Abstract
COVID-19 is a highly contagious disease that can cause severe pneumonia. Patients with pneumonia undergo chest X-rays (XR) to assess infiltrates that identify the infection. However, the radiographic characteristics of COVID-19 are similar to the other acute respiratory syndromes, hindering the imaging diagnosis. In this work, we proposed identifying quantitative/radiomic biomarkers for COVID-19 to support XR assessment of acute respiratory diseases. This retrospective study used different cohorts of 227 patients diagnosed with pneumonia; 49 of them had COVID-19. Automatically segmented images were characterized by 558 quantitative features, including gray-level histogram and matrices of co-occurrence, run-length, size zone, dependence, and neighboring gray-tone difference. Higher-order features were also calculated after applying square and wavelet transforms. Mann–Whitney U test assessed the diagnostic performance of the features, and the log-rank test assessed the prognostic value to predict Kaplan–Meier curves of overall and deterioration-free survival. Statistical analysis identified 51 independently validated radiomic features associated with COVID-19. Most of them were wavelet-transformed features; the highest performance was the small dependence matrix feature of “low gray-level emphasis” (area under the curve of 0.87, sensitivity of 0.85, ). Six features presented short-term prognostic value to predict overall and deterioration-free survival. The features of histogram “mean absolute deviation” and size zone matrix “non-uniformity” yielded the highest differences on Kaplan–Meier curves with a hazard ratio of 3.20 (). The radiomic markers showed potential as quantitative measures correlated with the etiologic agent of acute infectious diseases and to stratify short-term risk of COVID-19 patients.
Keywords: COVID-19, Radiomics, Coronavirus, Chest radiography, Medical image analysis
Introduction
By the end of 2019, a novel type of coronavirus, known as SARS-CoV-2, was discovered, causing several infections and pneumonia cases initially in Wuhan, China, and later on across the globe. The World Health Organization (WHO) defined the acute infectious disease caused by the SARS-CoV-2 as COVID-19 (Coronavirus Disease - 2019) [1, 2]. COVID-19 is a systemic infectious disease but mainly characterized by the inflammation of the human respiratory system and its high contagiousness. Currently, the diagnosis of COVID-19 is confirmed by real-time reverse transcription-polymerase chain reaction (RT-PCR) [3, 4]. However, the virus presence in the upper airways is transient, and the RT-PCR displays low sensitivity of 71% and requires dedicated instrumentation to be readily available, limiting its wide use during a pandemic [5, 6].
Most of the patients with suspected pneumonia are submitted to chest radiography (XR) and computed tomography (CT) to assess the infiltrates’ presence and patterns. The infection caused by COVID-19 typically presents bilateral lung infiltrates with patterns resembling ground-glass and consolidation [3, 6, 7]. These radiological characteristics are informative but are similar to signs from other acute respiratory syndromes like SARS (Severe Acute Respiratory Syndrome) and MERS (Middle East Respiratory Syndrome) [8]. Moreover, as those characteristics describe the internal structure of lung lesions subjectively, qualitatively, or semi-quantitatively, they can lead to intra- and inter-observer variability [9, 10]. Due to the limitations of the aforementioned methods (i.e., RT-PCR and visual/qualitative radiological assessment), a quantitative/computational approach may add to clinical routine.
Computer-aided diagnosis/detection (CAD) tools contribute to improve the interpretation of radiological findings and to identify diseases in early stages [11, 12]. The goal of CAD is to improve the accuracy and consistency of medical image diagnosis and interpretation using the suggestion provided by a computer. CAD tools traditionally provide a single answer (second opinion) to specialists, but not short-term prognostic information, limiting the applicability to the clinical routine [13, 14].
The field of radiomics has emerged as a promising quantitative approach to develop medical imaging biomarkers and support clinical decisions [12, 15] Radiomics is an extension of CAD that associates computer-extracted medical image features with clinical endpoints (e.g., genomics, staging, survival, recurrence, among others). This radiomic association allows a more comprehensive characterization of the underlying phenotype, ultimately increasing the power of decision support models [14, 16]. The recent advances in target therapies for precision medicine imperatively required an inexpensive and easily obtainable imaging approach for phenotyping diseases, and radiomics can provide it as it is a non-invasive, fast, low cost, and reproducible tool [9, 15]
Therefore, our goal in this work is to use radiomics to identify XR quantitative imaging biomarkers for COVID-19. For this purpose, we first segmented the lungs automatically from radiography images; then extracted quantitative features from the segmented regions of interest (ROIs); and finally associated them with COVID-19 endpoints, such as etiology and patient survival.
Materials and Methods
Patients
In this study, we used XR images of 227 patients from publicly available cohorts, and hence, no institutional review board approval was needed. At first, we used three cohorts to discover potential biomarkers for COVID-19, namely the discovery set, and the other two cohorts as an independent validation set.
The discovery set was composed of 195 patients:
29 patients with COVID-19 that had images and clinical data provided by the Italian Society of Medical and Interventional Radiology [17];
127 cases of pneumonia non-related to COVID-19 from the Spanish chest XR cohort (PadChest dataset) with image-associated reports from patients that attended the San Juan de Alicante Hospital, University of Alicante, Spain [18];
39 patients with pneumonia non-related to COVID-19 from the National Library of Medicine, National Institutes of Health (OpenI dataset), who attended various hospitals of the Indiana University School of Medicine, USA [19].
Physicians performed image labeling for the cases from PadChest and OpenI datasets. The validation set was composed of 32 patients from around the world. In this set, images and clinical data were initially collected from the literature by researchers of the University of Montreal [20]. Then, it was completed with follow-up data (survival time, imaging and event dates) by us to perform the radiomic analysis. From those 32 patients, 20 had the diagnosis confirmed for COVID-19 by RT-PCR, and 12 from a different etiology distributed as following: one caused by Pneumocystis fungal pathogen, two by the bacteria Streptococcus, four of them had ARDS, and five had SARS. Table 1 describes demographic data from the cohorts.
Table 1.
Description of the patients
| Discovery Set | Validation Set | ||||
|---|---|---|---|---|---|
| Italian cases of COVID-19 | Spanish cases of other pneumonia | American cases of other pneumonia | World cases of COVID-19 | World cases ofother pneumonia | |
| (n = 29) | (n = 127) | (n = 39) | (n = 20) | (n = 12) | |
| Age* | 61.1 ± 13.2 | 63.1 ± 18.1 | NA | 48.6 ± 14.8 | 49.5 ± 17.4 |
| (27–87) | (29–99) | (12–71) | (25–74) | ||
| Gender | |||||
| Female | 10 | 49 | NA | 8 | 5 |
| Male | 19 | 78 | NA | 9 | 5 |
| Chest abnormalities | |||||
| Airspace disease | - | - | 13 | 1 | - |
| Aortic changes | - | 24 | 2 | - | - |
| Cardiomegaly | - | 17 | - | - | 1 |
| Consolidation | 3 | 2 | 9 | 7 | 6 |
| Heart insufficiency | - | 10 | - | - | - |
| Hilar enlargement | 1 | 7 | - | - | - |
| Infiltrate | - | 6 | 4 | 2 | - |
| Pleural effusion | 2 | 11 | 4 | 1 | - |
| Pleural thickening | - | 6 | - | 1 | - |
| Pulmonary atelectasis | - | 11 | 5 | - | - |
| Pulmonary emphysema | - | 2 | 1 | - | - |
| Pulmonary fibrosis | - | 2 | - | - | - |
NA, not available
* Mean ± standard deviation (min–max)
Image Segmentation
We first automatically segmented the lung from the radiography using an algorithm based on an artificial intelligence model (i.e., convolutional neural network U-Net) [21, 22]. Although the model was previously trained and assessed for lung segmentation, yielding a performance (Dice coefficient) of 0.978 [22], we evaluated the performance of the model with the images from this work to enable a robust radiomic analysis. Two experienced medical image analysts (one with 12 years of experience in CT and XR imaging and one with 9 years of experience in XR, intravascular optical coherence tomography, and ultrasound imaging) manually segmented the lungs to be used as reference for the images segmented by the model. The Dice coefficient and the Jaccard index obtained from the automatic over the manual segmentation of all images were, respectively, 0.951 (± 0.031 of standard deviation) and 0.909 (± 0.053 of standard deviation).
The image segmentation algorithm created a binary mask of both lungs and then a ROI from the lungs mask extreme points to generate the segmented image. This segmentation step removed unnecessary anatomical structures for pneumonia assessment, such as head, neck, and arms, along with textual information relative to the exam (such as DICOM metadata).
Feature Extraction
Quantitative radiography-based features on all imaging levels (first order, second order, and higher order) were investigated as potential biomarkers for COVID-19. These radiomic features were extracted on each image segmented by the method described previously. The first-order features describe the gray-level distribution of an image without considering pixel locations. Second-order features describe the spatial relationships of gray levels inside the ROI. On the other hand, higher-order features simultaneously evaluate location and relationships between pixels without considering spatial properties by using image filtering [9, 11, 14]
For each patient, first-order features were extracted from the gray-level histogram of the segmented radiography (First-order histogram). Eighteen statistical measures were calculated from each image histogram. The second-order features were extracted from five gray-level matrices (Second-order texture): co-occurrence matrix (GLCM, 24 features), run-length matrix (GLRLM, 16 features), size zone matrix (GLSZM, 16 features), dependence matrix (GLDM, 14 features), and neighboring gray-tone difference matrix (NGTDM, 5 features). Higher-order features were obtained from wavelet transforms and a square filter (higher-order spectrum). Coiflet transforms were applied to decompose the image in four different frequency domain bands (HH, HL, LH, and LL). The square filter took the gray levels square and linearly scaled them back to the radiography’s original range. After filtering, the first- and second-order measures were calculated on the filtered image’s histogram or matrix. The radiomic features were extracted using PyRadiomics v3.0 package, in compliance with the Imaging Biomarker Standardization Initiative (IBSI) [16, 23]. A total of 558 radiomic features characterized each patient comprised in the analysis (Table 2).
Table 2.
List of all features extracted for the radiomic analysis
| Type | Features |
|---|---|
| Statistics (n = 18) | Energy, Total Energy, Entropy, Minimum, 10th Percentile, 90th Percentile, Maximum, Mean, Median, Range, Interquartile Range, Mean Absolute Deviation (MAD), Robust Mean Absolute Deviation (rMAD), Root Mean Squared (RMS), Skewness, Kurtosis, Variance, and Uniformity. |
| GLCM (n = 24) | Autocorrelation, Joint Average, Cluster Prominence, Cluster Shade, Cluster Tendency, Contrast, Correlation, Difference Average, Difference Entropy, Difference Variance, Joint Energy (or Angular Second Moment), Joint Entropy, two Informational Measures of Correlation (IMC), Inverse Difference Moment (IDM), Maximal Correlation Coefficient (MCC), Inverse Difference Moment Normalized (IDMN), Inverse Difference (ID), Inverse Difference Normalized (IDN), Inverse Variance, Maximum Probability (or Joint Maximum), Sum Average, Sum Entropy, and Sum of Squares (or Joint Variance). |
| GLRLM (n = 16) | Short Run Emphasis (SRE), Long Run Emphasis (LRE), Gray Level Non-Uniformity (GLN), Gray Level Non-Uniformity Normalized (GLNN), Run Length Non-Uniformity (RLN), Run Length Non-Uniformity Normalized (RLNN), Run Percentage (RP), Gray Level Variance (GLV), Run Variance (RV), Run Entropy (RE), Low Gray Level Run Emphasis (LGLRE), High Gray Level Run Emphasis (HGLRE), Short Run Low Gray Level Emphasis (SRLGLE), Short Run High Gray Level Emphasis (SRHGLE), Long Run Low Gray Level Emphasis (LRLGLE), and Long Run High Gray Level Emphasis (LRHGLE). |
| GLSZM (n = 16) | Small Area Emphasis (SAE), Large Area Emphasis (LAE), Gray Level Non-Uniformity (GLN), Gray Level Non-Uniformity Normalized (GLNN), Size-Zone Non-Uniformity (SZN), Size-Zone Non-Uniformity Normalized (SZNN), Zone Percentage (ZP), Gray Level Variance (GLV), Zone Variance (ZV), Zone Entropy (ZE), Low Gray Level Zone Emphasis (LGLZE), High Gray Level Zone Emphasis (HGLZE), Small Area Low Gray Level Emphasis (SALGLE), Small Area High Gray Level Emphasis (SAHGLE), Large Area Low Gray Level Emphasis (LALGLE), and Large Area High Gray Level Emphasis (LAHGLE). |
| GLDM (n = 14) | Small Dependence Emphasis (SDE), Large Dependence Emphasis (LDE), Gray Level Non-Uniformity (GLN), Dependence Non-Uniformity (DN), Dependence Non-Uniformity Normalized (DNN), Gray Level Variance (GLV), Dependence Variance (DV), Dependence Entropy (DE), Low Gray Level Emphasis (LGLE), High Gray Level Emphasis (HGLE), Small Dependence Low Gray Level Emphasis (SDLGLE), Small Dependence High Gray Level Emphasis (SDHGLE), Large Dependence Low Gray Level Emphasis (LDLGLE), and Large Dependence High Gray Level Emphasis (LDHGLE). |
| NGTDM (n = 5) | Coarseness, Contrast, Busyness, Complexity, and Strength. |
Statistical Analysis
A univariate analysis statistically evaluated the radiomic association between XR features and COVID-19 diagnosis, using the receiver operating characteristic (ROC) curve with sensitivity and specificity metrics. The Mann–Whitney U test evaluated the statistical difference between feature distributions from the groups of patients with pneumonia [9]. Each feature had the area under the ROC curve (AUC) and p-value calculated individually.
The short-term prognostic analysis was performed by correlating the radiomic features with overall and deterioration-free survival using the Kaplan–Meier time-to-event method. Higher and lower-risk groups of patients were split according to the median value of the quantitative features [12]. As the number of patients with follow-up data (survival time and outcome result) was relatively low for this analysis, we combined all cases with COVID-19 in a single set of 28 patients (14 from discovery and 14 from the validation set).
The mean follow-up time was 20.4 days (±7.1 of standard deviation). Overall survival analysis used death by any nature as event, and deterioration-free survival analysis used worsening on clinical/radiological conditions or death by any cause. Patients who survived or remained clinically stable or had loss of follow-up were censored. The log-rank test assessed the statistical difference between the survival curves from both stratified groups to identify features with potential prognostic value [15].
The SciPy v1.2.3 and R v3.4.4 packages were used to perform statistical analysis. Tests with p < 0.05 were considered statistically significant.
Results
Demographic Findings
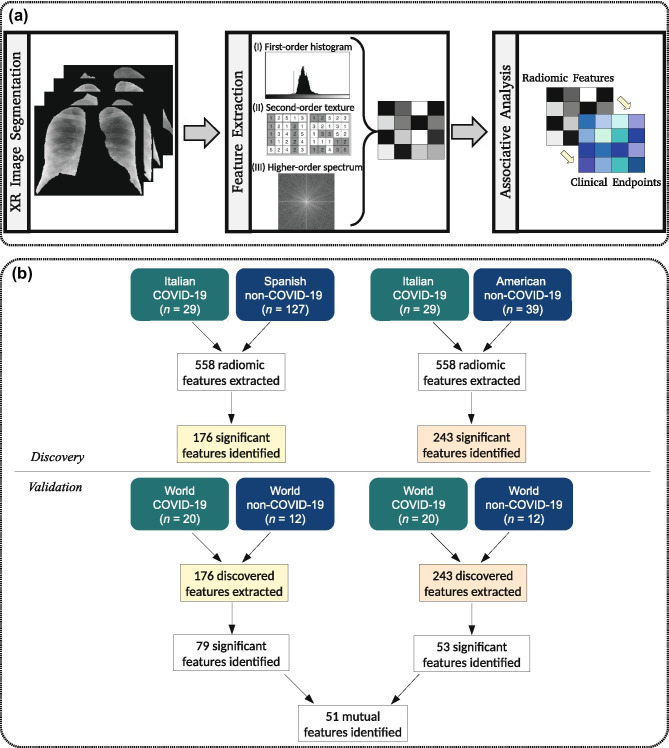
From the 49 patients later diagnosed with COVID-19 (29 from discovery and 20 from validation set), 44 patients displayed clinical data publicly available for analysis. All 44 patients attended a hospital after onset, mainly with fever (52% of the cases with symptoms data available), cough (27%), and dyspnea (25%). Twenty-six patients were men with a mean age of 53.1 years old (±16.2 of standard deviation), and 18 were women with a mean age of 61.4 years old (±12.1 of standard deviation). We divided the sample into two datasets for discovery and independent validation sets (Table 1). Figure 1 depicts the radiomic analysis performed in this study.
Fig. 1.
Workflow employed in this work: (a) radiomic pipeline for the association between radiographic features and COVID-19 endpoints; (b) radiomic analysis performed to identify potential biomarkers for the diagnosis of COVID-19
Diagnostic Biomarker Findings
Statistical analysis identified 176 radiomic features associated with COVID-19 in the Spanish discovery set. Seventy-nine of those obtained a significant correlation with SARS-CoV-2 in the validation set (p < 0.05). Moreover, we identified 243 radiomic features associated with COVID-19 in the American discovery set. Fifty-three of those also obtained a significant correlation with the novel coronavirus in the validation set (p < 0.05).
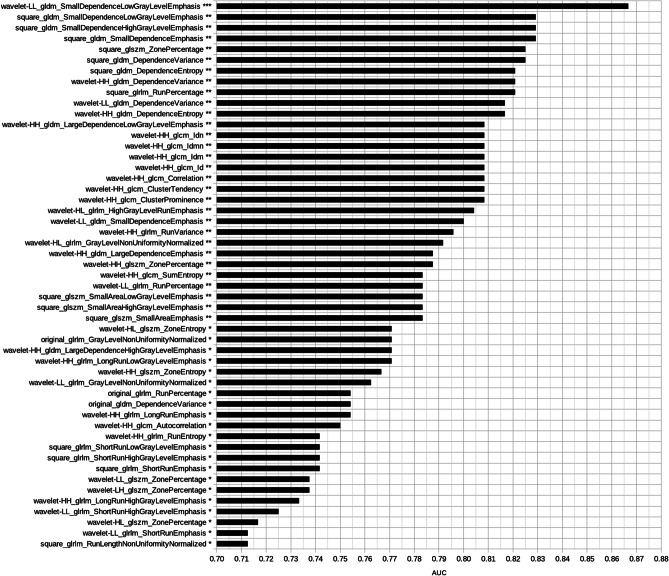
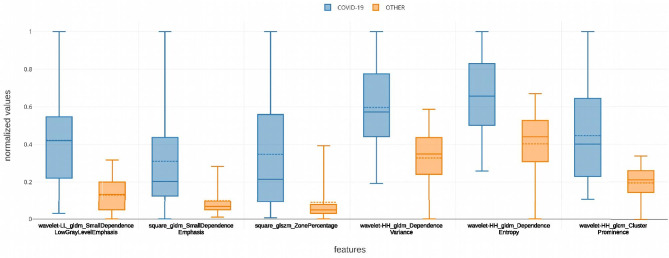
The intersection set between the features identified in the validation set resulted in 51 radiomic biomarkers for COVID-19 (Fig. 1b). Figure 2 shows the most significant radiomic biomarkers for COVID-19 and their respective AUC value. Most of them were higher-order features extracted after the wavelet (HH band) transform (41%). Figure 3 presents boxplots of the values from some features associated with pneumonia caused by the SARS-CoV-2 virus. One wavelet feature identified as f521, according to the supplementary material, obtained the best association with COVID-19. The feature f521 yielded an AUC of 0.867, sensitivity of 0.85, and specificity of 0.67 (Fig. 4). The significant features identified only in the American discovery set were f56, yielding an AUC of 0.775, and f54 with an AUC of 0.742. The most significant features identified only in the Spanish discovery set were f74, f76, and f246 with AUC of 0.821.
Fig. 2.
Most significant radiomic biomarkers for COVID-19. In the end of each feature name, there is a statistical significance symbol used according to the following notation: *** for p < 0.001, ** for 0.001 p < 0.01, and * for 0.01 p < 0.05
Fig. 3.
Distribution of some significant radiomic features associated with COVID-19. The dashed line depicts the mean value of the feature for the corresponding group
Fig. 4.
Performance of the feature f521 to recognize COVID-19 radiographic patterns: (a) ROC curve; (b) true positive XR of a 40-year-old woman with COVID-19 presented as a very discrete ground-glass opacity in the right lower lobe; (c) false negative XR of a 50-year-old woman with COVID-19 presented as multiple small bilateral patchy opacifications
Short-term Prognostic Biomarker Findings
There was a small number of patients with follow-up data for time-to-event analyses, which allowed to combine all patients with COVID-19 in a single set (n = 28). One radiomic feature was identified with short-term prognostic value to predict overall survival. The feature f287 yielded a significant difference in overall survival rates from the stratified risk groups of COVID-19 patients. The normalized threshold value used for stratification was 0.177509. High values (greater than the median) of the potential biomarker identified lower-risk patients with a mean survival time of 25 days. This group was composed of six women (58.5 ±14.2 years) and seven males (51.6 ±9.6 years). One patient did not have all the clinical data available. Furthermore, low values (less than the median) of the feature stratified patients with higher risk and mean survival time of 13 days. This group included four women (65.5 ±13.0 years) and ten males (61.2 ±11.7 years).
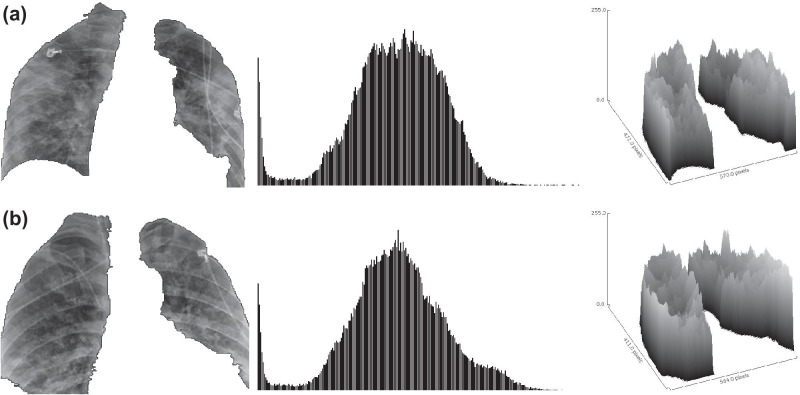
Figure 5 presents two examples of COVID-19 patients stratified by the radiomic feature f287 identified with prognostic potential. It is important to emphasize that both patients from Fig. 5 were correctly classified as COVID-19 cases by the radiomic biomarker f521, which yielded the highest performance to detect COVID-19, as previously reported.
Fig. 5.
Radiography image, gray-level histogram, and tridimensional surface plot of COVID-19 patients stratified by the radiomic biomarker f287: (a) 67-year-old woman with bilateral consolidation and 13 days of survival (no occurrence of an event of death on follow-up), classified as a lower-risk case by the biomarker; (b) 36-year-old man with scattered consolidation and nine days of survival until death, classified as a higher-risk case by the biomarker. Although both cases look visually very similar, as described by radiological assessment and gray-level distributions, the higher-order radiomic biomarker could stratify the risk of the patient according to spectral properties of the radiographic image
Five radiomic features (f143, f174, f294, f340, and f344) presented prognostic value to predict patient deterioration-free survival (Table 3). The biomarker f174 yielded the highest significant difference in survival curves, using the normalized median of 0.328535 as the threshold for stratification. High values of the feature identified lower-risk patients, while low values stratified patients with a higher risk of deterioration (12 events with mean deterioration-free survival time of 4.8 days and hazard ratio of 3.2). The lower risk patient group included five women (57.4 ±13.4 years) and nine males (55.3 ±13.1 years). The higher risk patient group included five women (65.2 ±13.8 years) and eight males (57.6 ±10.4 years). One patient did not have all the clinical data available. Figure 6 presents the Kaplan–Meier curves of risk groups of COVID-19 patients identified by the most significant radiography-based radiomic feature.
Table 3.
Radiomic features associated with deterioration-free survival of COVID-19 patients
| Radiomic Feature | Risk | Deterioration | Mean Survival Time in Days | Hazard Ratio | p |
|---|---|---|---|---|---|
| (value range) | Group | Events | (95% confidence interval) | (95% confidence interval) | |
| square_glszm_SizeZoneNonUniformity | Higher | 12 | 4.8 (3.5 to 6.2) | 3.198 (1.145 to 8.932) | 0.0265 |
| (3.750 to 11.423) | Lower | 7 | 12.1 (6.3 to 17.8) | - | |
| wavelet-HL_glrlm_LongRunEmphasis | Higher | 12 | 5.0 (3.3 to 6.6) | 3.049 (1.133 to 8.206) | 0.0273 |
| (1.428E+15 to 2.214E + 16) | Lower | 7 | 12.5 (6.8 to 18.1) | - | |
| wavelet-HL_firstorder_Skewness | Higher | 12 | 5.1 (3.3 to 6.9) | 2.823 (1.056 to 7.547) | 0.0386 |
| (-1.423 to 0.097) | Lower | 7 | 12.0 (6.6 to 17.5) | - | |
| square_gldm_LargeDependenceEmphasis | Higher | 10 | 4.5 (3.5 to 5.4) | 3.1443 (1.047 to 9.439) | 0.0411 |
| (7.804E+14 to 8.066E+15) | Lower | 9 | 11.2 (6.2 to 16.2) | - | |
| wavelet-HL_glrlm_RunEntropy | Higher | 11 | 5.3 (3.5 to 7.1) | 2.770 (1.012 to 7.582) | 0.0473 |
| (7.422E+14 to 3.776E+16) | Lower | 8 | 12.5 (7.2 to 17.9) | - |
Fig. 6.
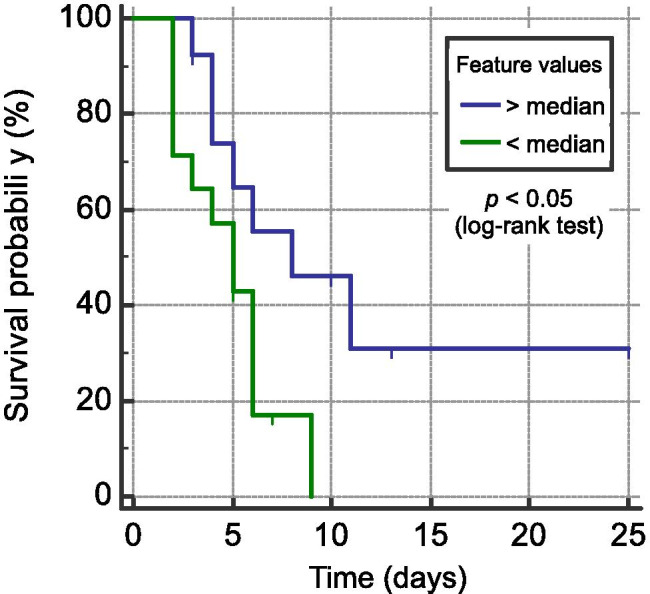
Kaplan–Meier deterioration-free survival curves of COVID-19 patients stratified by the radiomic biomarker f174
Discussion
In this study, we provided evidence that 57 radiomic features from chest radiographs can improve diagnostics’ specificity and determine the worst outcome in the short-term in COVID-19 patients. The early diagnosis of COVID-19 is crucial for the patient’s isolation to prevent virus spread and for rapid treatment decisions to improve the patient’s short-term prognosis [24]. Medical imaging plays a critical role in evaluating COVID-19, mainly on staging the disease’s extent and monitoring the progression after treatment (dexamethasone to critically ill patients on ventilators, for instance) [4, 25, 26]. In early stages, multiple small patchy shadows and interstitial changes emerge in the lungs, while in severe stages, the lesions aggravate, leading to massive infiltrating consolidations and ground-glass opacities, ultimately changing into fibrosis in a dissipative phase [2, 27].
Most of those radiological characteristics are subjectively evaluated with CT imaging as it has higher sensitivity on visual assessment than radiography [5, 7]. However, XR is more accessible and exposes the patient to less radiation. Therefore, chest radiographic biomarkers may have a significant impact on supporting clinical decisions. It is widely known that COVID-19 has a worse prognosis in older people and patients with chronic comorbidities (e.g., hypertension, diabetes, and cardiac diseases) due to their weaker immune system [1, 4].
In this work, we identified XR features associated with COVID-19 that can stratify the patient’s short-term risk even without comorbidity conditions and at an early stage of care (at hospital admission, for instance). These features could indicate the patient’s rapid worsening before the clinical condition deteriorates and when treatment is more likely to have greater benefit. Moreover, f287 and f174 identified patients at a higher/lower risk, confirming the worse short-term prognosis to men in comparison to women [28].
The COVID-19-correlated features of f521, f287, and f174 highlights the challenge of visually recognizing intricate XR patterns, as they were uncovered only after wavelet transform or square filtering. Thus, the radiomics of COVID-19 only identified the biomarkers from a higher-order imaging level with frequency domain analysis. These higher-order features traditionally describe different properties of spectral components from a ROI, characterizing image heterogeneity [13, 15], but the wavelet transforms enabled to capture higher textural heterogeneity on radiography from COVID-19 and not from other pneumonia etiologies.
Radiomic models have previously been developed to improve chest radiographic assessment of pneumonia cases. Sousa et al. [29] used wavelet-derived features as input to three different multivariate methods to detect childhood pneumonia. Chandra et al. [30] employed five different artificial intelligence techniques with first-order histogram features to detect adult pneumonia in XR. Deep-learning models have also been used to detect pediatric pneumonia [31, 32], but all of them were done prior to the COVID-19.
The small sampling of the cohorts precludes our findings’ generalization, which will require validation in future studies. That will be facilitated by the fact that the pandemic has not disappeared, and soon a large number of images will be available in the public domain to validate/improve these findings. Moreover, as sharing data policy is being heavily stimulated, we expect to access clinical data to enhance the number of candidate biomarkers for COVID-19. A prospective evaluation of the biomarkers will also be necessary to confirm the differences in texture and spectrum of images from similar visually identical radiological assessments. Further validation of these biomarkers may also be instrumental in teleradiology to reduce the gap from distant resource-limited places, where x-ray scanners are the only imaging healthcare option to assist diagnostics and predict outcomes of COVID-19 patients.
Conclusion
Altogether, we identified 57 radiomic biomarkers (51 diagnostics and 6 prognostics, p < 0.05) correlated with the etiologic agent of acute infectious diseases and short-term outcomes from COVID-19 patients. The biomarkers of f521, f287, and f174 have the potential to improve the clinical routine as it could automatically prioritize the exams from higher-risk patients with COVID-19 for further reading from a specialist and investigation. The feature f521 specifically distinguished pneumonia etiologies, which could be used as an initial biomarker to stratify the early identification of COVID-19. Finally, the biomarkers of f521, f287, and f174 could recommend the need for intensive care with mechanical ventilation, for instance, ultimately leading to better outcomes such as decreased deterioration and mortality.
Finally, it will be paramount to test the usefulness of these radiomics to predict or anticipate the critical cases, especially the ones requiring attention to thrombotic events.
Acknowledgements
We thank Zerbini Foundation and Foxconn Brazil for the financial support granted for the research project Machine Learning in Cardiovascular Medicine.
Funding
This work was supported by Foxconn Brazil and Zerbini Foundation as part of the research project "Machine Learning in Cardiovascular Medicine".
Declarations
Conflicts of Interest
The authors received research grants from Foxconn Brazil and Zerbini Foundation.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
José Raniery Ferreira Junior and Diego Armando Cardona Cardenas contributed equally to this work
References
- 1.Yuan M, Yin W, Tao Z, Tan W, Hu Y. Association of radiologic findings with mortality of patients infected with 2019 novel coronavirus in Wuhan. China. PloS One. 2020;15(3):e0230548. doi: 10.1371/journal.pone.0230548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li M, Lei P, Zeng B, Li Z, Yu P, Fan B, Wang C, Li Z, Zhou J, Hu S, et al. Coronavirus disease (COVID-19): spectrum of CT findings and temporal progression of the disease. Acad Radiol. 2020;27(5):603–608. doi: 10.1016/j.acra.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu K, Xu P, Lv WF, Qiu XH, Yao JL, Jin-Feng G, et al: CT manifestations of coronavirus disease-2019: a retrospective analysis of 73 cases by disease severity. Eur J Radiol, 108941, 2020 [DOI] [PMC free article] [PubMed]
- 4.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang Y, Zhang H, Xie J, Lin M, Ying L, Pang P, and Ji W: Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology, 200432, 2020 [DOI] [PMC free article] [PubMed]
- 6.Bai HX, Hsieh B, Xiong Z, Halsey K, Choi JW, Tran TML, Pan I, Shi LB, Wang DC, Mei J, et al: Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology, 200823, 2020 [DOI] [PMC free article] [PubMed]
- 7.Guan CS, Lv ZB, Yan S, Du YN, Chen H, Wei LG, Xie RM, Chen BD. Imaging features of coronavirus disease 2019 (COVID-19): evaluation on thin-section CT. Acad Radiol. 2020;7(5):609–613. doi: 10.1016/j.acra.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng MY, Lee EY, Yang J, Yang F, Li X, Wang H, Lui MMS, Lo CSY, Leung B, Khong PL, et al: Imaging profile of the COVID-19 infection: radiologic findings and literature review. Radiology: Cardiothoracic Imaging, 2(1):e200034, 2020 [DOI] [PMC free article] [PubMed]
- 9.Ferreira-Junior JR, Koenigkam-Santos M, Tenório APM, Faleiros MC, Cipriano FEG, Fabro AT, Näppi J, Yoshida H, de Azevedo-Marques PM. CT-based radiomics for prediction of histologic subtype and metastatic disease in primary malignant lung neoplasms. Int J Comput Assist Radiol Surg. 2020;15(1):163–172. doi: 10.1007/s11548-019-02093-y. [DOI] [PubMed] [Google Scholar]
- 10.Degnan AJ, Ghobadi EH, Hardy P, Krupinski E, Scali EP, Stratchko L, Ulano A, Walker E, Wasnik AP, Auffermann WF. Perceptual and interpretive error in diagnostic radiology causes and potential solutions. Acad Radiol. 2019;26(6):833–845. doi: 10.1016/j.acra.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Santos MK, Ferreira Júnior JR, Wada DT, Tenório APM, Barbosa MNH, and Marques PMDA: Artificial intelligence, machine learning, computer-aided diagnosis, and radiomics: advances in imaging towards to precision medicine. Radiologia Brasileira, 52(6):387–396, 2019 [DOI] [PMC free article] [PubMed]
- 12.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5(1):1–9. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolossváry M, Kellermayer M, Merkely B, Maurovich-Horvat P. Cardiac computed tomography radiomics. J Thorac Imaging. 2018;33(1):26–34. doi: 10.1097/RTI.0000000000000268. [DOI] [PubMed] [Google Scholar]
- 14.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferreira Junior JR, Koenigkam-Santos M, Machado CVB, Faleiros MC, Correia NSC, Cipriano FEG, Fabro AT, and de Azevedo-Marques PM: Radiomics analysis of lung cancer for patient prognosis and intratumor heterogeneity assessment. Radiologia Brasileira, Accepted for publication, 2020 [DOI] [PMC free article] [PubMed]
- 16.Van Griethuysen JJ, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RG, Fillion-Robin JC, Pieper S, and Aerts HJ: Computational radiomics system to decode the radiographic phenotype. Cancer Res, 77(21):e104–e107, 2017 [DOI] [PMC free article] [PubMed]
- 17.Italian Society of Medical and Interventional Radiology. COVID-19 Database. Online; last acess on March 24, 2020. Available at www.sirm.org/category/senza-categoria/covid-19/, 2020
- 18.Bustos A, Pertusa A, Salinas JM, and de la Iglesia-Vayá M: Padchest: A large chest x-ray image dataset with multi-label annotated reports. arXiv preprint arXiv:1901.07441, 2019. [DOI] [PubMed]
- 19.Demner-Fushman D, Kohli MD, Rosenman MB, Shooshan SE, Rodriguez L, Antani S, Thoma GR, McDonald CJ. Preparing a collection of radiology examinations for distribution and retrieval. J Am Med Inform Assoc. 2016;23(2):304–310. doi: 10.1093/jamia/ocv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen JP, Morrison P, and Dao L: COVID-19 image data collection. Online; last acess on March 24, 2020. Available at https://github.com/ieee8023/covid-chestxray-dataset, 2020
- 21.Ronneberger O, Fischer P, and Brox T: U-Net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, 234–24, 2020
- 22.Pazhitnykh I, and Petsiuk V: Lung segmentation (2D). Online; last acess on March 24, 2020. Available at https://github.com/imlab-uiip/lung-segmentation-2d, 2017
- 23.Zwanenburg A, Vallières M, Abdalah MA, Aerts HJ, Andrearczyk V, Apte A, Ashrafinia S, Bakas S, Beukinga RJ, Boellaard R, et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295(2):328–338. doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, Bai J, Lu Y, Fang Z, Song Q, et al: Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology, 200905, 2020 [DOI] [PMC free article] [PubMed]
- 25.Randomised Evaluation of COVid-19 thERapY (RECOVERY) Trial. Low-cost dexamethasone reduces death by up to one third in hospitalised patients with severe respiratory complications of covid-19. Online; last access on June 18, 2020. Available at https://www.recoverytrial.net/news/low-cost-dexamethasone-reduces-death-by-up-to-one-third-in-hospitalised-patients-with-severe-respiratory-complications-of-covid-19, 2020
- 26.Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, Tao Q, Sun Z, and Xia L: Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology, 200642, 2020 [DOI] [PMC free article] [PubMed]
- 27.Chung M, Bernheim A, Mei X, Zhang N, Huang M, Zeng X, Cui J, Xu W, Yang Y, Fayad ZA, et al: CT imaging features of 2019 novel coronavirus (2019-nCoV). Radiology, 295(1):202–207, 2020 [DOI] [PMC free article] [PubMed]
- 28.Moradi B, Ghanaati H, Kazemi MA, Gity M, Hashemi H, Davaritanha F, Chavoshi M, Rouzrokh P, and Kolahdouzan K: Implications of sex difference in CT scan findings and outcome of patients with COVID-19 pneumonia. Radiology: Cardiothoracic Imaging, 2(4):e200248, 2020 [DOI] [PMC free article] [PubMed]
- 29.Sousa RT, Marques O, Soares FAA, Sene II, Jr, de Oliveira LL, Spoto ES. Comparative performance analysis of machine learning classifiers in detection of childhood pneumonia using chest radiographs. Procedia Computer Science. 2013;18:2579–2582. doi: 10.1016/j.procs.2013.05.444. [DOI] [Google Scholar]
- 30.Chandra TB, and Verma K. Pneumonia detection on chest x-ray using machine learning paradigm. In Proceedings of 3rd International Conference on Computer Vision and Image Processing, Springer, 21-33, 2020
- 31.Kermany DS, Goldbaum M, Cai W, Valentim CC, Liang H, Baxter SL, McKeown A, Yang G, Wu X, Yan F, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 32.Liang G, and Zheng L: A transfer learning method with deep residual network for pediatric pneumonia diagnosis. Comput Methods Programs Biomed, 104964, 2020 [DOI] [PubMed]