Abstract
Background: Mutations of thyroid hormone receptor α1 (TRα1) cause resistance to thyroid hormone (RTHα). Patients exhibit growth retardation, delayed bone development, anemia, and bradycardia. By using mouse models of RTHα, much has been learned about the molecular actions of TRα1 mutants that underlie these abnormalities in adults. Using zebrafish models of RTHα that we have recently created, we aimed to understand how TRα1 mutants affect the heart function during this period.
Methods: In contrast to human and mice, the thra gene is duplicated, thraa and thrab, in zebrafish. Using CRISPR/Cas9-mediated targeted mutagenesis, we created C-terminal mutations in each of two duplicated thra genes in zebrafish (thraa 8-bp insertion or thrab 1-bp insertion mutations). We recently showed that these mutant fish faithfully recapitulated growth retardation as found in patients and thra mutant mice. In the present study, we used histological analysis, gene expression profiles, confocal fluorescence, and transmission electron microscopy (TEM) to comprehensively analyze the phenotypic characteristics of mutant fish heart during development.
Results: We found both a dilated atrium and an abnormally shaped ventricle in adult mutant fish. The retention of red blood cells in the two abnormal heart chambers, and the decreased circulating blood speed and reduced expression of contractile genes indicated weakened contractility in the heart of mutant fish. These abnormalities were detected in mutant fish as early as 35 days postfertilization (juveniles). Furthermore, the expression of genes associated with the sarcomere assembly was suppressed in the heart of mutant fish, resulting in abnormalities of sarcomere organization as revealed by TEM, suggesting that the abnormal sarcomere organization could underlie the bradycardia exhibited in mutant fish.
Conclusions: Using a zebrafish model of RTHα, the present study demonstrated for the first time that TRα1 mutants could act to cause abnormal heart structure, weaken contractility, and disrupt sarcomere organization that affect heart functions. These findings provide new insights into the bradycardia found in RTHα patients.
Keywords: zebrafish, TRα; mutations; bradycardia; heart defects; contractility; blood speed
Introduction
Thyroid hormone nuclear receptors (TRs) mediate the genomic actions of thyroid hormone (triiodothyronine [T3]) in growth, development, and differentiation, as well as in maintaining metabolic homeostasis. There are two TR genes in humans, THRA and THRB, encoding three major T3 binding TR isoforms (TRα1, TRβ1, and TRβ2). The expression of these TR isoforms is tissue dependent and developmentally regulated. The transcriptional activity of TR is modulated by a host of many nuclear coregulatory proteins, such as corepressors (e.g., nuclear receptor corepressor 1) and coactivators (e.g., steroid hormone receptor coactivators). For T3 positively regulated genes, TRs recruit the nuclear corepressors (NCOR1 and NCOR2) for transcriptional repression. In the presence of T3, TR undergoes conformational changes, resulting in the release of corepressors and allowing recruitment of a multiprotein coactivator complex for transcriptional activation (1–5).
TRs' effect on critical functions is evident in that mutations of THRB gene cause resistance to thyroid hormone (RTH) actions in target tissues (RTHβ). Patients with RTHβ manifest elevated serum thyroid hormones, elevated nonsuppressible thyrotropin (TSH), and other symptoms, including slow growth, hearing loss, and attention-deficit/hyperactivity disorder (4,6). Studies using mouse models of RTHβ have elucidated how TRβ mutants act to cause defects in growth and bone (7,8) and have clarified the molecular basis underlying clinical issues of heterogeneity in target tissue resistance of patients. Furthermore, an RTHβ mouse model expressing a mutant NCOR1 (ThrbPV/+NcorΔID/ΔID mice) was used to identify NCOR1 as a critical regulator of the dominant negative actions of TRβ mutants (9). These findings have advanced the understanding of in vivo molecular actions of TRβ mutants in RTHβ.
Still, whether mutations of the THRA gene could cause human diseases was not known until 23 years after the identification of THRB mutations in RTHβ patients. In 2012–2013, the first THRA mutations were reported (10–14). RTHα patients are clinically distinct from RTHβ patients and are characterized by nearly normal thyroid hormone and TSH levels, as well as growth retardation, delayed bone development, constipation, erythroid disorders, and bradycardia (10,13–16). The molecular actions of TRα1 mutants in growth and bone abnormalities have been studied in the Thra1PV/+ mouse, a model of RTHα. As found in patients, Thra1PV/+ mice have delayed closure of the skull sutures with enlarged fontanelles, as well as severe postnatal growth retardation with delayed bone age (17,18). Analysis of bone phenotypes in the Thra1PV/+ mouse led to the conclusion that TRα1 plays a major physiological role in skeletal development, linear growth, and the maintenance of adult skeletal integrity in vivo.
Recently, how TRα1 mutants act to cause anemia and constipation in RTHα patients has been studied in Thra1PV mice. TRα1 mutants act to suppress the clonogenic potential of progenitors in the erythrocytic lineage in bone marrow, leading to a reduction of mature erythrocytes (19,20). Furthermore, TRα1 mutants suppress the expression of the Gata-1 gene, a T3 directly regulated gene. TRα1 mutants impair erythropoiesis, via repression of the Gata-1 gene expression and its downstream regulated genes, causing anemia (19,20). Adult Thra1PV mice were also found to exhibit constipation as do RTHα patients (21). Further elucidation showed that TRα1 mutants act to decrease villi and reduce stem cell proliferation in the intestine crypts of Thra1PV mice, thereby impairing intestine functions (21).
While the Thra1PV/+ mouse is very useful to dissect the molecular actions of TRα1 mutants in adults and to understand how TRα1 mutants underpin the pathogenesis in patients, this model has limitations. It is not suitable for studying molecular actions during early development. The almost insurmountable difficulty is that female Thra1PV/+ mice are virtually infertile, and males have deficient fertility. Therefore, it is nearly impossible to get sufficient mutant embryos and neonates for phenotypic characterization during development.
Consequently, alternative animal models are needed to circumvent this difficulty, and therefore, we recently developed zebrafish models of RTHα (22). We chose zebrafish because they have been increasingly used as models for human diseases due to their high fecundity and rapid external embryonic development, as well as the easy visualization of transparent embryos.
In contrast to humans and mice, in zebrafish, the thra gene is duplicated as the thraa and thrab genes. Using CRISPR/Cas9 gene editing, we have created two mutant fish expressing C-terminal truncated mutations as found in RTHα patients (thraa 8-bp insertion and thrab 1-bp insertion mutations). As found in patients and in Thra1PV/+ mice, these mutant fish exhibit growth retardation, indicating that the in vivo mutant actions are conserved among humans, mice, and fish. Thus, these mutant zebrafish are valid models to study other RTHα phenotypes during development.
Characterization of the heart phenotype in an RTHα mouse model, Thra1R384C/+ mice, showed bradycardia, decreased blood pressure, and impaired contractions (23,24). However, in these studies, it was not known the stage of development at which the heart activities were affected in the mutant mice. Furthermore, how the structures of the heart might have changed in Thra1R384C/+ mice to cause these phenotypic manifestations was not clear. In the present study, we analyzed the heart phenotype of mutant zebrafish. We found that both homozygous thraa 8-bp ins and thrab 1-bp ins mutant fish exhibit abnormal heart shape and size, slow heart beats, and decreased contractions and blood flow speed. These changes were detected as early as in juveniles (31 days postfertilization [dpf]) and persisted until adulthood. Decreased expression of contractile genes and several genes related to sarcomere assembly was altered in mutant fish. Mutant fish exhibited abnormal sarcomere structures as revealed by transmission electron microscopy (TEM). Importantly, the zebrafish models allowed us to discern detailed internal sarcomere structures. With this histological information, it provides the foundation to explore whether these cardiac defects also exist in the mouse models of RTHα, and thereby gain insight into the molecular actions of mutant TRα1 in vivo.
Materials and Methods
Ethics statement
All zebrafish experiments were performed in compliance with the guidelines for animal handling and approved animal study protocols under the National Cancer Institute, Animal Care and Use Committee.
Zebrafish husbandry
The TAB5 mutant line of zebrafish was used for studies. The wild-type (WT) and thra mutant lines of fish and genotyping methods have been previously described (22). To generate thra-Tg line, thraa 8-bp ins (m/m) expressing gata1-DsRed, we outcrossed tg(gata1:dsRed) with thraa 8-bp ins (m/m) fish. The DsRed expression was imaged with a fluorescence microscope (Leica TL5000) at 48 hours postfertilization. Tg (gata1:DsRed) was obtained from Dr. Raman Sood (National Institutes of Health).
Measure of heart rate of zebrafish
Before measurements, fish were anesthetized with 1 × MS222 solution (0.2 g MS222/50 mL phosphate-buffered saline [PBS]). Heart rate was measured at two minutes after anesthesia 1 × MS222 at room temperature. Heart rates were measured every minute for 30 minutes. Heart rate (beats per minute) was calculated by counting the number of heart beats in 15 seconds and multiplying that number by four. All heart rates were measured on a LEICA GZ4 microscope.
Analysis of blood flow speed
The tg (gata1:DsRed) and tg [thraa 8-bp ins (m/m):gata1:DsRed] lines were used. Based on our results from measuring the heart rate of zebrafish, fish were anesthetized with 1 × MS222 solution (0.2 g MS222/50 mL PBS) for 20 minutes. Confocal images were acquired using a Nikon Ti2-E spinning disk confocal microscope equipped with a Yokogawa CSU-W1 50 μm pinhole disk, a 20 × C-Apochromat (N.A. 0.95) water immersion lens, and Photometrics BSI sCMOS camera for an effective x–y pixel size of 0.33 μm. For time-lapse imaging, images were collected every 20 ms for a duration of 30 seconds. Cell tracking analysis, including speed measurements, was done using the Spots tracking module of Imaris software (v.9.3.1).
Transmission electron microscopy
Zebrafish heart ventricle tissues, ∼1 mm3 in size, were fixed for 48 hours at 4°C in 2.5% glutaraldehyde and 1% paraformaldehyde in 0.1 M cacodylate buffer (pH 7.4), and washed with cacodylate buffer three times. The tissues were fixed with 1% OsO4 for two hours, washed again with 0.1 M cacodylate buffer three times, washed with water, and placed in 1% uranyl acetate for one hour. The tissues were subsequently serially dehydrated in ethanol and propylene oxide and embedded in EMBed 812 resin (Electron Microscopy Sciences, Hatfield, PA). Thin sections, ∼80 nm, were obtained by utilizing the Leica Ultracut-UCT ultramicrotome (Leica, Deerfield, IL), placed onto 300 mesh copper grids, and stained with saturated uranyl acetate in 50% methanol and then with lead citrate. The grids were viewed with a JEM-1200EXII electron microscope (JEOL Ltd., Tokyo, Japan) at 80 kV and images were recorded on the XR611M, midmounted, 10.5M pixel CCD camera (Advanced Microscopy Techniques Corp., Danvers, MA).
Zebrafish sample collection
Fish were euthanized with MS222 before heart collections. For real-time quantitative polymerase chain reaction (RT-qPCR) analysis, ∼7 to 11 hearts were obtained from adult male and female zebrafish at 10–13 months postfertilization (mpf). For Western blot analysis, ∼9 to 11 hearts were obtained from adult male and female zebrafish at 11 mpf. For histological analysis, ∼3 hearts per age group were obtained: from juveniles at 1.1 mpf and from adult zebrafish at 4–5 mpf and 11.9 mpf. In all cases, sample collection was performed with a LEICA GZ4 microscope.
Histological analyses
Zebrafish were euthanized and fixed in 4% formaldehyde at 4°C for a minimum of 24 hours, followed by decalcification in a 1:1 ratio of formic acid/sodium citrate for 24 hours at room temperature. The fish were then dehydrated through a series of ethanol and then xylene, and finally were embedded in paraffin. Five micrometer sections were prepared and stained with hematoxylin and eosin (H&E) (Histoserv, Germantown, MD). Histological section images for H&E were captured with a light microscope (Olympus LC 30 camera).
RNA isolation and RT-qPCR
Total RNA from hearts was isolated using TRIzol (Invitrogen) according to the manufacturer's protocol. RT-qPCR was performed with the one-step SYBR Green RT-qPCR Master Mix (Qiagen, Valencia, CA) on an ABI 7900HT system. In each genotype, samples in triplicates were tested for the target genes. Data were analyzed using Prism 8 software (GraphPad Software, Inc.). Primer sequences are shown in Supplementary Table S1. Ef1a was used as the housekeeping gene for controls.
Western blot analysis
The Western blot analyses were performed as described previously (25). The species specificity and other relevant information about the antibodies used in the present study are listed in Supplementary Table S2. Antibodies were used at the manufacturer's recommended concentration. For control of protein loading, the blot was probed with an antibody against GAPDH.
Statistical analysis
All data are expressed as mean ± standard deviation. All data analyses used two-tailed unpaired t-tests, and p < 0.05 was considered statistically significant. GraphPad Prism version 8 for Mac OS X was used to perform analyses of variances.
Results
The heart of thraa and thrab mutant zebrafish exhibits abnormal histology
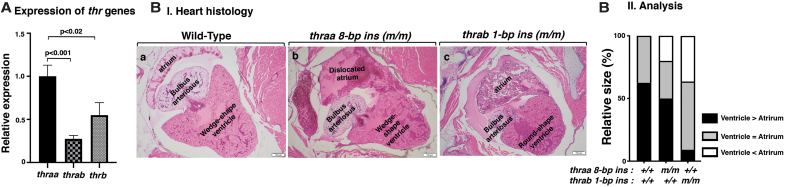
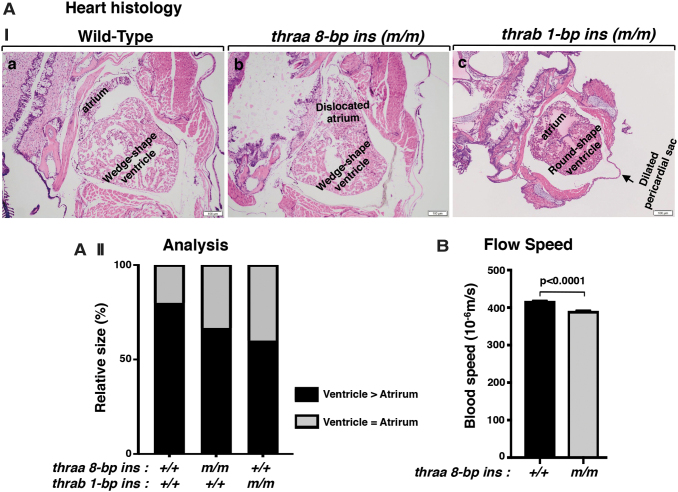
We first determined the expression of the thyroid hormone receptor α (thr) genes in the heart. Consistent with what has been reported for humans and mice (15,26,27), the thra gene was the major thr gene in the heart (Fig. 1A, first two bars). Between the two duplicated thra genes, the thraa gene expression level was 4.5-fold higher than that of the thrab gene. Analysis of adult heart showed abnormal shape and size in mutant fish (Fig. 1B[I]). In the heart of WT fish, the ventricle is usually wedge-shaped and larger than the atrium (ratio of ventricle vs. atrium >1). In contrast to the heart of WT fish, the size of the ventricle and atrium was similar (the ratio of ventricle vs. atrium was ∼1), and furthermore, the atrium was displaced in homozygous thraa 8-bp ins mutant fish (Fig. 1B[I-b]). In the heart of homozygous thrab 1-bp ins mutant fish, the ventricle was round, with the size close to that of atrium (ratio of ventricle vs. atrium was ∼1). Figure 1B(II) compares the extent of the abnormality in the size of ventricle and atrium among WT, thraa 8-bp ins, and thrab1-bp ins mutant fish. The greater extent in the distribution of enlarged atrium in the mutant fish suggested that the atrium was dilated. Furthermore, in the heart of thrab1-bp ins mutant fish, prominent pericardial effusion was also noted. These data showed that the mutations of the thra genes cause heart abnormalities in adults.
FIG. 1.
The mutation of thra genes results in cardiac atrial dilation in adult homozygous thra (m/m) mutant zebrafish. (A) The mRNA expression of thyroid hormone receptor genes (thraa, thrab, and thrb) in the heart of WT fish (3.7 mpf) was determined by RT-PCR as described in the Materials and Methods section. The number of WT fish analyzed is 14 (N = 14). The data are shown as mean ± SE (n = 3, three biological samples, each triplicate), with p-values to indicate significant changes. Two-tailed unpaired t-test, p-adjusted <0.05, was used for statistical analysis. GraphPad Prism version 8 for Mac OS X was used to perform analyses of variances. (B[I]) Representative histology features of zebrafish heart in WT (4 mpf) (B[a]); homozygous thraa 8-bp ins (m/m) (4.1 mpf) (B[b]); and homozygous thrab 1-bp ins (m/m) (5.1 mpf) (B[c]), sectioned and stained with H&E. The histology features of heart in thraa 8-bp ins (m/m) mutant fish show laterally displaced and dilated atrium; thrab 1-bp ins (m/m) mutant fish show a round-shaped ventricle and dilated atrium. (B[II]) Semiquantitative analysis of relative size of ventricle to atrium. Relative ventricle and atrium sizes at the aortic valve and atrioventricular valves are compared between WT (N = 6), thraa 8-bp ins (m/m) (N = 6), and thrab 1-bp ins (m/m) (N = 6) mutant fish. mpf, months postfertilization; RT-PCR, real-time polymerase chain reaction; SE, standard error; H&E, hematoxylin and eosin; WT, wild type.
Thraa 8-bp ins and thrab 1-bp ins mutations impair contractility of the heart
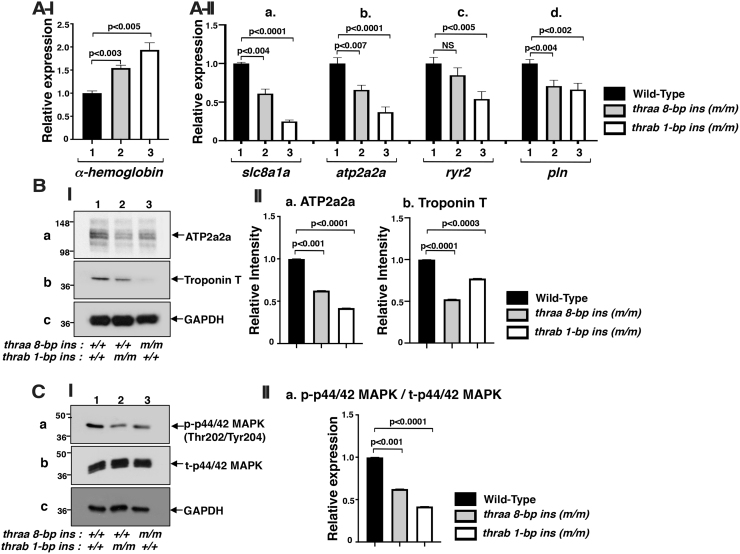
In addition to the abnormality in size and shape, the trabeculae, which are clearly apparent in the ventricle of WT fish (Fig. 1B[I-a]), were not visible in the ventricles of either thraa 8-bp ins mutant fish (Fig. 1B[I-b]) or thrab 1-bp ins mutant fish (Fig. 1B[I-c]). Instead, the ventricles were filled with erythrocytes, suggesting that blood flow out of the ventricles was impeded. To validate that there was more retention of erythrocytes in the mutant hearts, we determined the expression of the α-hemoglobin gene (hbaa1 gene) in the heart. We found that, compared with WT fish, the expressions of the hbaa1 gene were 1.5- and 1.9-fold higher in the hearts of thraa 8-bp ins and thrab 1-bp ins, respectively (Fig. 2A[I]), supporting that there were more erythrocytes retained in the mutant hearts than in the WT heart.
FIG. 2.
Increased gene expression of α-hemoglobin (A[I]) altered calcium handling regulator gene mRNA expression (A[II]), protein abundance (B), and attenuated ERK signaling (C) in adult homozygous thra (m/m) mutant zebrafish. (A[I]) The mRNA expression of α-hemoglobin gene in the heart of WT, homozygous thraa 8-bp ins (m/m), and homozygous thrab 1-bp ins (m/m) fish (6.2 mpf) (N = 15). (A[II]) The mRNA expression of Ca2+ handling regulator genes, (a) slc8a1a, (b) atp2a2a, (c) ryr2, and (d) pln, in the heart of WT, homozygous thraa 8-bp ins (m/m), and homozygous thrab 1-bp ins (m/m) fish (11–12 mpf) (N = 10–12) was determined by RT-PCR as described in the Materials and Methods section. (B[I]) Western blot analysis was carried out for (a) ATP2a2a, (b) troponin T, and (c) GAPDH using heart as described in the Materials and Methods section. (B[II]) Quantitative analysis of relative protein abundance of the ratios of (a) ATP2a2a and (b) troponin T using GAPDH as a loading control. (C[I]) Western blot analysis of (a) p-ERK, (b) total ERK, and (c) GAPDH from the heart of fish with genotypes indicated. (C[II]) Quantitative analysis of relative protein abundance of the ratio of p-ERK to total ERK using GAPDH as a loading control. For Western blot analysis, the number of fish used was 9–11 (11 mpf). The data are shown as mean ± SE (n = 3) with p-values to indicate significant changes. Two-tailed unpaired t-test, p-adjusted <0.05, was used for statistical analysis. GraphPad Prism version 8 for Mac OS X was used to perform analyses of variances. NS, not significant.
We next tested whether the retention of erythrocytes was due to the weakened contractility by the heart to pump out the blood to peripheral circulation in the mutant fish. Accordingly, we compared the expressions of the contractile-related genes in the heart of WT and mutant fish. As shown in Figure 2A(II), the expressions of solute carrier family 8 member a1 (slc8a1a) (Fig. 2A[II-a]), ATPase/sarcoplasmic/endoplasmic reticulum Ca+2 transporting 2a (atp2a2a) (Fig. 2A[II-b]), ryanodine receptor 2 (ryr2b) (Fig. 2A[II-c]), and phospholamban (pln) (Fig. 2A[II-d]) were suppressed, ranging from 34% to 75% in the heart of thraa 8-bp ins and thrab 1-bp mutant fish. SLC8A1A is involved in several processes, including calcium ion transport, cellular calcium homeostasis, and sarcomere organization (28–31). The sarcoplasmic reticulum (SR) plays an important role in the contraction and relaxation coupling in the myocardium. ATP2A2A pumps cytosolic Ca2+ into the SR lumen during relaxation of the cardiac myocytes. ATP2A2A regulates heart contraction (31–33). RYR2b mediates the release of sequestered calcium from the endoplasmic reticulum into the cytosol to regulate heart functions (34,35). Phospholamban (PLN) regulates the Ca2+ transport activity of ATP2a2a in heart muscle. PLN is involved in cardiac activity (36–38). It is known that the expression of atp2a2a gene in the myocardium is increased by thyroid hormone (T3) (39–42). However, the two THRA mutant proteins lose T3 binding activity (22), resulting in the suppression of the expression of the atp2a2a gene (Fig. 2A[II-b]). The expression of other critical contractile-related genes was also significantly lower in the mutant heart than the WT heart (Fig. 2A[II-a, c, d]).
We further analyzed the protein levels of the major contractile genes. Consistent with mRNA expression, we found that the protein levels of ATP2a2a were reduced by 37% and 58%, respectively, in the heart of thraa 8-bp ins and thrab 1-bp mutant fish (Fig. 2B[I-a]; quantitation shown in Fig. 2B[II-a]). Likewise, the protein levels of troponin T were reduced by 48% and 23%, respectively, in the heart of thraa 8-bp ins and thrab 1-bp mutant fish (Fig. 2B[I-b]; quantitation shown in Fig. 2B[II-b]). Troponin T is one of the three major components of the troponin complex, via calcium regulates excitation/contraction coupling in the heart. Troponin T attaches troponin to tropomyosin and to the myofibrillar thin filament to affect contractility (43). Taken together, these results indicate that both thraa 8-bp ins and thrab 1-bp ins mutations suppress the expression of contractile genes to weaken heart contractility.
The mitogen-activated protein kinase (MAPK) signaling pathway is essential for the proliferation of cardiomyocytes, especially during heart regeneration (44). We next analyzed whether cardiomyocyte proliferation was affected in the heart of mutant fish. We found that the phosphorylated p-44/42 MAPK was decreased in the heart of thraa 8-bp ins and thrab 1-bp ins mutant fish by 39% and 59%, respectively, suggesting that the mutations attenuated the signaling to decrease cardiomyocyte proliferation. The decreased cardiomyocyte proliferation could lead to defective activities of the heart.
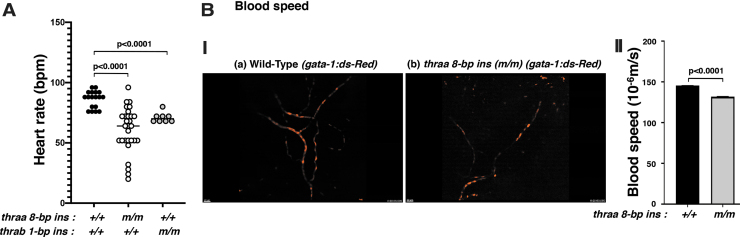
To demonstrate the functional consequences of the reduced expression of contractile genes of the heart, we compared the heart rates of WT and mutant fish. We found that the heart rates in thraa 8-bp ins and thrab 1-bp ins mutants were 32% and 22% lower, respectively, than in WT fish (Fig. 3A). These data indicate that these mutant fish exhibit bradycardia as found in RTHα patients (10,13) and in mutant TRα1R384C/+ mice, a model of RTHα (23,24).
FIG. 3.
thraa 8-bp ins (m/m) mutants show decreased heart rate (A) and decreased blood flow speed (B). (A) Heart rates in adult WT, thraa 8-bp ins (m/m), and thrab 1-bp ins (m/m) zebrafish were counted as described in the Materials and Methods section. For the measuring of heart rates, 8–29 fish (11–14 mpf) were used. (B[I]) Decreased flow speed of red blood cells in tg (thraa 8-bp ins(m/m):gata-1-dsRed) (b) compared with tg (WT: thraa+/+:gata-1-dsRed) (a) (2 mpf). The video can be viewed in Supplementary Figure S1. (B[II]) Comparison of the flow speed of red blood cells in WT and homozygous thraa 8-bp ins mutant fish. The images were captured from 3 to 4 individual WT or mutant fish; each fish was imagined 4–5 times. The data are shown as mean ± SE with p-values to indicate significant changes. Two-tailed unpaired t-test, p-adjusted <0.05, was used for statistical analysis. GraphPad Prism version 8 for Mac OS X was used to perform analyses of variances.
The decreased expression of contractile genes suggested that the heart could be deficient in pumping out the blood to circulate in the body. We therefore crossed transgenic fish expressing gata-1:DsRed with thraa 8-bp ins mutant fish and imaged the blood flow. The gata-1 gene is essential for terminal differentiation of erythrocytes. Transgenic fish expressing DsRed chromophore (45) driven by the gata-1 promoter have been used to study the ontogenic development of erythropoiesis noninvasively (46). As shown in Figure 3B(II) (dynamic video can be viewed in Supplementary Video S1A and S1B), the flow speed of the WT fish (thra+/+:gata-1-DsRed; Fig. 3B[I-a]) was faster than thraa 8-bp ins(m/m)gata-1:DsRed fish (Fig. 3B[I-b]). Quantitative analysis showed that the red cell flow speed of thraa 8-bp ins(m/m):gata-1:DsRed fish was 9.3% slower than that of WT fish (Fig. 3B[II]). Taken together, these data indicate that mutations of the thra gene could cause heart abnormalities.
Abnormal myofibril organization in the heart of mutant fish
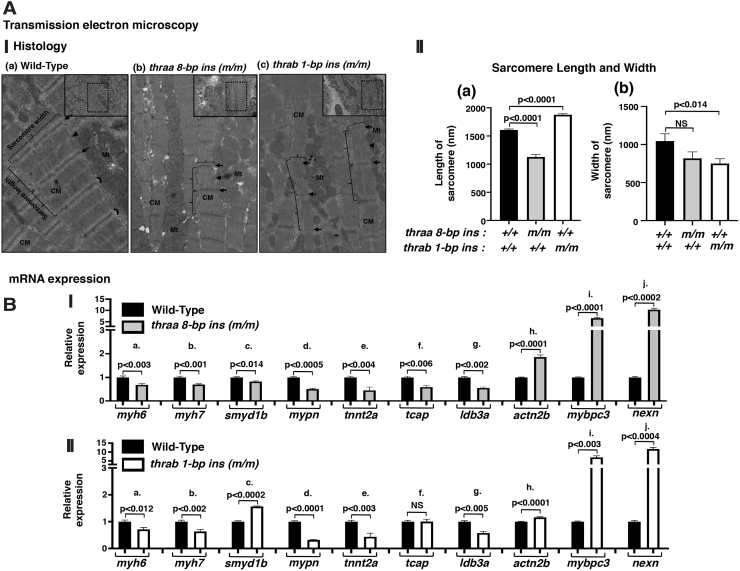
The dilation of the atrium (Fig. 1B[I, II]) and weakened heart contractility in the mutant fish prompted us to ascertain whether the sarcomere assembly could be affected in the thraa 8-bp ins and thrab 1-bp ins mutant fish. Using TEM, we analyzed the sarcomere organization in the heart of WT and two mutant fish. Figure 4A shows the representative micrographs. We found abnormal sarcomere assembly in the heart of mutant fish. WT sarcomeres were well organized with discrete sarcomere units separated by clearly visible Z-disc/I bands (Fig. 4A[I-a]). These sarcomeres unite to form individual myofibrils that align along the longitudinal axis of the cardiomyocytes. Myofibrils are highly ordered structures brought together by three components: actin and myosin filaments, accessory proteins of actin and myosin, and scaffolding proteins (Fig. 4A[I-a]) (47). As shown in Figure 4A(I-b), the myofibrils were thinner with narrower and shorter sarcomere units in the heart of thraa 8-bp ins mutant fish. Interestingly, the sarcomere units were more elongated, but narrower in the heart of thrab 1-bp ins mutant fish (Fig. 4A[I-c]) compared with WT sarcomeres (Fig. 4A[I-a]). The quantitative analysis shown in Figure 4A(II) indicates that the sarcomere length was reduced by 37% in thraa 8-bp ins mutant fish, but increased by 10% in thrab 1-bp ins mutant fish. Moreover, the width of sarcomeres was reduced by 28% and 33%, respectively, in the heart of thraa 8-bp ins and thrab 1-bp ins mutant fish (Fig. 4A[II-b]). These results indicate that mutations of the thraa and thrab genes impaired the assembly of sarcomeres to affect the contractility of the heart.
FIG. 4.
TEM shows myofilaments in ventricular sarcomere of adult zebrafish (A[I]). Representative micrographs of WT (a) and thraa 8-bp ins (m/m) (b) and thrab 1-bp ins (m/m) (c) mutants. Small lined rectangle indicates electron micrographs ( × 2000). Dotted rectangle areas were enlarged from each electron micrograph. Disrupted structure of ventricular sarcomere (length, width, and I-band) observed in thraa 8-bp ins (m/m) (b) and thrab 1-bp ins (m/m) (c) mutants compared with WT (a). Ventricular myocardium constituted of four to five overlapping layers of cardiac myocytes. Arrows point to Z-disc, crescent indicates I-band, arrow heads point to M-band, and brackets indicate representative sarcomere length (solid lines) and width (dotted lines), Mt; mitochondria, CM, cardiac myocytes. (A[II]). The length (a) and width (b) of individual sarcomeres were measured and graphed (n = 20) in randomly picked TEM images, the number of fish (N = 4 each genotype). The data are expressed as mean ± SE; the p-values are indicated. Two-tailed unpaired t-test, p-adjusted <0.05, was used for statistical analysis. (B) The mRNA expression of filaments and sarcomere structure genes in the heart of homozygous thraa 8-bp ins mutant fish (I) and homozygous thrab 1-bp ins (m/m) fish (11–12 mpf) (N = 10–12) (II) was determined by RT-PCR as described in the Materials and Methods section. The genes are as follows: (B[Ia] and B[IIa]) myh6, (B[Ib] and B[IIb]) myh7, (B[Ic] and B[IIc]) smyd1b, (B[Id] and B[IId]) mypn, (B[Ie] and B[IIe]) tnnt2, (B[If] and B[IIf]) tcap, (B[Ig] and B[IIg]) ldb3a, (B[Ih] and B[IIh]) actn2b, (B[Ii] and B[IIi]) mybpc3, and (B[Ij] and B[IIj]) nexn. The data are shown as mean ± SE (n = 3 biological samples, each with triplicates) with p-values to indicate significant changes. Two-tailed unpaired t-test, p-adjusted <0.05, was used for statistical analysis. GraphPad Prism version 8 for Mac OS X was used to perform analyses of variances. TEM, transmission electron microscopy.
To understand the molecular basis underlying the abnormal organization of myofibrils, we analyzed the expression of a battery of genes associated with sarcomeres. The expression of two cardiac myosin heavy chain genes, myh6 and myh7, which are the major components of thick filaments in a sarcomere unit (47), was lower in the heart of thraa 8-bp ins mutant fish (Fig. 4B[I]). The expression of smyd1b (SET and MYND domain containing 1b), which plays a key role in thick filament assembly during myofibrillogenesis, was lower (Fig. 4B[I-c]). In addition, the expression of genes encoding sarcomere's structural- and associated-proteins, mypn [myopalladin (33); tnnt2a [troponin T (48); tcap (telethonin (49)]; and idba3a [LIM domain binding 3a; cypher (50)] was suppressed in the heart of thraa 8-bp ins mutant fish (Fig. 4B[I-d–g]). Intriguingly, the expression of actn2b (actinin alpha 2b), mybpc3 (myosin binding protein C), and nexn (F actin binding protein) genes was elevated in the heart of thraa 8-bp mutant fish (Fig. 4B[I-h–j]). The human orthologs of these three genes are known to be involved in dilated cardiomyopathy and hypertrophic cardiomyopathy (51–54). It is known that many gene functions are conserved among humans, mice, and zebrafish. The genes affected by TRα1 mutations in the zebrafish identified in the present study could be used to explore the potential molecular defects in patients and mouse models of RTHα.
A similar expression profile of the genes encoding major sarcomere assembling proteins was also observed in the heart of thrab 1-bp ins mutant fish (Fig. 4B[II]). However, one major difference was the expression of the smyd1b gene. Instead of being suppressed in the heart of thraa 8-bp ins mutant fish, the expression of the smyd1b gene was activated (Fig. 4B[II-c]). Another small difference was the expression of the tcap gene. Instead of being suppressed in the heart of thraa 8-bp ins mutant fish, its expression was unchanged (Fig. 4B[II-c]). The differential expression of these two genes could account for the subtle differences observed in the sarcomere structures revealed by TEM (Fig. 4A).
Juvenile thraa 8-bp ins and thrab 1-bp ins mutant fish exhibit heart abnormalities
The finding that the adult mutant fish displayed the heart abnormalities shown above prompted us to ascertain the developmental stage at which the heart abnormality developed. At 31 dpf, the mutant fish developed abnormal heart shape and size in the atrium and ventricle as found in adult fish (Fig. 1B). In the juvenile heart of WT fish, the ventricle is wedge-shaped and larger than the atrium (ratio of ventricle vs. atrium >1; Fig. 5A[I-a]). In contrast to the heart of WT fish, the size of the ventricle and atrium in the mutant juveniles was similar (ratio of ventricle vs. atrium ∼1); moreover, the atrium was dislocated in homozygous thraa 8-bp ins mutant fish (Fig. 5A[I-b]). In the heart of homozygous thrab 1-bp ins mutant fish, the ventricle was round-shaped, with the size close to that of the atrium (ratio of ventricle vs. atrium ∼1). Figure 5A(II) compares the extent in the abnormality of the size of ventricle and atrium among WT, thraa 8-bp ins, and thrab 1-bp ins mutant fish. The higher extent in the distribution of enlarged atrium in the mutant fish suggested that the atrium was dilated as early as in juveniles. In line with the findings that atrium was dilated in the heart of juvenile mutant fish, the blood flow speed was also decreased by 7% in the thraa 8-bp ins mutant fish (Fig. 5B) as similarly found in the adult fish (Fig. 3B[II]). Taken together, these data showed that the mutations of the thra genes caused heart abnormalities detectable at the juvenile stage that persisted to adulthood.
FIG. 5.
Juveniles of homozygous thra mutant fish develop heart abnormalities. (A[I]) The histology features of zebrafish heart in WT (I[a]), homozygous thraa 8-bp ins (m/m) (I[b]), and homozygous thrab 1-bp ins (m/m) (I[c]) were sectioned and stained with H&E. The histology features of heart in thraa 8-bp ins (m/m) mutant fish show a dislocated and dilated atrium, and thrab 1-bp ins (m/m) mutant fish show a round-shaped ventricle and dilated atrium. (A[II]) Quantitative analysis of relative size of ventricle to atrium. Relative ventricle and atrium sizes at the aortic valve and atrioventricular valves are compared between WT (N = 3), thraa 8-bp ins (m/m) (N = 3), and thrab 1-bp ins (m/m) (N = 3) (1.1 mpf) mutant fish. GraphPad Prism version 8 for Mac OS X was used to perform analyses of variances. (B). Comparison of the flow speed of red blood cells in WT and homozygous thraa 8-bp ins mutant fish. The images were captured from 3 to 4 individual WT or mutant fish; each fish was imagined 4–5 times. Two-tailed unpaired t-test, p-adjusted <0.05, was used for statistical analysis. GraphPad Prism version 8 for Mac OS X was used to perform analyses of variances.
Discussion
Patients with mutations of the THRA gene have been reported to have low heart rates (10,13). The TRa1R384C/+ mutant mouse, a model of RTHα, was also reported to have slower heart rates than WT mice (23,24). In the mouse studies, no information was provided regarding the developmental stage at which the heart abnormalities were detectable. Furthermore, it was unclear how TRα1 mutants disrupted physiology at the structural and molecular levels to cause the heart defects. In the present study, we used the recently created zebrafish models of RTHα to address these issues. We found abnormal heart shape and size in mutant fish, beginning at the juvenile stage and persisting into adulthood, leading to bradycardia and reduced circulating blood speed. Analysis of internal structures of mutant fish ventricles by TEM revealed defective myofibrils with altered structures of sarcomeres. Gene expression analyses indicated that TRα1 mutants acted to alter critical genes involved in sarcomere organization, thereby resulting in defective heart functions. Of note, at present, besides bradycardia, no structural changes in the heart of RTHα patients have been reported so far. Nonetheless, the present study has provided new insights into the molecular actions of TRα1 mutants causing heart abnormalities.
The mutant fish used in the present study were created by CRISPR/Cas9-mediated targeted mutagenesis (22). The stable expression of mutant receptors made it possible to analyze actions of mutant receptors from embryos to adulthood. The first visible heart morphological abnormality was detected in juveniles (35 dpf; Fig. 5), which persisted to adulthood (Fig. 1B). We attempted to ascertain whether homozygous thraa 8-bp ins mutation could impact heart development in the embryos. In whole-mount in situ hybridization using the ventricular-specific myosin light chain-2 (cmlc2) gene as a probe, we did not find any apparent morphological differences between the WT and mutant embryos at 48 dpf. Furthermore, we analyzed the expression of two transcription factors, NK2 homeobox 5 (nkx2.5) and GATA binding protein 4 (gata4), which are involved in cardiac chamber formation, cardiac development, cardiac muscle tissue regeneration, and proper atrioventricular formation and function (55,56). We also evaluated the expression of transforming growth factor-beta 2 (tgfβ2), which is critical for cardiomyocyte proliferation and heart valve formation (57,58) as well as the expression of atrial natriuretic peptide (the nppa gene), important for the differentiating myocardium of the atrium and ventricles of the developing heart (59,60).
Of note, it is known that the expression of the nkx2.5 and the nppa genes is regulated by thyroid hormone. The expression of the nkx2.5 gene is increased by thyroid hormone in embryonic stem cell-derived cardiomyocytes (61). The expression of the nppa gene in neonatal cardiomyocytes was reported to be stimulated by the thyroid hormone (62,63). As shown in Supplementary Figure S1, there were no differences in the expression of these four genes between the WT and homozygous thraa 8-bp ins mutant embryos/early larvae at 3 and 5 dpf. Taken together, these results suggested that mutation of the thraa gene had no effect in the early development of the heart. These findings are consistent with our earlier observations of no discernible morphological abnormalities in the embryos and early larvae in homozygous thraa 8-bp ins mutant fish (22).
As reported earlier for zebrafish, total levothyroxine (LT4) levels were relatively low in the embryonic stage, suggesting that the WT TRs were most likely unliganded. Thus there was not sufficient ligand-dependent transcription activity of WT receptors to be interfered/antagonized by mutant receptors that could not bind thyroid hormones. As total T3 and LT4 began to rise on day 15 and peaked at the larva-juvenile transition stage, more WT TRs, as expected, are bound to thyroid hormones in WT fish (22). However, the homozygous thrab 1-bp ins mutant, which could not bind the thyroid hormone, acted dominantly negatively to interfere with the transcription activity on the T3-target genes in the heart, as shown in Figures 2A(II) and 4B. These findings suggest that mutations of the thra gene are detrimental in the postlarval development of the heart.
Unlike in humans and mice, in zebrafish the thra gene is duplicated. Previously, we have shown that the two duplicated genes have isoform-redundant and isoform-specific functions (22). Severe growth retardation was found in homozygous thrab-1-bp ins mutant fish, whereas only a very small growth defect was detected in homozygous thraa 8-bp mutant fish (22). Furthermore, hypoplasia of the epidermis was exhibited only in thrab 1-bp ins mutant fish, not in thraa 8-bp ins mutant fish (22). In the present study, a different pattern of functions in the duplicated genes emerged. Both homozygous thrab 1-bp ins and thraa 8-bp ins mutant fish exhibited prominent histological abnormalities in the heart, although with some minor differences (Fig. 1). Abnormal myofibrils were found in the heart of both mutant fish, but with some small isoform-dependent differences as revealed by TEM and profiling of genes critical in the assembling of sarcomeres (Fig. 4). These different phenotypic abnormalities displayed in these two lines of mutant fish clearly indicated the complexity in the regulation of the biological functions by the duplicated thra genes. The differential effects mediated by these two TRα1 mutant isoforms must await future studies.
Mutations identified in RTHα patients vary in the extent of the loss of thyroid hormone binding and the level of dominant negative activity (10–13,16,64,65). The efficacy in the treatment of RTHα patients with LT4 depends on the severity of clinical phenotypes mediated by the in vivo molecular actions of TRα1 mutants. A milder clinical manifestation seen in a 17-year-old patient with A263V mutation had a favorable response to LT4 treatment. In contrast, a more severe clinical manifestation found in another RTHα patient with L274P mutation was refractory to hormone therapy (66). Moreover, LT4 treatment may not ameliorate symptoms in all target tissues. The selective action of LT4 therapy is exemplified in an RTHα patient with Ala382ProfsX7 mutation whose diminished alertness and constipation responded to LT4 therapy. Despite these responses, the patient's cardiac parameters (heart rate, contractility) were relatively resistant to treatment. Furthermore, the patient's reduced red cell mass with macrocytosis was unresponsive to LT4 therapy (13). For hormone-treatment-refractory patients, other alternative treatments are needed.
Our zebrafish models would be valuable in exploring other treatment options. The present study showed that the blood flow speed of thraa 8-bp ins(m/m)gata-1:DsRed fish was slower than in WT fish. In this approach, the expression of fluorescence DsRed is driven by the promoter of the gata-1 gene, which is a critical regulator of erythropoiesis. Such thraa 8-bp ins mutant fish expressing a fluorescence tag can be used for rapid screening of small molecules for potential therapeutics to correct the heart abnormalities. Moreover, this approach can be extended to search for therapeutics for defects besides the heart. Expressing fluorescence probes driven by the promoters to target specific tissues in the mutant zebrafish, such as expressing brightness-enhanced green fluorescence protein driven by keratin genes, could be used to search for therapeutics to correct skin defects (22). The development of thra mutant zebrafish of RTHα expressing fluorescence proteins would facilitate identification of potential therapeutics for treatment of RTHα patients.
Supplementary Material
Acknowledgments
We thank Blake Carrington for the analysis of genotyping sequencing, and Kevin Bishop for advice during the course of this project. We are grateful to Ruth Woodword, Animal Program Director, NICHD, and Tania Clark, Animal Program Director, NHGRI, Michael Wisnieski, Aquatic Project Manager, Stephen Frederickson, Research Specialist, Charles River, NIH-Contractor, for their special assistance to make it possible to complete the revision experiments for this article during the unprecedented SARS-CoV-2 pandemic time.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This research was supported by the Intramural Research Program (Innovation Award) of the Center for Cancer Research, National Cancer Institute, National Institutes of Health.
Supplementary Material
References
- 1. Koenig RJ 1998. Thyroid hormone receptor coactivators and corepressors. Thyroid 8:703–713 [DOI] [PubMed] [Google Scholar]
- 2. Kim MK, Lee JS, Chung JH. 1999. In vivo transcription factor recruitment during thyroid hormone receptor-mediated activation. Proc Natl Acad Sci U S A 96:10092–10097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vella KR, Ramadoss P, Costa ESRH, Astapova I, Ye FD, Holtz KA, Harris JC, Hollenberg AN. 2014. Thyroid hormone signaling in vivo requires a balance between coactivators and corepressors. Mol Cell Biol 34:1564–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ortiga-Carvalho TM, Sidhaye AR, Wondisford FE. 2014. Thyroid hormone receptors and resistance to thyroid hormone disorders. Nat Rev Endocrinol 10:582–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mendoza A, Hollenberg AN. 2017. New insights into thyroid hormone action. Pharmacol Ther 173:135–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weiss RE, Dumitrescu A, Refetoff S. 2010. Approach to the patient with resistance to thyroid hormone and pregnancy. J Clin Endocrinol Metab 95:3094–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bassett JH, Williams GR. 2016. Role of thyroid hormones in skeletal development and bone maintenance. Endocr Rev 37:135–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Williams GR 2013. Thyroid hormone actions in cartilage and bone. Eur Thyroid J 2:3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fozzatti L, Kim DW, Park JW, Willingham MC, Hollenberg AN, Cheng SY. 2013. Nuclear receptor corepressor (NCOR1) regulates in vivo actions of a mutated thyroid hormone receptor alpha. Proc Natl Acad Sci U S A 110:7850–7855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bochukova E, Schoenmakers N, Agostini M, Schoenmakers E, Rajanayagam O, Keogh JM, Henning E, Reinemund J, Gevers E, Sarri M, Downes K, Offiah A, Albanese A, Halsall D, Schwabe JW, Bain M, Lindley K, Muntoni F, Vargha-Khadem F, Dattani M, Farooqi IS, Gurnell M, Chatterjee K. 2012. A mutation in the thyroid hormone receptor alpha gene. N Engl J Med 366:243–249 [DOI] [PubMed] [Google Scholar]
- 11. Demir K, van Gucht AL, Buyukinan M, Catli G, Ayhan Y, Bas VN, Dundar B, Ozkan B, Meima ME, Visser WE, Peeters RP, Visser TJ. 2016. Diverse genotypes and phenotypes of three novel thyroid hormone receptor-alpha mutations. J Clin Endocrinol Metab 101:2945–2954 [DOI] [PubMed] [Google Scholar]
- 12. Espiard S, Savagner F, Flamant F, Vlaeminck-Guillem V, Guyot R, Munier M, d'Herbomez M, Bourguet W, Pinto G, Rose C, Rodien P, Wemeau JL. 2015. A novel mutation in THRA gene associated with an atypical phenotype of resistance to thyroid hormone. J Clin Endocrinol Metab 100:2841–2848 [DOI] [PubMed] [Google Scholar]
- 13. Moran C, Schoenmakers N, Agostini M, Schoenmakers E, Offiah A, Kydd A, Kahaly G, Mohr-Kahaly S, Rajanayagam O, Lyons G, Wareham N, Halsall D, Dattani M, Hughes S, Gurnell M, Park SM, Chatterjee K. 2013. An adult female with resistance to thyroid hormone mediated by defective thyroid hormone receptor alpha. J Clin Endocrinol Metab 98:4254–4261 [DOI] [PubMed] [Google Scholar]
- 14. Tylki-Szymanska A, Acuna-Hidalgo R, Krajewska-Walasek M, Lecka-Ambroziak A, Steehouwer M, Gilissen C, Brunner HG, Jurecka A, Rozdzynska-Swiatkowska A, Hoischen A, Chrzanowska KH. 2015. Thyroid hormone resistance syndrome due to mutations in the thyroid hormone receptor alpha gene (THRA). J Med Genet 52:312–316 [DOI] [PubMed] [Google Scholar]
- 15. Moran C, Chatterjee K. 2015. Resistance to thyroid hormone due to defective thyroid receptor alpha. Best Pract Res Clin Endocrinol Metab 29:647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Mullem AA, Chrysis D, Eythimiadou A, Chroni E, Tsatsoulis A, de Rijke YB, Visser WE, Visser TJ, Peeters RP. 2013. Clinical phenotype of a new type of thyroid hormone resistance caused by a mutation of the TRalpha1 receptor: consequences of LT4 treatment. J Clin Endocrinol Metab 98:3029–3038 [DOI] [PubMed] [Google Scholar]
- 17. O'Shea PJ, Bassett JH, Sriskantharajah S, Ying H, Cheng SY, Williams GR. 2005. Contrasting skeletal phenotypes in mice with an identical mutation targeted to thyroid hormone receptor alpha1 or beta. Mol Endocrinol 19:3045–3059 [DOI] [PubMed] [Google Scholar]
- 18. O'Shea PJ, Bassett JH, Cheng SY, Williams GR. 2006. Characterization of skeletal phenotypes of TRalpha1 and TRbeta mutant mice: implications for tissue thyroid status and T3 target gene expression. Nucl Recept Signal 4:e011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park S, Han CR, Park JW, Zhao L, Zhu X, Willingham M, Bodine DM, Cheng SY. 2017. Defective erythropoiesis caused by mutations of the thyroid hormone receptor alpha gene. PLoS Genet 13:e1006991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Han CR, Park S, Cheng SY. 2017. NCOR1 modulates erythroid disorders caused by mutations of thyroid hormone receptor alpha1. Sci Rep 7:18080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bao L, Roediger J, Park S, Fu L, Shi B, Cheng SY, Shi YB. 2019. Thyroid hormone receptor alpha mutations lead to epithelial defects in the adult intestine in a mouse model of resistance to thyroid hormone. Thyroid 29:439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Han CR, Holmsen E, Carrington B, Bishop K, Zhu YJ, Starost M, Meltzer P, Sood R, Liu P, Cheng SY. 2020. Generation of novel genetic models to dissect resistance to thyroid hormone receptor alpha in zebrafish. Thyroid 30:314–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mittag J, Wallis K, Vennstrom B. 2010. Physiological consequences of the TRalpha1 aporeceptor state. Heart Fail Rev 15:111–115 [DOI] [PubMed] [Google Scholar]
- 24. Tinnikov A, Nordstrom K, Thoren P, Kindblom JM, Malin S, Rozell B, Adams M, Rajanayagam O, Pettersson S, Ohlsson C, Chatterjee K, Vennstrom B. 2002. Retardation of post-natal development caused by a negatively acting thyroid hormone receptor alpha1. EMBO J 21:5079–5087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhu X, Enomoto K, Zhao L, Zhu YJ, Willingham MC, Meltzer P, Qi J, Cheng SY. 2017. Bromodomain and extraterminal protein inhibitor JQ1 suppresses thyroid tumor growth in a mouse model. Clin Cancer Res 23:430–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wikstrom L, Johansson C, Salto C, Barlow C, Campos Barros A, Baas F, Forrest D, Thoren P, Vennstrom B. 1998. Abnormal heart rate and body temperature in mice lacking thyroid hormone receptor alpha 1. EMBO J 17:455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gloss B, Trost S, Bluhm W, Swanson E, Clark R, Winkfein R, Janzen K, Giles W, Chassande O, Samarut J, Dillmann W. 2001. Cardiac ion channel expression and contractile function in mice with deletion of thyroid hormone receptor alpha or beta. Endocrinology 142:544–550 [DOI] [PubMed] [Google Scholar]
- 28. Shimizu H, Langenbacher AD, Huang J, Wang K, Otto G, Geisler R, Wang Y, Chen JN. 2017. The calcineurin-FoxO-MuRF1 signaling pathway regulates myofibril integrity in cardiomyocytes. ELife 6:e27955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shih YH, Zhang Y, Ding Y, Ross CA, Li H, Olson TM, Xu X. 2015. Cardiac transcriptome and dilated cardiomyopathy genes in zebrafish. Circ Cardiovasc Genet 8:261–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ebert AM, McAnelly CA, Handschy AV, Mueller RL, Horne WA, Garrity DM. 2008. Genomic organization, expression, and phylogenetic analysis of Ca2+ channel beta4 genes in 13 vertebrate species. Physiol Genomics 35:133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ebert AM, Hume GL, Warren KS, Cook NP, Burns CG, Mohideen MA, Siegal G, Yelon D, Fishman MC, Garrity DM. 2005. Calcium extrusion is critical for cardiac morphogenesis and rhythm in embryonic zebrafish hearts. Proc Natl Acad Sci U S A 102:17705–17710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vassalle M, Lin CI. 2004. Calcium overload and cardiac function. J Biomed Sci 11:542–565 [DOI] [PubMed] [Google Scholar]
- 33. Hsieh FC, Lu YF, Liau I, Chen CC, Cheng CM, Hsiao CD, Hwang SL. 2018. Zebrafish VCAP1X2 regulates cardiac contractility and proliferation of cardiomyocytes and epicardial cells. Sci Rep 8:7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Michalak M, Campbell KP, MacLennan DH. 1980. Localization of the high affinity calcium binding protein and an intrinsic glycoprotein in sarcoplasmic reticulum membranes. J Biol Chem 255:1317–1326 [PubMed] [Google Scholar]
- 35. Wu HH, Brennan C, Ashworth R. 2011. Ryanodine receptors, a family of intracellular calcium ion channels, are expressed throughout early vertebrate development. BMC Res Notes 4:541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Traaseth NJ, Ha KN, Verardi R, Shi L, Buffy JJ, Masterson LR, Veglia G. 2008. Structural and dynamic basis of phospholamban and sarcolipin inhibition of Ca(2+)-ATPase. Biochemistry 47:3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bhupathy P, Babu GJ, Periasamy M. 2007. Sarcolipin and phospholamban as regulators of cardiac sarcoplasmic reticulum Ca2+ ATPase. J Mol Cell Cardiol 42:903–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hagemann D, Xiao RP. 2002. Dual site phospholamban phosphorylation and its physiological relevance in the heart. Trends Cardiovasc Med 12:51–56 [DOI] [PubMed] [Google Scholar]
- 39. Nagai R, Zarain-Herzberg A, Brandl CJ, Fujii J, Tada M, MacLennan DH, Alpert NR, Periasamy M. 1989. Regulation of myocardial Ca2+-ATPase and phospholamban mRNA expression in response to pressure overload and thyroid hormone. Proc Natl Acad Sci U S A 86:2966–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zarain-Herzberg A, Marques J, Sukovich D, Periasamy M. 1994. Thyroid hormone receptor modulates the expression of the rabbit cardiac sarco (endo) plasmic reticulum Ca(2+)-ATPase gene. J Biol Chem 269:1460–1467 [PubMed] [Google Scholar]
- 41. Holt E, Sjaastad I, Lunde PK, Christensen G, Sejersted OM. 1999. Thyroid hormone control of contraction and the Ca(2+)-ATPase/phospholamban complex in adult rat ventricular myocytes. J Mol Cell Cardiol 31:645–656 [DOI] [PubMed] [Google Scholar]
- 42. Dillmann WH 2002. Cellular action of thyroid hormone on the heart. Thyroid 12:447–452 [DOI] [PubMed] [Google Scholar]
- 43. Parmacek MS, Solaro RJ. 2004. Biology of the troponin complex in cardiac myocytes. Prog Cardiovasc Dis 47:159–176 [DOI] [PubMed] [Google Scholar]
- 44. Liu P, Zhong TP. 2017. MAPK/ERK signalling is required for zebrafish cardiac regeneration. Biotechnol Lett 39:1069–1077 [DOI] [PubMed] [Google Scholar]
- 45. Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY. 2000. The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci U S A 97:11990–11995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yaqoob N, Holotta M, Prem C, Kopp R, Schwerte T. 2009. Ontogenetic development of erythropoiesis can be studied non-invasively in GATA-1:DsRed transgenic zebrafish. Comp Biochem Physiol A Mol Integr Physiol 154:270–278 [DOI] [PubMed] [Google Scholar]
- 47. England J, Loughna S. 2013. Heavy and light roles: myosin in the morphogenesis of the heart. Cell Mol Life Sci 70:1221–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ferrante MI, Kiff RM, Goulding DA, Stemple DL. 2011. Troponin T is essential for sarcomere assembly in zebrafish skeletal muscle. J Cell Sci 124:565–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang R, Yang J, Zhu J, Xu X. 2009. Depletion of zebrafish Tcap leads to muscular dystrophy via disrupting sarcomere-membrane interaction, not sarcomere assembly. Hum Mol Genet 18:4130–4140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang J, Shih YH, Xu X. 2014. Understanding cardiac sarcomere assembly with zebrafish genetics. Anat Rec (Hoboken) 297:1681–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen Q, Smith CY, Bailey KR, Wennberg PW, Kullo IJ. 2013. Disease location is associated with survival in patients with peripheral arterial disease. J Am Heart Assoc 2:e000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hodatsu A, Konno T, Hayashi K, Funada A, Fujita T, Nagata Y, Fujino N, Kawashiri MA, Yamagishi M. 2014. Compound heterozygosity deteriorates phenotypes of hypertrophic cardiomyopathy with founder MYBPC3 mutation: evidence from patients and zebrafish models. Am J Physiol Heart Circ Physiol 307:H1594–H1604 [DOI] [PubMed] [Google Scholar]
- 53. Tu S, Chi NC. 2012. Zebrafish models in cardiac development and congenital heart birth defects. Differentiation 84:4–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bakkers J 2011. Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc Res 91:279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Colombo S, de Sena-Tomás C, George V, Werdich AA, Kapur S, MacRae CA, Targoff KL. 2018. Nkx genes establish second heart field cardiomyocyte progenitors at the arterial pole and pattern the venous pole through Isl1 repression. Development 145:dev161497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. 2010. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 464:601–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dogra D, Ahuja S, Kim HT, Rasouli SJ, Stainier DYR, Reischauer S. 2017. Opposite effects of activin type 2 receptor ligands on cardiomyocyte proliferation during development and repair. Nat Commun 8:1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Azhar M, Brown K, Gard C, Chen H, Rajan S, Elliott DA, Stevens MV, Camenisch TD, Conway SJ, Doetschman T. 2011. Transforming growth factor Beta2 is required for valve remodeling during heart development. Dev Dyn 240:2127–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Grassini DR, Lagendijk AK, De Angelis JE, Da Silva J, Jeanes A, Zettler N, Bower NI, Hogan BM, Smith KA. 2018. Nppa and Nppb act redundantly during zebrafish cardiac development to confine AVC marker expression and reduce cardiac jelly volume. Development 145:dev160739. [DOI] [PubMed] [Google Scholar]
- 60. Houweling AC, van Borren MM, Moorman AF, Christoffels VM. 2005. Expression and regulation of the atrial natriuretic factor encoding gene Nppa during development and disease. Cardiovasc Res 67:583–593 [DOI] [PubMed] [Google Scholar]
- 61. Lee YK, Ng KM, Chan YC, Lai WH, Au KW, Ho CY, Wong LY, Lau CP, Tse HF, Siu CW. 2010. Triiodothyronine promotes cardiac differentiation and maturation of embryonic stem cells via the classical genomic pathway. Mol Endocrinol 24:1728–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Stoykov I, van Beeren HC, Moorman AF, Christoffels VM, Wiersinga WM, Bakker O. 2007. Effect of amiodarone and dronedarone administration in rats on thyroid hormone-dependent gene expression in different cardiac components. Eur J Endocrinol 156:695–702 [DOI] [PubMed] [Google Scholar]
- 63. Ferdous A, Wang ZV, Luo Y, Li DL, Luo X, Schiattarella GG, Altamirano F, May HI, Battiprolu PK, Nguyen A, Rothermel BA, Lavandero S, Gillette TG, Hill JA. 2020. FoxO1-Dio2 signaling axis governs cardiomyocyte thyroid hormone metabolism and hypertrophic growth. Nat Commun 11:2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Moran C, Agostini M, Visser WE, Schoenmakers E, Schoenmakers N, Offiah AC, Poole K, Rajanayagam O, Lyons G, Halsall D, Gurnell M, Chrysis D, Efthymiadou A, Buchanan C, Aylwin S, Chatterjee KK. 2014. Resistance to thyroid hormone caused by a mutation in thyroid hormone receptor (TR)alpha1 and TRalpha2: clinical, biochemical, and genetic analyses of three related patients. Lancet Diabetes Endocrinol 2:619–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. van Gucht AL, Meima ME, Zwaveling-Soonawala N, Visser WE, Fliers E, Wennink JM, Henny C, Visser TJ, Peeters RP, van Trotsenburg AS. 2016. Resistance to thyroid hormone alpha in an 18-month-old girl: clinical, therapeutic, and molecular characteristics. Thyroid 26:338–346 [DOI] [PubMed] [Google Scholar]
- 66. Moran C, Agostini M, McGowan A, Schoenmakers E, Fairall L, Lyons G, Rajanayagam O, Watson L, Offiah A, Barton J, Price S, Schwabe J, Chatterjee K. 2017. Contrasting phenotypes in resistance to thyroid hormone alpha correlate with divergent properties of thyroid hormone receptor alpha1 mutant proteins. Thyroid 27:973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.