngeal samples by real-time reverse transcription polymerase chain reaction (rRT-PCR) is the gold standard for diagnosis of COVID-19 (Tahamtan and Ardebili, 2020)(Jamal et al., 2020).
Low rRT-PCR cycle thresholds (CT), are a relative indication for high viral titers and confer a higher risk for developing severe COVID-19 which may relate to increased mortality (Rao et al., 2020). High CT-values could either indicate an incipient (novel) infection, an evading (asymptomatic) infection (Drew et al., 2020)(Lan et al., 2020) or a nonspecific rRT-PCR reaction (Tahamtan and Ardebili, 2020). The amount of SARS-CoV-2 RNA in the upper respiratory tract peaks around symptoms onset (Walsh et al., 2020)(Cevik et al., 2020). Viral RNA may, however persist up to 3 months without the presence of transmissible virus or being infectious (Li et al., 2020)(Wang et al., 2020), which makes the interpretation of weak positive results challenging. Further, CT-values may inherently vary because of the patients’ tolerance to sampling, sampling skills of medical personnel and the use of different RNA extraction kits and PCR instrumentation (Dabouh et al., 2020)(Raymaekers et al., 2009).
Since limiting SARS-CoV-2 transmission is of vital importance to curb COVID-19 spread, we aimed to investigate at which ratio weak positive, further termed 'borderline' rRT-PCR test results, taking a minimum of >25 template copies per milliliter as a threshold for true positivity, ultimately culminate in true infections.
2. Methods
2.1. Ethical approval
Ethical approval was obtained from the ethics committee of the Jessa Hospital (number f/2020/166). The study is registered on ClinicalTrials.gov (NCT04636294).
2.2. Sample collection
After deep nasopharyngeal sampling, swabs (FLOQSwabs, Copan, Italy) were brought into 2 milliliter of cell lysis buffer (Promega). All samples were tested within 24 hours upon arrival in the laboratory.
2.3. rRT-PCR
All samples were analyzed with one of the following 3 methods:
The first method included RNA extraction on the Maxwell RSC Instrument (Promega) using the RSC Viral TNA kit (Promega) according to the manufacturers’ instructions. An extraction and amplification control (Phocine Distemper Virus, kindly provided by the Department of Viroscience, Erasmus Medical Centre Rotterdam) was added to each sample as amplification and extraction control. These extracts were analyzed on QuantStudio 7 flex (Thermo Fisher Scientific), with an in-house rRT-PCR test, based on the CDC oligonucleotide primers and probes for detection of the viral nucleocapsid (N) gene of SARS-CoV-2. The second method was performed on the m2000 (Abbott) using the m2000 RealTime SARS-CoV-2 assay, according to the manufacturers’ instructions. The last method comprised the fully-automated instrument Alinity (Abbott), using the Alinity m SARS-CoV-2 amp kit. Signals generated from the m2000 and Alinity platforms were derived from a combined RdRp/N-gene fluorophore. Viral loads were semiquantitatively calculated based on a standard curve from a viral suspension (Sciensano, Belgian Institute for Health, Brussels, Belgium).
2.4. Threshold for SARS-CoV-2 positivity
The threshold for SARS-CoV-2 positivity was set on the presence of more than 25 viral template copies per milliliter, which was calculated based on a SARS-CoV-2 dilution series of SCV2QC (Qnostics). The cut-off CT-values were determined for each platform separately. Results of samples with CT-values representing ≤25 viral copies per milliliter, but with a visible PCR signal, crossing the threshold, were assigned as ‘borderline’ test results. Samples were considered as being negative if no amplification occurred.
2.5. Inclusion criteria
All samples sent to the molecular department of the clinical laboratory of the Jessa Hospital (Hasselt, Belgium) for determination of SARS-CoV-2 between August 1, 2020 and November 20, 2020 were included. The inclusion criterion for the follow-up study was the presence of a borderline SARS-CoV-2 test result accompanied by a follow-up test within 96 hours.
2.6. Statistical analyses and data visualization
Statistics were performed using GraphPad Prism. Comparison of initial and follow-up viral loads was performed using a Wilcoxon signed rank test. Proportions were compared using a Fisher's exact test. A result was considered as significantly different when the P-value was lower than 0.05. Confidence intervals (CIs) for proportions were calculated using the modified Wald method. Figures were made using MS office PowerPoint and GraphPad Prism.
3. Results
3.1. Study population
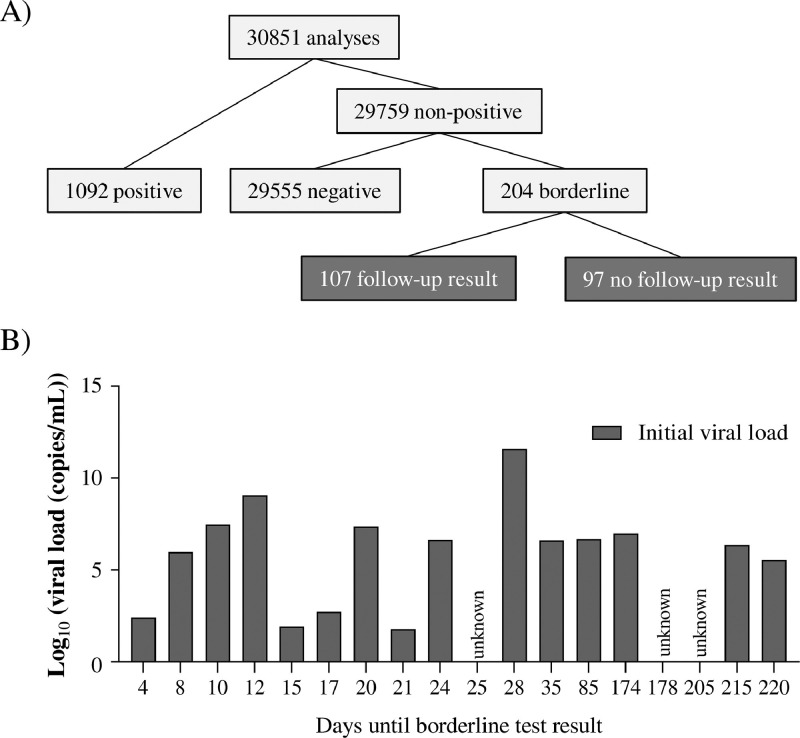
From a total of 30,851 analyses, 1,092 tests were found positive and 29,555 were found negative, resulting in a positivity ratio of 3.54%. Borderline test results were observed for 204 samples (i.e., 0.66% of all SARS-CoV-2 test results) (Fig. 1 A), indicating that 15.74% (95% CI [13.86; 17.83]) of the reported non-negative test results comprised inconclusive test results. From these 204 borderline test results, 18 samples were preceded by a positive test result, ranging from 4 to 220 days in the past (Fig. 1B). The majority of patients with a borderline SARS-CoV-2 rRT-PCR test result were ambulatory patients.
Fig. 1.
Description of the patient cohort. (A) Organigram of positive, negative and borderline SARS-CoV-2 rRT-PCR test results. (B) From a total of 204 SARS-CoV-2 borderline samples, 18 were preceded by a positive test result.
3.2. Follow-up testing of borderline SARS-CoV-2 patients
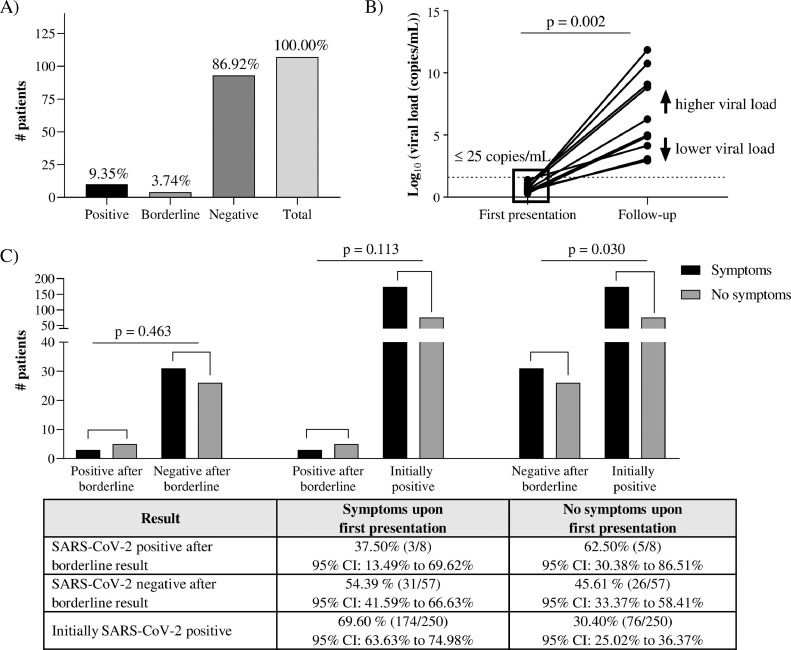
We retrospectively analyzed SARS-CoV-2 rRT-PCR results of patients with a borderline test result and who were re-tested in the same laboratory within a timeframe of 96 hours. Out of 107 follow-up samples, 10 samples (9.35%, 95% CI [4.99; 16.52]) showed conversion of the borderline viral load to a positive test result. Four borderline samples still showed a borderline test result (3.74%, 95% CI [1.16; 9.53]) and 93 samples tested negative (86.92%, 95% CI [79.11; 92.17]) (Fig. 2 A).
Fig. 2.
Follow-up of borderline SARS-CoV-2 rRT-PCR results. (A) Ten out of 107 patients (9.35%) tested positive within 96 hours after a borderline SARS-CoV-2 test result. (B) Follow-up viral loads were significantly higher in SARS-CoV-2-positive patients (Wilcoxon signed rank test). (C) Patients with a positive SARS-CoV-2 test result within 96 hours after a borderline test result, did not experience more often COVID-19 symptoms upon first presentation compared to patients that turned out SARS-CoV-2 negative in the follow-up test (Fischer's exact test).
For the 10 patients that became SARS-CoV-2 positive after follow-up testing, significantly higher viral titers were obtained at re-testing (Fig. 2B, P-value for Wilcoxon signed rank test = 0.002). The mean viral load at follow-up testing was 7 × 1010 copies/milliliter. As such, 9.35% of retested patients became true SARS-CoV-2 positive after follow-up testing (Fig. 2A).
To differentiate clinically between individuals with a positive and negative test result in the follow-up test, we investigated whether the first group might initially present more often with typical COVID-19 symptoms. Data regarding the presence of COVID-19 symptoms were available in 80% (8/10) of the positive follow-up cases and 61% (57/93) of the negative follow-up cases. Three out of 8 (37.50%, 95% CI [13.49; 69.62]) SARS-CoV-2 positive follow-up patients presented with typical COVID-19 symptoms, while 31 out of 57 (54.38%, 95% CI [41.59; 66.63]) SARS-CoV-2 negative patients were also symptomatic, resulting in no significant difference between both groups regarding the presence of typical COVID-19 symptoms upon first presentation (P = 0.463) (Fig. 2C). Patients that tested SARS-CoV-2 negative in the follow-up test, significantly experienced less COVID-19 symptoms upon first presentation compared to patients that tested positive without pre-existing borderline test result (P = 0.030). Patients that tested positive in the follow-up test did not experience significantly more or less symptoms than patients that tested positive without pre-existing borderline test result (P = 0.113). The fact that SARS-CoV-2 positivity was observed in 9.35% of the follow-up samples makes that 0.92% (95% CI 0.47–1.70) of all positive samples originated from previous borderline test results that would have been neglected without advising follow-up testing. In addition, the conversion ratio to positivity after a borderline test result is 2.64 times higher than the general positivity ratio in this cohort.
4. Discussion
SARS-CoV-2-infected patients show heterogenic clinical presentations ranging from asymptomatic patients to patients needing mechanical intubation and transfer to the intensive care unit (Boeckmans et al., 2020)(Botta et al., 2020). Screening programs, especially after high risk contacts, are important to prevent COVID-19 outbreaks (Cheng et al., 2020). Different CT-cutoff values for SARS-CoV-2 positivity may be used among different laboratories. This inconsistency triggered us to investigate the time course of patients exhibiting almost undetectable viral loads. We applied a testing strategy in which samples containing a viral titer of more than 25 template copies per milliliter were assigned as being positive. Samples in which ≤25 template copies per milliliter were present, but in which a signal was generated during the PCR reaction, were considered as “borderline” for which follow-up testing within 1 to 2 days was proposed to the treating physician.
We found that more than 9% of patients with borderline SARS-CoV-2 test results developed higher viral titers within a timeframe of 96 hours after initial sample collection, which implies that 0.92% of all positive samples originated from earlier borderline test results. The conversion rate to positivity after a borderline test result is 2.64 times higher than the general positivity ratio, which substantiates the relevance of reporting these low viral loads. Of note, positivity conversion could be substantially higher since not all borderline test results were accompanied by a follow-up test. Further, consequent follow-up testing after 5 or 10 days could as well result in a higher conversion ratio, since the mean incubation time was found to be 5.68 days (99% CI: 4.78–6.59 days) in a meta-analysis (Khalili et al., 2020).
Significantly higher viral loads were observed among the positive retested borderline patients, indicating viral shedding in the period between the initial and follow-up test. Borderline SARS-CoV-2 patients are hence potentially highly infectious (Drew et al., 2020) during the intermittent period before the follow-up test result is known, which implicates that follow-up testing should be strongly encouraged and isolation measures should be taken until proven negative. In addition, patients with a positive follow-up test result did not experience more often COVID-19 symptoms upon first presentation than patients that turned negative in the follow-up test, hampering symptom-based differentiation. It should be mentioned that results regarding the possible presence of symptoms at first presentation in this study could be underpowered due to the relative small-sample size of the follow-up SARS-CoV-2 positive patient group and the fact that governmental testing policies and test indications vary throughout time. In addition, not all clinical information was present for all patients which could be due to large screening programs. Nevertheless, a meta-analysis showed that about 20% of SARS-CoV-2-positive adults were never febrile and less than 60% developed a cough (Grant et al., 2020).
Another finding of this study is the persistence of borderline CT-values among 3.74% of the samples in this specific setting within 96 hours. Absence of evolution to higher viral loads was in these cases presumed as a sign of previous infection, in which residual viral particles were present in the nasopharynx (Lan et al., 2020). Further, 18 of 204 borderline test result were preceded by a positive test result, which likely also implies the persistence of virus particles without being infectious (D'Ardes et al., 2020)(Cento et al., 2020). Since reinfection cannot be ruled out, we speculate that reporting borderline test results might become less relevant upon attaining (partial) herd immunity since a large part of the population is likely to exhibit a borderline test result. Upon reaching a stadium of herd immunity, determination of IgM/IgG antibody titers might serve as a more appropriate diagnostic tool (Zhang et al., 2020), perhaps in combination with rRT-PCR. Antibody, memory B-cell, CD4+ T-cell, and CD8+ T-cell responses to SARS-CoV-2, however, all exhibit distinct kinetics (Dan et al., 2021) and the key triggers for developing humoral and/or cellular immunity remain unclear. The development of an efficient testing algorithm will therefore likely rely on specific epidemiological conditions and available diagnostic tools. An efficient testing strategy could, for example, be implemented by pooling samples for rRT-PCR to reduce costs and work load. Nonetheless, SARS-CoV-2 borderline patients might be missed due to sample dilution, leading to false negative test results (Lohse et al., 2020). Therefore we advocate consequent follow-up testing after a borderline SARS-CoV-2 rRT-PCR result when a recent infection is not documented. Further, emerging SARS-CoV-2 variants (Fiorentini et al., 2021) could possibly escape the current SARS-CoV-2 rRT-PCR primers, making testing strategies even more complicated.
Notably, CE-in vitro diagnostic medical devices marking of the PCR instrumentation used in our study states that every sample in which amplification occurs, should be reported as being positive. This strategy would result in a considerable number of false positives, seen that the majority of borderline follow-up tests turned out as being negative. It should, however, also be mentioned that nonspecific PCR reactions are as well a possible source of false positive results (Ruiz-Villalba et al., 2017).
To efficiently implement a SARS-CoV-2 borderline testing strategy, it is of utmost importance to maximize efforts in reducing sample to sample and amplicon contamination. Strict laboratory decontamination is performed at our site to reduce false positive PCR results. One particular method for laboratory decontamination is using decontamination solutions (bleach, RNAse and DNAse solutions for surfaces, instruments and biosafety cabinets) and separation of activities (Fischer et al., 2016), although this is not possible when using fully automated systems. Another strategy to avoid false weak positive results could be lowering the CT-threshold for positivity. This approach seems, however, not recommended because of the significantly higher viral loads present in the positive follow-up samples, indicating the relevance of the previously reported low borderline viral loads.
5. Conclusion
Reporting SARS-CoV-2 rRT-PCR borderline test results and follow-up testing of patients with borderline test results is crucial to early detect COVID-19 cases and prevent outbreaks, which is especially relevant for hospitals and nursing homes. Future studies should, however, clarify how testing strategies could be optimized taking these borderline results into account.
Conflict of interest
The authors declare that they have no conflict of interest.
Funding support
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Author statement
Joost Boeckmans: conceptualization, data curation, formal analysis, investigation, methodology, writing-original draft. Reinoud Cartuyvels: conceptualization, investigation, methodology, writing-original draft, supervision, writing-review & editing. Petra Hilkens: conceptualization, investigation, methodology, writing-review & editing. Liesbeth Bruckers: statistical analysis, methodology, writing-review & editing. Koen Magerman: conceptualization, investigation, methodology, supervision, writing-review & editing. Luc Waumans: conceptualization, investigation, methodology, supervision, writing-review & editing. Marijke Raymaekers: conceptualization, investigation, methodology, supervision, writing-review & editing.
Acknowledgments
We thank all medical laboratory technologists for their commitment during the COVID-19 pandemic.
References
- Boeckmans J, Rodrigues RM, Demuyser T, Piérard D, Vanhaecke T, Rogiers V. COVID‑19 and drug‑induced liver injury : a problem of plenty or a petty point ? Arch Toxicol. 2020;94:1367–1369. doi: 10.1007/s00204-020-02734-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botta M, Tsonas AM, Pillay J, Boers LS, Algera AG, Bos LDJ, et al. Ventilation management and clinical outcomes in invasively ventilated patients with COVID-19 (PRoVENT-COVID): a national, multicentre, observational cohort study. Lancet Respir Med. 2020;19:1–10. doi: 10.1016/s2213-2600(20)30459-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cento V, Colagrossi L, Nava A, Lamberti A, Senatore S, Travi G, et al. Persistent positivity and fluctuations of SARS-CoV-2 RNA in clinically-recovered COVID-19 patients. J Infect. 2020;81:90–92. doi: 10.1016/j.jinf.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. SARS-CoV-2, SARS-CoV-1 and MERS-CoV viral load dynamics, duration of viral shedding and infectiousness: a living systematic review and meta-analysis. Lancet Microbe. 2020;5247:1–10. doi: 10.2139/ssrn.3677918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HY, Jian SW, Liu DP, Ng TC, Huang WT, Lin HH. Contact tracing assessment of COVID-19 transmission dynamics in taiwan and risk at different exposure periods before and after symptom onset. JAMA Intern Med. 2020;180:1156–1163. doi: 10.1001/jamainternmed.2020.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DK, Akl EA, Duda S, Solo K, Yaacoub S, Schünemann HJ, et al. Physical distancing, face masks, and eye protection to prevent person-to-person transmission of SARS-CoV-2 and COVID-19: a systematic review and meta-analysis. Lancet. 2020;395:1973–1987. doi: 10.1016/S0140-6736(20)31142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ardes D, Boccatonda A, Rossi I, Pontolillo M, Cocco G, Schiavone C, et al. Long-term positivity to SARS-CoV-2: a clinical case of COVID-19 with persistent evidence of infection. Eur J Case Rep Intern Med. 2020;7 doi: 10.12890/2020_001707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabouh E, Lázaro-Perona F, Romero-gómez M, Mingorance J, Garciá-Rodriguez J. Ct values from SARS-CoV-2 diagnostic PCR assays should not be used as direct estimates of viral load. J Infect. 2020;21 doi: 10.1016/j.jinf.2020.10.017. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan JM, Mateus J, Kato Y, Hastie KM, Faliti CE, Ramirez SI, et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;4063:1–23. doi: 10.1101/2020.11.15.383323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew RJ, Donnell SO, Leblanc D, Mcmahon M, Natin D. The importance of cycle threshold values in interpreting molecular tests for SARS-CoV-2. Diagn Microbiol Infect Dis. 2020;98:1–3. doi: 10.1016/j.diagmicrobio.2020.115130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentini S, Messali S, Zani A, Caccuri F, Giovanetti M, Ciccozzi M, et al. First detection of SARS-CoV-2 spike protein N501 mutation in Italy in August, 2020. Lancet Infect Dis. 2021;3099:3099. doi: 10.1016/s1473-3099(21)00007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Renevey N, Thür B, Hoffmann D, Beer M, Hoffmann B. Efficacy assessment of nucleic acid decontamination reagents used in molecular diagnostic laboratories. PLoS One. 2016;11:1–9. doi: 10.1371/journal.pone.0159274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant MC, Geoghegan L, Arbyn M, Mohammed Z, McGuinness L, Clarke EL, et al. The prevalence of symptoms in 24,410 adults infected by the novel coronavirus (SARS-CoV-2; COVID-19): a systematic review and meta-analysis of 148 studies from 9 countries. PLoS One. 2020;15 doi: 10.1371/journal.pone.0234765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Yi GY, Zhu Y. Estimation of the basic reproduction number, average incubation time, asymptomatic infection rate, and case fatality rate for COVID-19: meta-analysis and sensitivity analysis. J Med Virol. 2020;92:2543–2550. doi: 10.1002/jmv.26041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal AJ, Mozafarihashjin M, Coomes E, Powis J, Li AX, Paterson A, et al. Sensitivity of nasopharyngeal swabs and saliva for the detection of severe acute respiratory syndrome Coronavirus 2. Clin Infect Dis. 2020;27:9–11. doi: 10.1093/cid/ciaa848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalili M, Karamouzian M, Nasiri N, Javadi S, Mirzazadeh A, Sharifi H. Epidemiological characteristics of COVID-19: a systematic review and meta-analysis. Epidemiol Infect. 2020;148:1–17. doi: 10.1017/S0950268820001430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L, Xu D, Ye G, Xia C, Wang S, Li Y, et al. Positive RT-PCR test results in patients recovered from COVID-19. J Am Med Assoc. 2020;323:1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Wang X, Lv T. Prolonged SARS-CoV-2 RNA shedding: not a rare phenomenon. J Med Virol. 2020;92:2286–2287. doi: 10.1002/jmv.25952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse S, Pfuhl T, Berkó-Göttel B, Rissland J, Geißler T, Gärtner B, et al. Pooling of samples for testing for SARS-CoV-2 in asymptomatic people. Lancet Infect Dis. 2020;20:1231–1232. doi: 10.1016/S1473-3099(20)30362-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotfi M, Hamblin MR, Rezaei N. COVID-19: Transmission, prevention, and potential therapeutic opportunities. Clin Chim Acta. 2020;508:254–266. doi: 10.1016/j.cca.2020.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SN, Manissero D, Steele VR, Pareja J. A narrative systematic review of the clinical utility of cycle threshold values in the context of COVID-19. Infect Dis Ther. 2020;9:573–586. doi: 10.1007/s40121-020-00324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymaekers M, Smets R, Maes B, Cartuyvels R. Checklist for optimization and validation of real-time PCR assays. J Clin Lab Anal. 2009;23:145–151. doi: 10.1002/jcla.20307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano M, Ruggiero A, Squeglia F, Maga G, Berisio R. A structural view of SARS-CoV-2 RNA replication machinery: rna synthesis, proofreading and final capping. Cells. 2020;9:1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Villalba A, van Pelt-Verkuil E, Gunst QD, Ruijter JM, van den Hoff MJ. Amplification of nonspecific products in quantitative polymerase chain reactions (qPCR) Biomol Detect Quantif. 2017;14:7–18. doi: 10.1016/j.bdq.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahamtan A, Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev Mol Diagn. 2020;20:453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh KA, Jordan K, Clyne B, Rohde D, Drummond L, Byrne P, et al. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J Infect. 2020;81:357–371. doi: 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hang X, Wei B, Li D, Chen F, Liu W, et al. Persistent SARS-COV-2 RNA positivity in a patient for 92 days after disease onset: A case report. Medicine (Baltimore) 2020;99:e21865. doi: 10.1097/MD.0000000000021865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZL, Hou YL, Li DT, Li FZ. Diagnostic efficacy of anti-SARS-CoV-2 IgG/IgM test for COVID-19: a meta-analysis. J Med Virol. 2020 doi: 10.1002/jmv.26211. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]