Abstract
Background
Gastric cancer (GC) is the third most common cause of cancer deaths worldwide. In the present study, we aimed to identify novel GC biomarkers by integrating isobaric tags of relative and absolute quantitation (iTRAQ) for aberrantly expressed proteins in GC patients.
Methods
Using stable isotope tags, we labeled an initial discovery group comprising four paired gastric cancer and adjacent gastric tissue samples, and subjected them to LC‐ESI‐MS/MS. We used a validation set comprising 129 paired gastric cancer and adjacent gastric tissues from patients and benign healthy controls to validate the candidate targets.
Results
We identified two proteins, NAD(P)‐dependent steroid dehydrogenase‐like (NSDHL) and neutral cholesterol ester hydrolase 1 (NCEH1), that were significantly overexpressed in GC tissues. The sensitivity and specificity of NSDHL were 80.6% and 74.4%, respectively, in GC compared with a sensitivity of 25.6% in adjacent tissues and 24% in benign healthy controls. The area under the ROC curve (AUC) for NSDHL was 0.810 for GC detection. Overexpression of NSDHL in GC was significantly correlated with local tumor invasion. The sensitivity and specificity of NCEH1 were 77.5% and 73.6%, respectively, in GC compared with a sensitivity of 26.4% in adjacent tissues and 20% in benign controls. The AUC for NSDHL was 0.792. Overexpression of NCEH1 was significantly associated with tumor histological classification and local invasion. Moreover, a combined analysis of NSDHL and NCEH1 achieved a sensitivity and specificity of 85.7% and 83%, respectively, and the AUC was 0.872. The combined analysis of NSDHL and NCEH1 was significantly correlated with histological grade and TNM Ⅱ‐Ⅳ staging.
Conclusions
iTRAQ‐labeled quantitative proteomics represents a powerful method to identify novel cancer biomarkers. The present study identified NSDHL and NCEH1 as useful biomarkers for screening, diagnosis, and prognosis of patients with gastric cancer.
Keywords: Cholesterol metabolism, Gastric cancer, iTRAQ, NCEH1, NSDHL
In the present study, The AUCs for NSDHL and NCEH1 were 0.810 and 0.792. A combined analysis of NSDHL and NCEH1 achieved a sensitivity and specificity of 85.7% and 83%, and the AUC was 0.872. The combined analysis of NSDHL and NCEH1 significantly improved the AUC for detecting gastric cancer.
![]()
1. INTRODUCTION
Gastric cancer (GC) is the third most common cause of cancer deaths worldwide. Because of a lack of early‐stage gastric cancer diagnostic and therapeutic technologies and targets, the majority of gastric cancer patients exhibit poor survival rates. 1 , 2 , 3 Although research in this area has steadily progress over the last several decades, the currently available gastric cancer diagnostic biomarkers, such as carbohydrate antigen 19.9 (CA 19.9), carcinoembryonic antigen (CEA), and pepsinogen I/II, have limited diagnostic sensitivity and specificity. 4 , 5 , 6 Therefore, it is important to identify novel diagnostic biomarkers that are responsible for gastric carcinoma etiology and progression. This would be beneficial for early‐stage detection of gastric cancer and for the development of effective therapeutic strategies.
Proteomics represents a powerful tool for the identification of novel biomarkers in malignant tumors. Compared with 2‐D electrophoresis, mass spectrometry is a superior technique for detecting the relative abundance of proteins in biological samples. A great number of methods have been developed that support relative quantification measurements. Stable isotope labeling of proteins in samples prior to analysis is one of the most popular methods for relative quantification by mass spectrometry. 7 Isobaric tags for relative and absolute quantification (iTRAQ) is now the most widely used method for high‐throughput protein quantification, and it is capable of simultaneously quantitating four to eight different biological samples. 8 , 9 Thus, iTRAQ provides an unbiased method to identify novel potential diagnostic and therapeutic targets for early cancer stage detection.
In this study, we used iTRAQ‐labeled quantitative proteomics as a gold standard technique to identify novel biomarkers in gastric cancer compared with adjacent tissues. We identified NAD(P)‐dependent steroid dehydrogenase‐like (NSDHL) and neutral cholesterol ester hydrolase 1 (NCEH1) as potential novel biomarkers, which were further validated in a separate series of surgical gastric cancer samples for early‐stage gastric cancer screening.
2. MATERIALS AND METHODS
2.1. Clinical specimens
The specimens comprised fresh stomach cancer tissues surgically removed from gastric cancer patients along with surrounding adjacent tissues, which were 5 cm from the edge of the tumor and had no obvious tumor cell content as determined by an experienced pathologist. All 129 patients with gastric cancer and 25 healthy control subjects with benign gastritis from January 2010 to December 2018 were included, and no patient underwent radiotherapy or chemotherapy prior to surgery. The study was approved by the Human Research Ethics Committee of North Sichuan Medical College.
2.2. Sample preparation and iTRAQ labeling
Frozen tissue samples representing four pairs of samples in the discovery group were quickly weighed, homogenized in lysis buffer, and centrifuged. The supernatant containing the proteins was collected and stored at −80°C. iTRAQ 8‐plex experiments were performed on the tissue extracts. Each iTRAQ channel contained 50 μg of protein per sample. The proteins were digested with trypsin (10% w/w, sequencing grade modified trypsin, Sigma). The iTRAQ 8‐plex labeling was performed according to the manufacturers’ instructions (iTRAQ labeled for 2.5 hours). iTRAQ labels 113 and 114 were used to label the first pair of gastric cancer and adjacent normal gastric tissue, respectively. Labels 115/116, 117/118, and 119/121 were used for the remaining three tissue pairs, respectively. After iTRAQ labeling, the samples were pooled, desalted on a 500‐mg Sep‐Pak C18 column (Millipore), dried in a Speed Vac Concentrator (Thermo Scientific), and subjected to peptide fractionation.
2.3. Strong cation exchange chromatography
The dried iTRAQ‐labeled sample was diluted 10‐fold in a loading buffer. Then, the samples were reconstituted in 4 mL of buffer A (25 mM NaH2PO4 in 25% ACN, pH 2.7) and loaded onto a 5‐μm particle size, 4.6 × 250 mm Ultremex SCX Column (Phenomenex). The column was eluted with a gradient of buffer A for 10 min, 5%–35% buffer B (25 mM NaH2PO4, 1 M KCl, 25% ACN, pH 2.7) for 11 min, and 35%–80% buffer B for 1 min. The peptides were pooled into 20 fractions, and each fraction was collected at 1‐min intervals. Finally, samples were dried under vacuum and stored at −20°C until the MS analysis.
2.4. LC‐ESI‐MS/MS analysis
The LC setup was equipped with a linear ion trap mass spectrometer, the LTQ‐Orbitrap Discovery (Thermo Fisher, San Jose, CA, USA). A data‐dependent acquisition protocol, in which one MS scan was followed by three MS/MS scans for the eight most abundant precursor ions in the MS survey scan, was applied. The sample volume was 10 μl per injection. A blank was introduced after each sample to clear the system. A 1.8 kV electrospray voltage was applied. The dynamic exclusion settings were 2 s for repeat counts, 30 s for repeat duration, and 120 s for exclusion duration. The automatic gain control to avoid overfilling of the ion trap and the PQD spectra was generated by 5 × 104 ions accumulated in the ion trap. The m/z scan range was 350‐2,000 Da.
2.5. Database search and bioinformatics
The MASCOT search engine (Matrix Science, London, UK; version 2.20) was used to compare MS/MS spectra with that of the International Protein Index (IPI) human database (https://www.ebi.ac.uk/IPI). A threshold of 10 ppm for intact peptide tolerance masses and that of 0.05 Da for PQD fragment ions were set for protein identification. For quantification of the identified proteins, we defined the threshold for a significant change as a 1.2‐fold increase or decrease in expression. We also searched the spectra against a decoy database and estimated that the false‐discovery rate (FDR) of our identified peptides was 1.2%. Gene ontology analysis was performed by Blast2GO software, and pathways associated with the candidate proteins were identified using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.
2.6. Immunohistochemical staining
Immunohistochemical (IHC) staining was performed as previously described. 10 Briefly, 5‐μm‐thick consecutive sections of formalin‐fixed, paraffin‐embedded specimens from gastric cancer patients were stained with antibodies using the Boster immunohistochemical kit (Boster Biology). IHC staining was performed using specific antibodies against NSDHL (Santa Cruz; diluted 1:50‐100) and NCEH1 (Abcam, MA, USA; diluted 1:50‐100). Two pathologists independently evaluated the intensities and percentages of the stained sections. The staining intensity was scored as 0 for a negative stain and 1, 2, or 3 to indicate weak, moderate, or strong staining, respectively. The percentages of positive tumor cells were scored as 0 (<5%), 1 (6%–25%), 2 (26%–50%), 3 (51%–75%), or 4 (76%–100%). The IHC scores were calculated by multiplying the percentage of positively stained cells by their corresponding intensities to obtain a final protein expression score. Slides with final scores ≥ 3 were considered positive, and those with scores <3 were considered negative.
2.7. Statistical analysis
The differences between experimental groups were analyzed with a chi‐square test. All analyses including chi‐square tests and area under the ROC curves (AUCs) were performed using SPSS 23.0 (SPSS, Chicago, IL, USA) software for Windows. p ≤ .05 (two‐sided) was considered significant.
3. RESULTS
3.1. NSDHL and NCEH1 are overexpressed in gastric cancer tissues
In this study, we used four technical replicates to demonstrate the reproducibility of the experimental results and to perform complementation. A total of 431 proteins were characterized as differentially expressed in gastric cancer tissues compared with adjacent tissues. 11 Functional clustering of the differentially expressed proteins revealed that the majority represented metabolic proteins. We found that two key enzymes, NSDHL and NCEH1, which are associated with cholesterol metabolism, were significantly overexpressed in gastric cancer tissues. NSDHL was upregulated 1.476‐fold, and NCEH1 was upregulated 1.303‐fold in gastric cancer tissues.
3.2. Validation of NSDHL and NCEH1 expression in gastric cancer tissues by immunohistochemistry
The expression of NSDHL (Figure 1A, B) and NCEH1 (Figure 1C, D) was further validated by immunohistochemistry. NSDHL was overexpressed in 80.6% of GC tissues compared with 25.6% of surrounding normal tissues (Figure 2A). The localization of NSDHL was mainly in the cytoplasm. NCEH1 was overexpressed in 77.5% of GC tissues compared with 26.4% in adjacent normal tissues (Figure 2A, Table 1). The NCEH1 was mainly localized to the cytoplasm. Furthermore, a combined analysis revealed that NSDHL and NCEH1 were overexpressed in 85.7% of gastric cancer tissue compared with those in 17% of adjacent gastric tissues and 22% of healthy controls (Figure 2B). The AUCs were 0.810 for NSDHL, 0.792 for NCEH1, and 0.872 for the combination of NSDHL and NCEH1 (Figure 3A, B, and C, respectively). These results suggest that the diagnostic value for gastric cancer was significantly increased by the combined analysis of NSDHL and NCEH1.
Figure 1.
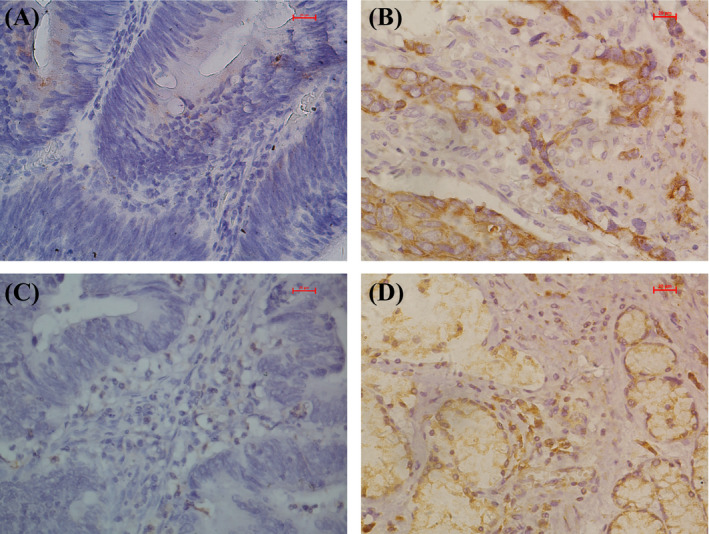
Immunohistochemical analysis of the expression of NSDHL and NCEH1 in gastric cancer tissues. NSDHL was primarily located in the cytoplasm (A, adjacent tissues; B, gastric cancer tissues), and NCEH1 was mainly located in the cytoplasm (C, adjacent tissues; D, gastric cancer tissues;)
Figure 2.
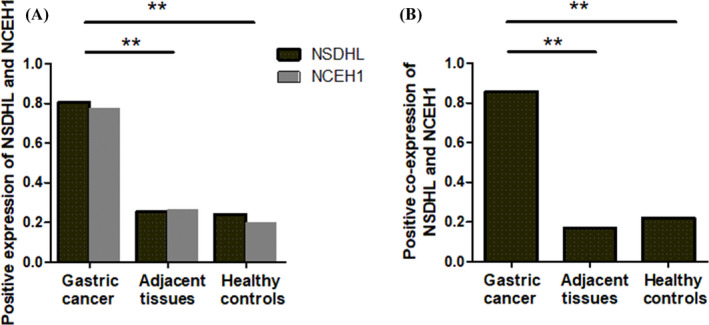
Expression of NSDHL (A) and NCEH1 (A) and both of them (B) in gastric cancer tissues, adjacent tissues, and benign healthy controls
Table 1.
Validation of NSDHL and NCEH1 expression in gastric cancer tissues, adjacent normal tissues, and benign healthy controls
| Adjacent normal tissues | Cancer tissues |
Healthy controls |
p‐values* | |
|---|---|---|---|---|
| NSDHL(‐) | 96 | 25 | 19 | |
| NSDHL(+) | 33 | 104 | 6 | <.001 |
| NCEH1(‐) | 95 | 29 | 20 | |
| NCEH1(+) | 34 | 100 | 5 | <.001 |
| NSDHL and NCEH1(‐) | 78 | 15 | 18 | |
| NSDHL and NCEH1(+) | 16 | 90 | 5 | <.001 |
The p‐values were determined by the chi‐square test.
Figure 3.
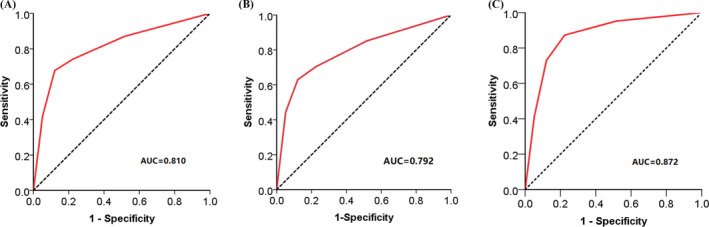
ROC curves for NSDHL and NCEH1. (A), The AUCs were 0.810 for NSDHL. (B), The AUC were 0.792 for NCEH1. (C), The AUC were 0.872 for the combination of NSDHL and NCEH1. p < .001
3.3. Clinicopathological features between NSDHL‐positive and NSDHL‐negative patient groups
Next, we performed a chi‐square test to analyze the relationship between NSDHL‐positive and NSDHL‐negative patient samples with clinicopathological factors including histological type, T staging, lymph node involvement, and distant metastasis. As shown in Table 2, NSDHL expression was significantly upregulated in the T3 and T4 group compared with the T1 and T2 group (p = .013).
Table 2.
Correlation of NSDHL expression with clinicopathological features in gastric cancer
| Clinicopathological features | NSDHL (‐) | NSDHL (+) | Case | p‐values* |
|---|---|---|---|---|
| Histology classification | ||||
| Adenocarcinoma | 20 (18.7%) | 87 (81.3%) | 107 | .663 |
| Signet‐ring cell carcinoma | 5 (22.7%) | 17 (77.3%) | 22 | |
| pT | ||||
| T1 + T2 | 17 (28.8%) | 42 (71.2%) | 59 | .013 |
| T3 + T4 | 8 (11.4%) | 62 (88.6%) | 70 | |
| pN | ||||
| pN0 | 13 (23.2%) | 43 (76.8%) | 56 | .402 |
| pN1 + pN2 | 8 (20.5%) | 31 (79.5%) | 39 | |
| pN3a + pN3b | 4 (11.8%) | 30 (88.2%) | 34 | |
| pM | ||||
| pM0 | 25 (20.3%) | 98 (79.7%) | 123 | ‐ |
| pM1 | 0 | 6 (‐) | 6 | |
The p‐values were determined by the chi‐square test.
3.4. The correlation of NCEH1 expression with clinicopathological features
To explore the clinical association of NCEH1 expression with clinicopathological features, we found that NCEH1 was highly expressed in gastric adenocarcinoma compared with that in signet‐ring cell carcinoma (Table 3, p = .023). Furthermore, patients with T3 and T4 TNM stage disease exhibited a higher expression of NCEH1 compared with patients in stage T1 or T2 (Table 3, p = .004).
Table 3.
Correlation of NCEH1 expression with clinicopathological features in gastric cancer
| Clinicopathological features | NCEH1 (‐) | NCEH1 (+) | Case | p‐values* |
|---|---|---|---|---|
| Histology classification | ||||
| Adenocarcinoma | 20 (18.7%) | 87 (81.3%) | 107 | .023 |
| Signet‐ring cell carcinoma | 9 (40.9%) | 13 (59.1%) | 22 | |
| pT | ||||
| T1 + T2 | 20 (33.9%) | 39 (66.1%) | 59 | .004 |
| T3 + T4 | 9 (12.9%) | 61 (87.1%) | 70 | |
| pN | ||||
| pN0 | 15 (26.8%) | 41 (73.2%) | 56 | .217 |
| pN1 + pN2 | 10 (25.6%) | 29 (74.4%) | 39 | |
| pN3a + pN3b | 4 (11.8%) | 30 (88.2%) | 34 | |
| pM | ||||
| pM0 | 28 (22.8%) | 95 (77.2%) | 123 | ‐ |
| pM1 | 1 (16.7%) | 5 (83.3%) | 6 | |
The p‐values were determined by the chi‐square test.
3.5. Clinical association of NSDHL and NCEH1 expression with pathological features in gastric cancer
As shown in Table 4, to further explore the combined diagnostic values of NSDHL and NCEH1 in gastric cancer, we found that NSDHL and NCEH1 expression was significantly associated with histological grade and TNM staging. Combined NSDHL and NCEH1 expression was significantly higher in patients with histological grade I compared with that in patients with histological grade Ⅱ and Ⅲ (p = .048). Compared with TNM stage I, gastric cancer patients with TNM stage Ⅱ to Ⅳ presented with higher NSDHL and NCEH1 expression (Table 4).
Table 4.
Correlation of NSDHL and NCEH1 expression with clinicopathological features in gastric cancer
| Clinicopathological features | NSDHL and NCEH1 (‐) | NSDHL and NCEH1 (+) | Case | p‐values* |
|---|---|---|---|---|
| Age (year) | ||||
| ≥60 | 10 (14.9%) | 57 (85.1%) | 67 | .804 |
| ≤59 | 5 (13.2%) | 33 (86.8%) | 38 | |
| Sex | ||||
| Male | 11 (14.9%) | 63 (85.1%) | 74 | .793 |
| Female | 4 (12.9%) | 27 (87.1%) | 31 | |
| Histology classification | ||||
| Adenocarcinoma | 10 (11.5%) | 77 (88.5%) | 87 | .072 |
| Signet‐ring cell carcinoma | 5 (27.8%) | 13 (72.2%) | 18 | |
| Histological grade | ||||
| I | 7 (9.7%) | 65 (90.3%) | 72 | .048 |
| II and III | 8 (24.2%) | 25 (75.8%) | 33 | |
| Lymph node metastasis | ||||
| Positive | 7 (11.5%) | 54 (88.5%) | 61 | .333 |
| Negative | 8 (18.2%) | 36 (81.8%) | 44 | |
| TNM Staging | ||||
| I | 10 (25.6%) | 29 (74.4%) | 39 | .011 |
| Ⅱ‐Ⅳ | 5 (7.6%) | 61 (92.4%) | 66 | |
The p‐values were determined by the chi‐square test.
4. DISCUSSION
Although there have been improvements in the treatment of gastric cancer in recent years, prognosis remains poor primarily because the disease is often diagnosed at advanced stages. Identification of reliable biomarkers is an essential for early detection and prognosis of gastric cancer. We are the first to discover that NSDHL and NCEH1 are significantly overexpressed in gastric cancer tissues compared with adjacent gastric tissues and healthy controls. Notably, the aberrantly high expression of NSDHL and NCEH1 in gastric cancer patients was significantly associated with clinicopathological histology classification, local invasion, histological grade, and TNM staging.
Cholesterol is a major component of cell membranes and a precursor for steroid hormones. The biosynthesis and catabolism reactions of cholesterol ester are central cholesterol metabolic pathways in animal cells. Comprising more than 20 reactions, cholesterol biosynthesis is strictly regulated within cells. Dysregulation or imbalance of cholesterol metabolic pathways contributes to many human diseases such as atherosclerosis, neuropathy, and cancer progression. 12 , 13 Several recent studies have demonstrated that dysregulated cholesterol metabolism is a salient feature in the progression of cancer. 14 , 15 In the present study, we found that NSDHL and NCEH1 were significantly overexpressed in patients with gastric cancer. These results suggest that patients with gastric cancer may experience dysregulated cholesterol metabolism because NSDHL and NCEH1 are key enzymes that contribute to cholesterol metabolism.
NAD(P)‐dependent steroid dehydrogenase‐like is an essential enzyme in the penultimate step of cholesterol biosynthesis. It catalyzes NAD+‐dependent oxidative decarboxylation of the C4 methyl groups of 4α‐carboxysterols. Previous studies have demonstrated that a loss‐of‐function mutation in the human NSDHL gene results in congenital hemidysplasia with ichthyosiform nevus and limb defect (CHILD) syndrome. This is an X‐linked human embryonic developmental disorder that is lethal in males and causes limb reduction in females. 16 Recently, several studies have revealed that NSDHL may also serve as an oncogene in cancer progression. 17 , 18 , 19
Xue et al found that NSDHL was overexpressed in 67NR, 168FARN, and 4T1 murine breast cancer cell lines and translocated to the plasma membrane from the intracellular compartment. The study suggested that the altered localization and differential expression of NSDHL may contribute to breast cancer cell metastasis and serve as a novel target for anti‐metastasis therapy. 17 Mechanistic studies revealed that NSDHL is a key regulator of growth and proto‐oncogene signaling in cancer cells. Gabitova et al demonstrated that NSDHL deficiency resulted in the accumulation of sterol metabolites, which inhibit tumor growth. 18 In addition, the loss of NSDHL gene expression sensitized the response of cancer cells to EGFR‐targeting inhibitors. 19 To develop a targeted compound, Kim et al analyzed the protein structure of NSDHL and identified a novel molecule (compound 9), which could inhibit NSDHL and its biological function in cells. The results revealed that compound 9 decreased EGFR expression and signal transduction. Moreover, when combined with an EGFR inhibitor, compound 9 enhanced its antitumor effect. 14
Consistent with the above studies, we found that NSDHL was overexpressed by 80.6% in gastric cancer tissues and more than 25.6% in surrounding normal gastric tissues. In addition, NSDHL expression in gastric cancer patients with T3 and T4 stage disease was significantly higher than that in patients with T1 and T2 stage (Table 2). The results suggest that NSDHL has the potential to regulate the local invasion of gastric cancer cells. NSDHL exhibited a sensitivity of 80.6% and a specificity of 74.4% in the detection of patients with gastric cancer. To further improve the sensitivity and specificity, we analyzed the combined expression of NSDHL and NCEH1 in gastric cancer.
Neutral cholesterol ester hydrolase 1 (NCEH1), also known as arylacetamide deacetylase‐like 1 (AADACL1) and KIAA1363, is a serine hydrolase, which has diverse biological activities and cellular functions. 20 , 21 , 22 , 23 , 24 NCEH1 hydrolyzes cholesterol esters to free cholesterol, which contributes to the development of atherosclerosis. NCEH1 also hydrolyzes 2‐acetyl monoalkylglycerol, a metabolic intermediate in the ether lipid pathway to form alkyl‐LPA. Moreover, NCEH1 may also hydrolyze exogenous organophosphorus compounds during organ detoxification. 25 NCEH1 is overexpressed in a variety of aggressive human cancer cell lines and primary tumors. 20 , 21 , 22 , 23 Although NCEH1 regulates the levels of neutral ether lipids and cholesterol ester metabolism, the manner in which these metabolites regulate the progression of cancer remains unclear.
Recent studies have demonstrated that after a single and repeated treatment of chlorpyrifos and chlorpyrifos‐oxon, NCEH1 is elevated in MCF‐7 and MDA‐MA‐231 cells and induces proliferation. The results indicated that NCEH1 may participate with chlorpyrifos as factors in the induction of breast cancer. 26 Chang et al reported that NCEH1 was highly expressed in prostate cancer cells. Using covalent PET probe imaging, they observed that NCEH1 was primarily localized to the outer edges of xenograft tissues, with less localization within the tumor. These results indicate that NCEH1 may contribute to the aggressiveness and metastasis of breast cancer cells. 21 By hydrolyzing the metabolic intermediate, 2‐acetylmonoalkylglycerol, NCEH1 may regulate the ether lipid signaling network that links platelet activating factor and lysophosphatidic acid metabolism. To develop a novel inhibitor of NCEH1, they identified JW480, an O‐aryl carbamate, which acts as a potent and selective inhibitor of NCEH1. JW480 reduced monoalkylglycerol ether and inhibited prostate cancer cell migration, invasion, and growth in vivo. 20
In addition to Chang's studies, other researchers have designed and developed compounds to detect and inhibit NCEH1 activity. Fan et al designed the first “off‐on” NIR probe, NB‐AX, which exhibits a high selectivity for NCEH1. This probe can rapidly distinguish breast cancer cells from normal cells with a detection limit of 0.58 μg/ml. 27 To develop novel inhibitors for NCEH1 in cancer, Shreder et al demonstrated that AX13057 inhibits NCEH1 activity in SK‐OV‐3 xenograft‐bearing mice based on protein profiling. 28
In the present study, we found that NCEH1 was highly expressed in 77.5% of gastric cancer tissues and was higher compared with the 26.4% expression observed in adjacent gastric tissues. Further analysis indicated that the high expression of NCEH1 was significantly associated with histological type and local invasion of T3 and T4 stage disease. Traditional biomarkers, such as CA 19.9, exhibit a sensitivity of 20%–56%. However, our study revealed that NCEH1 had a sensitivity of 77.5% and a specificity of 73.6% for the detection of gastric cancer. Our results suggest that NCEH1 may serve as a novel biomarker for gastric cancer detection and progression, particularly for patients with an adenocarcinoma histology type and T3 or T4 stage disease. To further improve the sensitivity and specificity of NCEH1, we combined NSDHL with NCEH1, another key enzyme in the cholesterol ester catabolism pathway, and analyzed their diagnostic potential in gastric cancer. We observed that the combination of NSDHL and NCEH1 exhibited a sensitivity of 85.7% and specificity of 83% in detecting gastric cancer. Moreover, the AUC value of 0.872 for combined NSDHL and NCEH1 was higher than either marker alone. In addition, the coexpression of NSDHL and NCEH1 was significantly associated with advanced TNM stage in gastric cancer patients. Our results suggest that the combined analysis of NSDHL and NCEH1 achieved a higher sensitivity and specificity for screening gastric cancer than that of a single marker and may be useful in the screening and follow‐up assessment of gastric cancer patients.
In conclusion, we used iTRAQ‐based quantitative proteomics to discover that NSDHL and NCEH1, key enzymes in cholesterol metabolism, were aberrantly expressed in gastric cancer. Dysregulated expression of NSDHL and NCEH1 in gastric cancer was significantly correlated with histological type, local invasion, and TNM staging. Moreover, our results indicate that the combined analysis of NSDHL and NCEH1 as coordinate markers was superior to a single marker in detecting gastric cancer. Therefore, our study indicates that NSDHL and NCEH1 may serve as novel biomarkers in the screening and follow‐up of patients with gastric cancer.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (81702093); the Sichuan Provincial Department of Science and Technology (2020YJ0379); the Special Foundation of Cooperation between Nanchong Government and North Sichuan Medical College (18SXHZ0281); and the National Training Program of Innovation and Entrepreneurship for Undergraduates (201910634023; 201910634021).
Xiao Y, Xie J, Liu L, et al. NAD(P)‐dependent steroid dehydrogenase‐like protein and neutral cholesterol ester hydrolase 1 serve as novel markers for early detection of gastric cancer identified using quantitative proteomics. J Clin Lab Anal.2021;35:e23652 10.1002/jcla.23652
DATA AVAILABILITY STATEMENT
Data are available in the article.
REFERENCES
- 1. Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635‐648. [DOI] [PubMed] [Google Scholar]
- 2. Baretton GB, Aust DE. Aktuelle Biomarker beim Magenkarzinom. [Current biomarkers for gastric cancer] Pathologe. 2017;38:93‐97. [DOI] [PubMed] [Google Scholar]
- 3. Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23:700‐713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu J, Li G, Wang Z, et al. Circulating MicroRNA‐21 is a Potential Diagnostic Biomarker in Gastric Cancer. Dis Markers. 2015;435656:1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gong X, Zhang H. Diagnostic and prognostic values of anti‐helicobacter pylori antibody combined with serum CA724, CA19‐9, and CEA for young patients with early gastric cancer. J Clin Lab Anal. 2020;34(e23268):1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jiang W, Sun G, Zhang Y, et al. Simultaneous determination of gastric cancer biomarkers pepsinogen PGI/PGII using element tagged immunoassay coupled with inductively coupled plasma mass spectrometry detection. J Clin Lab Anal. 2020;34(7):1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Swiatly A, Horala A, Matysiak J, Hajduk J, Nowak‐Markwitz E, Kokot ZJ. Understanding ovarian cancer: iTRAQ‐based proteomics for biomarker discovery. Int J Mol Sci. 2018;19:2240‐2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Voss BJ, Loh JT, Hill S, Rose KL, McDonald WH, Cover TL. Alteration of the Helicobacter pylori membrane proteome in response to changes in environmental salt concentration. Proteomics Clin Appl. 2015;9:1021‐1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Okano T, Seike M, Kuribayashi H, et al. Identification of haptoglobin peptide as a novel serum biomarker for lung squamous cell carcinoma by serum proteome and peptidome profiling. Int J Oncol. 2016;48:945‐952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hsu CH, Hsu CW, Hsueh C, et al. Identification and Characterization of Potential Biomarkers by Quantitative Tissue Proteomics of Primary Lung Adenocarcinoma. Mol Cell Proteomics. 2016;15:2396‐2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jiang Z, Shen H, Tang BO, Qin YU, Ji X, Wang LI. Quantitative proteomic analysis reveals that proteins required for fatty acid metabolism may serve as diagnostic markers for gastric cancer. Clin Chim Acta. 2017;464:148‐154. [DOI] [PubMed] [Google Scholar]
- 12. Luo J, Yang H, Song BL. Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol. 2020;21:225‐245. [DOI] [PubMed] [Google Scholar]
- 13. Riscal R, Skuli N, Simon MC. Even cancer cells watch their cholesterol!. Mol Cell. 2019;76:220‐231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim DG, Cho S, , , et al. Crystal structures of human NSDHL and development of its novel inhibitor with the potential to suppress EGFR activity. Cell Mol Life Sci. 2020; 1–19. [ahead of print]. 10.1007/s00018-020-03490-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bovenga F, Sabbà C, Moschetta A. Uncoupling nuclear receptor LXR and cholesterol metabolism in cancer. Cell Metab. 2015;21:517‐526. [DOI] [PubMed] [Google Scholar]
- 16. Beck TN, Georgopoulos R, Shagisultanova EI, et al. EGFR and RB1 as Dual Biomarkers in HPV‐Negative Head and Neck Cancer. Mol Cancer Ther. 2016;15:2486‐2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xue T, Zhang Y, Zhang L, Yao L, Hu X, Xu LX. Proteomic analysis of two metabolic proteins with potential to translocate to plasma membrane associated with tumor metastasis development and drug targets. J Proteome Res. 2013;12:1754‐1763. [DOI] [PubMed] [Google Scholar]
- 18. Gabitova L, Restifo D, Gorin A, et al. Endogenous sterol metabolites regulate growth of EGFR/KRAS‐dependent tumors via LXR. Cell Rep. 2015;12:1927‐1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sukhanova A, Gorin A, Serebriiskii IG, et al. Targeting C4‐demethylating genes in the cholesterol pathway sensitizes cancer cells to EGF receptor inhibitors via increased EGF receptor degradation. Cancer Discov. 2013;3:96‐111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chang JW, Nomura DK, Cravatt BF. A potent and selective inhibitor of KIAA1363/AADACL1 that impairs prostate cancer pathogenesis. Chem Biol. 2011;18:476‐484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chang JW, Bhuiyan M, Tsai HM, et al. In Vivo Imaging of the Tumor‐Associated Enzyme NCEH1 with a Covalent PET Probe. Angew Chem Int Ed. 2020;59(35):15161‐15165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chiang KP, Niessen S, Saghatelian A, Cravatt BF. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chem Biol. 2006;13:1041‐1050. [DOI] [PubMed] [Google Scholar]
- 23. Jessani N, Liu Y, Humphrey M, Cravatt BF. Enzyme activity profiles of the secreted and membrane proteome that depict cancer cell invasiveness. Proc Natl Acad Sci U S A. 2002;99:10335‐10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Okazaki H, Igarashi M, Nishi M, et al. Identification of neutral cholesterol ester hydrolase, a key enzyme removing cholesterol from macrophages. J Biol Chem. 2008;283:33357‐33364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ross MK, Pluta K, Bittles V, Borazjani A, Allen CJ. Interaction of the serine hydrolase KIAA1363 with organophosphorus agents: Evaluation of potency and kinetics. Arch Biochem Biophys. 2016;590:72‐81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moyano P, García J, García JM, et al. Chlorpyrifos‐induced cell proliferation in human breast cancer cell lines differentially mediated by estrogen and aryl hydrocarbon receptors and KIAA1363 enzyme after 24 h and 14 days exposure. Chemosphere. 2020;251:1‐13. [DOI] [PubMed] [Google Scholar]
- 27. Fan J, Guo S, Wang S, et al. Lighting‐up breast cancer cells by a near‐infrared fluorescent probe based on KIAA1363 enzyme‐targeting. Chem Commun (Camb). 2017;53:4857‐4860. [DOI] [PubMed] [Google Scholar]
- 28. Shreder KR, Lin EC, Wu J, et al. Synthesis and structure‐activity relationship of (1‐halo‐2‐naphthyl) carbamate‐based inhibitors of KIAA1363 (NCEH1/AADACL1). Bioorg Med Chem Lett. 2012;22:5748‐5751. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available in the article.


