SUMMARY
Macrophages reprogram their lipid metabolism in response to activation signals. However, a systems-level understanding of how different pro-inflammatory stimuli reshape the macrophage lipidome is lacking. Here, we use complementary “shotgun” and isotope tracer mass spectrometry approaches to define the changes in lipid biosynthesis, import, and composition of macrophages induced by various Toll-like receptors (TLRs) and inflammatory cytokines. “Shotgun” lipidomics data revealed that different TLRs and cytokines induce macrophages to acquire distinct lipidomes, indicating their specificity in reshaping lipid composition. Mechanistic studies showed that differential reprogramming of lipid composition is mediated by the opposing effects of MyD88- and TRIF-interferon-signaling pathways. Finally, we applied these insights to show that perturbing reprogramming of lipid composition can enhance inflammation and promote host defense to bacterial challenge. These studies provide a framework for understanding how inflammatory stimuli reprogram lipid composition of macrophages while providing a knowledge platform to exploit differential lipidomics to influence immunity.
In Brief
Using a combination of shotgun lipidomics and stable-isotope tracing, Hsieh et al. show that distinct pro-inflammatory stimuli reshape the macrophage lipid composition in a signal-specific manner and that targeting this change can increase immunity. Thus, the study provides an in-depth resource and framework for understanding this lipidomic response while suggesting approaches for future therapy.
Graphical Abstract
INTRODUCTION
Lipids are essential components of all cells, ensuring proper energetic, biochemical, and biophysical properties. Macrophages are a key component of the innate immune system that are critical for the clearance of infectious agents and cellular debris, generation of pro- and anti-inflammatory programs, and initiation of wound healing (Gordon and Taylor, 2005; Lawrence and Natoli, 2011; Okabe and Medzhitov, 2016). Activation of macrophages by pattern recognition receptors (PRRs; e.g., Toll-like receptors [TLRs]) or cytokines have been shown to rapidly induce changes to their lipid metabolic and energetic programs (Blanc et al., 2011; Liu et al., 2013; Oishi et al., 2017; York et al., 2015), leading to a model where reprogramming of lipid homeostasis is required to ensure proper macrophage inflammatory function. Consistent with this concept, genetic or pharmacologic interference of TLR3- or TLR4-meditated lipid metabolic reprogramming affect the ability of macrophages to mediate inflammatory responses and host defense to pathogens (Blanc et al., 2011; Oishi et al., 2017; York et al., 2015). Despite the importance of lipid metabolic reprogramming for macrophage function, there remains significant gap in our knowledge of the signaling and enzymatic pathways that coordinate reshaping of macrophage lipid composition with different pro-inflammatory signals.
To gain further insights as to how distinct pro-inflammatory signals regulate reprogramming of the lipid composition of macrophages, we took a systems-level approach using a combination of direct infusion “shotgun” lipidomics and stable-isotope labeling mass spectrometry (MS) on macrophages stimulated with a panel of TLR ligands. Informatics analysis of lipidomics datasets revealed that activation of macrophages with all pro-inflammatory stimuli induce significant remodeling of the macrophage lipidome. However, we found that changes to lipid composition occur in a TLR-specific manner. Mechanistic studies indicated that TLR-mediated reprogramming of the lipidome occurs, in large part, by the opposing influences of the Myeloid differentiation primary response protein 88 (MyD88)- and TIR-domain-containing adapter-inducing interferon-β (TRIF)-interferon-signaling pathways. We show that MyD88-dependent TLRs reprogram lipid composition through nuclear factor-E2-related factor 2 (NRF2) and sterol regulatory element binding proteins (SREBP)-transcriptional axes, but find no evidence for IRG1 signaling in this process. We also demonstrate that TRIF-dependent PRR signaling requires autocrine production of type I IFNs, but IFNγ signaling in trans can replace this requirement. Finally, we apply the knowledge gained from these MS studies to show that perturbing the ability of macrophages to reshape lipid composition in response to MyD88-dependent TLRs alters inflammatory responses and facilitates protection from Staphylococcus aureus challenge in vivo. Taken together, these studies provide a conceptual framework for understanding how distinct pro-inflammatory signals reshape macrophage lipid composition, and provide the proof-of-concept evidence that manipulating the ability of macrophages to adopt their preferred lipidomic state can be exploited to enhance host defense against bacterial challenge.
RESULTS
Distinct Reprogramming of the Macrophage Lipidome by Different TLRs
Activation of macrophages with TLR agonists (e.g., Poly[I:C], or LPS, or cytokines [e.g., IFNγ or IFNβ]) have been shown to reprogram components of lipid homeostasis within these cells (Araldi et al., 2017; Blanc et al., 2013; Liu et al., 2013; Oishi et al., 2017). However, a systems-level understanding of the influence that different pro-inflammatory stimuli have on reshaping the macrophage lipidome is lacking. To begin to address this gap in knowledge, C57BL/6 bone marrow-derived macrophages (BMDMs) were stimulated with TLR1/2, TLR3, TLR4, TLR7, or TLR9 agonists (Pam3CSK4, Poly(I:C), LPS, CL307, and ODN1668, respectively). Stimulation with a TLR5 agonist did not sufficiently activate BMDMs for inclusion in this study. Lipids were extracted at 24 and 48-h time points after activation, and then subjected to direct infusion “shotgun” MS. In these studies, we were able to quantify the pool size of approximately 750 individual lipid species in the macrophage lipidome, representing the glycerophospholipid (e.g., phospholipids and lysophospholipid species), glycerolipid (e.g., di-acyl and tri-acylglycerides), sphingolipid (e.g., sphingomyelins and ceramides), and cholesterol and fatty acids classes (Fahy et al., 2009; Liebisch et al., 2013).
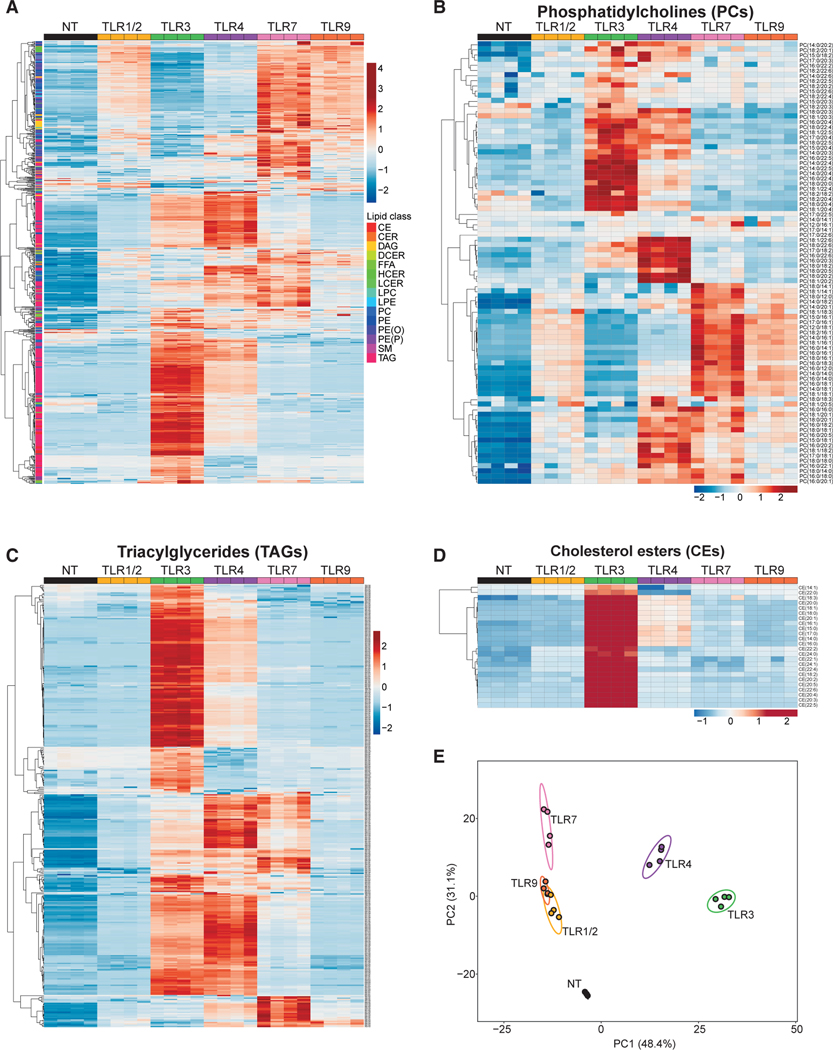
Analysis of the total pool size for each lipid class or subclass revealed that TLR-mediated activation of macrophages largely increased lipid content (Figure S1A). Cluster analysis of individual lipid species demonstrated that activation of TLRs resulted in marked changes to lipid composition at the 24 and 48-h time points in a TLR-dependent manner (Figures 1A and S1). Inspection of individual lipid (sub)classes revealed additional layers of complexity in the types of changes to the lipid composition induced by different TLRs. For example, examination of phosphatidylcholines (PCs), an abundant subclass of lipids in the macrophage lipidome, showed that TLR1/2, TLR7, or TLR9 activation resulted in the accumulation of largely overlapping PC species, whereas TLR3 and TLR4 activation resulted in the accumulation of PC species that were distinct from that of TLR1/2, TLR7, or TLR9 stimulation (Figure 1B). Inspection of the neutral lipids triacylglycerides (TAGs) species revealed a different pattern of remodeling. Activation of macrophages with TLR3, TLR4, or TLR7 agonists resulted in the accumulation of TAGs, although TLR7 signaling induced a restricted number of TAG species when compared with TLR4 and TLR3 (Figure 1C). In contrast, TLR1/2- and TLR9-activated macrophages did not accumulate TAGs, and were indistinguishable from unstimulated macrophages for this lipid class (Figure 1C). In other instances, we observed that activation of a specific TLR (e.g., TLR3) resulted in the marked accumulation of a single subclass of lipid species (e.g., cholesterol esters) (Figure 1D). This differential pattern of changes in macrophage lipid composition could be observed across nearly all lipid (sub)classes measured (Figures S1B–S1K).
Figure 1. Distinct Reprogramming of the Macrophage Lipidome by Different TLRs.
(A) Heat map of all individual lipids quantified by direct infusion MS analysis of quiescent BMDMs (indicated as NT) or BMDMs stimulated with TLR1/2, TLR3, TLR4, TLR7, or TLR9 agonists for 48 h.
(B) Heat map of individual PC species measured by direct infusion MS from BMDMs stimulated as above.
(C) Heat map of individual TAG species measured by direct infusion MS from BMDMs stimulated as above.
(D) Heat map of individual CE species measured by direct infusion MS from BMDMs stimulated as above.
(E) PCA of individual lipids quantified by MS from BMDMs stimulated as above. Percentage of total variance explained by individual principal components indicated in axis. Prediction ellipses are set at 95% probability.
All MS experiments and analysis shown are from four biologic replicates per experimental condition. Heatmap scales are Z score for number of deviations away from the row mean. Rows are clustered using correlation distance and complete linkage. Experiments shown are representative of more than three independent studies.
We applied principal component analysis (PCA) to the lipidomics data to better define the lipids contributing to the variance observed among different TLR stimulations. Principal component 1 (PC1) accounted for approximately 48% of the total variance in lipidomes for both the 24 and 48-h time points, and largely reflected changes to the lipidome driven by TLR3 signaling (Figure 1E). Principal component 2 (PC2) delineated approximately 26%–31% of the total variance, and reflected changes to the lipidome induced by TLR1/2, TLR7, or TLR9 signals (Figures 1E and S1M). TLR4-stimulated macrophages occupied a distinct space, resembling an admixture of PC1 and PC2 eigenvectors (Figure 1E). PC3 and PC4 contributed less than 9.0% and 4.0%, respectively, to the total variance, indicating that PC1 and PC2 captured major differences in the lipidome induced by TLR activation (Figure 1E). PCA of four independent lipidomic datasets from TLR-activated macrophages demonstrated a high degree of reproducibility in the differential reprogramming of the lipidome (Figure S1N). Taken together, these data reveal that activation of macrophages by various TLRs reprogram lipid composition of macrophages in distinct but partially overlapping manners.
Lipid Loading Perturbs TLR-Mediated Reprogramming of the Lipidome
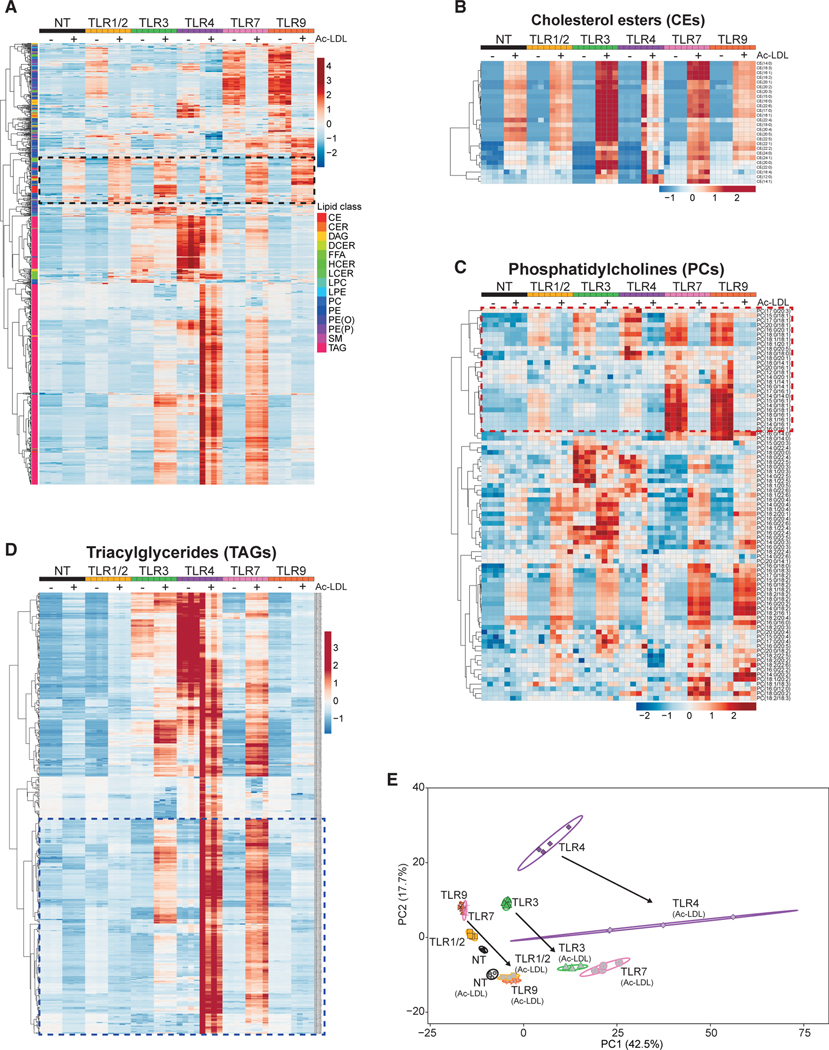
Macrophages play a key role in clearing excess lipids from tissues under normal and pathologic conditions (Moore et al., 2013). Thus, we asked if lipid loading of macrophages would interfere with TLR-mediated reprogramming of the lipidome. To address this, acetylated-LDL (Ac-LDL) was added to wildtype (WT) BMDMs cultures for 2 h before stimulation with TLR1/2, TLR3, TLR4, TLR7, or TLR9 agonists. After 48 h of stimulation, lipids were extracted and then subjected to shotgun MS. Cluster analysis of individual lipid species revealed a distinct group of lipids that accumulated in macrophages as a consequence of Ac-LDL loading, irrespective of stimulation (indicated by the box with the black dashed outline in Figures 2A and S2A). Not unexpectedly, these species were dominated not only by cholesterol esters (Figure 2B), but also included a select number of other lipids, such as PCs (Figure 2C), DAGs, PEs, SMs, and FFAs (Figure S2).
Figure 2. Lipid Loading Perturbs TLR-mediated Reprogramming of the Lipidome.
(A) Heat map of all individual lipids quantified by direct infusion MS analysis of BMDMs pretreated with acetylated-LDL or stimulated with TLR1/2, TLR3, TLR4, TLR7, or TLR9 agonists for 48 h. Black dashed box denotes changes induced by Ac-LDL loading.
(B) Heat map of individual CE species measured by direct infusion MS from BMDMs stimulated as above.
(C) Heat m ap of individual PC species measured by direct infusion MS from BMDMs stimulated as above. Red dashed box denotes changes induced by Ac-LDL loading.
(D) Heat map of individual TAG species measured by direct infusion MS from BMDMs stimulated as above. Blue dashed box denotes changes induced by Ac-LDL loading.
(E) PCA of individual lipids quantified by MS from BMDMs stimulated as above. Percentage of total variance explained by individual principal components indicated in axis. Prediction ellipses are set at 95% probability. Arrows indicate the shift in PCA in response to Ac-LDL loading.
All MS experiments and analysis are from four biologic replicates per experimental condition. Heatmap scales are Z score for number of deviations away from the row mean. Rows are clustered using correlation distance and complete linkage.
Importantly, we observed that Ac-LDL loading markedly altered TLR-driven remodeling of lipid composition (Figures 2A–2D and S2). For example, Ac-LDL treatment inhibited the accumulation of phospholipid species that contained saturated and monounsaturated fatty acyl tails (Figure 2C, denoted by the box with the red dashed outline, and Figures S2D–S2F). We also found that TLR3, TLR4, or TLR7 activation of Ac-LDL-loaded macrophages resulted in the accumulation of a large number of distinct TAG lipid species (denoted by the box with the blue dashed outline in Figure 2D). Accumulation of TAG species was not observed in TLR1/2 or TLR9-activated macrophages despite lipid loading (Figure 2D), reinforcing the concept that TAG accumulation is a distinct and tightly regulated program in response to some, but not all pro-inflammatory signals. PCA and cluster analysis further highlighted how lipid loading of macrophages altered reprogramming of lipid composition in response to inflammatory signals (Figures 2E, see arrows; and S2). Taken together, these data show that lipid loading of macrophages does perturb TLR-induced reprogramming of the lipidome, and underscore the concept that the basal metabolic state of a macrophage will be an important determinant of how macrophages reshape their lipid composition in response to activation signals.
MyD88 and TRIF-IFN Signaling Drive Distinct Aspects of Lipidomic Reprogramming
MyD88 and TRIF adaptor proteins are essential for acquisition of pro-inflammatory immune effector functions (Takeda and Akira, 2004). MyD88 is required for TLR1/2, TLR5, TLR7, and TLR9 signaling, whereas TRIF is used by TLR3. TLR4 engages both the MyD88 and TRIF signaling pathways. Thus, we asked if the selective loss of these pathways would influence reprogramming of the lipid composition in response to TLR-mediated activation.
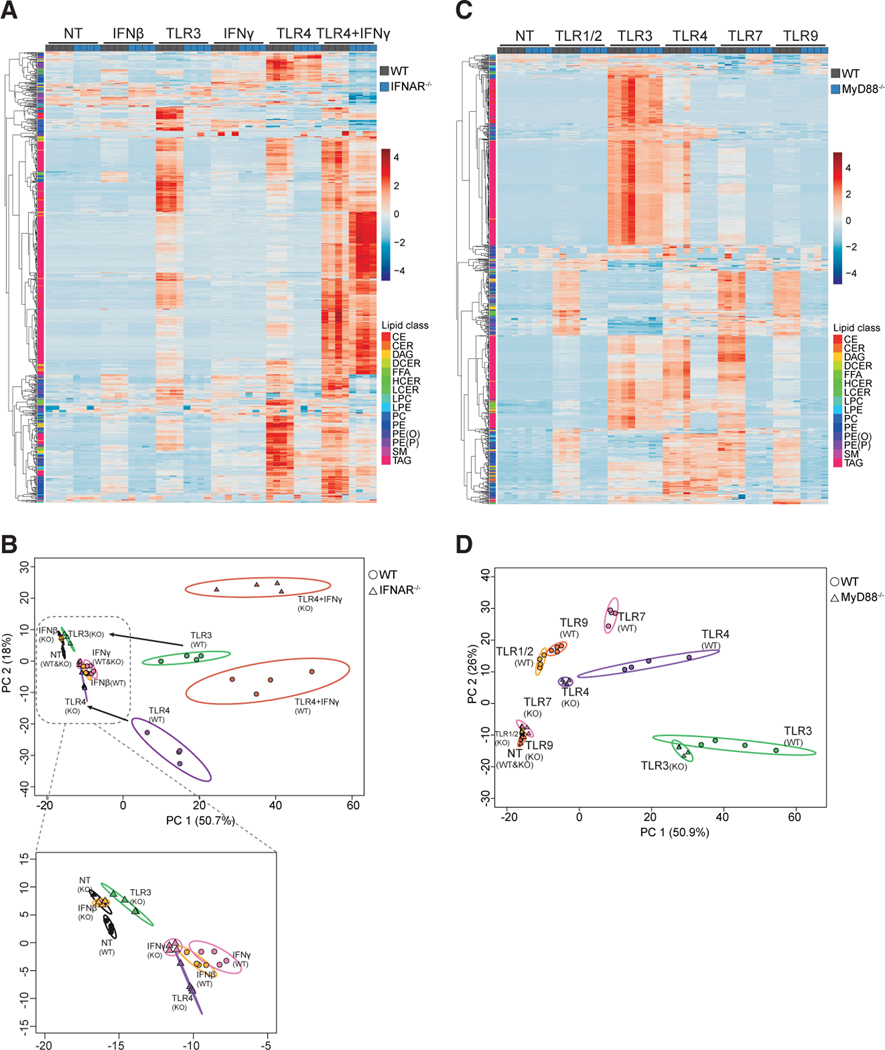
To address this question, we stimulated Ticam1Lps2 (TRIF-deficient) and control BMDMs with TLR1/2, TLR3, or TLR4 agonists (representative of MyD88- and TRIF-dependent TLRs) for 48 h, followed by shotgun lipidomics. No differences in lipid composition of quiescent (unstimulated) TRIF-deficient and control macrophages were observed (Figures S3A and S3B). However, loss of TRIF impacted both TLR3- and TLR4-mediated reprogramming of the lipidome (Figures S3A and S3B). As expected, loss of TRIF did not influence TLR1/2-induced changes to the lipidome (Figures S3A and S3B). PCA revealed that loss of TRIF shifted the lipidome of TLR3- or TLR4-activated macrophages to more closely resemble that of their TLR1/2-activated counterparts (Figure S3B; see arrows). A key effector function of TLR3- and TLR4-mediated activation is the generation of type I IFNs (Yamamoto et al., 2003), leading us to ask if the changes to the lipidome observed in response to TLR3 or TLR4 stimulation is dependent on autocrine production of type I IFNs. To that end, control and type I IFN receptor-deficient (IFNAR−/−) BMDMs were stimulated with TLR3 or TLR4 agonists, followed by shotgun lipidomics. Deletion of IFNAR abrogated the ability of TLR3 signaling to drive lipid reprogramming, largely phenocopying the results observed in TRIF-deficient macrophages (Figures 3A and 3B; see arrows in PCA plot). Analysis of TLR4-stimulated macrophages also showed that IFNAR signaling is required for a component of reprogramming. However, we still observed marked changes to the lipid composition in the absence of IFNAR, presumably as a function of TLR4-mediated MyD88 signaling (Figures 3A and 3B). The addition of IFNβ or IFNγ alone to cultures also induced changes to lipid composition, although these changes were modest compared with that of TLR3 or TLR4 stimulation (Figures 3A, 3B, and S3C–S3O). However, loss of IFNAR blocked IFNγ-mediated changes for some lipid species (for example TAGs; Figure S3O). The combination of IFNγ and TLR4 agonist further enhanced reprogramming of the lipidome, predominated by the accumulation of the neutral lipids cholesterol esters (CEs) and TAGs (Figures 3A, S3J, and S3O). The addition of exogenous IFNγ to cultures was sufficient to overcome the loss of IFNAR in TLR4-stimulated macrophages for most lipid species (Figures 3A and 3B). This observation demonstrates a largely overlapping role for both type I and II IFNs in reprogramming lipid composition in the context of TLR signaling. Together, these studies support a model where IFN-mediated inflammation is required to drive remodeling of the lipidome in response to TLR3 or TLR4 through autocrine or paracrine sources.
Figure 3. Type I IFN and MyD88 Signaling Are Required to Drive Distinct Aspects of Lipidomic Reprogramming.
(A) Heat map of all individual lipids quantified by direct infusion MS analysis of WT control or IFNAR−/− BMDMs stimulated with TLR3, TLR4 agonists, or IFNβ, IFNγ, or IFNγ + TLR4 agonist for 48 h.
(B) PCA of individual lipids quantified by MS from WT control or IFNAR−/− BMDMs (designated as O or Δ, respectively) stimulated with indicated TLR agonists and/or cytokine for 48 h. Arrows indicate the shift of IFNAR−/− samples from controls in PCA stimulated with indicated TLR agonist. Inset of PCA provided to show the small changes induced by cytokines or TLR3 agonist. Percentage of total variance explained by individual principal components indicated in axis. Prediction ellipses are set at 95% probability.
(C) Heat map of all individual lipids quantified by direct infusion (“shotgun”) MS analysis of WT control or MyD88−/− BMDMs stimulated with TLR1/2, TLR3, TLR4, TLR7, or TLR9 agonists for 48 h.
(D) PCA of individual lipids quantified by MS from WT control or MyD88−/− BMDMs (designated as O or Δ, respectively) stimulated with TLR agonists as indicated for 48 h. Arrows indicate the shift of MyD88−/− samples from WT controls in PCA stimulated with indicated TLR agonist. Percentage of total variance explained by individual principal components indicated in axis. Prediction ellipses are set at 95% probability.
All MS experiments and analysis are from four independent biologic replicates per experimental condition. Heatmap scales are Z score for number of deviations away from the row mean. Rows are clustered using correlation distance and complete linkage.
Next, we determined the contribution of MyD88 to reprogramming of the lipidome in response to TLR activation. Thus, we stimulated MyD88-deficient (MyD88−/−) or control macrophages with various TLR agonists. No differences in the lipidome of unstimulated control and MyD88−/−macrophages were observed (Figures 3C and 3D). However, loss of MyD88 abrogated the ability of TLR1/2, TLR7, and TLR9 to reprogram the lipidome (Figures 3C, 3D, and S4A–S4M). MyD88 deficiency had little impact on changes to the lipidome induced by TLR3 signaling (Figures 3C and 3D). Given that TLR4 signals through both TRIF and MyD88, we predicted that loss of MyD88 in TLR4-activated macrophages would result in a lipidome that resembled that of TLR3-stimulated macrophages. However, clustering and PCA of TLR4-stimulated macrophages revealed that loss of MyD88 results in a lipidome that more closely resembles suboptimal stimulation, rather than changes induced by unopposed TRIF/IFNAR signaling (Figures 3D and S4A–S4M). Together, these data support a model where MyD88 and TRIF signaling are required to drive distinct remodeling programs, and that IFNs can overwrite the changes to the lipidome induced by MyD88.
TLR Signals Promote Divergent Fatty Acid Synthetic Programs
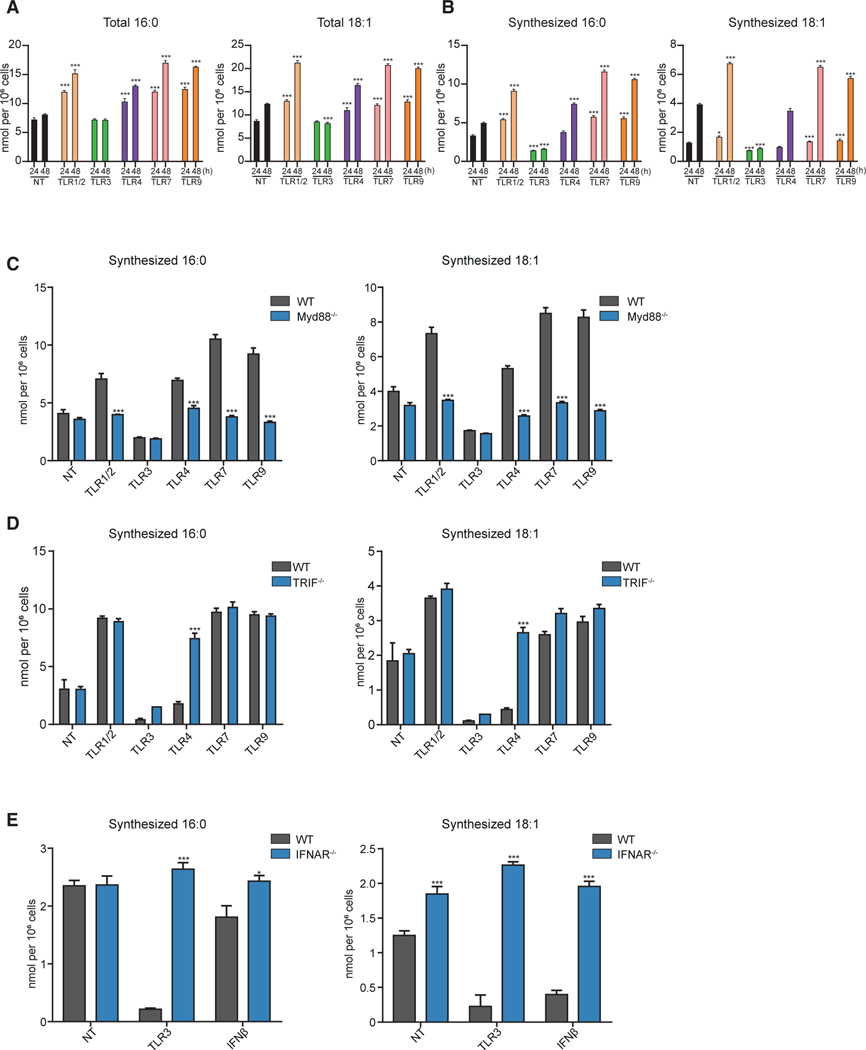
Long chain fatty acids (LCFAs) are a major component of the complex lipids in the lipidome. LCFAs are synthesized de novo or imported from extracellular sources (Argus et al., 2018), leading us to ask if TLR signals differentially reprogram the source of fatty acids used to remodel the macrophage lipidome. To answer this question, we stimulated BMDMs with TLR agonists in complete media containing [U-13C]glucose for up to 48 h, followed by lipid extraction, derivatization, and GC/MS. Total and isotope labeled LCFA pools (e.g., 16:0, 16:1, 18:0, 18:1) were determined, and the contribution of synthesis to the total LCFA pool was modeled using isotopic spectral analysis (ISA) (Argus et al., 2018; Kelleher and Nickol, 2015). MS data showed that activation of macrophages with all TLRs maintained or increased the total pools of the saturated LCFAs (termed SFAs hereafter), and monounsaturated LCFAs (termed MUFAs) (Figures 4A and S5A). As expected, TLR3 signaling decreased de novo synthesis of SFAs and MUFAs (Figures 4B and S5B) (Argus et al., 2018) confirming that the increase in LCFAs during TLR3-induced reprogramming is derived from extracellular sources. In contrast, activation of MyD88-dependent TLRs (i.e., TLR1/2, TLR7, or TLR9) increased de novo synthesis of SFA and MUFAs (Figures 4B and S5B). TLR4 activation resulted in a more complex phenotype, with an increase in the synthesized pool for saturated fatty acid species observed (e.g., 16:0), while little or no changes in the pool of synthesized 16:1, 18:1, and 18:0 observed (Figures 4B and S5B). Changes in lipid synthesis were mirrored by changes in lipid synthesis gene expression (Figure S5C), indicating that the observed changes in lipid composition are, in part, transcriptionally programed in response to different TLR signals.
Figure 4. TLRs Promote Divergent Fatty Acid Synthetic Programs.
(A) Total palmitic acid (16:0) and oleic acid (18:1) from quiescent BMDMs (indicated as NT) and BMDMs stimulated with TLR1/2, TLR3, TLR4, TLR7, or TLR9 agonists for either 24 or 48 h.
(B) Net synthesized palmitic acid (16:0) and oleic acid (18:1) quiescent BMDMs (NT) or BMDMs stimulated with TLR1/2, TLR3, TLR4, TLR7, or TLR9 agonists for either 24 or 48 h.
(C) Net synthesized palmitic acid (16:0) and oleic acid (18:1) from quiescent control or MyD88−/− BMDMs and BMDMs stimulated with TLR1/2, TLR3, TLR4, TLR7, or TLR9 agonists for 48 h.
(D) Net synthesized of palmitic acid (16:0) and oleic acid (18:1) from quiescent WT control or TRIF−/− BMDMs, and stimulated with TLR1/2, TLR3, TLR4, TLR7, or TLR9 agonists for 48 h.
(E) Net synthesized palmitic acid (16:0) and oleic acid (18:1) in control or IFNAR−/− BMDMs stimulated with TLR3 agonist or IFNβ for 48 h.
All isotope labeling experiments are from four biologic replicates per experimental condition. All data are presented as mean ± SEM. *p < 0.05; **p < 0.01, ***p < 0.001. Experiments shown are representative of more than three independent studies.
MyD88 and TRIF-IFNAR Drive Opposing Fatty Acid Synthesis Programs
Next, we assessed the importance of TRIF, IFNAR, and MyD88 on LCFA synthesis in response to TLR agonists. In line with our shotgun lipidomics studies, we observed that loss of MyD88 abrogated TLR1/2, TLR4, TLR7, or TLR9-induced upregulation of de novo LCFAs synthesis (i.e., 16:0, 16:1, 18:0, and 18:1) (Figures 4C, S5D, and S5E). As expected, MyD88-deficiency did not influence repression of LCFA synthesis in response to TLR3 activation (Figures 4C and S5E). We also found that TRIF has an important role in TLR4-mediated reprogramming of LCFA synthesis. We observed that TRIF-deficient macrophages upregulated LCFA synthesis in response to LPS, indicating that TRIF signaling opposes MyD88 signaling to establish the balance of LCFA synthetic flux in TLR-stimulated macrophages (Figures 4D, S5F, and S5G). Notably, no difference in the downregulation of LCFA synthesis between control and TRIF-deficient macrophages was observed in response to TLR3 activation (Figures 4D, S5F, and S5G). Presumably this is because Poly(I:C) can generate type I IFN production via other PRR pathways (e.g., RIG-I), independent of TLR3/TRIF signaling (Kato et al., 2008). Consistent with this hypothesis, genetic deletion of the type I IFN receptor (IFNAR) abrogated the ability of Poly(I:C) to decrease de novo synthesis of 16:0, 16:1, 18:0, and 18:1 fatty acids (Figures 4E, S5H, and S5I). Together, these studies reinforce the notion of the opposing roles that the MyD88 and IFN signaling pathways have on reprogramming of lipid metabolism of macrophages.
The NRF2-SREBP Axis Is Required for MyD88-Inducible LCFA Synthesis
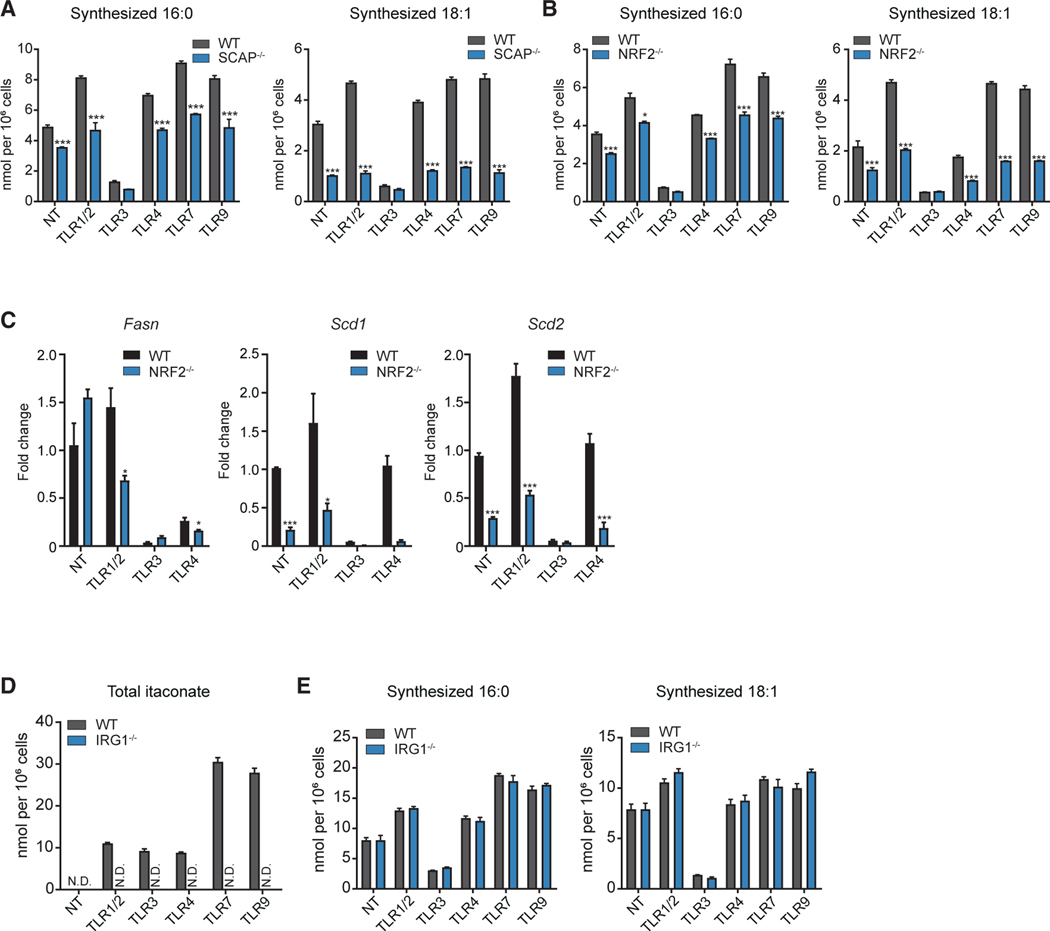
The SREBPs are transcription factors with well-defined roles in regulating lipogenesis, leading us to ask if SREBPs are required for the upregulation of LCFA synthesis in response to TLR/MyD88 signaling. Thus, we stimulated SREBP loss-of-function macrophages (LysMCre/SCAPfl/fl; designated SCAP−/−) with various TLR agonists and LCFA synthesis assessed using stable-isotope tracing. ISA demonstrated that SREBP activity is required for the upregulation of LCFA synthesis in response to MyD88-driven activation (Figures 5A, S6A, and S6B). As previously reported, no significant difference in the repression of LCFA synthesis in response to TLR3 signaling was noted between control and SCAP-deficient macrophages (Figure 5A) (York et al., 2015). The transcription factor NRF2 has also been shown to influence SREBP activity in metabolic tissues (Huang et al., 2010). Thus, we also asked if NRF2-deficiency impacted LCFA synthesis in TLR-stimulated macrophages. ISA showed that NRF2 is also required for the increase in SFA (16:0, 18:0) and MUFA (16:1 and 18:1) synthesis (Figures 5B, S6C, and S6D). Examination of gene expression demonstrated that NRF2 is required for the upregulation of LCFA synthesis genes (e.g., Fasn, Scd1, Scd2) in response to MyD88-dependent TLRs (Figure 5C). Similar to loss of SREBP, deletion of NRF2 did not influence the repression of de novo LCFA synthesis in response to TLR3 activation (Figures 5B, S6C, and S6D). Thus, we conclude that NRF2 and SREBP transcriptionally link LCFA synthetic flux with MyD88-dependent TLRs.
Figure 5. NRF2/SREBP Axis Sets Basal and TLR Inducible Lipogenesis in Macrophages.
(A) Net synthesized palmitic acid (16:0) and oleic acid (18:1) from WT control and SCAP−/− BMDMs stimulated with TLR1/2, TLR3, TLR4, TLR7, TLR9 agonists, or not treated (NT) for 48 h.
(B) Net synthesized palmitic acid (16:0) and oleic acid (18:1) from WT control and NRF2−/− BMDMs stimulated with TLR1/2, TLR3, TLR4, TLR7, TLR9 agonists, or not treated (NT) for 48 h.
(C) qPCR analysis of Fasn, Scd1, and Scd2 gene expression from WT control or NRF2−/− BMDMs stimulated with TLR1/2, TLR3, TLR4 agonists or not-treated (NT) for 24 h.
(D) MS analysis of itaconate from WT control or IRG1−/− BMDMs stimulated with TLR1/2, TLR3, TLR4, TLR7, TLR9 agonists or not treated (NT) for 24 h.
(E) Net synthesized palmitic acid (16:0) and oleic acid (18:1) in WT control or IRG1−/− BMDMs stimulated with TLR1/2, TLR3, TLR4 agonists or not-treated (NT) for 48 h.
All isotope labeling experiments are from four independent replicates per experimental condition and are representative of greater than three experiments. Gene expression studies are from three biologic replicates per experimental condition and are representative of greater than three experiments. All data are presented as mean ± SEM. *p < 0.05; **p < 0.01, ***p < 0.001.
Recent work has implicated immune-responsive gene 1 (Irg1) in the regulation of NRF2 in TLR4-stimulated macrophages, through the enzymatic generation of the metabolite itaconate (Bambouskova et al., 2018; Michelucci et al., 2013; Mills et al., 2018). MS showed that macrophages accumulate itaconate in response to both MyD88- and TRIF-dependent TLR signals in an IRG1-dependent manner (Figure 5D). These data led us to explore if IRG1 activity was also required for the upregulation of LCFA synthesis in response to TLR signaling. However, IRG1-deficiency had little impact on the ability of macrophages to regulate LCFA synthesis in response to TLR activation (Figure 5E). Thus, neither TLR-mediated activation of IRG1, nor the accumulation of the metabolite itaconate, are required to link NRF2-dependent upregulation of LCFA synthesis with MyD88-dependent TLR signals.
Abrogating MUFA Synthesis Prolongs MyD88-Mediated Inflammation
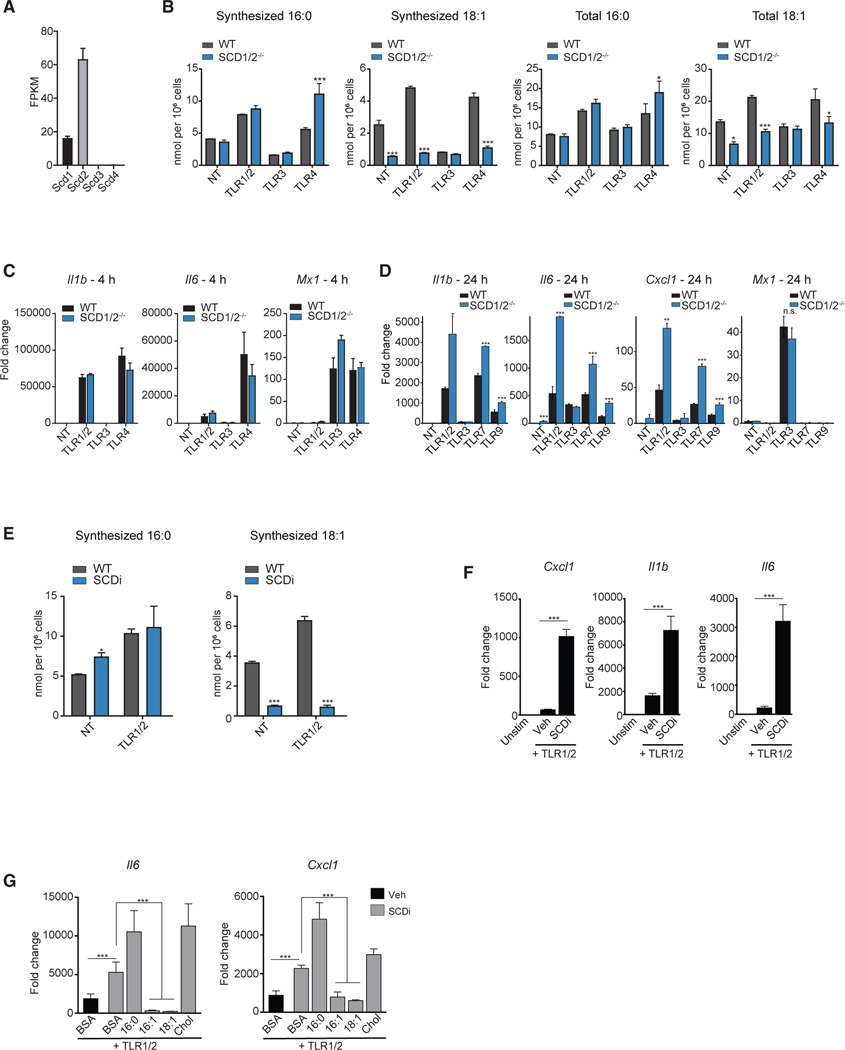
Our MS data indicate that MyD88 signaling leads to mono-desaturation of the lipidome by increasing MUFA synthesis (Figures 1A, 4B, and 4C). The stearoyl-CoA desaturases (SCDs) are Δ−9 desaturase enzymes that are responsible for the synthesis of MUFAs from SFA precursors. In mice, there are four isoforms of SCD that share up to 85% sequence homology with some degree of overlap in substrate utilization (Paton and Ntambi, 2009). Analysis of RNA-seq data showed that Scd1 and Scd2 are expressed in BMDMs, with Scd2 expressed at a markedly higher level (Figure 6A). Scd3 and Scd4 were not detectable in BMDMs. Thus, we explored if the combined loss of SCD1 and SCD2 (LysMCre/SCD1/2fl/fl) would influence MUFA synthesis in response to TLR activation. To that end, SCD1/2-deficient BMDMs were stimulated with TLR1/2, TLR3 or TLR4 agonist, and synthesis of SFAs and MUFAs were determined. As predicted, loss of SCD1/2 resulted in a marked reduction in the pool size of synthesized MUFAs in response to TLR1/2 and TLR4 activation and decreased the total MUFA pool (Figures 6B, S6E, and S6F).
Figure 6. Perturbing MUFA Synthesis Prolongs MyD88-Mediated Inflammation.
(A) FPKM values of Scd transcripts in quiescent BMDMs.
(B) Net synthesized and total palmitic acid (16:0) and oleic acid (18:1) from WT control or SCD1/2−/− BMDMs stimulated with TLR1/2, TLR3, TLR4, TLR7, TLR9 agonists or not treated (NT) for 48 h.
(C) qPCR analysis of Il1b, Il6, and Mx1 gene expression in WT control or SCD1/2−/− BMDMs stimulated with TLR1/2, TLR3, TLR4 agonists or not-treated for 4 h.
(D) qPCR analysis of CXCL1, Il1b, Il6, and Mx1 gene expression in WT control or SCD1/2−/− BMDMs stimulated with TLR1/2, TLR3, TLR7, TLR9 agonists or not-treated for 24 h.
(E) Net synthesized palmitic acid (16:0) and oleic acid (18:1) in BMDMs stimulated with TLR1/2 ± SCDi for 48 h.
(F) qPCR analysis of CXCL1, Il1b, and Il6 gene expression in BMDMs stimulated with TLR1/2 ± SCDi for 24 h.
(G) qPCR analysis of Il6, and CXCL1 gene expression in BMDMs stimulated with TLR1/2 ± SCDi (10 nM) ± BSA-conjugated 16:0, 16:1, or 18:1 fatty acids, or cholesterol for 24 h.
FPKM values of Scd transcripts from five independent RNA-sequencing experiments. All isotope labeling experiments are from four biologic replicates per experimental condition and are representative of greater than three experiments. Gene expression studies are from three biologic replicates per experimental condition and are representative of greater than three experiments. Data are presented as mean ± SEM. *p < 0.05; **p < 0.01, ***p < 0.001.
Gene expression studies revealed little difference between control and SCD1/2-deficient macrophages in the induction of inflammatory genes at early time points (i.e., 4 h) in response to TLR activation (Figure 6C). However, we observed that SCD1/2-deficient macrophages exhibited prolonged inflammatory gene expression at a later time point (i.e., 24 h [Figure 6D]). Moving forward, we focused our studies on TLR1/2 as a representative of MyD88-driven inflammation. Treatment of macrophages with the SCD inhibitor Cay10566 (termed SCDi hereafter) abrogated the synthesis of 16:1 and 18:1 MUFAs in response to TLR1/2 activation, with little impact on SFA pools (Figures 6E, S6G, and S6H). Similar to SCD1/2-deficiency, SCDi treatment promoted inflammatory responses to MyD88-dependent TLR activation at later time points with no influence on inflammation at early time points (Figures 6F, S6I, and S6J). The impact of SCDi on chemokine and cytokine production could be reduced by providing exogenous 16:1 or 18:1, but not by the addition of palmitate (16:0) or cholesterol (Figure 6G). Together, these data support a model where the upregulation of SCD activity is critical for negatively controlling the MyD88-driven inflammation by ensuring the accumulation of MUFAs.
SREBP1c Attenuates Inflammation through Regulating MUFA Homeostasis
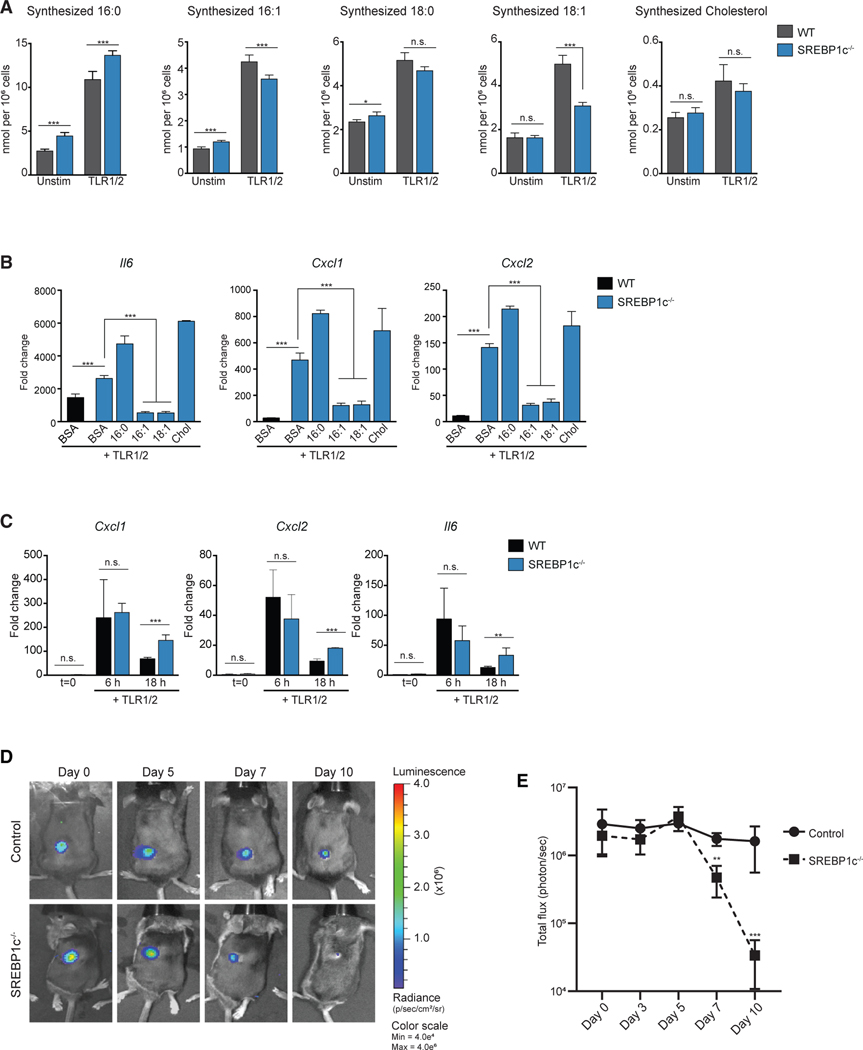
Gene expression studies showed that Srebf1c is upregulated in response to MyD88-dependent TLR signals (Figure S7A). Given the well-defined role of SREBP1c in regulating Fasn and Scd expression (Horton et al., 2002), we next explored if loss of SREBP1c would influence LCFA homeostasis and inflammation. SREBP1c-deficient macrophages were stimulated with TLR1/2 agonist and LCFA synthesis was evaluated using stable-isotope labeling. Loss of SREBP1c resulted in modest alterations in fatty acid homeostasis, with increases in the accumulation of synthesized SFA (16:0) pools and decreases in synthesized MUFAs (16:1 and 18:1) in TLR1/2-stimulated macrophages (Figure 7A). No difference in synthesized 18:0 and cholesterol were noted between the two genotypes (Figure 7A), but modest increases in total LCFA and cholesterol pools were noted in SREBP1c-deficient macrophages (Figure S7B). Consistent with SCD loss-of-function studies, we observed that SREBP1c-deficient macrophages exhibit sustained inflammatory chemokine and cytokine gene expression in response to TLR1/2 activation (Figure 7B). Supplementing cultures with exogenous 16:1 or 18:1 returned inflammatory gene expression to control levels (Figure 7B). In contrast, providing exogenous palmitate (16:0) increased the pro-inflammatory phenotype of SREBP1c−/− macrophages (Figure 7B). Taken together, these studies support a model where NRF2-SREBP1c-SCD metabolically attenuates MyD88-driven inflammation in macrophages by controlling MUFA homeostasis.
Figure 7. Upregulation of SREBP1c Is Required for MUFA Flux to Control Inflammation.
(A) Net synthesized palmitic acid (16:0), palmitoleic acid (16:1), stearic acid (18:0), oleic acid (18:1), and cholesterol from WT control or SREBP1c−/− BMDMs stimulated with TLR1/2 agonist.
(B) qPCR analysis of inflammatory gene expression from WT control or SREBP1c−/− BMDMs stimulated with TLR1/2 agonist ± BSA-conjugated 16:0, 16:1, or 18:1 fatty acids, or cholesterol for 24 h.
(C) qPCR analysis of inflammatory gene expression from cells collected by peritoneal lavage from WT control or SREBP1c−/− mice injected (intraperitoneal) with TLR1/2 agonist for 6 h and 18 h.
(D) Time course bioluminescence images from a representative WT control and SREBP1c−/− mouse from day 0 (immediately post-infection) through day 10 post-infection challenged with the bioluminescent strain of Staphylococcus aureus (Xen36).
(E) Time course quantification of total flux (photons/sec) from WT control or SREBP1c−/− mice infected with bioluminescent S. aureus.
All isotope labeling experiments are from four biologic replicates per experimental condition and representative of greater than three experiments. Gene expression studies are from three biologic replicates per experimental condition and are representative of greater than three experiments. In vivo experiments with S. aureus infections or TLR1/2 agonists are representative of four independent experiments.
All data are presented as mean ± SEM. *p < 0.05; **p < 0.01, ***p < 0.001.
Loss of SREBP1c Facilitates Host Defense to S. aureus In Vivo
Finally, we asked if the loss of SREBP1c would influence inflammatory responses in vivo. Initially, we examined the inflammatory response to i.p. injection of TLR1/2 ligand (Pam3CSK4). Peritoneal lavage was collected at 6 and 18 h post-injection and inflammatory gene expression was assessed from the collected cells. Similar to our in vitro studies, no difference in inflammation was observed at an early (6 h) time point (Figure 7C), but cells in lavage fluid exhibited heightened inflammatory gene expression at 18 h post-injection (Figure 7C).
These data led us to hypothesize that loss of SREBP1c would influence host defense to pathogens that are known to trigger TLR2-dependent responses. Stimulation of BMDMs in vitro with S. aureus resulted in increased Srebp1c and Scd2 expression, as well as increased the pool of synthesized MUFAs (Figures S7C–S7G), phenocopying our TLR2 ligand studies. To test this in vivo, we employed a skin infection model of S. aureus where pathogen burden can be monitored using bioluminescent imaging (Fournier and Philpott, 2005; Scumpia et al., 2017). Control and SREBP1c−/− mice were injected with 2.5 × 106 CFUs (intradermal) of S. aureus, an inoculum that is cleared by C57BL/6 mice over approximately 2 weeks (Scumpia et al., 2017). Bioluminescence analysis immediately after injection (designated day 0) confirmed that both control and SREBP1c−/− mice received equivalent bacterial inoculums (Figures 7D and 7E). Minimal difference in bacterial load in the skin was observed at early time points (Figures 7D and 7E). However, SREBP1c−/− animals had lower bacterial burden by day 7 post-infection when compared with control mice, with bacteria becoming minimally detectable by day 10 in SREBP1c−/− animals (Figures 7D and 7E).
We considered the possibility that accelerated clearance of bacteria by SREBP1c−/− mice was a function of heightened phagocytosis. However, we observed that loss of SREBP1c decreased phagocytosis of fluorescently labeled S. aureus (Figure S7H). Moreover, the addition of palmitate (16:0) or oleate (18:1) did not restore this difference in phagocytic capacity (Figure S7H), consistent with the notion that the prolonged inflammatory responses observed in SREBP1c−/− macrophages underlie protection from S. aureus challenge. Taken together, these data indicate that limiting the ability of cells to undergo SREBP1c-dependent metabolic programing of fatty acid metabolism can enhance resistance to bacteria, and suggest that pharmacologically targeting LCFA homeostasis could be a viable approach in controlling bacterial infections.
DISCUSSION
In this study, we sought to better understand how the macrophage lipidome is reshaped in response to pro-inflammatory signals generated by different TLRs. By employing shotgun lipidomics, we were able to show that activation of macrophages by TLRs and cytokines induces changes to glycerolipid, glycerophospholipid, sphingolipid, cholesterol, and fatty acid composition. These studies also revealed that activation by different pro-inflammatory stimuli results in the acquisition of distinct lipidomes, revealing specificity in this process. We think it likely that other classes of PRRs also induce similar changes. For example, activation of PRRs that induce a robust type I IFN response (e.g., STING) will likely resemble that of TLR3 activation. PCA of combined lipidomic data from TLR1/2, TLR3, or TLR4-activated macrophages (four in total) showed that the major changes to lipid composition induced by the various TLRs was consistent. Thus, we conclude that TLR-mediated remodeling of the lipidome is a regulated process that ensures activated macrophages acquire a specific lipidome. Having said that, we also found that loading of macrophages with Ac-LDL had a profound effect on TLR-induced reshaping of the lipidome. Thus, the lipid composition or metabolic state of a macrophage at the time of stimulation appears to be an important determinant for TLR-mediated alterations to lipid composition. Based on this observation, we also think it likely that the availability of other metabolites (e.g., glucose or amino acids) or oxygen influence reprogramming of the lipidome.
Using loss-of-function models, we were able to show that TLR-induced remodeling of the lipidome are driven, in large part, through the opposing effects of the MyD88 and TRIF/Type I interferon-signaling pathways. Stable-isotope enrichment MS studies showed that MyD88-dependent TLRs upregulate de novo LCFA synthesis (SFAs and MUFAs), and that the increase in LCFA synthesis is dependent on NRF2 and SREBP1 transcriptional activity. Whereas, TRIF-dependent TLRs attenuate de novo LCFA synthesis through autocrine type I IFNs signaling. We also found that providing IFNγ in trans supplants the requirement for type I IFNs in TLR-stimulated macrophages, indicating that IFNγ and IFNβ have largely overlapping roles. Our studies also show that IFN signals can overwrite the lipid metabolic program induced by MyD88-dependent TLRs, suggesting that the presence of IFNs in the inflammatory milieu conveys important information to macrophages regarding the appropriate lipidome to support effector function. Interestingly, treatment of naive macrophages with IFNβ or IFNγ alone resulted in far more modest changes to the lipidome, indicating that TLR signaling is required to “prime” macrophages to ensure maximal IFN-induced reshaping of lipid composition. Taken together, these studies provide a basis for understanding how reprogramming of the macrophage lipidome is achieved in the context of distinct pro-inflammatory signals, and it will be of interest to see if these “rules” apply to other immune cell types.
As to why different TLRs would induce macrophages to reprogram their lipid composition in distinct manners remains an important open question. One idea is that changes in lipid composition are required to ensure appropriate inflammatory and host defense functions in response to different categories of pathogens. Given the important role for lipids in regulating membrane function, we think it likely that reprogramming lipid composition of plasma and intracellular membranes facilitates cellular host defense through multiple effector pathways. Changes in membrane lipid composition have been shown to alter membrane curvature and fluidity, thereby influencing a wide variety of membrane-dependent processes, including phagocytosis, viral envelope fusion, viral egress, microbial invasion and lifecycle, to name a few (Araldi et al., 2017; Blanc et al., 2013; Lee et al., 2018; Liu et al., 2013; Mulvihill et al., 2015; Vromman and Subtil, 2014). Another important reason for remodeling lipid composition is to facilitate subcellular distribution or function of key immune effector molecules. Elegant mechanistic studies on the IL-1β inflammatory axis revealed that the pore forming protein Gasdermin D, responsible for allowing IL-1β to move through the plasma membrane (Heilig et al., 2018), is recruited to the inner leaflet via interactions with acidic phospholipids (Ding et al., 2016; Liu et al., 2016; Mulvihill et al., 2018). Importantly, artificially inducing changes to membrane lipid composition can impact oligomerization of Gasdermin D. Likewise, it has been shown that Cyclic GMP-AMP synthase (cGAS), a nucleotidyltransferase that plays a key role in recognition of viral intracellular DNA, is directed to distinct subcellular locations through interactions with PIP lipid species (Barnett et al., 2019). Thus, remapping lipid composition in a PRR-or cytokine-specific manner becomes a central process for regulating and coordinating multiple aspects of cellular immunity.
The opposing role of MyD88 and TRIF-IFN signaling in controlling reprogramming of the lipidome also leads to the hypothesis that changes in lipid composition will be important for entraining specific types of pro-inflammatory responses. Studies on the IFN signaling axis have shown that these cytokines profoundly attenuate de novo cholesterol biosynthesis (Blanc et al., 2011, 2013; Liu et al., 2013; York et al., 2015), and that change in the synthesis of cholesterol facilitates the generation of type I IFNs while negatively regulating inflammasome activation, and the generation of IL-1β (Dang et al., 2017; Reboldi et al., 2014). In this way, control over cellular cholesterol homeostasis becomes a regulatory node for balancing IFN and IL-1β inflammatory programs. Likewise, flux through the fatty acid synthetic enzyme fatty acid synthase (FASN) has been shown to mediate inflammation in the context of diabetogenic diet and LPS challenge (Carroll et al., 2018; Wei et al., 2016). In the absence of saturated fatty acid synthesis, it has been shown that macrophages are unable to efficiently transduce pro-inflammatory signals at the plasma membrane. This deficiency could not be rescued by the addition of exogenous saturated fatty acids, but rather, exogenous cholesterol was necessary to restore TLR signaling (Wei et al., 2016). Given that biophysical and biochemical actions of membranes are sensitive to stoichiometry of different lipid species, we think it is likely that the impact of reprogramming specific lipid metabolic pathways (e.g., fatty acid synthesis) in response to inflammatory signals will influence inflammation and cellular immunity through secondary interactions with other lipid species in the plasma or intracellular membranes.
Finally, our data indicate that increased synthetic flux through the Δ9-desaturases Stearoyl-CoA desaturases (SCDs) in response to MyD88-dependent TLR signaling is required to control inflammation. It remains unclear how accumulation of newly synthesized MUFAs promote resolution of MyD88-driven inflammation. As discussed above, deletion of FASN was found to influence inflammatory response by altering membrane composition and TLR4 signaling (Wei et al., 2016). It may be that a similar disruption of membrane function could be operative in macrophages that have SCD activity attenuated. Although we found that providing exogenous cholesterol or saturated fatty acids was not able to reduce inflammation in our studies, we cannot rule out the possibility that loss of MUFA homeostasis perturbs plasma membrane function resulting in persistent TLR, MyD88, or cytokine receptor signaling. An alternative hypothesis is that desaturated fatty acids modulate ER membrane function and ER stress. It has been shown that accumulation of saturated fatty acids can activate NF-κB signaling through engagement of the ER stress pathways (Ackerman et al., 2018; Fu et al., 2011; Halbleib et al., 2017; Lancaster et al., 2018). In this way, decreasing MUFA synthesis results in the accumulation of saturated fatty acid containing complex lipids. This model would align with studies where perturbations in PUFA metabolism also results in heightened inflammatory responses downstream of the ER stress response (Giannakis et al., 2019; Oishi et al., 2017; Rong et al., 2015).
In conclusion, the studies detailed here advance our understanding of how distinct activation signals shape the lipidome of macrophages and show that targeting these changes can be exploited to positively affect host defense. It is our expectation that the data in this manuscript will serve as a resource for ongoing research efforts, and that future systems-level studies will build upon these data to provide a comprehensive understanding of the crosstalk between lipid metabolism and inflammation.
Limitations of Study
This study examined the impact of different TLR signals on reprogramming of fatty acid synthetic fluxes and the lipidome of BMDMs from C57BL/6 mice. Here, the data reflect changes induced in a highly reductionist system that specifically examines one macrophage type that is derived in vitro. It will be important to determine the extent to which the changes in lipid metabolism is observed in other genetic backgrounds of mice, as well as human-derived monocytes and macrophages. An additional limitation of this study is that we performed a targeted shotgun MS approach. Although we quantitatively measure upwards of 750 lipid species in this study, we have not exhaustively examined the macrophages lipidome, and are missing key classes (e.g., eicosanoids). We believe that the data serve as a strong foundation, and as new studies are performed, we anticipate that there will be identification of additional lipid species that change in response to different pro-inflammatory signals. An additional limitation of our study is that we have not examined how reprogramming of the lipidome in response to TLRs or other cytokines can be observed in macrophages in vivo. Macrophages alter their function in response to specific cues within different tissue microenvironment (Glass and Natoli, 2016). It is anticipated that local environmental differences in lipid metabolite availability will influence the extent to which macrophages undergo reprogramming of their lipidome in response to activation signals (Giannakis et al., 2019; Okabe and Medzhitov, 2016). One final limitation to the current study is the use of germline SREBP1c deletion for in vivo studies. Macrophages have a key role in the clearance of bacteria through their ability to phagocytose extracellular bacteria, generate inflammation, release antimicrobial products generate, and oxidative bursts (Okabe and Medzhitov, 2016). Our studies did not exhaustively characterize the function of SREBP1c-deficient macrophages, and as such it may be that the increased clearance of bacteria observed in SREBP1c-deficient mice may not be dependent specifically on their ability to generate increased inflammation. Likewise, it is anticipated that loss of SREBP1c in additional cell types beyond that of macrophages, such as neutrophils and skin resident immune cells, will contribute to the increased clearance of S. aureus in the skin in SREBP1c-deficient mice (Kabashima et al., 2019). Finally, comprehensive analysis of how MUFA homeostasis influences the antimicrobial functions of other innate immune cells will be a key future objective.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Steven J. Bensinger (SBensinger@mednet.ucla.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
These data are available at the NIH Common Fund’s National Metabolomics Data Repository (NMDR) website, the Metabolomics Workbench (https://www.metabolomicsworkbench.org) where it has been assigned Project ID: PR000914. The data can be accessed directly via the Project DOI: https://doi.org/10.21228/M8JH69. This work is supported by NIH grant U2CDK119886.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal housing and all the experimental procedures were authorized by the UCLA Animal Research Committee under protocol 2005135-41B. Mice were housed 4 per cage in a temperature (22 – 24 °C) and humidity controlled colony room, maintained on a 12 h light/dark cycle (07:00 to 19:00 light on), with standard chow diet (LabDiet 5053) and water provided ad libitum and environmental enrichments. General health of the animal was assessed weekly by UCLA DLAM veterinarians.
Isolation of Bone Marrow-Derived Macrophages (BMDMs)
Bone marrow cells were isolated from femurs of male WT control or knockout mice between the age of 8–12 weeks. Cells were treated with RBC lysis buffer for 3 min to remove red blood cells, centrifuged at 365 ×g for 5 min, and resuspended in 10% BMDM medium. Cells were maintained at 37 °C in a humidified 5% CO2 incubator. BMDMs were differentiated for 6 days prior to experiments, and medium was changed at day 4 of differentiation.
LysMCre SCD1/2−/− BMDMs
LysMCre SCD1/2−/− BMDMs were generated from crossing Scd1/2 flox/flox mice (generously provided by Dr. Miyazaki; (Miyazaki et al., 2005)) with B6.129P2-Lyz2tm1(cre)Ifo/J (LysM-Cre) mice and maintained on a C57BL/6 genetic background.
S. aureus Infection
All procedures were approved by UCLA Animal Research Committee under protocol 2015–021. Experiments were performed as previously described in (Scumpia et al., 2017) with minor modifications. Briefly, the mice were shaved on the back and inoculated intradermally with 100 μL of mid logarithmic growth phase S. aureus strain Xen36 (~2.5×106 CFUs/100 μL) after overnight culture in sterile pharmacy grade saline (0.9%) by a 27-gauge needle and a tuberculin syringe (Abbott Laboratories; Chicago, IL). Actual bacterial concentrations were confirmed with conventional bacteriological plating of serial dilutions.
METHOD DETAILS
Macrophage Media
For all experiments involving BMDMs, cells were cultured in either DMEM supplemented with 5% or 10% v/v heat-inactivated fetal bovine serum (FBS) (designated as 5% or 10% BMDM media) plus 2 mM L-glutamine, 100 units/mL, 100 μg/mL penicillin/streptomycin, 500 μM sodium pyruvate, and 5% v/v conditioned media containing macrophage colony stimulating factor (M-CSF) (Takeshita et al., 2000) produced by CMG cells to induce differentiation to BMDMs.
Shotgun Lipidomics Analysis
Day 6 BMDMs were seeded at a density of 9×105 cell/well in 6 well dishes (Fisher 08-772-1B). On day 8, BMDM were mock-treated (media change) or co-treated with ligands as described below in 5% BMDM media for 48 h. Prior to collection, cell number was assessed by imaging as described below. After removal of culturing media, 1 mL of ice-cold PBS was added into each well and cells were scraped with cell lifters and spun down in glass tubes at 365 ×g for 5 min at 4°C. Two wells of cells were combined as a single replicate. A modified Bligh and Dyer extraction (Bligh and Dyer, 1959) was carried out on samples. Prior to biphasic extraction, a 13-lipid class Lipidyzer Internal Standard Mix is added to each sample (AB Sciex, 5040156). Following two successive extractions, pooled organic layers were dried down in a Genevac EZ-2 Elite. Lipid samples were resuspended in 1:1 methanol/dichloromethane with 10 mM Ammonium Acetate and transferred to robovials (Thermo 10800107) for analysis. Samples were analyzed on the Sciex Lipidyzer Platform for targeted quantitative measurement of 1100 lipid species across 13 classes. Differential Mobility Device on Lipidyzer was tuned with SelexION tuning kit (Sciex 5040141). Instrument settings, tuning settings, and MRM list available upon request. Data analysis performed on Lipidyzer software. Quantitative values were normalized to cell counts. For informatics analysis, heat maps and PCA plots were generated with Clustvis (Metsalu and Vilo, 2015). Rows were centered and unit variance scaling is applied to rows (Z score). Rows were clustered using correlation distance and complete linkage. For PCA, unit variance scaling was applied to rows, Nipals PCA is used to calculate principal components. X and Y axis show principal component 1 and principal component 2 that explain percentage of the total variance, respectively. Prediction ellipses are such that with probability 0.95, a new observation from the same group will fall inside the ellipse.
Isotope Labeling Experiments of BMDMs
All isotope labeling experiments from BMDMs were performed as previously described (Williams et al., 2013; York et al., 2015). Briefly, day 6 BMDMs were plated at 1×105 cells/well in 24-well black plates in 10% BMDM media. On day 8, BMDM were mock-treated (media change) or co-treated with ligands in DMEM (no glucose) supplemented with 10 mM [13C] glucose and 10 mM unlabeled glucose (i.e. 50% [13C6]glucose), 5% v/v FBS, 2 mM L-glutamine, 100 units/mL penicillin, 100 mg/mL streptomycin, and 5% v/v conditioned media for 48 h. Prior to collection, cell number was assessed by imaging as described below. Isotopomer spectral analysis (ISA) was conducted using an Agilent 5975C mass spectrometer coupled to a 7890 Gas Chromatograph as previously described (Williams et al., 2013). Data were normalized by cell count. For collection with guanidine HCl, cells were treated with 50 μL of 6 M aqueous guanidine HCl and transferred to glass tubes for derivatization with an additional 100 μL of 3 M methanolic guanidine HCl. Samples were prepared alongside internal standard curve samples made up of FAMEs (Nu-Chek Prep).
For all lipidomics experiments, 50 μg/mL acetylated-LDL, 50 ng/mL Pam3CSK4, 1 μg/mL Poly(I:C), 50 ng/mL LPS, 100 nM CL307, 100 nM ODN1668, 20 ng/mL IFNβ, 20 ng/mL IFNγ, or 50 ng/mL LPS + 20 ng/mL IFNγ were used.
For isotope labeling and gene expression experiments, 50 ng/mL Pam3CSK4, 1 mg/mL Poly(I:C), 50 ng/mL LPS (for Figures 4D, S5F, S5G, S6I, and S6J), 10 ng/mL LPS (for Figures 4A–4C, 5, 6B–6D, S5A, S5B, S5D, S5E, and S6A–S6D), 100 nM CL307, 100 nM ODN1668, 20 ng/mL IFNβ, or 10 nM of SCDi were used.
For isotope labeling experiments involving S. aureus, BMDMs were mock-infected (media change) or infected with either live or heat killed (98 °C, 10 min) S. aureus (strain 8325–4) for 1 h in antibiotic free 10% macrophage media. Inoculum is subsequently removed and BMDMs were cultured in labelling media as described above.
Cell Counts and Normalization
For both isotope labeling and lipidomics experiments, prior to sample collection, 1.25 mM Calcein-AM dissolved in glucose-free DMEM (final concentration 1.45 μM) was added to each well and the plates were then imaged on a Molecular Devices ImageXpress XL. 20 high magnification fluorescence images were captured for each well (21.83% of total well surface area for 24 well plates, or 24% for 6 well plates) using a 10x objective (Nikon Plan Fluor, 0.3 NA). Cell number was assessed using MetaXpress Software with Powercore using the Multi-wavelength cell scoring module.
Itaconate Measurements
Itaconate was measured as in Cordes and Metallo (Cordes and Metallo, 2019) using an Agilent 7890B gas chromatograph coupled to a 5977B mass spectrometer with an Agilent J&W DB-35ms column (30 m × 0.25 mm i.d. × 0.25 μm). Day 6 BMDMs were seeded at 0.33×105 cells/well in 12-well tissue culture dishes. After 48 h., cells were mock-treated or treated with TLR ligands (media composition and concentrations as in “Isotope labeling experiments”) for 24 h. At the time of harvest and extraction, wells were quickly washed twice with ice-cold 0.9% (w/v) saline. Polar metabolites were extracted with a Folch-like extraction of 40% MeOH, 40% CHCl3, and 20% H2O with 1 μg norvaline as an internal standard. The polar phase was collected and dried overnight in a refrigerated benchtop vacuum concentrator. Dried samples were prepared for GC/MS analysis by first treating with 20 μL of 2% (w/v) methoxyamine hydrochloride (“MOX”) in pyridine and incubated for 45 minutes at 37 °C, followed by addition of 20 μL N-tert-butyldimethylsiyl-nmethyltrifluoroacetamide with 1% tert-butyldimethylchlorosilane for 45 minutes at 37 °C to form methoxyamine-tertbutyldimethylsiyl (MOX-tBDMS) derivatives.
The GC oven was held at 100 °C for 2 min, increased to 255 °C at 3.5 °C min−1, then immediately increased to 320 °C at 15 °C min−1 and held for 3 min. Itaconate was measured using selected ion monitoring (SIM) mode and integrating fragment ions (m/z 301–308) at a retention time of 23.6 min. Samples were calibrated against a standard curve of itaconic acid (0.07–50 nmol). Cell numbers were quantified using identically treated matched plates. At the time of harvest for GC/MS, the matched plates were fixed with 4% paraformaldehyde for 20 min at room temperature and refrigerated. Two days later, cells were stained with Hoescht 33342 at 10 ng/mL overnight at 4°C. The following day, cell counts were obtained using the Operetta High Content Imaging System (Perkin Elmer).
Gene Expression Analysis
For all experiments involving gene expression analysis, day 6 BMDMs were seeded at 3×105 cells/well in 12-well plates in 10% BMDM media. On day 8, BMDMs were mock-treated, co-treated, or sequentially treated with ligands in 5% BMDM media as indicated in the figure legend. Cells were collected in TRIzol and RNA was extracted according to manufacturer’s protocol. cDNA was synthesized using 700 ng RNA per reaction with high-capacity cDNA reverse transcription kit. KAPA SYBR FAST qPCR Master Mix (2X) kit and a LightCycler 480 were used for quantitative RT-PCR using 0.5 μmol/L primers. Fold change related to the control group was calculated using 2ΔΔCP method with 36b4 as the reference gene.
Quantification of S. aureus In Vivo
In vivo bioluminescence was performed with the Xenogen IVIS imaging system (Xenogen Corporation; Alameda, CA) at the Crump Institute for Molecular Imaging at UCLA as previously described (Scumpia et al., 2017). Mice were anesthetized via isoflurane injection. Data are presented on color scale overlaid on a gray-scale photograph of mice and quantified as total flux and average radiance (photons/s) within a circular region of interest (1×103 pixels) with Living Image software (Xenogen) (lower limit of detection: 1 × 104 photons/s). Data shown is the combination of three separate experiments performed on different days using the same aliquot of S. aureus each day on age and sex matched control mice (from het × het breeding) and SREBP1c−/− mice from the same colony.
Macrophage Phagocytosis Assay
Day 6 BMDMs were plated at 2.5 × 105 cells/well in 24-well plates in 10% media. The next day, cells were pretreated with TLR1/2 ligand with or without free fatty acids in 10% BMDM media for 24 h. pHrodo red S. aureus Bioparticles Conjugate (0.4 mg/mL) were added to macrophage cultures in fresh BMDM media and incubated for 25 min at 37 °C. Macrophages cultures were vigorously washed twice with room temp PBS to reduce non-phagocytosed S. aureus. Cells were then scraped off the plate, and subject to flow cytometry analysis.
Preparation of 24% BSA and BSA-Conjugated Free Fatty Acids
FFA-free, endotoxin-low BSA was prepared by adding 6 g of fatty-acid-free BSA to 17.5 mL 150 mM NaCl in a beaker at 37 °C while stirring. 24% BSA pH was adjusted to 7.4 with 1N NaOH. Free fatty acids (16:0, 16:1 and 18:1) were dissolved in 150 mM NaCl pH = 7.4 to achieve a final concentration of 25 mM. Mixture was shaken vigorously, and heated at 65 °C until free fatty acids dissolved in solution. 24% BSA solution (ice cold) is finally combined with 25 mM fatty acid solution at a 54:46 ratio to yield approximated 12.5 μM final concentration with pH = 7.4. The final mixture was mixed for 10 min at room temperature on shaker before use. All BSA-conjugated fatty acids are stored at −80 °C. FFAs were diluted into cell culture medium at 1:500 dilution for 25 mM final concentration. FFAs were added 4 h post-TLR stimulation.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical parameters, including the number of replicates (n), can be found in the figure legends. Statistical analyses were performed using Graph Pad Prism 8 software. Data are presented as the mean ± SEM. For all gene expression analysis, two-tailed unpaired Student’s t-test was used. For flux analysis involving only WT BMDMs, statistical significance determined using ordinary oneway ANOVA followed by Dunnett post-hoc multiple comparisons tests. For flux analysis involving TRIF−/−, MyD88−/−, NRF2−/−, IFNAR−/−, NRF2−/−, IRG1−/−, SCAP−/−, SCD1/2−/− (Miyazaki et al., 2005), and SCDi for each fatty acid measured, statistical significance determined using the multiple Student’s t-tests corrected for multiple comparison using Holm-Sidak method, with alpha = 0.05. Computations assume that all rows are sample from populations with the same scatter (SD). For flux analysis involving S. aureus infection, two-tailed unpaired Student’s t-test was used. For in vivo bacteria skin infection experiments, mice were injected with S. aureus in a blinded fashion. Numbers of mice used in cohorts were predetermined based on prior literature (Scumpia et al., 2017) and no exclusion criteria were used.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| RBC lysis buffer | Sigma-Aldrich | Cat# R7757 |
| DMEM (High glucose) | Gibco | Cat# 11965 |
| DMEM (No Glucose) | ThermoFisher | Cat# 11966–025 |
| Penicillin/Striptomycin | Gibco | Cat# 15140122 |
| Sodium Pyruvate | Gibco | Cat# 11360070 |
| L-Glutamine | Gibco | Cat# A2916801 |
| HyClone Characterized FBS | GE | Cat# SH30071.03 |
| Recombinant M-CSF generated from CMG14–12 culture supernatant | Takeshita et al., 2000 | N/A |
| Pam3CSK4 | InvivoGen | Cat# tlrl-pms |
| Poly(I:C) | InvivoGen | Cat# tlrl-picw |
| LPS | InvivoGen | Cat# tlrl-smlps |
| CL307 | InvivoGen | Cat# tlrl-c307 |
| ODN1668 | InvivoGen | Cat# tlrl-1668 |
| Recombinant Mouse IFN-β1 (carrier-free) | BioLegend | Cat# 581302 |
| Recombinant Murine IFN-γ | PeproTech | Cat# 315–05 |
| Palmitic Acid (16:0) | Nu-Chek Prep | Cat# N-16-A |
| Palmitoleic Acid (16:1) | Nu-Chek Prep | Cat# U-40-A |
| Oleic Acid (18:1) | Nu-Chek Prep | Cat# U-46-A |
| Cholesterol | Sigma-Aldrich | Cat# C4951 |
| Trizol | ThermoFisher | Cat#15596-018 |
| D-Glucose (U-13C6, 99%) | Cambridge Isotope Laboratories | Cat# CLM-1396-MPT-PK |
| Low Density Lipoprotein from Human Plasma, Acetylated (AcLDL) | ThermoFisher | Cat# L35354 |
| Calcein-AM | Santa Cruz Biotech | Cat# sc-203865 |
| Methoxyamine hydrochloride | Sigma-Aldrich | Cat# 89803 |
| N-fert-Butyldimethylsilyl-N-methyltrifluoroacetamide with 1% tert-Butyldimethylchlorosilane | Sigma-Aldrich | Cat# 00942 |
| Itaconic acid | Sigma-Aldrich | Cat# I29204 |
| Hoescht 33342 | ThermoFisher | Cat# 62249 |
| Endotoxin-low BSA | Sigma-Aldrich | Cat# A8806 |
| pHrodo Red S. aureus Bioparticles Conjugate | Invitrogen | Cat# A10010 |
| SCDi CAY10566 | Cayman Chemical | Cat# NC0493687 |
| Critical Commercial Assays | ||
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | Cat#4368813 |
| KAPA SYBR FAST qPCR Master Mix (2X) optimized for LightCycler 480 | KAPA Biosystems | Cat#KK4601 |
| Deposited Data | ||
| Raw and analyzed lipidomics data | This paper | https://www.metabolomicsworkbench.org/ (Project ID PR000914; https://doi.org/10.21228/M8JH69) |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | RRID:IMSR_JAX:000664 |
| Mouse: B6.129P2(SJL)-Myd88tm1.1Defr/J (MyD88−/−) | The Jackson Laboratory | RRID:IMSR_JAX:009088 |
| Mouse: C57BL/6J-Ticam1Lps2/J (TRIF’/_)) | The Jackson Laboratory | RRID:IMSR_JAX:005037 |
| Mouse: B6.129X1-Nfe2l2tm1Ywk/J (NRF2”/_) | The Jackson Laboratory | RRID:IMSR_JAX:017009 |
| Mouse: C57BL/6NJ-Acod1em1(IMPC)J/J (IRG1”/_) | The Jackson Laboratory | RRID:IMSR_JAX:029340 |
| Mouse: B6;129-Scaptm1Mbjg/J (SCAP’/_) | The Jackson Laboratory | RRID:IMSR_JAX:004162 |
| Mouse: B6;129S6-Srebf1tm1Mbr/J (SREBP1C’/_) | The Jackson Laboratory | RRID:IMSR_JAX:004365 |
| Mouse: SCD1/2 | Miyazaki et al., 2005 | N/A |
| Mouse: B6(Cg)-Ifnar1tm1.2Ees/J (IFNAR’/_) | The Jackson Laboratory | RRID:IMSR_JAX:028288 |
| Oligonucleotides | ||
| Murine Il1b primer: Fwd: GCCCATCCTCTGTGACTCAT Rev: AGGCCACAGGTATTTTGTCG | This paper | N/A |
| Murine Il6 primer: Fwd: AGTTGCCTTCTTGGGACTGA Rev: TCCACGATTTCCCAGAGAAC | This paper | N/A |
| Murine Nos2 primer: Fwd: CACCTTGGAGTTCACCCAGT Rev: ACCACTCGTACTTGGGATGC | This paper | N/A |
| Murine Mx1 primer: Fwd: GACCATAGGGGTCTTGACCAA Rev: AGACTTGCTCTTTCTGAAAAGCC | This paper | N/A |
| Murine Cxcl1 primer: Fwd: CTGGGATTCACCTCAAGAACATC Rev: CAGGGTCAAGGCAAGCCTC | This paper | N/A |
| Murine Cxcl5 primer: Fwd: TGCGTTGTGTTTGCTTAACCG Rev: CTTCCACCGTAGGGCACTG | This paper | N/A |
| Murine Cxcl10 primer: Fwd: CCAAGTGCTGCCGTCATTTTC Rev: GGCTCGCAGGGATGATTTCAA | This paper | N/A |
| Murine Irf1 primer: Fwd: GGCCGATACAAAGCAGGAGAA Rev: GGAGTTCATGGCACAACGGA | This paper | N/A |
| Murine Irf7 primer: Fwd: TCCAGTTGATCCGCATAAGGT Rev: CTTCCCTATTTTCCGTGGCTG | This paper | N/A |
| Murine Tnfa primer: Fwd: TGCCTATGTCTCAGCCTCTTC Rev: GAGGCCATTTGGGAACTTCT | This paper | N/A |
| Murine 36b4 primer: Fwd: CTGTGCCAGCTCAGAACACTG Rev: TGATCAGCCCGAAGGAGAAG | This paper | N/A |
| Murine Scd1 primer: Fwd: GCTCTACACCTGCCTCTTCG Rev: GCCGTGCCTTGTAAGTTCTG | This paper | N/A |
| Murine Scd2 primer: Fwd: TGGTTTCCATGGGAGCTG Rev: TTGATGTGCCAGCGGTACT | This paper | N/A |
| Murine Srebf1c primer: Fwd: GAGCCATGGATTGCACATTT Rev: GGGAAGTCACTGTCTTGGTTG | This paper | N/A |
| Murine Fasn primer: TGGGTTCTAGCCAGCAGAGT Rev: ACCACCAGAGACCGTTATGC | This paper | N/A |
| Software and Algorithms | ||
| Prism 8 | GraphPad Software | https://www.graphpad.com/ |
| Adobe Illustrator CC | Adobe | http://www.adobe.com/products/illustrator.html |
| Agilent MassHunter Software | Agilent | http://www.agilent.com/en/products/software-informatics/masshunter-suite/masshunter/masshunter-software |
Highlights.
A quantitative profiling of the mouse macrophage lipidome activated by immune stimuli
Macrophages alter lipid composition in a TLR-specific manner
MyD88-dependent TLRs alter lipid composition by increasing de novo MUFA synthesis
Inhibiting MUFA synthesis increases inflammation generated by MyD88-dependent TLRs
Context and Significance.
Macrophages can recognize various components of pathogens to clear microbial and viral invaders. Although it is known that macrophages rapidly change their lipid metabolism to support their function, a systems-level understanding of “how” and “why” macrophages reshape their lipid composition is lacking. Here, Hsieh et al. performed detailed lipid profiling of macrophages activated by various stimuli that mimic different types of pathogens and found pathogen class-specific changes. They also showed that blocking the metabolic pathways underlying some of these changes in lipid composition enhance immunity to bacteria. These results provide a resource that can be mined for insight into the potential development of new therapies that rely on selective targeting of the lipid metabolism of macrophages to fight infections better.
ACKNOWLEDGMENTS
A.G.Y. was supported in part by California HIV/AIDS Research Program training fellowship (D12-LA-351), and is currently supported by the Howard Hughes Medical Institute Hanna H. Gray Fellowship; S.J.B. was supported by NIH AI093768, and HL146358; P.O.S. was supported by NIH AR066545 and AR073940; J.S. was supported by NIH AG047156 and HL126647. The research described was also supported by NIH/National Center for Advancing Translational Sciences (NCATS) UCLA CTSI grant Number UL1TR001881.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.cmet.2020.05.003.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Ackerman D, Tumanov S, Qiu B, Michalopoulou E, Spata M, Azzam A, Xie H, Simon MC, and Kamphorst JJ (2018). Triglycerides promote lipid homeostasis during hypoxic stress by balancing fatty acid saturation. Cell Rep. 24, 2596–2605.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araldi E, Fernández-Fuertes M, Canfrán-Duque A, Tang W, Cline GW, Madrigal-Matute J, Pober JS, Lasunción MA, Wu D, Fernández-Hernando C, and Suárez Y (2017). Lanosterol modulates TLR4-mediated innate immune responses in macrophages. Cell Rep. 19, 2743–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argus JP, Wilks MQ, Zhou QD, Hsieh WY, Khialeeva E, Hoi XP, Bui V, Xu S, Yu AK, Wang ES, et al. (2018). Development and application of FASA, a model for quantifying fatty acid metabolism using stable isotope labeling. Cell Rep. 25, 2919–2934.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bambouskova M, Gorvel L, Lampropoulou V, Sergushichev A, Loginicheva E, Johnson K, Korenfeld D, Mathyer ME, Kim H, Huang LH, et al. (2018). Electrophilic properties of itaconate and derivatives regulate the IκBz-ATF3 inflammatory axis. Nature 556, 501–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett KC, Coronas-Serna JM, Zhou W, Ernandes MJ, Cao A, Kranzusch PJ, and Kagan JC (2019). Phosphoinositide interactions position cGAS at the plasma membrane to ensure efficient distinction between self- and viral DNA. Cell 176, 1432–1446.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc M, Hsieh WY, Robertson KA, Kropp KA, Forster T, Shui G, Lacaze P, Watterson S, Griffiths SJ, Spann NJ, et al. (2013). The transcription factor STAT-1 couples macrophage synthesis of 25-hydroxycholesterol to the interferon antiviral response. Immunity 38, 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc M, Hsieh WY, Robertson KA, Watterson S, Shui G, Lacaze P, Khondoker M, Dickinson P, Sing G, Rodríguez-Martín S, et al. (2011). Host defense against viral infection involves interferon mediated down-regulation of sterol biosynthesis. PLoS Biol. 9, e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, and Dyer WJ (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917. [DOI] [PubMed] [Google Scholar]
- Carroll RG, Zas1ona Z, Galván-Peña S, Koppe EL, Sévin DC, Angiari S, Triantafilou M, Triantafilou K, Modis LK, and O’Neill LA (2018). An unexpected link between fatty acid synthase and cholesterol synthesis in proinflammatory macrophage activation. J. Biol. Chem. 293, 5509–5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes T, and Metallo CM (2019). Quantifying intermediary metabolism and lipogenesis in cultured mammalian cells using stable isotope tracing and mass spectrometry. Methods Mol. Biol. 1978, 219–241. [DOI] [PubMed] [Google Scholar]
- Dang EV, McDonald JG, Russell DW, and Cyster JG (2017). Oxysterol restraint of cholesterol synthesis prevents AIM2 inflammasome activation. Cell 171, 1057–1071.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Wang K, Liu W, She Y, Sun Q, Shi J, Sun H, Wang DC, and Shao F (2016). Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 535, 111–116. [DOI] [PubMed] [Google Scholar]
- Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CRH, Shimizu T, Spener F, van Meer G, Wakelam MJO, and Dennis EA (2009). Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 50, S9–S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier B, and Philpott DJ (2005). Recognition of Staphylococcus aureus by the innate immune system. Clin. Microbiol. Rev. 18, 521–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S, Yang L, Li P, Hofmann O, Dicker L, Hide W, Lin X, Watkins SM, Ivanov AR, and Hotamisligil GS (2011). Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature 473, 528–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakis N, Sansbury BE, Patsalos A, Hays TT, Riley CO, Han X, Spite M, and Nagy L (2019). Dynamic changes to lipid mediators support transitions among macrophage subtypes during muscle regeneration. Nat. Immunol. 20, 626–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, and Natoli G (2016). Molecular control of activation and priming in macrophages. Nat. Immunol. 17, 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S, and Taylor PR (2005). Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 5, 953–964. [DOI] [PubMed] [Google Scholar]
- Halbleib K, Pesek K, Covino R, Hofbauer HF, Wunnicke D, Hänelt I, Hummer G, and Ernst R (2017). Activation of the unfolded protein response by lipid bilayer stress. Mol. Cell 67, 673–684.e8. [DOI] [PubMed] [Google Scholar]
- Heilig R, Dick MS, Sborgi L, Meunier E, Hiller S, and Broz P (2018). The gasdermin-D pore acts as a conduit for IL-1b secretion in mice. Eur. J. Immunol. 48, 584–592. [DOI] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, and Brown MS (2002). SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Tabbi-Anneni I, Gunda V, and Wang L (2010). Transcription factor Nrf2 regulates SHP and lipogenic gene expression in hepatic lipid metabolism. Am. J. Physiol. Gastrointest. Liver Physiol. 299, G1211–G1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashima K, Honda T, Ginhoux F, and Egawa G (2019). The immunological anatomy of the skin. Nat. Rev. Immunol. 19, 19–30. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, and Akira S (2008). Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid–inducible gene-I and melanoma differentiation–associated gene 5. J. Exp. Med. 205, 1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher JK, and Nickol GB (2015). Isotopomer spectral analysis: Utilizing Nonlinear Models in Isotopic Flux Studies. Methods Enzymol. 561, 303–330. [DOI] [PubMed] [Google Scholar]
- Lancaster GI, Langley KG, Berglund NA, Kammoun HL, Reibe S, Estevez E, Weir J, Mellett NA, Pernes G, Conway JRW, et al. (2018). Evidence that TLR4 is not a receptor for saturated fatty acids but mediates lipid-induced inflammation by reprogramming macrophage metabolism. Cell Metab. 27, 1096–1110.e5. [DOI] [PubMed] [Google Scholar]
- Lawrence T, and Natoli G (2011). Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat. Rev. Immunol. 11, 750–761. [DOI] [PubMed] [Google Scholar]
- Lee JH, Phelan P, Shin M, Oh BC, Han X, Im SS, and Osborne TF (2018). SREBP-1a–stimulated lipid synthesis is required for macrophage phagocytosis downstream of TLR4-directed mTORC1. Proc. Natl. Acad. Sci. USA 115, E12228–E12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebisch G, Vizcaíno JA, Köfeler H, Trötzmuller M, Griffiths WJ, Schmitz G, Spener F, and Wakelam MJO (2013). Shorthand notation for lipid structures derived from mass spectrometry. J. Lipid Res. 54, 1523–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SY, Aliyari R, Chikere K, Li G, Marsden MD, Smith JK, Pernet O, Guo H, Nusbaum R, Zack JA, et al. (2013). Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity 38, 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, and Lieberman J (2016). Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsalu T, and Vilo J (2015). ClustVis: A web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 43, W566–W570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelucci A, Cordes T, Ghelfi J, Pailot A, Reiling N, Goldmann O, Binz T, Wegner A, Tallam A, Rausell A, et al. (2013). Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl. Acad. Sci. USA 110, 7820–7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EL, Ryan DG, Prag HA, Dikovskaya D, Menon D, Zaslona Z, Jedrychowski MP, Costa ASH, Higgins M, Hams E, et al. (2018). Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556, 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki M, Dobrzyn A, Elias PM, and Ntambi JM (2005). Stearoyl-CoA desaturase-2 gene expression is required for lipid synthesis during early skin and liver development. Proc. Natl. Acad. Sci. USA 102, 12501–12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KJ, Sheedy FJ, and Fisher EA (2013). Macrophages in atherosclerosis: a dynamic balance. Nat. Rev. Immunol. 13, 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvihill E, Sborgi L, Mari SA, Pfreundschuh M, Hiller S, and Muller DJ (2018). Mechanism of membrane pore formation by human gasdermin-D. EMBO J. 37, e98321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvihill E, Van Pee K, Mari SA, Muller DJ, and Yildiz Ö (2015). Directly observing the lipid-dependent self-assembly and pore-forming mechanism of the cytolytic toxin listeriolysin O. Nano Lett. 15, 6965–6973. [DOI] [PubMed] [Google Scholar]
- Oishi Y, Spann NJ, Link VM, Muse ED, Strid T, Edillor C, Kolar MJ, Matsuzaka T, Hayakawa S, Tao J, et al. (2017). SREBP1 contributes to resolution of pro-inflammatory TLR4 signaling by reprogramming fatty acid metabolism. Cell Metab. 25, 412–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe Y, and Medzhitov R (2016). Tissue biology perspective on macrophages. Nat. Immunol. 17, 9–17. [DOI] [PubMed] [Google Scholar]
- Paton CM, and Ntambi JM (2009). Biochemical and physiological function of stearoyl-CoA desaturase. Am. J. Physiol. Endocrinol. Metab. 297, E28–E37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboldi A, Dang EV, McDonald JG, Liang G, Russell DW, and Cyster JG (2014). Inflammation. 25-Hydroxycholesterol suppresses interleukin-1driven inflammation downstream of type I interferon. Science 345, 679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong X, Wang B, Dunham MM, Hedde PN, Wong JS, Gratton E, Young SG, Ford DA, and Tontonoz P (2015). Lpcat3-dependent production of arachidonoyl phospholipids is a key determinant of triglyceride secretion. eLife 4, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scumpia PO, Botten GA, Norman JS, Kelly-Scumpia KM, Spreafico R, Ruccia AR, Purbey PK, Thomas BJ, Modlin RL, and Smale ST (2017). Opposing roles of toll-like receptor and cytosolic DNA-STING signaling pathways for Staphylococcus aureus cutaneous host defense. PLoS Pathog. 13, e1006496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, and Akira S (2004). TLR signaling pathways. Semin. Immunol. 16, 3–9. [DOI] [PubMed] [Google Scholar]
- Takeshita S, Kaji K, and Kudo A (2000). Identification and characterization of the new osteoclast progenitor with macrophage phenotypes being able to differentiate into mature osteoclasts. J. Bone Miner. Res. 15, 1477–1488. [DOI] [PubMed] [Google Scholar]
- Vromman F, and Subtil A (2014). Exploitation of host lipids by bacteria. Curr. Opin. Microbiol. 17, 38–45. [DOI] [PubMed] [Google Scholar]
- Wei X, Song H, Yin L, Rizzo MG, Sidhu R, Covey DF, Ory DS, and Semenkovich CF (2016). Fatty acid synthesis configures the plasma membrane for inflammation in diabetes. Nature 539, 294–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KJ, Argus JP, Zhu Y, Wilks MQ, Marbois BN, York AG, Kidani Y, Pourzia AL, Akhavan D, Lisiero DN, et al. (2013). An essential requirement for the SCAP/SREBP signaling axis to protect cancer cells from lipotoxicity. Cancer Res. 73, 2850–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, and Akira S (2003). Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301, 640–643. [DOI] [PubMed] [Google Scholar]
- York AG, Williams KJ, Argus JP, Zhou QD, Brar G, Vergnes L, Gray EE, Zhen A, Wu NC, Yamada DH, et al. (2015). Limiting cholesterol biosynthetic flux spontaneously engages Type i IFN signaling. Cell 163, 1716–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
These data are available at the NIH Common Fund’s National Metabolomics Data Repository (NMDR) website, the Metabolomics Workbench (https://www.metabolomicsworkbench.org) where it has been assigned Project ID: PR000914. The data can be accessed directly via the Project DOI: https://doi.org/10.21228/M8JH69. This work is supported by NIH grant U2CDK119886.