Abstract
Acquired mutations are pervasive across normal tissues. However, our understanding of the processes that drive transformation of certain clones to cancer is limited. Here we study this phenomenon in the context of clonal hematopoiesis (CH) and the development of therapy-related myeloid neoplasms (tMN). We find mutations are selected differentially based on exposures. Mutations in ASXL1 are enriched in current or former smokers, whereas cancer therapy with radiation, platinum and topoisomerase II inhibitors preferentially selects for mutations in DNA damage response (DDR) genes (TP53, PPM1D, CHEK2). Sequential sampling provides definitive evidence that DDR clones outcompete other clones when exposed to certain therapies. Among cases where CH was previously detected, the CH mutation was present at tMN diagnosis. We identify the molecular characteristics of CH that increase risk of tMN. The increasing implementation of clinical sequencing at diagnosis provides an opportunity to identify patients at risk of tMN for prevention strategies.
MAIN
The multistage model of carcinogenesis suggests that the successive acquisition of somatic mutations predates cancer development1. Each mutation contributes to a clone’s fitness advantage, resulting in clonal expansions that culminate in malignant transformation, in a process that parallels Darwinian evolution2. This evolutionary process results from a complex interplay between the mechanisms that drive mutagenesis, the genetic targets of selection and the contexts in which these mutations contribute to differential clonal fitness.
Systematic cancer sequencing studies have delivered a detailed understanding of the processes that lead to mutations, the resulting mutation signature, and the genetic drivers of malignant disease.3,4 However, our understanding of the evolutionary trajectories that underlie cancer development is primarily based on retrospective modeling of clonal structures observed at diagnosis5 or disease progression6. Such approaches do not allow characterization of the genetic and clonal dynamics of early oncogenesis. Recent sequencing studies of normal tissues show that acquisition of somatic mutations is pervasive with aging7–16. Our understanding of the environmental factors that drive a subset of these mutated clones towards malignant transformation is limited and largely based on in vitro and animal studies17–19. Progress in this regard has been challenged by the paucity of longitudinal genetic and clonal studies with detailed annotation of intervening exposures.
Studies of clonal hematopoiesis (CH) present a unique opportunity to study the evolutionary process underlying malignant transformation in blood. Non-invasive sampling enables acquisition of statistically powered cohorts and longitudinal samples that permit assessment of the transition from normal to transformed disease. Population studies show that individuals with CH are at increased risk of transformation to myeloid neoplasms (MN)20,21. However, only a small proportion of CH subjects progress to MN. Cancer patients are at heightened risk of subsequent therapy-related myeloid neoplasms (tMN) such as AML and MDS22,23. tMN was traditionally thought to develop from the mutagenic effects of cancer therapy23. However, recent studies show that tMN-initiating mutations can predate cancer therapy19, consistent with CH24. Here, we sought to characterize the relationships between CH and environmental exposures and determine how cancer therapy shapes patterns of selection that contribute towards progression to overt leukemia.
Molecular characteristics and clinical determinants of CH
Utilizing prospective targeted sequencing data (MSK-IMPACT) from 24,146 cancer patients representing a wide range of primary tumor types (n=56) and ages (Extended Data Table 1), we established a stringent variant calling and filtration workflow to detect CH variants in blood, with a minimum variant allele frequency (VAF) of 2% (see Methods and Supplementary Notes). We identified 11,076 unique CH mutations in 7,216 individuals, representing 30% of patients in our cohort. The median VAF of CH mutations was 5.0% (range, 2–78%). Among individuals with CH, 69% (n=4952) had one mutation and 31% (2264) had two or more. The spectrum of CH mutations followed expected patterns of positive selection for truncating variants and missense mutations in tumor suppressors and oncogenes, respectively (Supplementary Figure 1). As the design of our panel limits interrogation to bona fide cancer genes, we annotated each mutation on the basis of its putative role in cancer pathogenesis using OncoKB25 and recurrence in an in-house dataset of myeloid neoplasms26–28 (see Methods). Over half of the CH mutations that we detected were classified as putative cancer-driver mutations (CH-PD, 52%, n=5810). Almost all CH-PD variants (91%, n=5301) were recurrent mutations in myeloid neoplasms (CH-myeloid PD) (Supplementary Figure 2).
Overall, mutations in myeloid driver genes (median=0.047) and CH-PD (0.050) showed higher VAFs than non-myeloid (0.038) and non-PD (0.038) mutations, respectively (Supplementary Figure 3a-b, Extended Data Table 2). Similarly, hotspot mutations at R882 within DNMT3A had higher VAFs compared to non-hotspot mutations, even after accounting for total number of mutations (Supplementary Figure 4). The VAF of mutations within individuals who harbored multiple mutations were higher compared to individuals with one mutation (Extended Data Table 2, Supplementary Figure 3c). Consistent with prior literature13,14,24, CH mutations were most frequently identified in DNMT3A, TET2 and ASXL1. Overall, 48% of CH mutations identified were in myeloid driver genes, while only 20% of genes on the MSK-IMPACT panel are myeloid driver genes. The strong enrichment of myeloid variants highlights the strength of the fitness advantage imparted on hematopoietic stem and progenitor cells (HSPCs) by mutations in genes implicated in myeloid pathogenesis as compared to bona fide oncogenic mutations in other cancer driver genes (Supplementary Figure 2).
To assess the role of cancer therapy alongside other factors in driving selection of CH clones, we extracted and curated detailed clinical data for 10,138 patients who had received all their cancer care at Memorial Sloan Kettering (MSK) (see Supplementary Notes). These patients’ demographic characteristics and solid tumor primary site did not differ from those who received treatment outside of MSK or whose treatment information was unavailable (n=14,008) (Supplementary Table 1). As previously reported24, older age strongly correlated with the presence of CH clones in cancer patients (OR=1.9, p<10−6) (Extended Data Table 3). CH was less common in patients of Asian ancestry relative to Caucasian descent (OR=0.7, p=1×10−3) (Extended Data Table 3), consistent with recent reports 29.
Overall, a total of 5,978 patients (59%) were exposed to cancer therapy (including cytotoxic therapy, radiation therapy, targeted therapy and immunotherapy) prior to blood draw (Extended Data Figure 1), whereas 4,160 (41%) were treatment-naive. Patients who had received prior cancer treatment were more likely to have CH compared to treatment-naive patients at time of testing (OR=1.3, p=1×10−6). The same was true for current and former smokers (OR=1.1, p=5×10−3), and effect sizes were similar between current (n=729, OR=1.2, p=0.10) and former smokers (n=4260, OR=1.1, p=8×10−3). The number of CH mutations in each patient was positively associated with cancer therapy and smoking, and clone size was also positively associated with smoking (Extended Data Tables 2, 4). The association between age, therapy and CH was stronger for CH-PD compared to mutations not known to be putative cancer drivers (Extended Data Table 2). All subsequent analyses were limited to CH-PD.
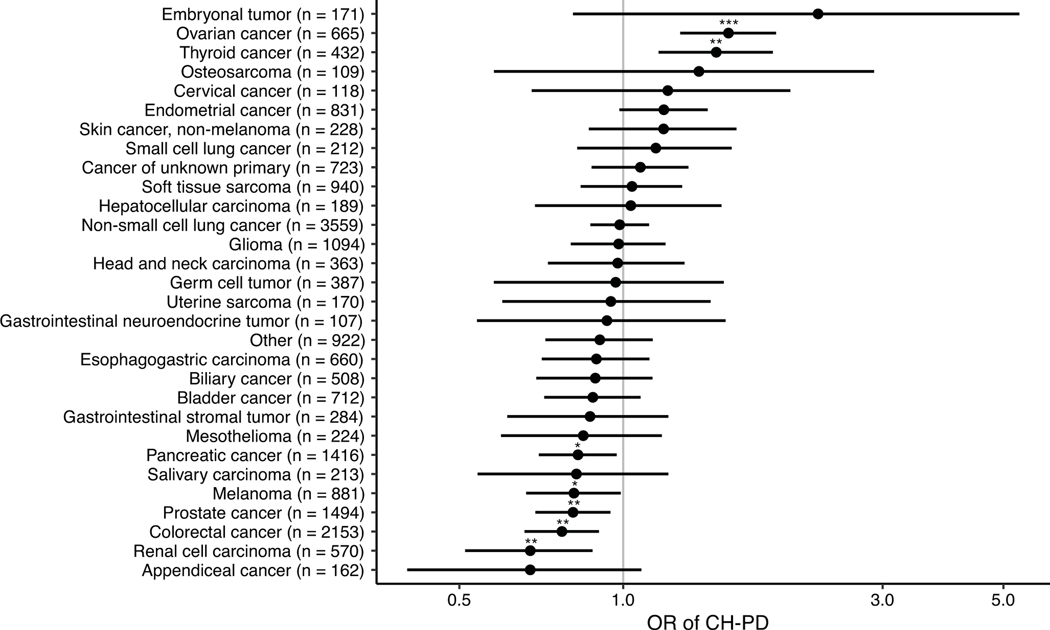
The odds of having CH among cancer patients differed by primary tumor type even after adjustment for age (Extended Data Figure 2). The overall mutational spectrum of CH was similar across cancer types, with the exception of DDR gene mutations being more frequent in patients with ovarian and endometrial cancers. This enrichment was most striking for mutations in PPM1D, which were found in 13% of patients with ovarian cancer and 7% of patients with endometrial cancer as compared to <5% in other cancer subgroups (Extended Data Figure 3). However, among patients who received no cancer therapy prior to blood draw, 8% of women with ovarian cancer and 0% of women with endometrial cancer had CH in PPM1D, suggesting that differences in the spectrum of CH mutations across tumor type could be explained by interactions with specific classes of cancer therapy and/or specific oncologic context.
Clinical parameters shape the fitness landscape of CH
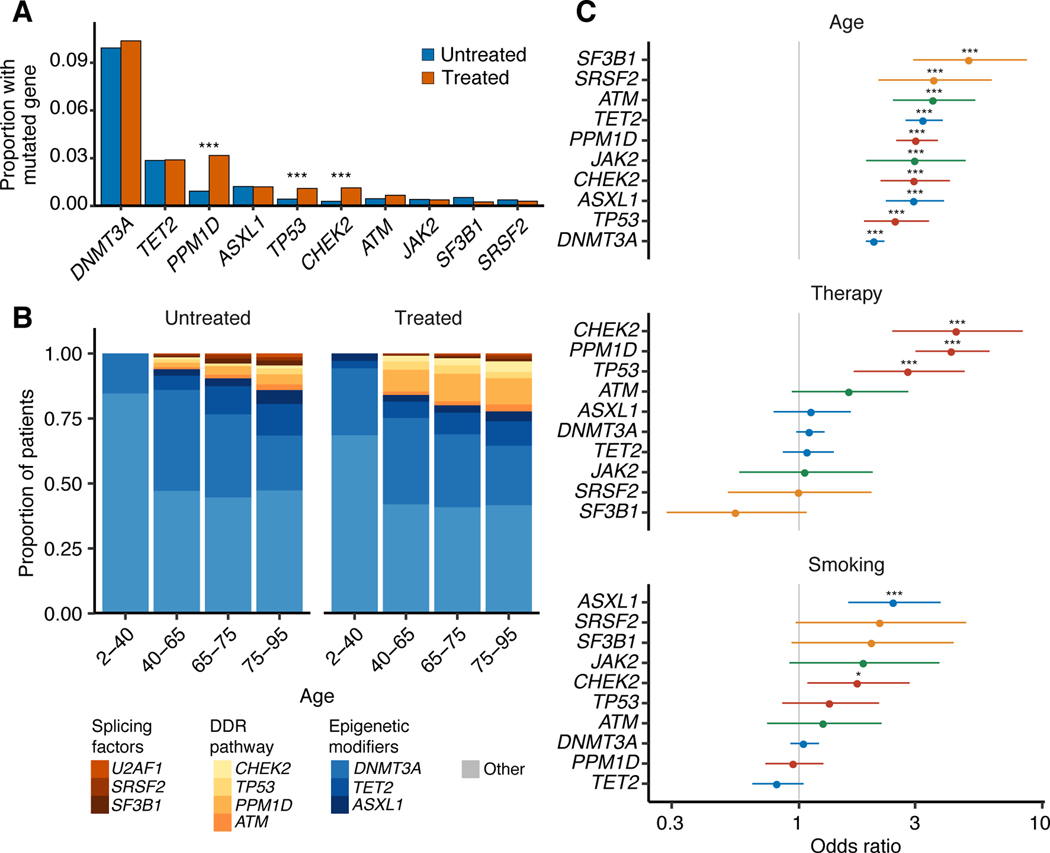
We next sought to determine how specific external exposures might influence the fitness landscape of CH mutations and found that age, treatment and smoking correlated with specific molecular subtypes of CH (Figure 1a-b, Supplementary Figure 5). For example, mutations in the spliceosome genes SRSF2 and SF3B1 were less common in our cohort relative to other CH mutations, but showed the strongest association with age (ORSRSF2 = 3.6, q (FDR-corrected p-value)=7×10−6; ORSF3B1 = 5.0, q=<10−6) (Figure 1b-c). Overall, in tests of heterogeneity, DNMT3A showed significantly weaker associations with age than other mutations, including spliceosome genes (Supplementary Figure 5). CH mutations in the DDR genes TP53, PPM1D and CHEK2 were most strongly associated with prior exposure to cancer therapy (ORTP53 = 2.8, q=2×10−4; ORPPM1D = 4.3, q=<10−6; ORCHEK2 =4.5, q= 6×10−6, Figure 1c). Besides differences in the frequency of DDR mutations, CH mutational features were otherwise similar between treated and untreated individuals (Supplementary Figure 6). Mutations in ASXL1 were significantly associated with smoking history (OR=2.5, q=1×10−4, Figure 1c). Current smokers had a stronger association with CH in ASXL1 (OR=3.1, p=1×10−3) compared to former smokers (OR=2.4, p=1×10−4) although the OR did not significantly differ (p=0.4). While CH was more frequent overall among patients who received cancer-specific therapy, CH defined by mutations in epigenetic modifiers (DNMT3A, TET2) or splicing regulators (SRSF2, SF3B1, U2AF1) was not strongly affected by exposure to therapy (Figure 1b-c). Together, these observations provide evidence that the relative fitness of acquired mutations in HSPCs is modulated by environmental factors such as cancer treatment, smoking or the aging microenvironment in a gene-dependent manner.
Figure 1. Specific molecular subtypes of CH-PD correlate with age, prior therapy exposure and smoking history.
(A) Proportion of patients with CH-PD mutations in specific genes among treated and untreated patients. Multivariable logistic regression was used to test whether the odds of having a specific gene mutated differed between treated (n=5,978) and untreated (n=4,160) patients after adjustment for age, gender, smoking and ethnicity. * p<0.05, ** p<0.01, *** p<0.001 (B) Among patients with CH-PD, the proportion with mutations in specific genes, by age group and treatment status. (C) Odds ratio with 95% confidence interval for CH-PD mutation in the ten most commonly mutated genes with top, increasing age (n=10,138); middle, for patients previously exposed to cancer therapy (n=5,978) compared to those with no exposure (n=4160); bottom, for current/former smokers (n=4,989) compared to non-smokers (n=5,145) in multivariable logistic regression models adjusted for therapy, smoking, ethnicity, age, gender and time from diagnosis to blood draw. *, q-value (FDR-corrected p-value) <0.05, ** q<0.01, *** q<0.001. Age is expressed as the mean centered values.
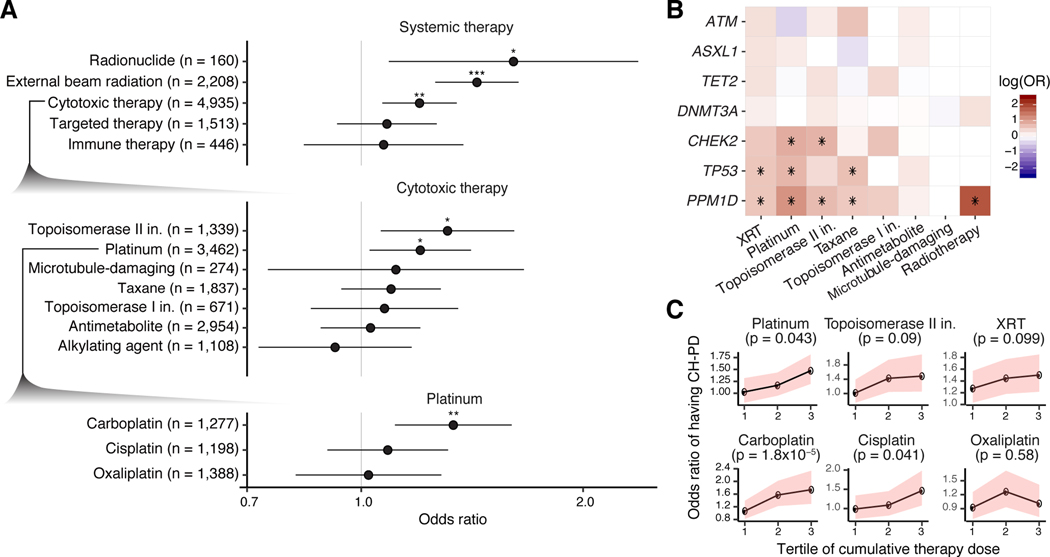
Given the variety of cancer therapies, different therapeutic classes may impart distinct effects on CH. In our study, subjects were exposed to 490 different agents (Supplementary Notes and Supplementary Table 2). To this point, we found evidence of heterogeneity in the strength of association between class agent and CH gene mutations. For example, of all treatment modalities, external beam radiation therapy (OR=1.4, p<10−6), cytotoxic chemotherapy (OR=1.2, p=2×10−3) and radionuclide therapy (OR=1.6, p=0.01) were most strongly associated with CH-PD (global test of heterogeneity phet=0.03). With respect to subclasses of cytotoxic therapy, CH-PD was most strongly associated with prior exposure to topoisomerase II inhibitors (OR=1.3, p=0.01) and platinum agents (OR=1.2, p=0.02), and of the platinum agents, carboplatin (OR=1.4, 0.001) was associated with CH, unlike cisplatin (OR=1.1, p=0.10) and oxaliplatin (OR=0.98, p=0.88) (Figure 2a). Targeted therapies and immunotherapeutic agent exposure were not significantly associated with CH (Figure 2a).
Figure 2. Association between CH-PD and prior exposure to cancer therapy.
(A) Odds ratios (OR) and 95% confidence intervals for CH-PD and specific classes of cancer therapy in multivariable logistic regression adjusted for each other, smoking, ethnicity, gender and time from diagnosis to blood draw. Top, OR for broad classes of cancer therapy; middle. OR between CH-PD and prior exposure to subclasses of cytotoxic therapy; bottom, OR between CH-PD and exposure to specific platinum-based drugs. (B) OR between prior receipt of cancer therapy and CH-PD stratified by tertile of cumulative exposure for the agent. Multivariable logistic regression was used adjusted as in (A) but with cumulative weight-adjusted dose of systemic therapy classes and cumulative radiation dose (as expressed in EQD2. The p-trend was calculated to test for association between CH and increasing tertiles of cumulative cancer therapy exposure among those who received the therapy in the multivariable model. Shaded bands indicate 95% confidence intervals. (C) Heatmap showing the log(OR) between CH-PD in specific genes and prior exposure to the major classes of cytotoxic therapy and radiation therapy in logistic regression models adjusted for therapy subclass, smoking, ethnicity, gender and time from diagnosis to blood draw. * q (FDR-corrected p-value) <0.05, ** q<0.01, *** q<0.001.
Associations with therapy exposure also varied by gene.
Mutations in PPM1D were most strongly associated with prior exposure to platinum (OR=3.2, q<10−6) or radionuclide therapy (OR=6.2, q=7×10−6) and also showed associations with topoisomerase II inhibitors (OR=2.0, q=0.002), taxanes (OR=1.8, q=0.003), topoisomerase I inhibitors (OR=1.7, q=0.002) and external beam radiation therapy (OR=1.8, q=0.04) (Figure 2b). Mutations in TP53 were associated with prior platinum (OR=2.1, q=0.03), radiation therapy (OR=1.8, q=0.04) and taxane (OR=1.9, q=0.05) exposure, whereas CHEK2 was associated with platinum (OR=2.4, q=0.02) and topoisomerase II inhibitors (OR=2.2, q=0.02) (Figure 2b). The strength of the association between DDR CH and cytotoxic therapy differed by cytotoxic therapy subclass (p=4×10−6) and platinum subclass (p=0.03).
To evaluate whether treatment dose modulated these relationships, we calculated each patient’s relative cumulative exposure to specific therapy classes (see Supplementary Notes and Supplementary Figure 7). Increasing exposure to platinum chemotherapy was associated with CH-PD (p-trend=0.04). Among platinum agents, CH-PD was associated with higher cumulative doses of carboplatin (p-trend=3×10−5) and cisplatin (p-trend=0.04) (Figure 2c). Evidence of dose-response further supports a possible causal relationship between the associated exposures and CH.
Clonal dynamics of CH in response to cancer therapy
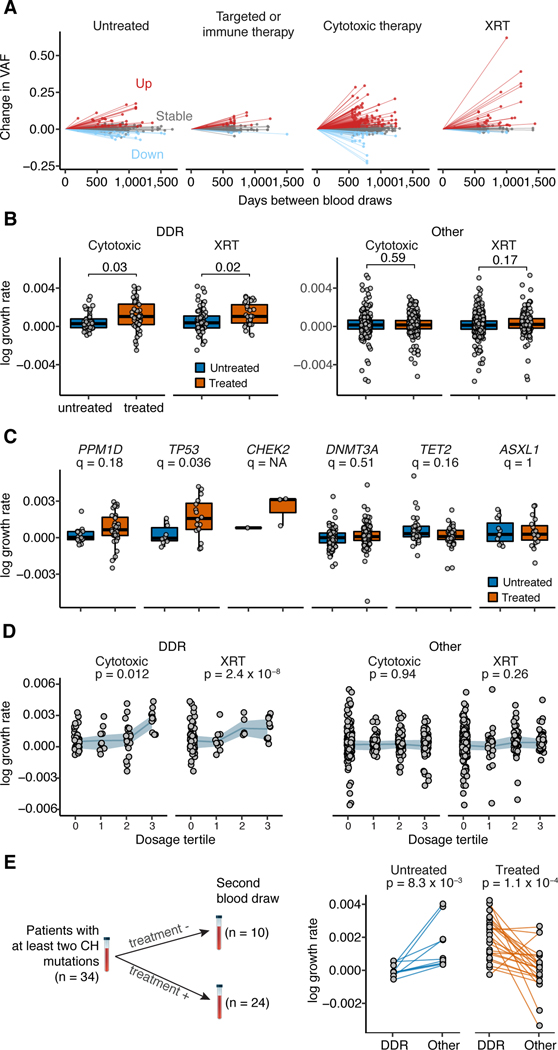
Our retrospective analysis suggests that exposure to cancer therapy results in a higher likelihood of CH, particularly in patients with mutations in DDR genes, following exposure to specific therapies. To definitively characterize how treatment affects mutational presentation and clonal dominance of CH across time, we collected sequential blood samples from 525 patients with solid tumors (median sampling interval time = 23 months, range: 6–53 months), of whom 61% received cytotoxic therapy or external beam radiation therapy and 39% received either targeted or immunotherapy or were untreated (see Methods and Supplementary Figure 8). None of these patients developed secondary hematologic malignancies during follow-up. Of these patients, 389 (74%) had CH, defined as a mutation present at a VAF of ≥2%, at the time of first sampling. The majority of CH mutations were present at both time points (n=590/620, 95%), allowing us to examine how clones evolved in the presence or absence of therapy and whether the clone-defining mutations influenced these trajectories.
We found evidence of both positive and negative changes in clone size across treatment modalities (Figure 3a). Among mutations detected at both time-points, the majority (62% (n=367) of CH mutations remained stable, 28% (n=164) had evidence of growth, and 10% (n=59) decreased in clonal size. Among patients receiving external beam radiation therapy or cytotoxic therapy, growth was most pronounced for CH with mutations in DDR genes TP53, CHEK2 and PPM1D (Figure 3b-c). Similar to our retrospective series, increasing cumulative exposure to these therapies resulted in faster clone growth in patients whose CH was defined by DDR mutations (Figure 3d). We did not see evidence of a significant association between change in VAF and time from end of cytotoxic therapy to the second blood sampling. Future studies with sequential sampling before, during and after therapy will be needed to characterize the kinetics of CH. Patients with multiple mutations exhibited faster CH growth30 as compared to those with one mutation (p=0.03) irrespective of mutation type and treatment status (Supplementary Figure 9). This likely reflects the greater competitive advantage of a subset of clones harboring multiple mutations, although this cannot be determined with certainty in the absence of single-cell sequencing. The proportion of patients with newly detected mutations among those who received interval cytotoxic/radiation therapy (4%, n=13) was non-significantly higher as compared to those who did not (1%, n=2, p=0.06) (Supplementary Figure 10). Thus, in addition to therapy selecting for CH, therapy may have mutagenic effects on HSPCs.
Figure 3. Clonal evolution of CH mutations under the selective pressure of cancer therapy.
(A) Change in VAF for CH mutations from initial to follow-up sequencing for patients stratified by type of therapy received during the follow-up period. XRT, external beam radiation. (B) Change in growth rate for DDR and non-DDR CH mutations among those who received XRT (n=167) or cytotoxic therapy (n=285) during the follow-up period. Shown are the p-values generated from t-tests comparing the growth rate of CH mutations among patients exposed to either of these therapies compared to untreated patients. (C) Change in growth rate for specific CH mutations stratified by whether patients received cytotoxic or radiation therapy (n=268) or no therapy (n=177) during the follow-up period. Shown are the FDR-corrected p-values (q-value) from a t-test comparing the growth rate of mutations in treated and untreated patients. (D) Change in growth rate for DDR and non-DDR CH mutations stratified by tertile of cumulative exposure to cytotoxic therapy and XRT. Shown are the p-values for a trend test for increasing growth rate of CH with increasing tertile of therapy exposure using generalized linear regression adjusted for age, gender and smoking. Shaded bands indicate interquartile ranges. Intra-subject competition between DDR and non-DDR CH mutations. Connecting lines show the difference in growth rate between DDR vs. other genes in patients who received XRT or cytotoxic therapy vs. those who did not receive such therapy during the follow-up period. A paired t-test was used to test for significance in the difference between growth rates of DDR and non-DDR CH mutations within individuals. All p-values are two-sided.
Many parameters likely influence evolutionary trajectories of emerging CH clones. To study competing clonal dynamics in patients, we identified 34 subjects in our prospective serial sampling series with one mutation in a DDR gene and one in a non-DDR gene (Figure 3e). The presence of these distinct classes of gene mutations within the same patient controls for any confounding parameters. In patients receiving interval cytotoxic therapy or radiation therapy, CH clones with DDR mutations grew faster compared to clones with other CH mutations in the same patient. However, the reverse was true in untreated patients: clones with mutations in non-DDR CH genes (e.g. DNMT3A) outcompeted clones with DDR mutations (Figure 3e). In summary, our serial sampling data provide direct evidence in patients that cancer therapy selects for clones with mutations in the DDR genes TP53, PPM1D and CHEK2 and that these clones have lower competitive fitness relative to non-DDR gene mutations in the absence of cytotoxic or radiation therapy.
Genetic and clonal evolution to tMN
Recent studies have shown that tMN-initiating mutations can predate cancer therapy19, challenging the traditional hypothesis that tMN develops from the mutagenic effects of cancer therapy31 and suggesting a relationship with CH. We hypothesized that tMN development is at least in part mediated by therapeutic selection of mutant clones in a gene-dependent manner.
To study the molecular events defining progression of CH to tMN, we analyzed 35 cases for which paired samples were available at the time of molecular profiling for primary cancer and at time of leukemic transformation for tMN (median inter-sampling time of 24 months, range:5–90 months) (Supplementary Table 3). We called mutations present at a VAF of ≥2% in at least one time-point. We detected disease-defining events at time of tMN in 34 patients. Strikingly at least one of these mutations was present at the time of CH (with at least one supporting read) in 19 patients (59%), with 13 (41%) harboring two or more. In all of these cases, the CH mutation was present at the time of tMN diagnosis (Extended Data Figure 4). However, these mutations are unlikely sufficient for leukemic transformation. In 91% of cases, transformation was associated with acquisition of additional somatic mutations, including chromosomal aneuploidies or mutations in genes (e.g. FLT3, KRAS, NRAS) known to drive late progression to myeloid disease27,32–34 (Supplementary Figure 11).
Nearly half (n=14, 40%) of the tMN patients had mutations in TP53. Overall, 10/14 TP53 mutations were detectable at time of CH testing. Of these, four cases had a concomitant TP53 mutation and another non-DDR mutation at time of CH. In agreement with prospective serial sequencing, in the presence of therapy the TP53 clone had consistently attained dominance by the time of tMN (Extended Data Figure 4). At transformation, in 12/13 (92%) cases with available karyotype, TP53 mutations co-occurred with isolated chromosomal aneuploidies or complex karyotype. This provides a direct mechanistic link, whereby cells carrying mutations in TP53 are positively selected when exposed to oncologic therapy and attain clonal dominance with further genetic diversification, such as the acquisition of chromosomal aneuploidies.
Clinical implications of CH in cancer patients
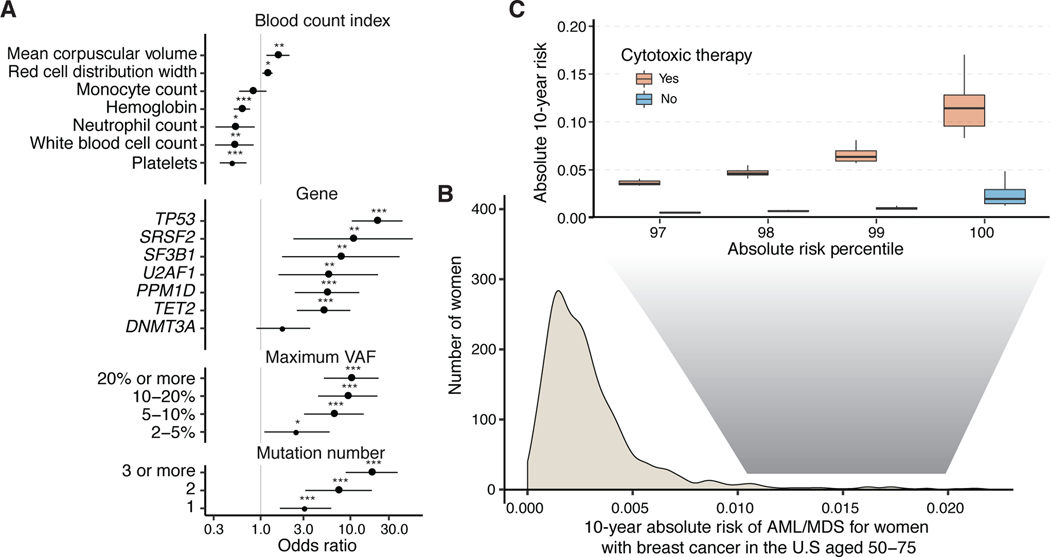
Based on the direct evidence that CH mutations lead to tMN transformation in our paired sample data, we sought to identify risk factors associated with tMN. By combining patient data from our cohort with detailed clinical histories and three previously published studies35–37, we created a cohort of 9,437 cancer patients exposed to cancer therapy, of whom 75 developed tMN (Supplementary Table 2, see Supplementary Notes). Cause-specific Cox proportional hazards analysis (Supplementary Table 2) showed that CH present at a VAF of >2% was positively associated with tMN risk (HR=6.9, p<10−6), and increased with the total number of mutations and clone size (Figure 4a). The strongest associations were observed for mutations in TP53, further validating the relevance of TP53 in tMN, and for mutations in spliceosome genes (SRSF2, U2AF1 and SF3B1). Future studies using error-corrected sequencing methods will clarify the relationship between CH and tMN at VAFs <2%. Comparison of HRs for tMN and AML risk showed similar effect sizes (Supplementary Figure 12) in our cohort as in recent studies of healthy individuals30,38. These data suggest that the relative risk of myeloid neoplasms associated with CH and related parameters (gene, VAF and mutation number) is similar between healthy individuals and cancer patients.
Figure 4. Risk of AML or MDS by clinical and CH-PD mutational characteristics in patients with solid tumors.
(A) Hazard ratio and 95% confidence intervals from Cox regression for blood count indexes, and CH-PD mutational characteristics for therapy-related myeloid neoplasms (tMN; AML or MDS, n=75). All models were adjusted for age and gender and stratified by study center. Blood counts are expressed as the mean centered score (the OR is per 1 SD of the blood count). * p<0.05, ** p<0.01, *** p<0.001. (B) Projected distribution of absolute 10-year risk of AML or MDS for women after a breast cancer diagnosis in the United States aged 50–75 at presentation based on our synthetic model. (C) Comparison of distribution of absolute 10-year risk of AML or MDS among women at the top percentiles of risk between those who go on to receive adjuvant cytotoxic chemotherapy and those who receive surgery only. n=9,437.
We next sought to evaluate how CH, in combination with clinical parameters such as age and peripheral blood counts, might help stratify tMN risk for cancer patients. For example, in solid tumor patients undergoing surgical resection, adjuvant cancer therapy can improve overall survival by reducing cancer recurrence. However, in some situations, the absolute survival benefit of adjuvant therapy is modest and is countered, at least in part, by the risk for subsequent tMN, which is almost universally fatal, with a 5-year survival of 10%39. In the absence of prospective clinical studies, we performed an exploratory analysis using a synthetic model to quantify the absolute risk of AML/MDS following a breast cancer diagnosis. Using previously established methodology40,41, we combined estimates of HR parameters obtained from our multivariable analysis with the distribution of CH mutational features and blood count parameters from untreated patients at MSK and external sources to model the 10-year cumulative absolute AML/MDS risk distribution for women with breast cancer aged 50–75 in the United States. This risk model assumes a multiplicative effect of CH mutational features and cancer therapy on risk of tMN, based on the similarity between risk estimates for CH mutational features in AML that develops in individuals never exposed to therapy and tMN (Supplementary Figure 12). We determined how the risk distribution would change with receipt of adjuvant therapy by shifting the population between receiving and not receiving adjuvant chemotherapy.
In our model, the majority (96%) of breast cancer patients have a low 10-year absolute risk (< 1%) for MN (Figure 4b) and for these patients, deferment of adjuvant chemotherapy would not impact their absolute MN risk (Figure 4c). However, for women at the highest risk of MN based on CH and blood count parameters in our synthetic model (top 1%), adjuvant chemotherapy increased the absolute risk of MN by approximately 9%. This would exceed the predicted absolute benefit in overall survival of chemotherapy in many women with early-stage breast cancer42. While not appropriate for clinical implementation, our findings may inform the design and provide a rationale for future studies to formally estimate the benefits of risk-adapted treatment decisions in cancer patients with CH.
DISCUSSION
Longitudinal studies of CH present a unique opportunity to study the patterns of early mutagenesis and the dynamics of clonal selection in the progression towards malignant transformation. Here, by combining epidemiologic and genetic approaches, we provide insights into the mechanisms that drive the transition of a normal HSPC to a cell with a considerably stronger proliferation advantage, and study how the ensuing trajectories are shaped by host and environmental exposures including age, ethnicity, smoking and cancer therapy. We provide evidence that the fate of CH mutations is dictated by a complex interplay between the inherent fitness advantage of the mutation(s) in HSPCs and parameters that preferentially select for specific mutations, i.e. aging for spliceosome mutations, smoking for mutations in ASXL1, and cancer therapy for specific genes involved in DDR (Extended Data Figure 5). These relationships provide insight into disease biology and may inform early detection and prevention strategies in cancer. We refine the relevance of CH as a predictor and precursor of tMN in cancer patients and show that CH mutations detected prior to tMN diagnosis were consistently part of the dominant clone at transformation. We demonstrate that cancer therapy directly favors growth of clones with mutations in genes such as TP53, which is associated with chemo-resistant disease and is strongly enriched in tMN. This provides a direct mechanistic link between genetic subtypes of CH, receipt of subsequent cancer therapy, and how these modulate the transition from CH to attainment of clonal dominance and, for a subset of cases, development of tMN.
Previous murine and in vitro modelling studies have provided evidence supporting an association between cancer therapy and increased fitness of DDR clones in CH. However, these observations have not been verified in human subjects, nor do they define how therapy enables the transition of CH to MN. Here we show that clones with DDR mutations are positively selected in the presence of cancer therapy but not in its absence. We also show that beyond clonal dominance the transition to tMN is most parsimoniously associated with the acquisition of further genetic lesions. Our detailed treatment information including agent class, dose and mechanism of action allowed us to refine the specificity and strength of the association between cancer therapy and CH and characterize distinct gene-treatment effects. We show that radiation therapy and cytotoxic therapy are significantly associated with CH, with regimens containing platinum and topoisomerase II inhibitors most strongly correlating with CH in specific DDR pathway genes including TP53, PPM1D and CHEK2. Serial sampling before and after therapy provided clear, definitive evidence that therapy induces gene-specific clonal expansion, whereby clones with mutations in DDR genes outcompete other clones in the setting of cancer therapy, but not in its absence. Last, the dose-response relationships observed in both our cross-sectional arm and longitudinal study further support a causal relationship between platinum and CH and the cumulative effect of therapy on selection.
The specificity of the associations at a genetic and exposure level (i.e. therapeutic subclasses and agents such as carboplatin) sets a framework for future correlative and mechanistic studies into early oncogenesis for blood disorders. The specific mechanisms and pathways through which chemotherapeutic agents induce HSC injury may be agent-specific43,44. Further work will be needed to elucidate the mechanisms responsible for the differential fitness effects of cancer therapy and other environmental exposures such as smoking on CH both during and after exposure, and how this relates to tMN risk. Beyond the most frequent cancer genes surveyed here, comprehensive genome studies such as deep whole exome or whole genome analyses in cohorts linked to detailed registries of environmental exposures are warranted to uncover the full repertoire of selection in CH.
We find overlap in the types of cancer therapy associated with selection of DDR CH and those linked to tMN risk (carboplatin, topoisomerase II inhibitors and radiation). Selection of TP53 is only one mechanism driving tMN and may be distinct from the processes driving initiation and selection for other tMN-associated alterations including chromosomal aneuploidies and genomic rearrangement (i.e. MLL fusion genes). Our work adds to early evidence45,46 that external stressors are critical in shaping gene-dependent selection of clonal mosaicism. Characterization of the complex interplay between genotype, fitness challenges, and environmental factors will be key to understanding age-associated clonal mosaicism and the associated exposures that result in malignant transformation. These insights would provide the premise for risk stratification and prevention strategies.
Our observations provide a rationale for clinical therapeutic intervention, including the development of therapies aimed to target high-risk CH clones and modulation of the use of adjuvant systemic cancer therapy in patients at highest risk of subsequent myeloid neoplasm. The latter could entail deferring adjuvant cytotoxic therapy or substituting therapies shown to promote high-risk CH with alternative agents when clinically appropriate. We showcase this with a prototype synthetic model; however, development and validation of risk prediction models for specific clinical scenarios are needed prior to implementation. The realization of precision medicine is reliant upon the development of evidence-based guidelines that consider molecular biomarkers alongside standard clinical criteria to inform clinical care. The decreasing cost of prospective clinical sequencing assays and the high frequency of CH in cancer patients suggest that screening for CH prior to initiation of cancer therapy may be feasible, and may enable molecularly based early detection and interception.
METHODS
MSK-IMPACT Cohort
The study population included patients with non-hematologic cancers at MSKCC that underwent matched tumor and blood sequencing using the MSK-IMPACT panel on an institutional prospective tumor sequencing protocol (ClinicalTrials.gov number, NCT01775072) before July 1st, 2018; all patients enrolled on this protocol provided informed consent. This study was approved by the MSKCC Institutional Review Board (IRB). A subset of patients that underwent tumor-genomic profiling as standard of care were not directly consented, in which case an IRB waiver was obtained to allow for inclusion into this study.
We extracted data on ethnicity, smoking, date of birth and cancer history through the MSK cancer registry. Subjects who had a hematologic malignancy diagnosed within three years prior to blood collection for MSK-IMPACT testing or who had an active hematologic malignancy at the time of blood draw were excluded. Subjects who were diagnosed with a hematologic malignancy less than three months following MSK-IMPACT were considered to have an active hematologic malignancy at the time of MSK-IMPACT and were also excluded. When unavailable through the cancer registry, we extracted data on ethnicity and smoking through structured fields in clinician medical notes if available. Subjects for which age was not available were excluded. Blood indices were taken from clinical labs closest to the date of blood collection for MSK-IMPACT, within one year before or after blood collection (median 0 days). The 8,810 individuals included in the previous MSK-IMPACT publication studying CH are included in the current manuscript. A major difference between the two studies, in addition to an expanded sample size, is the comprehensiveness of the clinical data, including therapeutic exposure data, that was obtained as detailed in the supplementary notes section.
Serial Sampling Cohort
In order to study the growth rate of clonal hematopoiesis mutations over time we collected additional blood samples on patients sequenced using MSK-IMPACT for repeat CH mutation testing. These came from three sources: first, from 372 patients with CH in whom we obtained a second blood sample at least 18 months after initial MSK-IMPACT blood collection, second, from 21 samples from patients with clonal hematopoiesis on MSK-IMPACT who had a blood sample banked at least 12 months prior to MSK-IMPACT testing, and third, from 132 samples that were taken for repeat MSK-IMPACT testing for clinical purposes at least six months after the first MSK-IMPACT testing irrespective of clonal hematopoiesis status (Supplementary Figure 8). For all patients who had sequential sampling data, we manually reviewed their medical records to capture receipt of cancer therapy received at outside institutions during the follow-up period. If subjects received therapy outside MSK during the follow-up period, we excluded them from analyses of dose-response relationships since cumulative dose of therapy could not be consistently collected from outside records. This study was approved by the MSKCC IRB.
Targeted Capture-Based Sequencing
Subjects had a tumor and blood sample (as a matched normal) sequenced using MSK-IMPACT, a FDA-authorized hybridization capture-based next-generation sequencing assay encompassing all protein-coding exons from the canonical transcript of 341, 410, or 468 cancer-associated genes (Supplementary Table 4). MSK-IMPACT is validated and approved for clinical use by New York State Department of Health Clinical Laboratory Evaluation Program and is used to sequence cancer patients at Memorial Sloan Kettering. Genomic DNA is extracted from formalin-fixed paraffin-embedded (FFPE) tumor tissue and patient matched blood sample, sheared and DNA fragments were captured using custom probes47. MSK-IMPACT contains most of the commonly reported CH genes with few exceptions. Earlier versions of the panel did not contain PPM1D or SRSF2. Additionally, three genes commonly reported to be observed in patients with malignancies, SRCAP, BRCC3 and ZNF318 were not included, the former two belonging to the DNA damage response pathway.
The blood samples in the serial sampling cohort that were obtained for repeat CH testing were sequenced using a comparable capture-based custom panel using 163 genes implicated in myeloid pathogenesis, which included the most commonly mutated genes in our MSK-IMPACT study, with the exception of ATM. The median sequencing depth was 665X (range=111–1987X) which was comparable to that obtained in the blood using MSK-IMPACT. For all subsequent analyses using the serial sampling cohort we only considered mutations that were present in both the initial and follow-up panel.
Variant Calling
Pooled libraries were sequenced on an Illumina HiSeq 2500 with 2×100bp paired-end reads. Sequencing reads were aligned to human genome (hg19) using BWA (0.7.5a). Reads were re-aligned around indels using ABRA (0.92), followed by base quality score recalibration with Genome Analysis Toolkit (GATK) (3.3–0). Median coverage in the blood samples was 497x, and median coverage in the tumors was 790x. Variant calling for each blood sample was performed unmatched, using a pooled control sample of DNA from 10 unrelated individuals as a comparator. Single nucleotide variants (SNVs) were called using Mutect and VarDict. Insertions and deletions were called using Somatic Indel Detector (SID) and VarDict. Variants that were called by two callers were retained. Dinucleotide substitution variants (DNVs) were detected by VarDict and retained if any base overlapped a SNV called by Mutect. All called mutations were genotyped in the patient matched tumor sample. Mutations were annotated with VEP (version 86) and OncoKb.
Post-Processing Filters for Clonal Hematopoiesis Calling
We applied a series of post-processing filters to further remove false positive variants caused by sequencing artifacts and putative germline polymorphisms. We removed variants that were found (with a VAF of >2% at least once) in a panel of sequencing data from 300 blood samples obtained from persons under 20 years of age and without evidence of clonal hematopoiesis. We further filtered single nucleotide deletions within a homopolymer stretch of (≥3 base repetition) of the same deleted base pair, single nucleotide substitutions completing a stretch of a ≥5 bp-long homopolymer (E.g. GGCGG -> GGGGG) in-frame deletions or insertions in a highly repetitive region (DUST48 algorithm score of ≥5), and variants with unequal proportions of forward/reverse direction supporting reads based on a fisher test. We performed manual review in IGV of recurrent mutations not previously reported in public databases. We required a variant allele fraction of at least 2% and at least 10 supporting reads. All genotypes were calculated using sequencing reads and bases with a quality value of at least 20. Because somatic mutations in the blood would be expected to be detected in the blood but not other tissue compartments, we compared the variant allele fraction (VAF) of mutations in the blood compared to the matched tumor. Variant calls that were present in the blood with a VAF of at least twice that in the tumor or 1.5 times the VAF if the tumor biopsy site was a lymph node were considered somatic. This ratio was chosen based on minimizing sensitivity and specificity of CH calls through simulations of leukocyte contamination in the tumor (see Supplementary Notes and Supplementary Figures 11 and 12). To further filter putative germline polymorphisms that passed the blood/tumor solid tissue ratio due to allelic imbalance in the tumor specimen, we removed any variant reported in any population in the gnomAD database at a frequency greater than 0.005.
Validation of Calls
To test the reproducibility of our clonal hematopoiesis mutation calling, we compared the mutational calling results from 1,173 samples, where the same DNA library for a blood sample was sequenced and analyzed twice using MSK-IMPACT. We detected 91% of variants in both samples using our calling criteria with a correlation coefficient of 0.98 for the variant allele fraction between the two calls indicating that the reproducibility of our calls was high. In 10 cases with CH, we obtained a second blood sample and re-sequenced using a custom capture based panel with unique molecular identifiers and found that this independent method confirmed all 18 of our CH calls using MSK-IMPACT.
Variant Annotation
Variants were annotated according to evidence for functional relevance in cancer (putative driver or CH-PD) and for relevance to myeloid neoplasms specifically (CH-myeloid-PD). We annotated variants as oncogenic in myeloid disease (CH-myeloid-PD) if they were in a gene hypothesized to drive myeloid/hematologic malignancies (Supplementary Table 5) and if they fulfilled any of the following criteria: 1) truncating variants in NF1, DNMT3A, TET2, IKZF1, RAD21, WT1, KMT2D, SH2B3, TP53, CEBPA, ASXL1, RUNX1, BCOR, KDM6A, STAG2, PHF6, KMT2C, PPM1D, ATM, ARID1A, ARID2, ASXL2, CHEK2, CREBBP, ETV6, EZH2, FBXW7, MGA, MPL, RB1, SETD2, SUZ12, ZRSR2 or in CALR exon 9; 2) translation start site mutations in SH2B3; 3) TERT promoter mutations; 4) FLT3-ITDs; 5) in-frame indels in CALR, CEBPA, CHEK2, ETV6, EZH2; 6) any variant occurring in the COSMIC “haematopoietic and lymphoid” category greater than or equal to 10 times; 7) any variant noted as potentially oncogenic in an in-house dataset of 7,000 individuals with myeloid neoplasm greater than or equal to 5 times. We annotated variants as oncogenic (CH-PD) if they fulfilled any of the following criteria: 1) any variant noted as oncogenic or likely oncogenic in OncoKB25; 2) any truncating mutations (nonsense, essential splice site or frameshift indel) in known tumor suppressor genes as per the Cancer Gene Census, OncoKB, or the scientific literature; 3) any variant reported as somatic at least 20 times in COSMIC49; 4) any variant meeting criteria for CH-Myeloid-PD as above. All missense variants not meeting the above criteria were individually reviewed for potential oncogenicity as previously described50.
Calculation of dN/dS Ratios
We used the dNdScv (https://github.com/im3sanger/dndscv) package to quantify the dN/dS ratios for missense and truncating mutations at the gene level as well as on the panel level. Due to the difference in the gene panel between different MSK-IMPACT panel versions, we excluded all MSK-IMPACT-341 samples and only included genes that were present on both MSK-IMPACT-410 and MSK-IMPACT-468 panels in the analysis. Finally, to generate the overall dN/dS landscape in CH, we only presented genes that reached a significance level of q<0.1 after multiple testing correction and contained more than 25 variants.
Modeling the Association Between CH and Prior Exposure to Cancer Therapy
We used multivariable logistic regression to evaluate for an association between clonal hematopoiesis (including gene and variant specific factors) and therapy, age, gender and smoking history. In addition to these variables, we also adjusted for time from cancer diagnosis to blood draw for MSK-IMPACT testing because trends in preferred oncologic agents vary over time and CH is known to associate with survival. We did not adjust for primary tumor type since we hypothesized that most of the difference in CH-PD rates reflected differences in treatment regimens. Indeed, among untreated patients, a global Wald test for differences in CH-PD prevalence by tumor type was not significant (p=0.98). Analyses stratified by the time since start and by completion of external beam radiation and chemotherapy showed no clear evidence of a time-dependence/latency between CH-PD and cumulative exposure to therapy. Thus, the time from start or stop of therapy was not adjusted for. While considering exploratory analyses, we performed multiple hypothesis correction using the false discovery rate (FDR) q-values for gene-specific analyses to control for inflation of type I error. We did not perform multiple hypothesis correction for analyses testing an association between subclasses of cancer therapy and CH because the association between cancer therapy and CH is known and our goal was to define the relative strength of these associations with subtypes of therapy rather than hypothesis testing. Heterogeneity p-values to test for differences in the strength of the association between subclasses of CH and clinical variables were calculated through logistic regression models limited to CH-positive individuals testing for a difference in the odds of having CH with the mutational feature of interest (e.g. CH-PD) vs. having CH without the mutational feature (e.g. non-CH-PD). Generalized estimating equations were used to test for an association between CH VAF and selected clinical and mutational features among CH positive individuals accounting for correlation between the VAF of mutations in the same person. Ordinal logistic regression among CH positive individuals was used to test for an association between clinical characteristics and increasing CH mutation number. A test for trend between increasing cumulative exposure to cancer therapy and the odds of CH-PD was performed using multivariable logistic regression limited to individuals exposed to the therapy of interest.
Modeling the Effect of Cancer Therapy on Mutation Growth Rate
For each mutation in each individual with sequential sequencing data available, we modeled the growth rate of the mutation between the two time points according to the following formula:
Where T and T0 indicates the age of the individual (in days) at the two measurement time points and V and V0 correspond to the VAF at T and T0 respectively. We also classified mutations as having increased, decreased or remained constant during the follow-up period based on a binomial test comparing the two VAFs. Generalized estimating equations were used to test for an association between exposure to cytotoxic therapy and external beam radiation therapy and CH growth rate adjusting for age, gender and smoking status accounting for correlation between the growth rate of mutations in the same person. Among patients with at least one mutation in a DDR CH gene and another non-DDR CH gene, we calculated the difference in the growth rate between mutations. When patients had more than two mutations in the same gene category, we used the highest growth rate for that category. A paired t-test was used to test for significance in the difference between growth rates of DDR mutations compared to non-DDR mutations within individuals who received cytotoxic therapy and/or external beam radiation therapy and within those who were untreated during the follow-up period.
Combined Analysis for AML/MDS Risk
We combined data from MSK and three previously published studies, Gillis et al., abbreviated MOF (n=68), Takahashi et al., abbreviated MDA (n=67), Gibson et al., abbreviated DFC (n=401) studying the effect of CH on tMN risk in cancer patients. We defined tMN as an MDS or AML diagnosed following exposure to therapeutic radiation or cytotoxic therapy as per the WHO criteria51. For all samples, uniform post processing filters were applied to ensure retention of variants in accordance with the QC standards of the MSK cohort including a universal 2% minimum VAF cutoff. We only included mutations within genes that are present on the panel from all centers and on all panel versions from each center (Supplementary Table 6). The only exceptions were SRSF2 which the IMPACT-341 sequencing panel did not cover and PPM1D which was not sequenced in IMPACT-341, MDA or MOF. We performed mean imputation of missing clinical data for blood counts. Only mutations that we classified as CH-PD were included in analyses. We performed univariate cause-specific Cox proportional hazards regression for the effect of maximum VAF, total number of CH mutations, CH in specific genes and blood count parameters adjusted for age and gender and stratified by study site. Interaction terms between study and CH were used to test for heterogeneity between studies on the effect of CH on tMN risk. The proportional hazards assumption was tested through visual inspection of residual plots and through the inclusion of time-varying covariates. We performed a multivariable analysis including age, gender and all variables that were significant in the univariate analysis with the exception of the genes not included in all studies to prevent reduction of sample size, PPM1D and SRSF2. Because our sample set was limited to individuals who received cancer therapy, we were unable to study gene-treatment interactions in the risk of myeloid neoplasm. Thus, in our combined model CH and cancer therapy are modeled as having multiplicative effects, i.e. no multiplicative interaction on myeloid neoplasm risk. We think this is a reasonable assumption for an exploratory analysis such as the one presented in our study. Much larger studies (including solid tumor patients who did and did not receive any cancer therapy besides surgery) would be needed to define the magnitude of CH-treatment interactions.
We also combined data from two studies investigating the effect of CH on AML risk in healthy individuals, Abelson et al., abbreviated PMC (n=969) and Young et al., abbreviated WSU (n=103), with data from MSK and applied uniform processing to mutation data from different centers. As in the solid tumor combined analysis, the same post processing filters used in the main MSK cohort including a universal 2% minimum VAF cutoff were applied to these studies and only mutations that we classified as CH-PD were included in analyses. We performed a multivariable Cox regression adjusted for age and gender including the variables used in the multivariable tMN risk analysis in solid tumor patients.
Modeling Absolute Risk of AML/MDS
We used the iCARE R package40,41 to build a model for absolute risk of AML/MDS in women with breast cancer aged 50–75 in the United States (U.S) by combining 1) the multivariate HR estimates from our study that were significant in the univariate model including maximum VAF of CH, gene specific effects and peripheral blood count indexes (RDW, hemoglobin); 2. Age-specific AML/MDS rates in breast cancer using data provided by the National Comprehensive Cancer Network (NCCN)52; 3. Competing hazards for mortality in women with breast cancer in the U.S aged 50–75 as reported in SEER53; 4. Previously published HR estimates for chemotherapy on the risk of tMN in women with breast cancer from the NCCN52; 5. The distribution of CH VAF, number of mutations, CH gene and peripheral blood count indexes using our cohort of MSK solid tumor cancer patients aged 50–75 who were untreated prior to blood draw; 6. The proportion of women who receive adjuvant chemotherapy for breast cancer in the U.S from SEER53. While our IMPACT cohort is not representative of the general breast cancer population in the U.S, since the distribution of CH mutational features is largely driven by age and since we do not see major differences in rates of CH between gender or untreated tumor types, we believe that the distribution of CH mutational features in untreated solid tumor patients sequenced on IMPACT reasonably approximates an age-matched untreated breast cancer population. While blood count indexes are known to differ by sex and we chose to use the distribution of blood counts from the entire treatment-naive IMPACT population (both male and female) to capture the inter-relationship between blood count indexes and CH mutational features. Sensitivity analyses using the distribution of blood count parameters from female IMPACT patients only produced similar results. This risk model assumes an additive association on the log scale of CH mutational features and cancer therapy for risk of tMN. This assumption is supported by the similarity between risk estimates for CH mutational features between AML in healthy individuals never exposed to therapy and tMN (Supplementary Figure 10).
All the statistical analyses were performed using the R statistical package (www.r-project.org). The code used in statistical analysis is provided in the Supplementary Notes.
Extended Data
Extended Data Figure 1. Distribution of cancer therapy received prior to blood collection for sequencing.
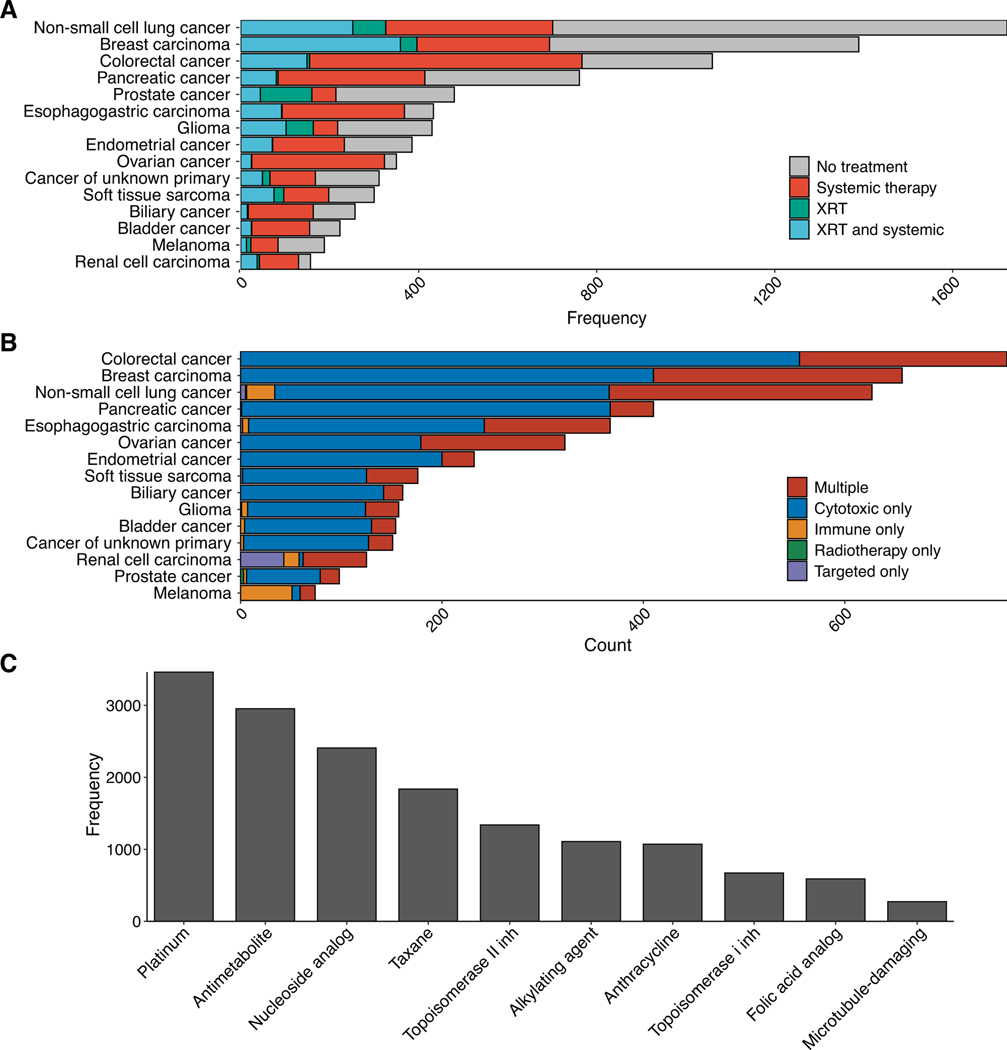
A) Frequency of patients receiving systemic therapy or external beam radiation therapy by primary tumor type. B) Frequency of patients receiving specific classes of systemic therapy by primary tumor type. C) Frequency of patients receiving top ten subclasses of cytotoxic therapy. Most patients (91%) who received at least one of these cytotoxic therapy classes received multiple classes.
Extended Data Figure 2. Association between primary tumor site and CH-PD.
Odds ratios (circle) and 95% confidence intervals for CH-PD in selected primary tumor types with at least 100 subjects compared to breast cancer (n=3540) in a logistic regression model adjusted for age. * p<0.05, ** p<0.01, *** p<0.001.
Extended Data Figure 3.
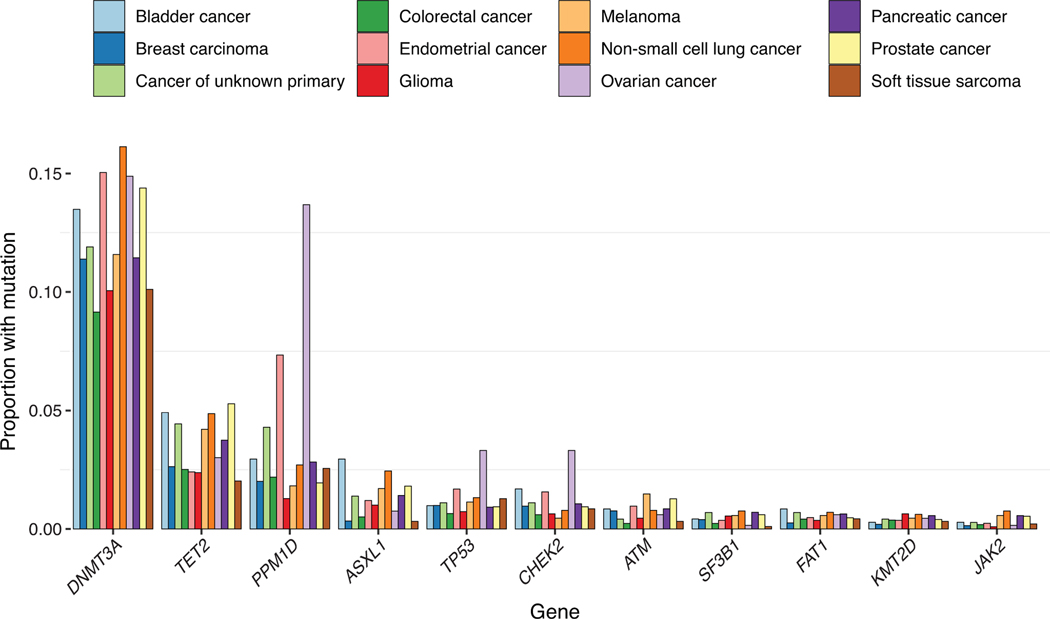
Proportion of patients with common CH-PD mutations by primary tumor sites. Genes mutated in at least 75 individuals and the top 12 primary tumor sites are shown.
Extended Data Figure 4. Variant frequencies (VAF) at time of pre-tMN testing and tMN diagnosis.
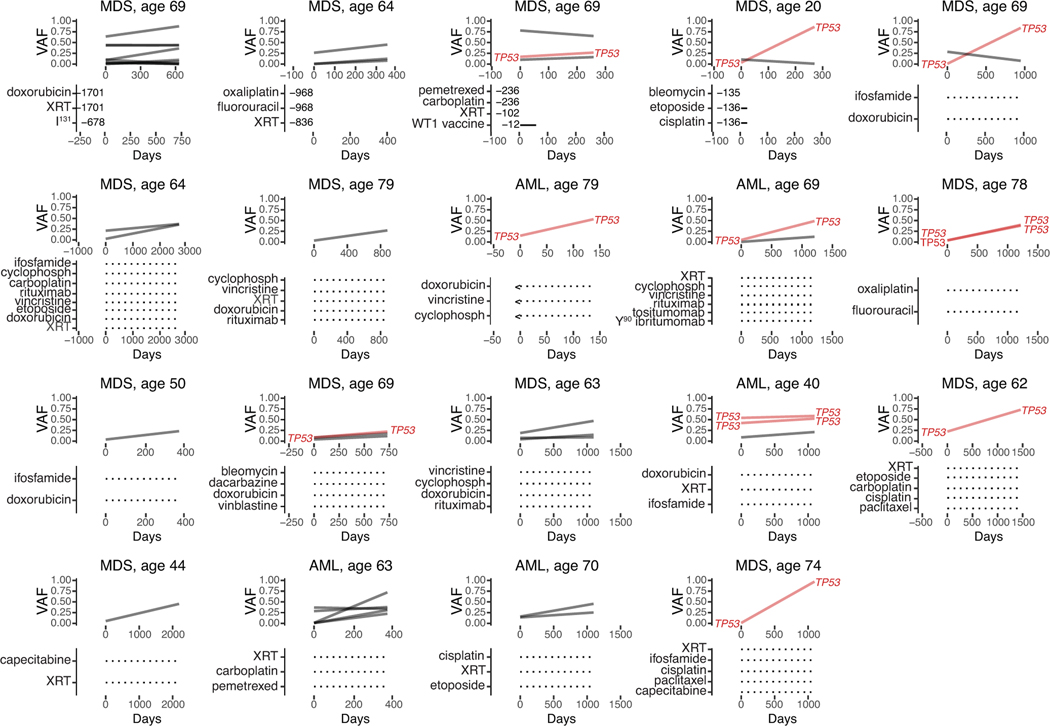
Plots show changes in mutational frequencies in relation to cancer therapy exposure in 19 CH cases. Below each graph are listed treatments received prior to pre-tMN testing and the number of days between the end of treatment and the pre-tMN sample.
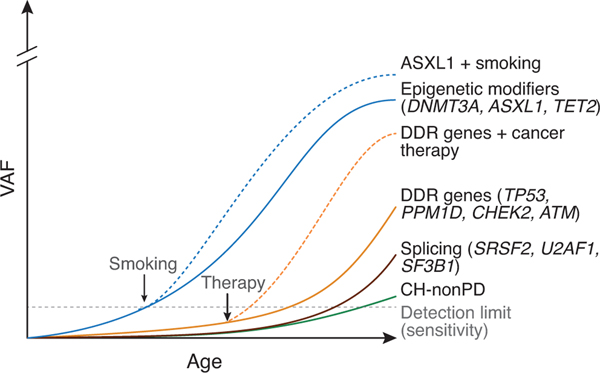
Extended Data Figure 5. Differences in the fitness effect of CH mutations and the environment shape clonal dominance over an individual’s lifetime.
Conceptual graph illustrating how associations between specific exposures and CH mutations may shape clonal dominance over an individual’s lifetime. AML, acute myeloid leukemia; cyclophosph, cyclophosphamide; MDS, myelodysplastic syndrome.
Extended Data Table 1.
Clinical characteristics of solid tumor patients assessed for CH.
| CH− | CH+ | |
|---|---|---|
| Total | 16930 (70%) | 7216 (30%) |
| Smoking status | ||
| Non-smoker | 8979 (74%) | 3086 (26%) |
| Current/former | 7255 (65%) | 3894 (35%) |
| Missing | 696 (75%) | 236 (25%) |
| Gender | ||
| Male | 7710 (70%) | 3315 (30%) |
| Female | 9220 (70%) | 3901 (30%) |
| Age | ||
| 0–10 | 324 (96%) | 13 (3.9%) |
| 10–20 | 284 (96%) | 13 (4.4%) |
| 20–30 | 672 (95%) | 36 (5.1%) |
| 30–40 | 1398 (92%) | 121 (8%) |
| 40–50 | 2757 (87%) | 420 (13%) |
| 50–60 | 4490 (78%) | 1298 (22%) |
| 60–70 | 4499 (64%) | 2575 (36%) |
| 70–80 | 2127 (50%) | 2092 (50%) |
| 80–90 | 379 (37%) | 648 (63%) |
| Ethnicity | ||
| White | 12628 (69%) | 5802 (31%) |
| Asian | 1274 (78%) | 356 (22%) |
| Black | 1081 (73%) | 410 (27%) |
| Other | 1175 (77%) | 355 (23%) |
| Unknown | 772 (72%) | 293 (28%) |
| Therapy | ||
| Treated | 4193 (70%) | 1785 (30%) |
| Untreated | 3027 (73%) | 1133 (27%) |
| Unknown | 9710 (69%) | 4298 (31%) |
| Primary tumor subtype | ||
| Ampullary carcinoma | 47 (76%) | 15 (24%) |
| Anal cancer | 38 (67%) | 19 (33%) |
| Appendiceal cancer | 128 (79%) | 34 (21%) |
| Biliary cancer | 351 (69%) | 157 (31%) |
| Bladder cancer | 445 (62%) | 267 (38%) |
| Breast carcinoma | 2610 (74%) | 930 (26%) |
| Cancer of unknown primary | 484 (67%) | 239 (33%) |
| Cervical cancer | 91 (77%) | 27 (23%) |
| Chondroblastoma | 1 (100%) | 0 (0%) |
| Chondrosarcoma | 42 (78%) | 12 (22%) |
| Chordoma | 27 (75%) | 9 (25%) |
| Choroid plexus tumor | 3 (100%) | 0 (0%) |
| Colorectal cancer | 1625 (75%) | 528 (25%) |
| Embryonal tumor | 153 (89%) | 18 (11%) |
| Endometrial cancer | 510 (61%) | 321 (39%) |
| Ependymomal tumor | 26 (90%) | 3 (10%) |
| Esophagogastric carcinoma | 464 (70%) | 196 (30%) |
| Ewing sarcoma | 66 (89%) | 8 (11%) |
| Gastrointestinal neuroendocrine tumor | 73 (68%) | 34 (32%) |
| Gastrointestinal stromal tumor | 200 (70%) | 84 (30%) |
| Germ cell tumor | 352 (91%) | 35 (9%) |
| Gestational trophoblastic disease | 10 (77%) | 3 (23%) |
| Glioma | 834 (76%) | 260 (24%) |
| Head and neck carcinoma | 252 (69%) | 111 (31%) |
| Hepatocellular carcinoma | 134 (71%) | 55 (29%) |
| Melanoma | 612 (69%) | 269 (31%) |
| Meningothelial tumor | 52 (79%) | 14 (21%) |
| Mesothelioma | 146 (65%) | 78 (35%) |
| Miscellaneous brain tumor | 22 (85%) | 4 (15%) |
| Miscellaneous neuroepithelial tumor | 11 (65%) | 6 (35%) |
| Nerve sheath tumor | 43 (88%) | 6 (12%) |
| Non-small cell lung cancer | 2235 (63%) | 1324 (37%) |
| Osteosarcoma | 98 (90%) | 11 (10%) |
| Ovarian cancer | 411 (62%) | 254 (38%) |
| Pancreatic cancer | 964 (68%) | 452 (32%) |
| Penile cancer | 7 (78%) | 2 (22%) |
| Pheochromocytoma | 6 (86%) | 1 (14%) |
| Pineal tumor | 1 (25%) | 3 (75%) |
| Prostate cancer | 971 (65%) | 523 (35%) |
| Renal cell carcinoma | 445 (78%) | 128 (22%) |
| Retinoblastoma | 38 (95%) | 2 (5%) |
| Salivary carcinoma | 161 (76%) | 52 (24%) |
| Sellar tumor | 53 (88%) | 7 (12%) |
| Sex cord stromal tumor | 29 (81%) | 7 (19%) |
| Skin cancer, non-melanoma | 137 (60%) | 91 (40%) |
| Small bowel cancer | 66 (77%) | 20 (23%) |
| Small cell lung cancer | 128 (60%) | 84 (40%) |
| Soft tissue sarcoma | 751 (76%) | 233 (24%) |
| Thymic tumor | 35 (70%) | 15 (30%) |
| Thyroid cancer | 267 (62%) | 165 (38%) |
| Uterine sarcoma | 124 (73%) | 46 (27%) |
| Vaginal cancer | 10 (67%) | 5 (33%) |
| Wilms tumor | 23 (96%) | 1 (4.2%) |
| Unknown | 75 (69%) | 34 (31%) |
Extended Data Table 2. Association between variant allele fraction (VAF) of CH mutations and clinical characteristics.
Generalized estimating equations were used to test for association between VAF of CH mutations (among those with a mutation) and selected clinical and mutational features, accounting for correlation between the VAF of mutations in the same person. Age expressed in decile.
| Variable (ref) | OR | 95% CI | p | |
|---|---|---|---|---|
| Age | - | 1 | 1–1.1 | 0.0011 |
| Ethnicity (white) | Asian | 1 | 0.94–1.2 | 0.42 |
| Black | 0.9 | 0.82–1 | 0.053 | |
| Other | 0.93 | 0.83–1 | 0.24 | |
| Unknown | 0.92 | 0.8–1.1 | 0.22 | |
| Smoking status (non-smoker) | Smoker | 1.1 | 1.1–1.2 | 0.000023 |
| Therapy (untreated) | Treated | 1 | 0.96–1.1 | 0.8 |
| PD status (Non-PD non-myeloid) | Myeloid PD | 1.3 | 1.3–1.4 | < 1 × 10−6 |
| Non-myeloid PD | 1.3 | 1.2–1.5 | 0.000052 | |
| Non-PD myeloid | 0.99 | 0.92–1.1 | 0.8 | |
| Number of mutations (1) | ≥ 2 | 1.1 | 1.1–1.2 | 0.0000038 |
Extended Data Table 3. Association among clinical characteristics and CH mutational characteristics.
Myeloid PD, genes mutated in myeloid neoplasms; non-myeloid, genes not linked to myeloid neoplasms; myeloid PD, variants known to be myeloid drivers or putative somatic driver mutations in myeloid neoplasms; myeloid non-PD, mutations within genes linked to myeloid neoplasms but that are not putative drivers; non-myeloid PD, mutations that are putative somatic driver mutations of cancer in genes not linked to myeloid neoplasms; non-myeloid non-PD, mutations within genes not linked to myeloid neoplasms that are not putative drivers of cancer. Associations were evaluated using multivariable logistic regression models to generate heterogeneity p-values. Sensitivity analyses restricted to individuals with only one mutation yielded similar results. Age expressed in decile.
| Variable (reference) | OR | 95% CI | p | |
|---|---|---|---|---|
| Age | - | 1 | 1–1.1 | 0.0011 |
| Ethnicity (white) | Asian | 1 | 0.94–1.2 | 0.42 |
| Black | 0.9 | 0.82–1 | 0.053 | |
| Other | 0.93 | 0.83–1 | 0.24 | |
| Unknown | 0.92 | 0.8–1.1 | 0.22 | |
| Smoke (non-smoker) | Smoker | 1.1 | 1.1–1.2 | 0.000023 |
| Therapy (untreated) | Treated | 1 | 0.96–1.1 | 0.8 |
| PD status (non-PD non-myeloid) | Myeloid PD | 1.3 | 1.3–1.4 | < 1 × 10−6 |
| Non-myeloid PD | 1.3 | 1.2–1.5 | 0.000052 | |
| Non-PD myeloid | 0.99 | 0.92–1.1 | 0.8 | |
| Number of mutations (1) | ≥ 2 | 1.1 | 1.1–1.2 | 0.0000038 |
Extended Data Table 4. Association between CH mutation number and clinical characteristics.
Ordinal logistic regression was used to test for association between clinical characteristics and mutation number in patients with clonal hematopoiesis in a multivariable model. Age expressed in decile.
| Variable (reference) | OR | 95% CI | p | |
|---|---|---|---|---|
| Age (0–10) | > 10 | 2.3 | 2–2.6 | < 1 × 10−6 |
| Gender (male) | Female | 1.1 | 0.94–1.3 | 0.2 |
| Ethnicity (white) | Non-white | 0.83 | 0.67–1 | 0.087 |
| Smoke (non-smoker) | Smoker | 1.2 | 1–1.4 | 0.027 |
| Therapy (untreated) | Treated | 1.2 | 1.1–1.5 | 0.011 |
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health (K08 CA241318 to K.B., K12 CA120780 to C.C., P50 CA172012 to L.B., P50 CA172012 to J.F., UG1 HL069315 to V.K.), American Society of Hematology (K.B. and Elli Papaemmanuil), EvansMDS Foundation (K.B.), European Hematology Association (Elli Papaemmanuil), Gabrielle’s Angels Foundation (Elli Papaemmanuil), V Foundation (Elli Papaemmanuil), Geoffrey Beene Foundation (Elli Papaemmanuil), UNC Oncology Clinical Translational Research Training Program (C.C.), Cycle for Survival (V.K.), Starr Cancer Consortium (to R.L., A.Z., M.F.B., R.P.), and the Cancer Colorectal Cancer Dream Team Translational Research Grant (SU2C-AACR-DT22-17 to L.D.). Elli Papaemmanuil is a Josie Robertson Investigator. C.C. is a recipient of the Conquer Cancer Foundation Young Investigator Award and the Prostate Cancer Foundation Young Investigator Award. K.S. is a recipient of the Defense Early Investigator Research Award (W81XWH-18-1-0330), Prostate Cancer Foundation Young Investigator Award and the Prostate Cancer Foundation Challenge Award. C.L., M.G. and L.M. are supported by funds from the Intramural Research Program of the National Cancer Institute, National Institutes of Health. Work performed at Memorial Sloan Kettering Cancer Center was supported in part by the Cancer Center Support Grant (P30 CA008748). N.G.’s work was supported in part by the Tissue Core and Genomic Core Facilities at the H. Lee Moffitt Cancer Center & Research Institute, an NCI-designated Comprehensive Cancer Center (P30 CA076292). The University of Cambridge has received salary support in respect of PDPP from the NHS in the East of England through the Clinical Academic Reserve.
The authors declare the following competing interests: K.L.B. has received research funding from GRAIL. C.C.C. has received honoraria from AbbVie, Loxo, H3 Biomedicine, Medscape, Octapharma, and Pharmacyclics; has served as a consultant for AbbVie, Covance, Cowen & Co., and Dedham Group and has received institutional research funding from AROG, Gilead, Loxo, H3 Biomedicine, and Incyte. Z.S. has an immediate family member who holds consulting/advisory roles within the field of ophthalmology with Allergan, Adverum Biotechnologies, Alimera Sciences, Biomarin, Fortress Biotech, Genentech, Novartis, Optos, Regeneron, Regenxbio, and Spark Therapeutics. E.B. receives research funding from Celgene. D.G. is a consultant of MNM Diagnostics and has received honoraria for speaking and scientific advisory engagements with Celgene, Prime Oncology, Novartis, Illumina and Kyowa Hakko Kirin. S.L. is an employee of GRAIL. M.E.R. holds an uncompensated advisory role with AstraZeneca, Daiichi-Sankyo, Merck, and Pfizer and receives institutional research funding from AstraZeneca, AbbVie, Medivation, and Pfizer. B.L.E. has received research funding from Celgene and Deerfield. T.D. is the Chief Medical Officer, ArcherDX, Inc. and receives salary from and holds an ownership stake in the company. K.T. receives consultancy fees from Symbio Pharmaceuticals. D.M.H. has consulted for Fount, Chugai, Boehringer Ingelheim, AstraZeneca, Pfizer, Bayer, and Genentech/Roche; has equity in Fount; and has received research grants from Loxo, Bayer, Puma, and AstraZeneca. J.B. is an employee of AstraZeneca; is on the Board of Directors of Foghorn and is a past board member of Varian Medical Systems, Bristol‐Myers Squibb, Grail, Aura Biosciences and Infinity Pharmaceuticals; has performed consulting and/or advisory work for Grail, PMV Pharma, ApoGen, Juno, Eli Lilly, Seragon, Novartis, and Northern Biologics; has stock or other ownership interests in PMV Pharma, Grail, Juno, Varian, Foghorn, Aura, Infinity Pharmaceuticals, ApoGen, Northern Biologics as well as Tango and Venthera, for which is a co‐founder; and has previously received honoraria or travel expenses from Roche, Novartis, and Eli Lilly. M. Ladanyi serves on the advisory boards for Astra-Zeneca, Bristol Myers Squibb, Takeda, Bayer, and Merck, and has received research support from Loxo Oncology and Helsinn Therapeutics. D.B.S. has served as a consultant or received honoraria from Pfizer, Loxo Oncology, Lilly Oncology, Illumina and Vivideon Therapeutics. M.F.B. is on the advisory board for Roche and receives research support from Illumina. M.S.T. receives research funding from AbbVie, Cellerant, Orsenix, ADC Therapeutics, and Biosight; serves on the advisory boards of Daiichi-Sankyo, KAHR, Rigel, Nohla, Delta Fly Pharma, Tetraphase, Oncolyze, and Jazz Pharma; has received royalties from UpToDate; and has received research funding from Incyte, Kura Oncology, and Celgene. L.A.D. is a member of the board of directors of Personal Genome Diagnostics (PGDx) and Jounce Therapeutics; is a paid consultant to PGDx and Neophore; is an uncompensated consultant for Merck (with the exception of travel and research support for clinical trials); is an inventor of multiple licensed patents related to technology for circulating tumor DNA analyses and mismatch repair deficiency for diagnosis and therapy from Johns Hopkins University, some of which are associated with equity or royalty payments directly to Johns Hopkins and L.A.D.; and holds equity in PGDx, Jounce Therapeutics, Thrive Earlier Detection and Neophore; his wife holds equity in Amgen. The terms of all these arrangements are being managed by Johns Hopkins and Memorial Sloan Kettering in accordance with their conflict of interest policies. R.L.L. is on the supervisory board of Qiagen and is a scientific advisor to Loxo, Imago, C4 Therapeutics and Isoplexis, which include equity interest; receives research support from and has consulted for Celgene and Roche and has consulted for Lilly, Janssen, Astellas, Morphosys and Novartis; and has received honoraria from Roche, Lilly and Amgen for invited lectures and from Gilead for grant reviews. A.Z. received honoraria from Illumina. E. Papaemmanuil receives research funding from Celgene and is a co-founder in Isabl Technologies, a software analytics company for high-throughput clinical whole-genome and RNA-sequencing analyses.
Footnotes
CODE AVAILABILITY
The code to replicate the findings in the article are publicly available, except those shown in Extended Data Figure 5 and Supplementary Figure 12, on Github: https://github.com/papaemmelab/bolton_NG_CH. The code used to generate the excepted figures are not included because the data cannot be shared (see above).
DATA AVAILABILITY
The minimal clinical and mutational data necessary to replicate the findings in the article, except those shown in Extended Data Figure 5 and Supplementary Figure 12, are publicly available on Github: https://github.com/papaemmelab/bolton_NG_CH. Data for the excepted figures (individual drug names and start and stop dates, and combinations of mutations at tMN diagnosis, respectively) cannot be made public to preserve patient anonymity. Raw sequencing data cannot be publicly deposited for legal and privacy reasons, as sequencing was performed for clinical purposes. Mutation calls are available on cBioPortal: http://www.cbioportal.org/study/summary?id=msk_ch_2020
COMPETING INTEREST DECLARATION
The remaining authors declare no competing interests.
REFERENCES
- 1.Armitage P & Doll R The Age Distribution of Cancer and a Multi-stage Theory of Carcinogenesis. British Journal of Cancer vol. 8 1–12 (1954). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greaves M & Maley CC Clonal evolution in cancer. Nature 481, 306–313 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexandrov LB et al. The Repertoire of Mutational Signatures in Human Cancer. doi: 10.1101/322859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabarinathan R et al. The whole-genome panorama of cancer drivers. doi: 10.1101/190330. [DOI] [Google Scholar]
- 5.Yates LR & Campbell PJ Evolution of the cancer genome. Nature Reviews Genetics vol. 13 795–806 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding L et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481, 506–510 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blokzijl F et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martincorena I et al. Somatic mutant clones colonize the human esophagus with age. Science 362, 911–917 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martincorena I, Jones PH & Campbell PJ Constrained positive selection on cancer mutations in normal skin. Proceedings of the National Academy of Sciences of the United States of America vol. 113 E1128–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martincorena I et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yokoyama A et al. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature 565, 312–317 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Suda K et al. Clonal Expansion and Diversification of Cancer-Associated Mutations in Endometriosis and Normal Endometrium. Cell Reports vol. 24 1777–1789 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Jaiswal S et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371, 2488–2498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genovese G et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371, 2477–2487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKerrell T et al. Leukemia-Associated Somatic Mutations Drive Distinct Patterns of Age-Related Clonal Hemopoiesis. Cell Reports vol. 10 1239–1245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie M et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nature Medicine vol. 20 1472–1478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandez-Antoran D et al. Outcompeting p53-Mutant Cells in the Normal Esophagus by Redox Manipulation. Cell Stem Cell 25, 329–341.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu JI et al. PPM1D Mutations Drive Clonal Hematopoiesis in Response to Cytotoxic Chemotherapy. Cell Stem Cell 23, 700–713.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong TN et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature 518, 552–555 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abelson S et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 559, 400–404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desai P et al. Somatic Mutations Predict Acute Myeloid Leukemia Years Before Diagnosis. doi: 10.1101/237941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morton LM et al. Evolving risk of therapy-related acute myeloid leukemia following cancer chemotherapy among adults in the United States, 1975–2008. Blood 121, 2996–3004 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNerney ME, Godley LA & Le Beau MM Therapy-related myeloid neoplasms: when genetics and environment collide. Nature Reviews Cancer vol. 17 513–527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coombs CC et al. Therapy-Related Clonal Hematopoiesis in Patients with Non-hematologic Cancers Is Common and Associated with Adverse Clinical Outcomes. Cell Stem Cell 21, 374–382.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chakravarty D et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol 2017, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papaemmanuil E et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med. 365, 1384–1395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papaemmanuil E et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 374, 2209–2221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grinfeld J et al. Classification and Personalized Prognosis in Myeloproliferative Neoplasms. N. Engl. J. Med. 379, 1416–1430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bick AG et al. Inherited Causes of Clonal Hematopoiesis of Indeterminate Potential in TOPMed Whole Genomes. bioRxiv 782748 (2019) doi: 10.1101/782748. [DOI] [Google Scholar]
- 30.Abelson S et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 559, 400–404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNerney ME, Godley LA & Le Beau MM Therapy-related myeloid neoplasms: when genetics and environment collide. Nature Reviews Cancer vol. 17 513–527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindsley RC et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 125, 1367–1376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Welch JS et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 150, 264–278 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cancer Genome Atlas Research Network et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gillis NK et al. Clonal haemopoiesis and therapy-related myeloid malignancies in elderly patients: a proof-of-concept, case-control study. Lancet Oncol. 18, 112–121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi K Germline polymorphisms and the risk of therapy-related myeloid neoplasms. Best Pract. Res. Clin. Haematol. 32, 24–30 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Gibson CJ et al. Clonal Hematopoiesis Associated With Adverse Outcomes After Autologous Stem-Cell Transplantation for Lymphoma. J. Clin. Oncol. 35, 1598–1605 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young AL, Tong RS, Birmann BM & Druley TE Clonal haematopoiesis and risk of acute myeloid leukemia. Haematologica (2019) doi: 10.3324/haematol.2018.215269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fianchi L et al. Characteristics and outcome of therapy-related myeloid neoplasms: Report from the Italian network on secondary leukemias. Am. J. Hematol. 90, E80–5 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Choudhury PP et al. iCARE: R package to build, validate and apply absolute risk models. doi: 10.1101/079954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maas P et al. Breast Cancer Risk From Modifiable and Nonmodifiable Risk Factors Among White Women in the United States. JAMA Oncol 2, 1295–1302 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Candido Dos Reis FJ et al. An updated PREDICT breast cancer prognostication and treatment benefit prediction model with independent validation. Breast Cancer Res. 19, 58 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meng A, Wang Y, Van Zant G & Zhou D Ionizing radiation and busulfan induce premature senescence in murine bone marrow hematopoietic cells. Cancer Res. 63, 5414–5419 (2003). [PubMed] [Google Scholar]
- 44.Hu W et al. Mechanistic Investigation of Bone Marrow Suppression Associated with Palbociclib and its Differentiation from Cytotoxic Chemotherapies. Clin. Cancer Res. 22, 2000–2008 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Meisel M et al. Microbial signals drive pre-leukaemic myeloproliferation in a Tet2-deficient host. Nature 557, 580–584 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu M et al. Somatic Mutations Increase Hepatic Clonal Fitness and Regeneration in Chronic Liver Disease. Cell 177, 608–621.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 47.Cheng DT et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J. Mol. Diagn. 17, 251–264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmieder R & Edwards R Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tate JG et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 47, D941–D947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Papaemmanuil E et al. Identification of Novel Somatic Mutations in SF3B1, a Gene Encoding a Core Component of RNA Splicing Machinery, in Myelodysplasia with Ring Sideroblasts and Other Common Cancers. European Journal of Cancer vol. 47 7 (2011). [Google Scholar]
- 51.Campo E et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. (IARC Who Classification of Tum, 2017). [Google Scholar]
- 52.Wolff AC et al. Risk of marrow neoplasms after adjuvant breast cancer therapy: the national comprehensive cancer network experience. J. Clin. Oncol. 33, 340–348 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Website. Surveillance, Epidemiology, and End Results (SEER) Program Populations (1969–2017) (www.seer.cancer.gov/popdata), National Cancer Institute, DCCPS, Surveillance Research Program, released December 2018. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.