Abstract
Patients with signs of COVID-19 were tested through diagnostic RT-PCR for SARS-CoV-2 using RNA extracted from the nasopharyngeal/nasal swabs. To determine the variants of SARS-CoV-2 circulating in the state of Nevada, specimens from 200 COVID-19 patients were sequenced through our robust sequencing platform, which enabled sequencing of SARS-CoV-2 from specimens with even very low viral loads, without the need of culture-based amplification. High genome coverage allowed the identification of single and multi-nucleotide variants in SARS-CoV-2 in the community and their phylogenetic relationships with other variants present during the same period of the outbreak. We report the occurrence of a novel mutation at 323aa (314aa of orf1b) of nsp12 (RNA-dependent RNA polymerase) changed to phenylalanine (F) from proline (P), in the first reported isolate of SARS-CoV-2, Wuhan-Hu-1. This 323F variant was present at a very high frequency in Northern Nevada. Structural modeling determined this mutation in the interface domain, which is important for the association of accessory proteins required for the polymerase. In conclusion, we report the introduction of specific SARS-CoV-2 variants at very high frequency in distinct geographic locations, which is important for understanding the evolution and circulation of SARS-CoV-2 variants of public health importance, while it circulates in humans.
Keywords: SARS-CoV-2, COVID-19, Genome enrichment, nsp12, RdRp, orf1b 314
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the cause of coronavirus disease 2019 (COVID-19), was first reported outbreak in Wuhan, Hubei, China, during December 2019 (Coronaviridae Study Group of the International Committee on Taxonomy of 2020; Petrosillo et al., 2020; Zhou et al., 2020). RNA sequencing and phylogenetic analysis of specimens taken during the initial outbreak in Wuhan determined that the virus is most closely related (89.1% nucleotide similarity) to a group of SARS-like coronaviruses (genus Betacoribavirus, subgenus Sarbecovirus), which had previously been identified in bats in China (Hu et al., 2018; Petrosillo et al., 2020). Coronaviruses have a recent history as emerging infections, first SARS-CoV in 2002–2003, and Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012, both zoonotic infections that cause severe respiratory illness in humans (Peiris et al., 2003; Zhong et al., 2003; Zaki et al., 2012; De Wit et al., 2016; Petrosillo et al., 2020). Unlike SARS-CoV and MERS-CoV, which displayed limited global spread, SARS-CoV-2 has spread around the world within a few months. There are specific characteristics of SARS-CoV-2, which have facilitated the transmission, including infections, that result in asymptomatic or mild disease, allowing for under-characterized transmission.
SARS-CoV-2 is an enveloped, positive single-stranded RNA virus. Detection of SARS-CoV-2 in patients has primarily occurred using RT-qPCR to detect viral RNA from respiratory specimens (primarily nasal and nasopharyngeal swabs). While RT-PCR determines a cycle threshold (Ct) value for each sample based on viral load, it does not yield sequence data leading to the description of genomic variants. To further study such variants and to better understand the epidemiology of the virus in the state of Nevada, we developed a workflow that allowed us to sequence SARS-CoV-2 genomic RNA from patient swabs containing a broad range of viral loads. Of the sequences of SARS-CoV-2 currently submitted to common databases (GenBank and GISAID), several were obtained after the virus had been passed in Vero cells (Kim et al., 2020; Licastro et al., 2020), and others came directly from patient specimens (Holland et al., 2020; Wu et al., 2020). Certain data have suggested a potential for lab-acquired mutations following passage in cell culture (Davidson et al., 2020; Kim et al., 2020). Specifically, a report of SARS-CoV-2 passage in Vero cells resulted in a spontaneous nine amino acid deletion within the spike (S) protein that overlaps with the furin cleavage site (Davidson et al., 2020). The loss of this site is suggested to increase the viral entry into Vero cells (Walls et al., 2020). For both research and epidemiological purposes, sequencing of SARS-CoV-2 directly from patient specimens not only reduces the possibility of laboratory-acquired mutations following passages in cell culture but also reduces the time that would be spent growing the virus from the patient specimens and subsequently also reduces handling larger amounts of infectious virus. A compounding factor for the specimens with high Ct values is that they would not have had enough infectious material to propagate virus in cell culture (Jaafar et al., 2020). Additionally, one of the goals in developing an optimized SARS-CoV-2 NGS protocol was to be able to generate the adequate depth of coverage of the viral genome while minimizing the sequencing of non-viral RNA, which would allow for more specimens to be multiplexed together during sequencing.
Our workflow employs a combination of RNA amplification, conversion into Illumina-compatible sequencing libraries, and enrichment of SARS-CoV-2 library molecules prior to sequencing. Using this methodology, we sequenced SARS-CoV-2 from a total of 200 patient specimens collected over a 3-month period originating from Nevada. Of the 200 selected, 173 were sequenced with enough quality to be used for determining SARS-CoV-2 nucleotide variants to perform further phylogenetic analysis and study the viral epidemiology within the state of Nevada. Analysis of these data suggests a specific epidemiological course for the local epidemic within Northern Nevada. This was characterized by an initial observation of variants closely resembling isolates identified from China or Europe. Subsequent to the government-mandated period of restrictions on business and social activity, we observed that a viral isolate not seen elsewhere in the world emerged within Northern Nevada cases (nucleotide 14,407 and 14,408). This isolate contains an amino acid change in residue P323 L/F of RdRp (nsp12). Furthermore, we found that sampled viral isolates in Southern Nevada, unlike those in Northern Nevada, closely resembled the makeup of the United States in general.
Results
RNA-seq workflow and assembly of SARS-CoV-2 genomes
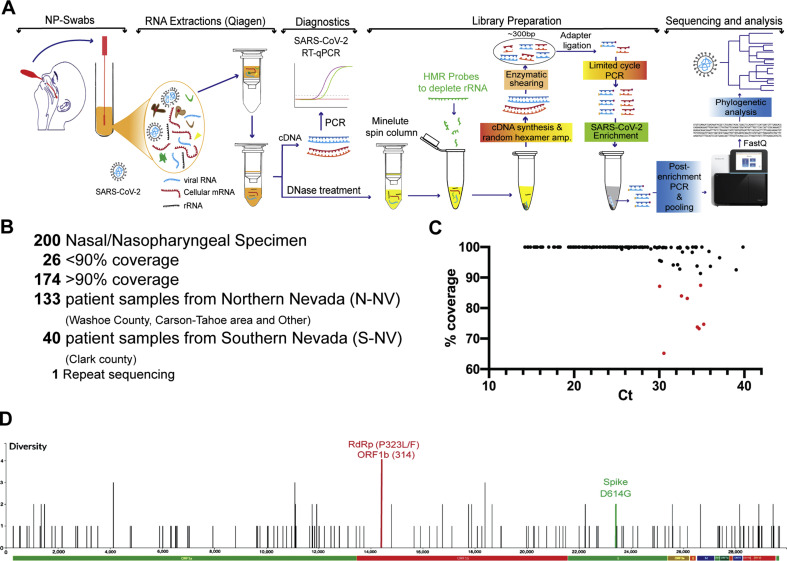
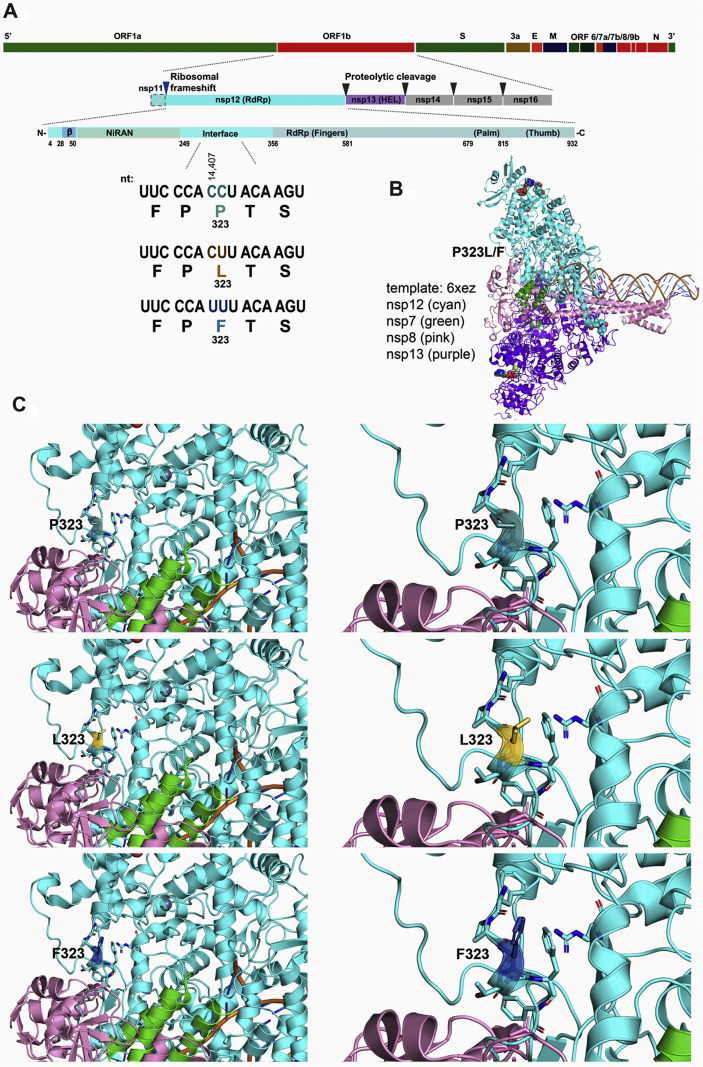
A total of 200 SARS-CoV-2 positive specimens collected in Nevada from March 6 to June 5 were randomly selected to have their viral genomes sequenced for variant analysis and subsequent epidemiological studies (Materials and methods; Fig. 1 A). Of the sequenced specimens, 173 had > 90% coverage and sufficient depth to accurately call those genomic positions with variants (Fig. 1B). An alignment of SARS-CoV-2 genomes from these specimens is presented as supplemental data (Fig. S1) showing 173 specimens with over 90% coverage. The number of sequences reads and the depth of coverage for each specimen is presented as a supplemental data (Table S1). These 173 specimens represented 133 patient specimens from Northern Nevada (including Washoe County of which Reno is the major city, the Carson-Tahoe area, and other northern, rural counties), 40 patient specimens from Southern Nevada (Clark County, which encompasses Las Vegas and surrounding cities). Nucleotide similarity and variants were determined and used to measure the phylogenetic relationships (Fig. S2). The combined nucleotide diversity across the entire SARS-CoV-2 genome for the Nevada specimens is shown in Fig. 1D, along with the genomic areas that were assessed for change in frequency corresponding to amino acids D614G, P323 L/F and nucleotide 379.
Fig. 1.
Workflow of SARS-CoV-2 genome sequencing and analysis from nasopharyngeal patient specimens in Nevada. A: RNA was extracted from nasal or nasopharyngeal (NP) swabs taken from patients in Nevada and first used to determine the presence of SARS-CoV-2 genomes by RT-qPCR. Next-generation sequencing (NGS) libraries were prepared from positive specimens; this included steps for ribosomal RNA depletion and SARS-CoV-2 enrichment. Subsequent libraries were pooled and used for whole genome sequencing at the Nevada Genomics Center on the Illumina NextSeq 500 instrument. FASTQ files were aligned to the reference genome and analyzed to determine nucleotide variation and phylogenetic relationship. B: A total of 200 specimens were sequenced, of which 173 had over 90% coverage of the SARS-CoV-2 genome. This included 133 patient specimens from Northern Nevada, 40 from Southern Nevada, and 1 specimen that was re-sequenced. C: Correlation between RT-qPCR Ct value and the percentage of coverage in the whole genome sequencing after trimming and alignment. D: Nucleotide variants across the SARS-CoV-2 genome in the 173 specimens from Nevada from March 6 to June 5.
During the sequencing analysis, we also examined the correlation between Ct values from the diagnostic RT-PCR and percentage coverage of the viral genome to determine the performance and robustness of our sequencing method in relation to available viral RNA in a specimen of a given Ct (Fig. 1C). On average, a Ct value less than 37 resulted in at least 90% coverage to the SARS-CoV-2 genome. Importantly, our in-house developed method for viral genome enrichment and sequencing directly from the patient’s specimens (nasal and nasopharyngeal swabs) was robust and yielded sequences covering over 90% of the genome even in samples having very high Ct (∼40) of viral genome detection. This is highly significant and shows the power of our workflow in sequencing of SARS-CoV-2 genome from a spectrum of samples, including the ones having inadequate amounts of specimen (due to the variability in collection) or lower viral loads in nasal secretions. Consequently, our sequencing protocol avoids any molecular epidemiological bias, which may get acquired through cell culture-based amplification, especially in those specimens with high Ct (low viral load) as our method eliminates the need of virus culture.
Prevalence of amino acid variant D614G of SARS-CoV-2 spike protein in specimens collected in Nevada
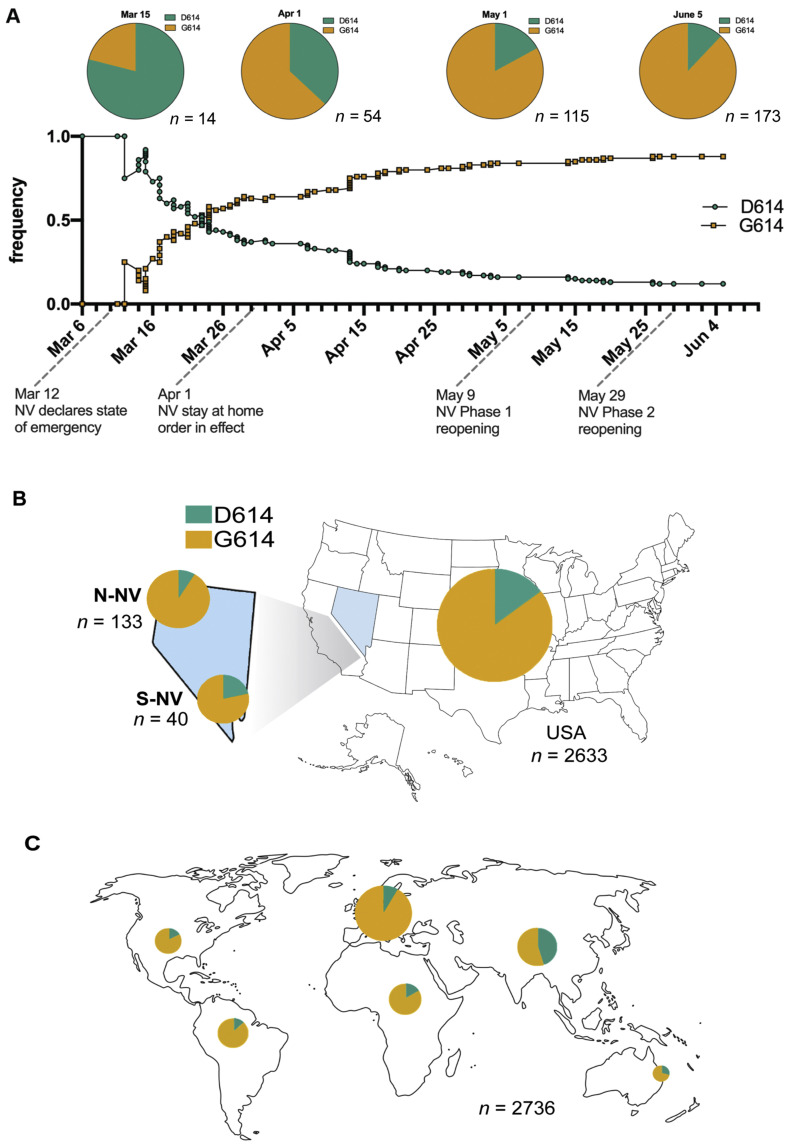
Earlier studies have revealed the emergence, spread, and potential importance of an alteration, D614G (genomic change at 23403A>G), of the spike protein (Korber et al., 2020). This missense mutation has become a clade-distinguishing locus that differentiates viral isolates originating in Asia from those that have emerged from Europe. A total of 173 cases were analyzed to determine the number and relative proportion of the specimens that carried the D614G spike protein variant in Nevada. The cumulative frequencies for D614 and G614 were plotted from March 6 to June 5 (Fig. 2 A). Specimens from the beginning of March represent the earliest known cases in Nevada, and of the 14 specimens sequenced during this time period (March 6–March 15), D614 was the predominant variant. This shifted from March to June with an increasing frequency of the G614 allele. The trend for specimens originating from either Northern Nevada (N-NV) and Southern Nevada (S-NV) both show a higher frequency of G614 (Fig. 2B). We used a subsampling of sequence data from Nextstrain.org to assess the frequency of D614G in the United States and globally during the same time period (March 6 to June 5) (Fig. 2B and 2C). The global trend by continent of D614G is also similar, with G614 at a higher frequency, the one noted exception is in Asia, where D614 and G614 continue to exist in equal proportions (Fig. 2C).
Fig. 2.
Distribution of D614G in Nevada and comparison with the United States and global proportion. A: Cumulative frequency of D614G in 173 patient specimens from Nevada from March 6 to June 5, 2020 (D614 is indicated by teal, G614 is indicated by yellow). Pie charts depict the cumulative proportion up to the indicated time point (March 15, April 1, May 1, June 5). The total number of specimens included at each time point is specified below each pie chart. Effective dates of emergency orders and regulatory responses to SARS-CoV-2 spread in Nevada are indicated on the frequency graph time axis. B: Proportion of D614G in the United States from March 6 to June 5, specimens from Nevada are divided in the geographic area that they originated from, Northern Nevada (N-NV) includes 133 specimens from Washoe County, Carson-Tahoe, and other northern counties, and Southern Nevada (S-NV) includes 40 specimens from Clark County. C: Global proportion of D614G in the shown regions during the same time period from a subsampling of sequences deposited in Nextstrain.org. The size of the pie chart corresponds to the relative specimen number for each region.
Frequency of SARS-CoV-2 clades in Nevada
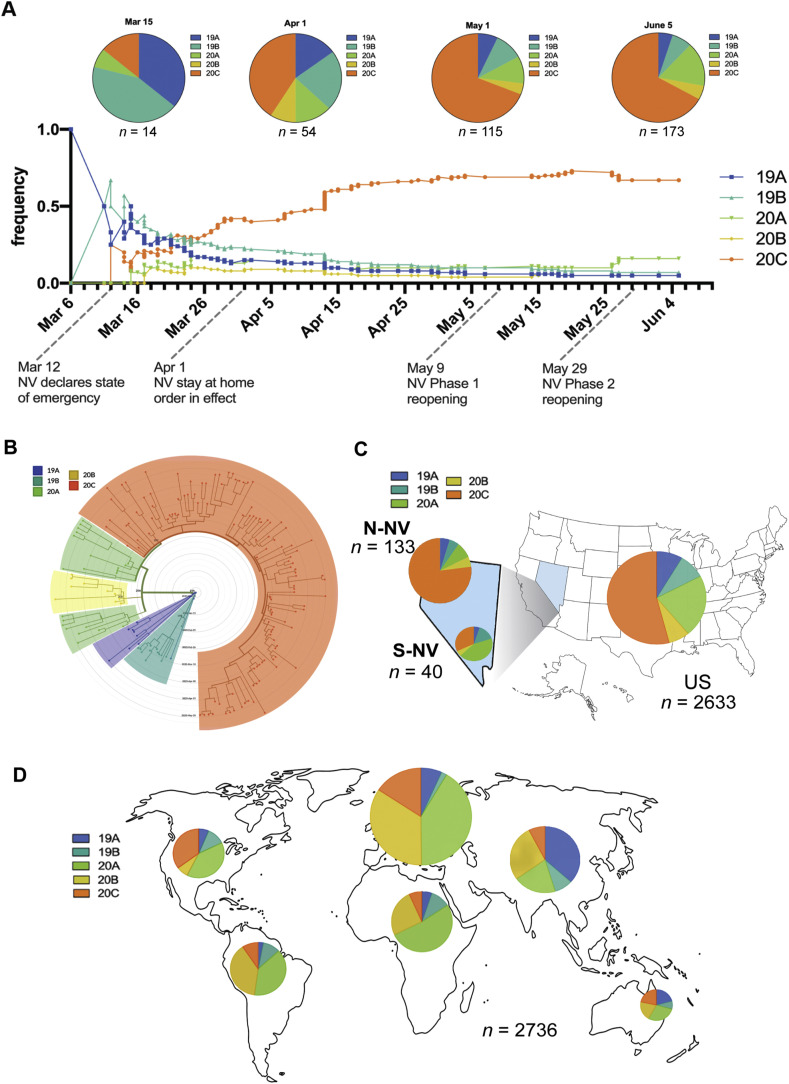
Worldwide, there are currently 5 main clades (19A, 19B, 20A, 20B, 20C) of SARS-CoV-2 differentiated based on specific nucleotide profiles in the Year-letter scheme of https://clades.nextstrain.org. Clades 19A and 19B are defined by C8782T and T28144C, respectively. 20A is a derivative of 19A and contains mutations C3037T, C14408T, and A23403G (resulting in D614G). 20B is defined by mutations G28881A, G28882A, and G28883C, and 20C contains C1059T and G25563T (Nextclade, 2020).
To assess the introduction and spread of the clades in Nevada, the cumulative frequencies for the clades were plotted from March 6 to June 5 (Fig. 3 A). The earliest sequenced specimens from Nevada were collected in the beginning of March (March 6–March 15) and are predominantly from Clades 19A and 19B. Additional sequenced specimens collected from March to June revealed a shift to a higher frequency of 20C (Fig. 3A). We performed phylogenetic reconstruction of the Nevada specimens and differentiated the clades on the circular dendrogram by color (Fig. 3B). There were discordant trends in the dominant clade for specimens originating from either Northern Nevada (N-NV) and Southern Nevada (S-NV) (Fig. 3C and 3D). Specimens from Northern Nevada (Washoe County, Carson-Tahoe, and other counties) showed a prevalence of 20C, while the Southern Nevada specimens from Clark County had a larger proportion of 20A (Figs. 3C and S3). We used a subsampling of Nextstrain.org data to assess the frequency of clades in the United States and globally by continent during the same time period (March 6 to June 5). The dominant clade in the United States was 20C, similar to the frequency seen in the total Nevada samples (N-NV and S-NV) (Fig. 3A and 3C). The global clade distributions were variable in areas outside of Asia, while Clades 19A and 19B are noted to be more prevalent in Asia (Fig. 3D). Clade distribution of SARS-CoV-2 among different countries are presented in Table S2.
Fig. 3.
Distribution of SARS-CoV-2 clades in Nevada. A: Cumulative frequency of SARS-CoV-2 clades in 173 patient specimens from Nevada during March 6 to June 5. The 5 clades are colored 19A (blue), 19B (teal), 20A (green), 20B (yellow), and 20C (orange). Pie charts depict the cumulative proportion up to the indicated time point (March 15, April 1, May 1, June 5). The total number of specimens included at each time point is specified below each pie chart. Dates of emergency orders and regulations meant to slow the spread of SARS-CoV-2 in Nevada are indicated on the time scale of the frequency graph. B: Circular dendrogram depicting clades from Nevada specimens. C: Pie chart of the clades from Northern Nevada (N-NV), Southern Nevada (S-NV), and the United States. D: Pie charts show the proportion of clades from global regions during the same time period from a subsampling of sequences deposited in Nextstrain.org. The size of the pie chart corresponds to the relative specimen number for each region.
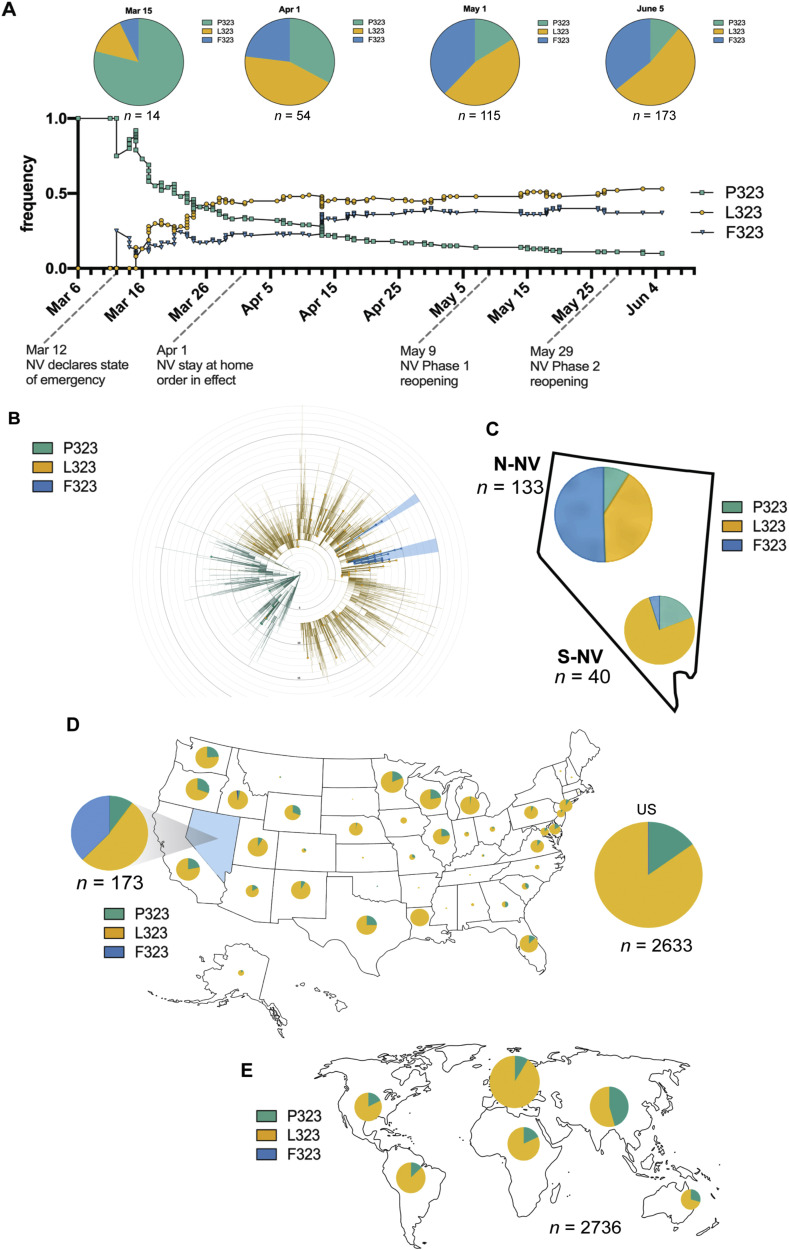
Prevalence of amino acid variant P323 L/F of SARS-CoV-2 nsp12 (RdRp) in Nevada
Analysis of sequencing data revealed a novel observation for our specimens at bases 14,407 and 14,408, which results in a change at residue 323 in nsp12 (RdRp). For the Wuhan isolate at 14,407 and 14,408, there is CC for proline (P), the variants have CT for leucine (P323L), and TT for phenylalanine (P323F). To assess the introduction and spread of P323 L/F in Nevada, the cumulative frequencies of P323, L323, and F323 were plotted from March 6 to June 5 (Fig. 3A). Nevada specimens from the beginning of March (March 6-March 15) showed P323 to be the predominant variant. As additional specimens were collected and sequenced from March to June, there was a shift to a higher frequency of L323 and F323 (Fig. 4 A). We performed phylogenetic reconstruction of the Nevada specimens and noted the P323 L/F variants on the circular dendrogram with the indicated colors (Fig. 4B). Interestingly, analysis of the Northern Nevada and Southern Nevada specimen showed very different dominant variants (Fig. 4B). In Northern Nevada the, F323 was more prevalent, while in Southern Nevada, L323 was more prevalent. We used a subsampling of Nextstrain.org data to assess the frequency of P323 L/F in the United States and globally during the same time period (March 6 to June 5). P323 was the predominant variant in Asia, while L323 was more prevalent in other areas of the world, and F323 was only appreciably noted in North America (Fig. 4D and 4E).
Fig. 4.
Distribution of P323 L/F (nsp12, RdRp) in Nevada. A: Cumulative frequency of P323 L/F (nsp12, RdRp) in 173 patient specimens from Nevada during March 6 to June 5. The amino acid at position 323 is indicated by teal for proline (P), yellow for leucine (L), and blue for phenylalanine (F). Pie charts depict the cumulative proportion up to the indicated time point (March 15, April 1, May 1, June 5). The total number of specimens included at each time point is specified below each pie chart. Dates of emergency orders and regulations meant to slow the spread of SARS-CoV-2 in Nevada are indicated on the time scale of the frequency graph. B: Circular dendrogram representing the distribution of amino acid change at residue 323 of nsp12 from a global subsampling of sequences deposited in Nextstrain.org from March 6 to June 5, the larger dots indicate specimens from Nevada. C: Pie chart indicating the ratio of P/L/F in Northern NV and Southern NV specimens from this study. D: Proportion of P323 L/F from a subsampling of sequences deposited in Nextstrain.org for the United States and E: global regions from March 6 to June 5. The size of the pie chart corresponds to the relative specimen number for each region.
While we are investigating the phenotypic significance of this predominant variant of RdRp, we performed in-silico structural modeling of RdRp to determine the spatio-temporal location of this 323aa on RdRp in complex with replisome proteins, nsp7, nsp8 and nsp13. Our data showed the location of 323aa in the interface domain of RdRp and variation of P323 to L or F did not significantly change the conformation of the protein (Fig. 5 ). As the interface domain (aa 251–398) acts as a protein interaction junction for the finger domain of the polymerase and the second subunit of nsp8 (accessory protein), required for the polymerase activity, we anticipate this mutation to have phenotypic effect on the RdRp activity. It remains to be seen if there is any altered phenotype associated with the P323 L/F RdRp variant, and a pending investigation will provide confirmatory results on this highly prevalent mutation.
Fig. 5.
Structure of SARS-CoV-2 nsp12 (RdRp) P323 L/F. A: Diagram depicting ORF1b genomic location and encoded proteins. Below the linear protein schematic of nsp12, specific nucleotide variants at position 14,407 and 14,408 and the resulting amino acid changes are indicated. B: SARS-CoV-2 replicase complex modeled from 6XEZ template. This model includes nsp12, nsp7, nsp8, nsp13, ligands (ZN, Mg), the template, and product strand of RNA. P323L/F within the interface domain of nsp12 is located at the left side of the complex. C: Model nsp12 showing P323 L/F within the interface domain. Residue 323 is shown with either P, L, or F, and amino acids with side chains within 5 Å of residue 323 are depicted as sticks in the cartoon model.
Discussion
We have developed a unique method, which combines low-abundance RNA amplification and targeted enrichment strategies, that results in efficient SARS-CoV-2 RNA-seq with high genome coverage and depth. There have been other reports that have successfully used similar targeted enrichment strategies for SARS-CoV-2 (Wen et al., 2020). An advantage of this protocol is that it generates sequence data directly from swab specimens without the need to passage the virus in cell culture, thereby reducing the handling of infectious material and induction of culture-acquired mutations. Another obstacle in sequencing directly from swab specimens is that most FDA-approved commercially available RNA extraction kits are specifically optimized to recover low amounts of total nucleic acids, include carrier polyA RNA that could be convertible into sequence able molecules, as has been observed previously with RNA-seq of Lassa- or Ebola-positive clinical specimens (Matranga et al., 2014).
The workflow incorporates amplification of low-abundance RNA into micrograms of DNA, followed by conversion from a fraction of the DNA into Illumina-compatible sequencing libraries and enrichment of these libraries for SARS-CoV-2 sequences. In addition, during the reverse transcription step, a reagent was incorporated to reduce the subsequent amplification of host ribosomal RNA. This approach is robust, in that it converts low amounts of RNA into microgram quantities of DNA representative of all the RNA species (aside of rRNA) present in the specimen. This DNA can be stored indefinitely to be interrogated by multiple techniques at a later date. Additionally, RNA amplification is likely less sensitive to low viral abundance compared to RT-PCR. Finally, the use of probes to enrich for coronavirus-specific sequencing library molecules is less sensitive to variants compared to tiling PCR amplicon approaches (Briese et al., 2015; O’Flaherty et al., 2018; Paskey et al., 2019; Paden et al., 2020; Xiao et al., 2020).
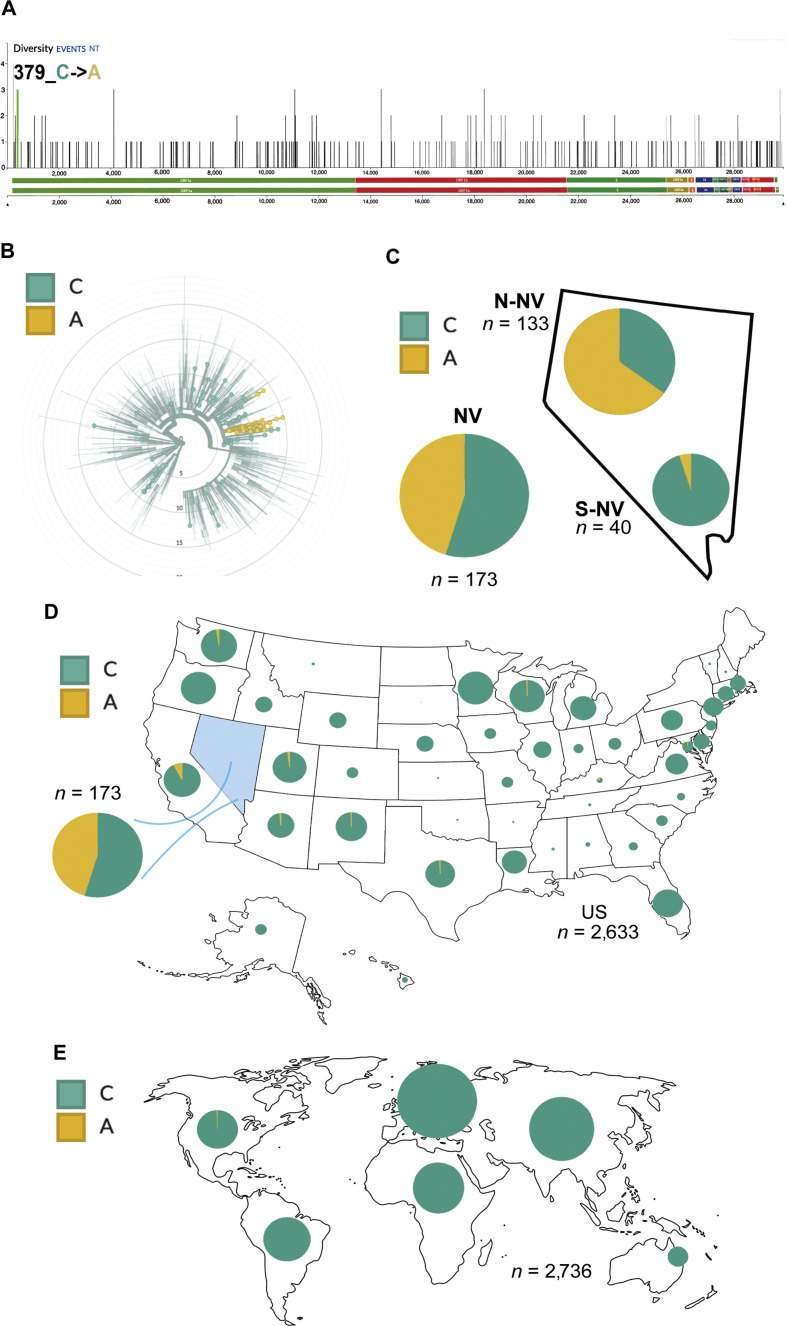
The data herein implicate that early in the pandemic, before the ‘stay-at-home’ order on April 1st, there were multiple introductions of SARS-CoV-2 into the state of Nevada. From April 1st to the beginning of June, Nevada experienced a period of semi-isolation, as the casinos and most hotels shut down, tourism and travel to the state essentially stopped. Because of the stay-at-home order and social distancing measures put in place, there was less mobility of people within and between states. It is possible that these measures, compounded by potential inherent transmission variability of some viral isolates, influenced the change in the frequency of D614G, clades and P323 L/F that we noted during this time period within Nevada. In addition, we also found 379C>A with a high prevalence in our study specimens compared to the subsampling of sequences from the United States and globally (Fig. 6 ). This is a synonymous mutation in nsp1; hence, the biological relevance of this nucleotide variant remains to be elucidated.
Fig. 6.
Distribution of nucleotide variant 379C>A. A: Green line at the far-left end of the genome denotes nucleotide position 379 of nsp1. B: Circular dendrogram of global subsample of sequences from Nextstrain.org with NV specimens indicated by larger dots. C: Pie chart indicating the proportion of sequences with either the cytidine (C) or adenosine (A) at position 379 from the Nevada specimens. Subsamples of sequences from Nextstrain.org are used to generate the proportion of 379C>A in D: the indicated states within the United States and E internationally. The size of the pie chart corresponds to the relative specimen number for each region.
We found the overall trend of D614G in Nevada during this time period to be similar with what was observed in other states and internationally, with the exception of within Asia where the D614 allele was first reported. We noted that there were differences between Northern and Southern Nevada. In Northern Nevada, Clades 20C and F323 were more frequent, while during this same time period, in Southern Nevada, Clades 20A and L323 were more prevalent. These data indicate that there were distinct genomic profiles of the SARS-CoV-2 viruses that were circulating in these populations during the initial months of the pandemic while the stay-at-home order was in place to help prevent transmission of the virus.
As of August 14, 2020, there were only 6 extant SARS-CoV-2 genomes submitted to NCBI, which had the P323F mutation, of the then 14,885 complete or near-complete > 27,000 nt searchable records. By January 9, 2021, the number of SARS-CoV-2 genome submissions with the P323F mutation increased to 24 (accession numbers: MW446823.1, MW403604.1, MW342060.1, MW276486.1, MW276484.1, MW276483.1, MW276482.1, MW245981.1, MW242671.1, MW217332.1, MW206559.1, MT345877.1, MT627429.1, MT706208.1, LR860619.1, MT810889.1, MT811171.1, MW064774.1, MW064823.1, MW067737.1, MW134169.1, MW134350.1, MW134357.1, MW181565.1) out of 47,196 SARS-CoV-2 genomes. This suggests that P323F has not resulted in an explosive growth and global spread seen in other variant lineages, such as B.1.1.7 (Tang et al., 2020; Kirby, 2021). Intriguingly, we had found 62 of the 133 analyzed specimens from Northern Nevada to carry P323F mutation. That is 46% of the specimens from Northern Nevada contained P323F compared to 0.04% of NCBI-deposited SARS-CoV-2 isolates. This was a significant accumulation of one specific SARS-CoV-2 variant in Northern Nevada, which could have been because of the circulation of this unique variant in the community without the introduction of new variants restricted by the shelter in place orders. However, regardless of the confined spread, P323F variant may have altered phenotypic characteristics, which have contributed to its increased prevalence and thus an active area of investigation.
In an attempt to understand the role of this amino acid, our structural modeling of RdRp showed that 323aa is located in the interface domain, which acts as the junction for the interaction of accessory protein (nsp8), required for the polymerase activity (Kirchdoerfer and Ward, 2019). Importantly, P323 of the SARS-CoV-2 (Wuhan-Hu-1) isolate has mutated to leucine (P323L) in all D614 variant of spike glycoprotein, which supposedly has higher transmission (Korber et al., 2020). Although the role of D61G in combination with P323L of RdRp on viral transmission has not been investigated, but the co-existence of these mutational changes (D614G and P323L) in almost all predominantly detected variants of SARS-CoV-2 reflects their importance in transmission and pathogenicity. Consequently, higher prevalence of P323 mutated to 323F (P323F) in variants circulating in the patients of Northern Nevada may suggest the importance of this specific amino acid in virus replication or transmission.
Materials and methods
SARS-CoV-2 specimen and library preparation
Nasal and nasopharyngeal swabs were received at the Nevada State Public Health Lab (NSPHL) or Southern Nevada Public Health Lab (SNPHL) for SARS-CoV-2 diagnostic testing, following a RNA extraction using either a QIAamp Viral RNA Mini Kit (QIAGEN) or Mag-Bind Viral DNA/RNA kit (Omega Biotek). Specimens were tested for the presence of coronaviral RNA using FDA-approved kits that employed RT-PCR to detect SARS-COV-2 RNA. RNA samples from SARS-CoV-2 positive deidentified specimens were used for our studies in accordance with the guidelines of the Institutional Review Board (IRB) of the University of Nevada, Reno, which determined this study to be EXEMPT FROM IRB REVIEW according to federal regulations and university policy.
A set of 200 coronavirus-positive specimens were selected for genome sequencing. Specimens were treated with DNase I (QIAGEN) for 30 min at room temperature and concentrated using RNeasy Minelute spin columns (QIAGEN) based on the manufacturer-supplied protocol. These concentrated samples were converted into Illumina-compatible sequencing libraries with a QIAseq FX Single Cell RNA Library kit (QIAGEN). RNA samples were annealed to a 1:12.5 dilution of QIAseq FastSelect – HMR probes (QIAGEN) to reduce subsequent amplification of human ribosomal RNA. After treatment to remove trace DNA from the samples, a reverse transcription reaction was carried out using random hexamers to generate cDNA. This cDNA was ligated to one another, followed by isothermal linear amplification. Amplified DNA (1 μg) was enzymatically sheared to an average insert size of 300 bp, and Illumina-compatible dual-indexed sequencing adapters were ligated to the ends. Next, about 300 ng of adapter-ligated sample was amplified with 6 cycles of PCR with KAPA HiFi HotStart polymerase (Roche Sequencing Solutions). Enrichment of library molecules containing SARS-CoV-2 sequence was conducted with a myBaits kit and coronavirus-specific biotinylated probes (Arbor Biosciences). Each enrichment used 500 ng of PCR-amplified DNA, was carried out based on manufacturer instructions at a hybridization temperature of 65°C for 16 h, and was completed with 8–16 cycles of PCR using KAPA HiFi HotStart polymerase. Samples were sequenced using an Illumina NextSeq 500 mid-output (2 × 75) kit. The generated FASTQ files were analyzed as described below. The sequences are available at GISAID: hCoV-19/USA/NV-NSPHL-A (0004–0210)/2020.
Computational analysis
Sequence pair libraries were trimmed using Trimmomatic, version 0.39 and adapter-clipping setting ‘2:30:10:2:keepBothReads’ (Bolger et al., 2014). Read pairs were aligned against the SARS-CoV-2 (Wuhan-Hu-1) reference genome (NC_045512.2) by Bowtie 2, version 2.3.5, local alignment (Langmead and Salzberg, 2012). PCR optical duplicates were removed via Picard MarkDuplicates (Picard-Toolkit, 2019).
Variants were called using Freebayes, version 1.0.2, with ploidy set to 1, minimum allele frequency 0.75, and minimum depth of 4 (Garrison and Marth, 2012). No variants were called in the first 200 bp and final 63 bp of the COVID-19 genome. High-quality variant sites were selected where site ‘QUAL >20’ using vcffilter, VCFlib version 1.0.0_rc2 (Garrison, 2019). Individual genomes were reconstructed by their filter-passing variants using bcftools consensus and only where aligned coverage depth ≥ 4; bases with coverage below 4 are reported as unknown (Ns) (Li, 2011).
A set of 3,644 complete, high-coverage SARS-CoV-2 genomes, subsample of data reported in the July 15, 2020, Nextstrain.org global, were used to obtain the metadata from GISAID and combined with our own samples to determine their phylogenetic placement (Elbe and Buckland-Merrett, 2017; Nextstrain, 2020). Four of the global samples were set aside after screening for unexpected FASTA characters. The combined sets of global and Nevadan samples were aligned together, with metadata, by the augur phylodynamic pipelines of the ncov build of the nextstrain command-line tool, version 2.0.0.post1 (Hadfield et al., 2018).
Cumulative frequencies of D614G, clades (19A, 19B, 20A, 20B, 20C), and P323 L/F were calculated at each time point based on the total number of specimens up to the indicated date. Plots and pie charts were generated using GraphPad Prism (version 8).
nsp12 protein modeling
Sequence of nsp12 (RdRp) protein for SARS-CoV-2 (YP_009725307.0) was retrieved from NCBI protein database, and a 3D model was structured based on a previously published report (PDB ID: 6XEZ (Chen et al., 2020). In addition to nsp12 (chain A), the model also contains nsp7 (chain C), nsp8 (Chains B and D), nsp13 (Chains E and F), ligands (Zn2+, Mg2+), and RNA template and product strands. Mutational changes to residue 323 within nsp12 were performed using PyMol Molecular Graphics System (version 2.0, Schrödinger LLC). The original proline (P) was mutated to either leucine (L) or phenylalanine (F) as indicated, and these residues along with residues containing side chains within 5 Å of P323 L/F are shown as sticks. The rotamers for each P323 L/F were assessed, and those with the least rotational strain and steric hindrance were used to generate the final image. To determine any NCBI-deposited sequences, which contain the P323F variant, standard protein BLAST from the BLASTp suite was used to find nsp12 protein sequences, which contained FSTVFPFTSFGP (P323F is bold and underlined) from full length SARS-CoV-2 genomes. The P323F amino acid changes were confirmed with the NCBI-deposited nucleotide sequences.
Ethics approval
Deidentified human specimens (nasal and nasopharyngeal swabs) used for the extraction of viral RNA in all the experiments were done in accordance with guidelines of the University of Nevada, Reno. The University of Nevada, Reno IRB reviewed this project and determined this study to be EXEMPT FROM IRB REVIEW according to federal regulations and university policy. The Environmental and Biological Safety committee of the University of Nevada, Reno, approved methods and techniques used in this study.
Data availability
All sequences hCoV-19/USA/NV-NSPHL-A (0004–0210)/2020 and the reported data are deposited, which is available at GISAID with the following accession numbers:
EPI_ISL_515409, EPI_ISL_515407, EPI_ISL_515408, EPI_ISL_515375, EPI_ISL_515376, EPI_ISL_515373, EPI_ISL_515374, EPI_ISL_515371, EPI_ISL_515372, EPI_ISL_515370, EPI_ISL_515416, EPI_ISL_515417, EPI_ISL_515414, EPI_ISL_515415, EPI_ISL_515379, EPI_ISL_515412, EPI_ISL_515413, EPI_ISL_515377, EPI_ISL_515410, EPI_ISL_515378, EPI_ISL_515411, EPI_ISL_515364, EPI_ISL_515365, EPI_ISL_515362, EPI_ISL_515363, EPI_ISL_515360, EPI_ISL_515361, EPI_ISL_515405, EPI_ISL_515406, EPI_ISL_515403, EPI_ISL_515404, EPI_ISL_515368, EPI_ISL_515401, EPI_ISL_515369, EPI_ISL_515402, EPI_ISL_515366, EPI_ISL_515367, EPI_ISL_515400, EPI_ISL_515353, EPI_ISL_515354, EPI_ISL_515351, EPI_ISL_515352, EPI_ISL_515350, EPI_ISL_515359, EPI_ISL_515357, EPI_ISL_515358, EPI_ISL_515355, EPI_ISL_515356, EPI_ISL_515342, EPI_ISL_515343, EPI_ISL_515340, EPI_ISL_515461, EPI_ISL_515341, EPI_ISL_515462, EPI_ISL_515460, EPI_ISL_515348, EPI_ISL_515349, EPI_ISL_515346, EPI_ISL_515347, EPI_ISL_515344, EPI_ISL_515345, EPI_ISL_515331, EPI_ISL_515298, EPI_ISL_515452, EPI_ISL_515332, EPI_ISL_515299, EPI_ISL_515453, EPI_ISL_515296, EPI_ISL_515450, EPI_ISL_515330, EPI_ISL_515297, EPI_ISL_515451, EPI_ISL_515294, EPI_ISL_515295, EPI_ISL_515293, EPI_ISL_515339, EPI_ISL_515337, EPI_ISL_515458, EPI_ISL_515338, EPI_ISL_515459, EPI_ISL_515335, EPI_ISL_515456, EPI_ISL_515336, EPI_ISL_515457, EPI_ISL_515333, EPI_ISL_515454, EPI_ISL_515334, EPI_ISL_515455, EPI_ISL_515319, EPI_ISL_515320, EPI_ISL_515441, EPI_ISL_515321, EPI_ISL_515442, EPI_ISL_515440, EPI_ISL_515328, EPI_ISL_515449, EPI_ISL_515329, EPI_ISL_515326, EPI_ISL_515447, EPI_ISL_515327, EPI_ISL_515448, EPI_ISL_515324, EPI_ISL_515445, EPI_ISL_515325, EPI_ISL_515446, EPI_ISL_515322, EPI_ISL_515443, EPI_ISL_515323, EPI_ISL_515444, EPI_ISL_515308, EPI_ISL_515429, EPI_ISL_515309, EPI_ISL_515390, EPI_ISL_515397, EPI_ISL_515430, EPI_ISL_515310, EPI_ISL_515398, EPI_ISL_515431, EPI_ISL_515395, EPI_ISL_515396, EPI_ISL_515393, EPI_ISL_515394, EPI_ISL_515391, EPI_ISL_515392, EPI_ISL_515317, EPI_ISL_515438, EPI_ISL_515318, EPI_ISL_515439, EPI_ISL_515315, EPI_ISL_515436, EPI_ISL_515316, EPI_ISL_515437, EPI_ISL_515313, EPI_ISL_515434, EPI_ISL_515314, EPI_ISL_515435, EPI_ISL_515311, EPI_ISL_515399, EPI_ISL_515432, EPI_ISL_515312, EPI_ISL_515433, EPI_ISL_515418, EPI_ISL_515419, EPI_ISL_515386, EPI_ISL_515387, EPI_ISL_515420, EPI_ISL_515384, EPI_ISL_515385, EPI_ISL_515382, EPI_ISL_515383, EPI_ISL_515380, EPI_ISL_515381, EPI_ISL_515306, EPI_ISL_515427, EPI_ISL_515307, EPI_ISL_515428, EPI_ISL_515304, EPI_ISL_515425, EPI_ISL_515305, EPI_ISL_515426, EPI_ISL_515302, EPI_ISL_515423, EPI_ISL_515303, EPI_ISL_515424, EPI_ISL_515300, EPI_ISL_515388, EPI_ISL_515421, EPI_ISL_515301, EPI_ISL_515389, EPI_ISL_515422.
Credit authorship contribution statement
Paul D. Hartley: Conceptualization, Formal analysis, Methodology, Writing - Original draft - Review & Editing. Richard L. Tillett: Conceptualization, Formal analysis, Methodology, Writing - Original draft - Review & Editing. David P. AuCoin: Conceptualization, Review & Editing. Joel R. Sevinsky: Formal analysis, Review & Editing. Yanji Xu: Formal analysis. Andrew Gorzalski: Methodology. Mark Pandori: Conceptualization, Specimen procurement, Diagnostic testing, Formal analysis, Writing - Review & Editing. Erin Buttery: Specimen procurement, Diagnostic testing. Holly Hansen: Specimen procurement, Diagnostic testing. Michael A. Picker: Diagnostic testing, Original draft - Review & Editing. Cyprian C. Rossetto: Conceptualization, Formal analysis, Methodology, Writing - Original draft - Review & Editing. Subhash C. Verma: Conceptualization, Formal analysis, Methodology, Project administration, Writing - Review & Editing.
Conflict of Interest
The authors declare that they have no competing interests with the contents of this article.
Acknowledgments
We thank the staff at the Nevada State Public Health Lab (NSPHL) and Southern Nevada, Public Health Laboratory (SNPHL) for providing the RNA extracts from the patient’s specimen. This work was supported by University of Nevada, Reno, Vice President for Research and Innovation (VPRI), Department of Microbiology & Immunology, UNR School of Medicine, Nevada IDeA Network of Biomedical Research Excellence (INBRE) from the National Institute of General Medical Sciences (GM 103440 and GM 104944) and from the National Institutes of Health. The authors wish to acknowledge the support of research and innovation and the Office of Information Technology at the University of Nevada, Reno, for computing time on the Pronghorn High-Performance Computing Cluster.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgg.2021.01.004.
Supplementary data
The following are the supplementary data to this article:
References
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese T., Kapoor A., Mishra N., Jain K., Kumar A., Jabado O.J., Lipkin W.I. Virome capture sequencing enables sensitive viral diagnosis and comprehensive virome analysis. mBio. 2015;6 doi: 10.1128/mBio.01491-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T. Structural basis for helicase-polymerase coupling in the SARS-cov-2 replication-transcription complex. Cell. 2020;182 doi: 10.1016/j.cell.2020.07.033. 1560–1573.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of V The species severe acute respiratory syndrome-related coronavirus: classifying 2019-ncov and naming it SARS-cov-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson A.D., Williamson M.K., Lewis S., Shoemark D., Carroll M.W., Heesom K.J., Zambon M., Ellis J., Lewis P.A., Hiscox J.A. Characterisation of the transcriptome and proteome of SARS-cov-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020;12:68. doi: 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E. Vcflib: a C++ library for parsing and manipulating VCF files. 2019. https://githubcom/vcflib/vcflib
- Garrison E., Marth G. Haplotype-based variant detection from short-read sequencing. Arxiv [q-biogn] 2012 http://arxiv.org/abs/1207.3907 Available. [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland L.A., Kaelin E.A., Maqsood R., Estifanos B., Wu L.I., Varsani A., Halden R.U., Hogue B.G., Scotch M., Lim E.S. An 81-nucleotide deletion in SARS-cov-2 ORF7a identified from sentinel surveillance in Arizona (january to March 2020) J. Virol. 2020;94 doi: 10.1128/JVI.00711-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D., Zhu C., Ai L., He T., Wang Y., Ye F., Yang L., Ding C., Zhu X., Lv R. Genomic characterization and infectivity of a novel SARS-like coronavirus in Chinese bats. Emerg. Microb. Infect. 2018;7:154. doi: 10.1038/s41426-018-0155-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaafar R., Aherfi S., Wurtz N., Grimaldier C., Hoang V.T., Colson P., Raoult D., La Scola B. Correlation between 3790 qpcr positives samples and positive cell cultures including 1941 SARS-cov-2 isolates. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1491. [DOI] [Google Scholar]
- Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-cov-2 transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby T. New variant of SARS-cov-2 in UK causes surge of COVID-19. Lancet Respir Med. 2021;9:e20–e21. doi: 10.1016/S2213-2600(21)00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Ward A.B. Structure of the SARS-cov nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B. Tracking changes in SARS-cov-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licastro D., Rajasekharan S., Dal Monego S., Segat L., D’Agaro P., Marcello A. Isolation and full-length genome characterization of SARS-cov-2 from COVID-19 cases in northern Italy. J. Virol. 2020;94 doi: 10.1128/JVI.00543-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga C.B., Andersen K.G., Winnicki S., Busby M., Gladden A.D., Tewhey R., Stremlau M., Berlin A., Gire S.K., England E. Enhanced methods for unbiased deep sequencing of Lassa and Ebola RNA viruses from clinical and biological samples. Genome Biol. 2014;15:519. doi: 10.1186/s13059-014-0519-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nextclade 2020. https://cladesnextstrainorg
- Nextstrain . 2020. Genomic epidemiology of novel coronavirus - global subsampling.https://nextstrain.org/ncov/global/2020-07-15?D=tree&l=clock&legend=closed [Google Scholar]
- O’Flaherty B.M., Li Y., Tao Y., Paden C.R., Queen K., Zhang J., Dinwiddie D.L., Gross S.M., Schroth G.P., Tong S. Comprehensive viral enrichment enables sensitive respiratory virus genomic identification and analysis by next generation sequencing. Genome Res. 2018;28:869–877. doi: 10.1101/gr.226316.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paden C.R., Tao Y., Queen K., Zhang J., Li Y., Uehara A., Tong S. Rapid, sensitive, full-genome sequencing of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26 doi: 10.3201/eid2610.201800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paskey A.C., Frey K.G., Schroth G., Gross S., Hamilton T., Bishop-Lilly K.A. Enrichment post-library preparation enhances the sensitivity of high-throughput sequencing-based detection and characterization of viruses from complex samples. BMC Genom. 2019;20:155. doi: 10.1186/s12864-019-5543-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosillo N., Viceconte G., Ergonul O., Ippolito G., Petersen E. COVID-19, SARS and MERS: are they closely related? Clin. Microbiol. Infect. 2020;26:729–734. doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard-Toolkit . 2019. http://broadinstitute.github.io/picard [Google Scholar]
- Tang J.W., Tambyah P.A., Hui D.S. Emergence of a new SARS-cov-2 variant in the UK. J. Infect. 2020;82:e27–e28. doi: 10.1016/j.jinf.2020.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., mcguire A.T., Veesler D. Structure, function, and antigenicity of the SARS-cov-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen S., Sun C., Zheng H., Wang L., Zhang H., Zou L., Liu Z., Du P., Xu X., Liang L. High-coverage SARS-cov-2 genome sequences acquired by target capture sequencing. J. Med. Virol. 2020;92:2221–2226. doi: 10.1002/jmv.26116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M., Liu X., Ji J., Li M., Li J., Yang L., Sun W., Ren P., Yang G., Zhao J. Multiple approaches for massively parallel sequencing of SARS-cov-2 genomes directly from clinical samples. Genome Med. 2020;12:57. doi: 10.1186/s13073-020-00751-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhong N.S., Zheng B.J., Li Y.M., Poon, Xie Z.H., Chan K.H., Li P.H., Tan S.Y., Chang Q., Xie J.P. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences hCoV-19/USA/NV-NSPHL-A (0004–0210)/2020 and the reported data are deposited, which is available at GISAID with the following accession numbers:
EPI_ISL_515409, EPI_ISL_515407, EPI_ISL_515408, EPI_ISL_515375, EPI_ISL_515376, EPI_ISL_515373, EPI_ISL_515374, EPI_ISL_515371, EPI_ISL_515372, EPI_ISL_515370, EPI_ISL_515416, EPI_ISL_515417, EPI_ISL_515414, EPI_ISL_515415, EPI_ISL_515379, EPI_ISL_515412, EPI_ISL_515413, EPI_ISL_515377, EPI_ISL_515410, EPI_ISL_515378, EPI_ISL_515411, EPI_ISL_515364, EPI_ISL_515365, EPI_ISL_515362, EPI_ISL_515363, EPI_ISL_515360, EPI_ISL_515361, EPI_ISL_515405, EPI_ISL_515406, EPI_ISL_515403, EPI_ISL_515404, EPI_ISL_515368, EPI_ISL_515401, EPI_ISL_515369, EPI_ISL_515402, EPI_ISL_515366, EPI_ISL_515367, EPI_ISL_515400, EPI_ISL_515353, EPI_ISL_515354, EPI_ISL_515351, EPI_ISL_515352, EPI_ISL_515350, EPI_ISL_515359, EPI_ISL_515357, EPI_ISL_515358, EPI_ISL_515355, EPI_ISL_515356, EPI_ISL_515342, EPI_ISL_515343, EPI_ISL_515340, EPI_ISL_515461, EPI_ISL_515341, EPI_ISL_515462, EPI_ISL_515460, EPI_ISL_515348, EPI_ISL_515349, EPI_ISL_515346, EPI_ISL_515347, EPI_ISL_515344, EPI_ISL_515345, EPI_ISL_515331, EPI_ISL_515298, EPI_ISL_515452, EPI_ISL_515332, EPI_ISL_515299, EPI_ISL_515453, EPI_ISL_515296, EPI_ISL_515450, EPI_ISL_515330, EPI_ISL_515297, EPI_ISL_515451, EPI_ISL_515294, EPI_ISL_515295, EPI_ISL_515293, EPI_ISL_515339, EPI_ISL_515337, EPI_ISL_515458, EPI_ISL_515338, EPI_ISL_515459, EPI_ISL_515335, EPI_ISL_515456, EPI_ISL_515336, EPI_ISL_515457, EPI_ISL_515333, EPI_ISL_515454, EPI_ISL_515334, EPI_ISL_515455, EPI_ISL_515319, EPI_ISL_515320, EPI_ISL_515441, EPI_ISL_515321, EPI_ISL_515442, EPI_ISL_515440, EPI_ISL_515328, EPI_ISL_515449, EPI_ISL_515329, EPI_ISL_515326, EPI_ISL_515447, EPI_ISL_515327, EPI_ISL_515448, EPI_ISL_515324, EPI_ISL_515445, EPI_ISL_515325, EPI_ISL_515446, EPI_ISL_515322, EPI_ISL_515443, EPI_ISL_515323, EPI_ISL_515444, EPI_ISL_515308, EPI_ISL_515429, EPI_ISL_515309, EPI_ISL_515390, EPI_ISL_515397, EPI_ISL_515430, EPI_ISL_515310, EPI_ISL_515398, EPI_ISL_515431, EPI_ISL_515395, EPI_ISL_515396, EPI_ISL_515393, EPI_ISL_515394, EPI_ISL_515391, EPI_ISL_515392, EPI_ISL_515317, EPI_ISL_515438, EPI_ISL_515318, EPI_ISL_515439, EPI_ISL_515315, EPI_ISL_515436, EPI_ISL_515316, EPI_ISL_515437, EPI_ISL_515313, EPI_ISL_515434, EPI_ISL_515314, EPI_ISL_515435, EPI_ISL_515311, EPI_ISL_515399, EPI_ISL_515432, EPI_ISL_515312, EPI_ISL_515433, EPI_ISL_515418, EPI_ISL_515419, EPI_ISL_515386, EPI_ISL_515387, EPI_ISL_515420, EPI_ISL_515384, EPI_ISL_515385, EPI_ISL_515382, EPI_ISL_515383, EPI_ISL_515380, EPI_ISL_515381, EPI_ISL_515306, EPI_ISL_515427, EPI_ISL_515307, EPI_ISL_515428, EPI_ISL_515304, EPI_ISL_515425, EPI_ISL_515305, EPI_ISL_515426, EPI_ISL_515302, EPI_ISL_515423, EPI_ISL_515303, EPI_ISL_515424, EPI_ISL_515300, EPI_ISL_515388, EPI_ISL_515421, EPI_ISL_515301, EPI_ISL_515389, EPI_ISL_515422.