Abstract
Background
This study was aimed at investigating the effects of long noncoding RNA (lncRNA) LINC02323 in ovarian cancer and its possible mechanism.
Methods
Microarray analysis and QPCR were utilized to identify lncRNA LINC02323 expression in patients with ovarian cancer. MTT assay was used for analysis of ovarian cancer cell proliferation. Western blot was utilized to investigate its possible mechanism.
Results
In patients with ovarian cancer, lncRNA LINC02323 expression was up‐regulated and miR‐1343‐3p expression was down‐regulated. Over‐expression of lncRNA LINC02323 promoted cell growth and reduced LDH activity levels in vitro model by suppression of miR‐1343‐3p expression. Down‐regulation of lncRNA LINC02323 reduced cell growth and increased LDH activity levels in vitro model by induction of miR‐1343‐3p expression. Over‐expression of miR‐1343‐3p reduced cell growth and reduced LDH activity levels in vitro model by suppression of TGF‐β receptor. Down‐regulation of miR‐1343‐3p promoted cell growth and reduced LDH activity levels in vitro model by induced of TGF‐β receptor.
Conclusion
Our findings show that Novel long noncoding RNA LINC02323 promotes cell growth of ovarian cancer via TGF‐β receptor 1 by miR‐1343‐3p.
Keywords: LINC02323, miR‐1343‐3p, ovarian cancer, TGF‐β
Serum levels of lncRNA LINC02323 in patients with ovarian cancer (A) and the risk of developing cancer (B), OS, and DFS (C and D).

1. INTRODUCTION
Ovarian cancer seriously endangers the health of women. Due to the atypical early symptoms and a lack of specific screening indicators of ovarian cancer, most patients are initially diagnosed in advanced stages. 1 Therefore, ovarian cancer is highly lethal and has poor prognosis. At present, the treatment of ovarian cancer is mainly based on cisplatin‐based combined chemotherapy after surgery. 2 However, due to the emergence of drug resistance of residual cancer cells, the 5‐year survival rate of patients with ovarian cancer is still at a low level. 3 Therefore, it is of great significance to explore effective approaches to inhibit drug resistance of ovarian cancer cells.
Long noncoding RNAs (LncRNAs) are a type of noncoding RNA molecules with over 200 nucleotides in length and without specific open reading frame. 4 , 5 LncRNAs not only affect gene expression regulation during cell growth, but also play an important role in the tumorigenesis, tumor progression, and drug resistance of malignant tumors. 6 At present, a series of studies have shown that different lncRNAs play sponge activity by combining with specific microRNAs (miRNAs) to inhibit miRNA functions, thereby affecting relevant pathways and affecting tumorigenesis and tumor progression. 7 , 8 , 9
High expression of TGF‐β1 is observed in a variety of tumors. 10 The role of TGF‐β1 in tumor progression has been investigated in vivo, in vitro, and in transgenic mice. Some results are contradictory and difficult to explain, which reflects the complex roles of TGF‐β1 on carcinogenesis. 11 , 12 Many tumor cells are no longer sensitive to the growth inhibitory effect mediated by TGF‐β1, indicating that increased expression of TGF‐β1 may change the microenvironment of tumor tissues, including induction of local vascular proliferation, production of extracellular matrix, and suppression of immune surveillance, all of which provide favorable environments for tumor cell proliferation. 11 In the early stage of tumor development, TGF‐β1 may act as a tumor suppressor; but in the advanced stage of tumor development, TGF‐β1 can promote tumor development as the synthesis of TGF‐β1 increases. 11 This study was aimed at investigating the effects of long noncoding RNA (lncRNA) LINC02323 in ovarian cancer and its possible mechanism.
2. MICROARRAY DATA ANALYSIS
2.1. Clinical patients with melanoma
Serum samples were obtained from patients with ovarian cancer patients or normal volunteer in Cangzhou Center Hospital. Written informed consent was obtained from all participants, and the research protocols were approved by the Ethics Committee of Cangzhou Center Hospital.
2.2. Cell culture and cell transfection
The human ovarian cancer cell line OVCAR3 was obtained from the Shanghai Institutes for Biological Sciences of Chinese Academy of Sciences (Shanghai, China) and cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc.) supplemented with 10% (v/v) heat‐inactivated fetal bovine serum (Thermo Fisher Scientific, Inc.) at 37°C in a humidified atmosphere of 5% CO2%‐95% air.
2.3. Quantitative real‐time polymerase chain reaction
RNA was isolated from SERUM and cells using TRIzol (Invitrogen), and cDNA was synthesized using a TaqMan microRNA Reverse Transcription Kit (Applied Biosystems of Thermo Fisher Scientific) according to the manufacturer's instructions. The TaqMan Universal Master Mix II (Thermo Fisher Scientific) was used for cDNA, and were quantified using SYBR Green real‐time PCR master mix (Applied Biosystems). The relative expressions of miRNA were calculated using the 2−ΔΔCt method.
2.4. Cell viability assay
Cell viability was enacted by MTT (Yeasen, Shanghai, China). After 4 h of transfection, cells were seeded in 96‐well plate. Cells were maintained at 37°C in humidified air with 5% CO2 for 24, 48, and 72 h. 20 μl of MTT substrate (5 mg/ml) was added to each well, and the plates were incubated for 4 h. The medium was then removed, and the cells were solubilized in 150 μl of DMSO for 15 min. The optical absorbance values were read at 490 nm using a Microplate Reader (Bio‐Rad).
2.5. Caspase‐3/9 and LDH activity levels
After 48 h of transfection, cell was collected, total proteins were obtained using PBS and BCA Protein Assay Kit (Pierce) was used for determining proteins concentration. Caspase‐3/9 and LDH activity levels were measured using ELISA assay, and optical absorbance values were read at 405 or 450 nm using a Microplate Reader (Bio‐Rad).
2.6. Transwell assay
100 μl (2 × 105 cells/ml) was added to upper Transwell without FBS and 600 μl culture medium with 10% FBS was added in the lower Transwell at 37°C with 95% air and 5% CO2 for 48 h. After incubation, filter was fixed with methanol for 5 min and stained by crystal violet for 10 min. Cells were counted under microscope.
2.7. Western blotting analysis
Western blot was to detect all the proteins expression. Total proteins were obtained using RIPA lysis buffer, and BCA Protein Assay Kit (Pierce) was used for determining proteins concentration. Protein were subjected to SDS‐PAGE and then electrophoretically transferred to a PVDF membrane (Millipore). Primary antibodies (CREB1, p53, Bax, and GAPDH) were prepared in 5% BSA and diluted according to the product instruction after blocking with 5% nonfat dry milk. Membranes were incubated with a horseradish peroxidase‐conjugated secondary antibody and were detected by ECL Detection Systems (Thermo Scientific).
2.8. Dual‐luciferase assay
The LINC02323 3′‐UTR or TGF‐β 3′‐UTR was amplified from human genomic DNA and subcloned into the pGL3‐basic luciferase reporter plasmid. LINC02323 3′‐UTR or TGF‐β 3′‐UTR luciferase reporter plasmids was co‐transfected with miR‐1343‐3p mimics using lipofectamine 2000 reagent. Dual‐Luciferase Reporter Assay was performed according to the manufacturer's instructions after 48 h of transfection.
2.9. Statistical analysis
All data are expressed as the mean ± SD and were analyzed by the SPSS 19.0 statistical software. The p‐values were calculated using a one‐way analysis of variance (ANOVA) or Student's t test. Results were considered statistically significant with p < 0.05.
3. RESULTS
3.1. Serum levels of lncRNA LINC02323 in patients with ovarian cancer
Our results revealed that serum levels of lncRNA LINC02323 in patients with ovarian cancer was up‐regulated (Figure 1A). The risk of developing cancer increased with serum levels of lncRNA LINC02323 in patients with ovarian cancer (Figure 1B). OS and DFS of lncRNA LINC02323 high expressions were lower than those of lncRNA LINC02323 low expression (Figure 1C,D).
Figure 1.
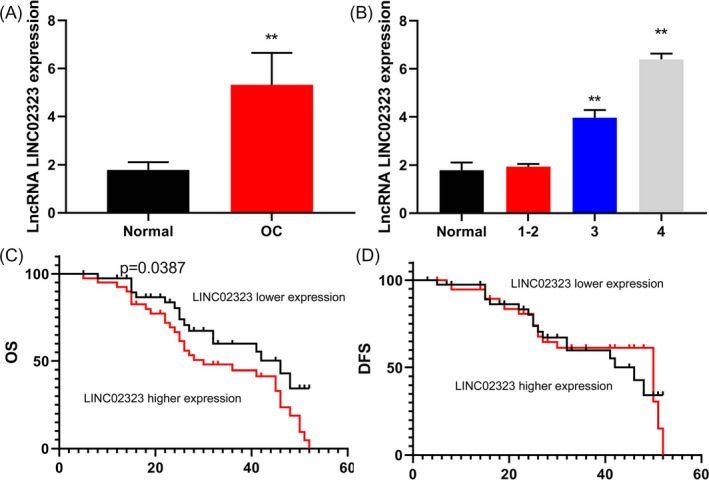
Serum levels of lncRNA LINC02323 in patients with ovarian cancer. Serum levels of lncRNA LINC02323 in patients with ovarian cancer (A) and the risk of developing cancer (B), OS, and DFS (C and D). All data were analyzed by the SPSS 19.0 statistical software. Normal, control normal group; OC, patients with ovarian cancer group. **p < 0.01 compared with healthy volunteers group
3.2. LncRNA LINC02323 regulated ovarian cancer cell proliferation and migration
To explore the underlying roles of LncRNA LINC02323 in ovarian cancer cell proliferation and migration, we used lncRNA LINC02323 plasmid and silncRNA LINC02323 plasmid to up‐regulate or down‐regulate the expression of lncRNA LINC02323 in vitro model (Figure 2A,B). Over‐expression of lncRNA LINC02323 promoted cell proliferation and migration rate, and reduced caspase‐3/caspase‐9 activity levels and LDH activity levels in vitro model (Figure 2C‐H). Down‐regulation of lncRNA LINC02323 reduced cell proliferation and migration rate, and increased caspase‐3/caspase‐9 activity levels and LDH activity levels in vitro model (Figure 2I‐N).
Figure 2.
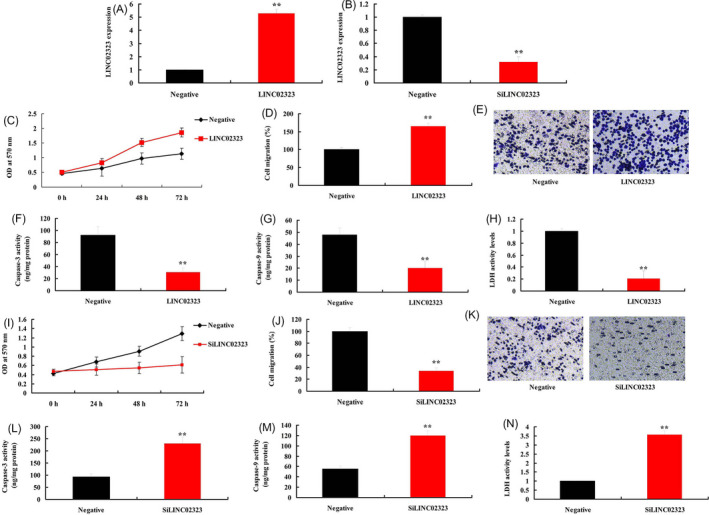
LncRNA LINC02323 regulated ovarian cancer cell proliferation and migration. LncRNA LINC02323 (A and B); cell proliferation (C), cell migration (D and E), caspase‐3/caspase‐9 activity levels (F and G), LDH activity levels (H) in vitro model by over‐expression of lncRNA LINC02323; cell proliferation (I), cell migration (J and K), caspase‐3/caspase‐9 activity levels (L and M), LDH activity levels (N) in vitro model by down‐regulation of lncRNA LINC02323. All data are expressed as the mean ± SD and were analyzed by the SPSS 19.0 statistical software. Negative, negative group; LINC02323, over‐expression of LINC02323 group; siLINC02323, down‐regulation of LINC02323 group. **p < 0.01 compared with negative group
3.3. LncRNA LINC02323 plays a crucial role in TGF‐β of ovarian cancer by targeting miR‐1343‐3p
Next, we explored the mechanism of lncRNA LINC02323 in ovarian cancer. We found that over‐expression of lncRNA LINC02323 reduced miR‐1343‐3p expression levels in vitro model, and negative correlations lncRNA LINC02323 and miR‐1343‐3p in patients with ovarian cancer (Figure 3A,B). Meanwhile, over‐expression of lncRNA LINC02323 reduced miR‐1343‐3p expression levels and down‐regulation of lncRNA LINC02323 promoted miR‐1343‐3p expression levels in vitro model (Figure 3C,D). LncRNA LINC02323 targeted miR‐1343‐3p and luciferase assay activity were reduced in over‐expression of LINC02323 group (Figure 3E,F).
Figure 3.
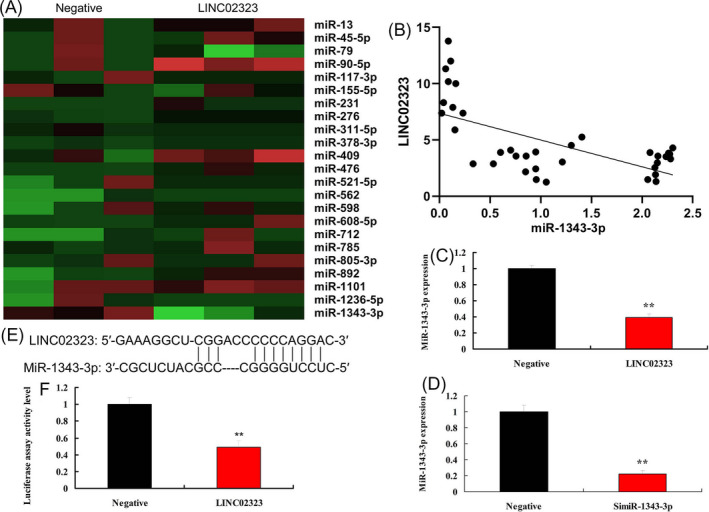
LncRNA LINC02323 regulated miR‐1343‐3p in vitro model. Heat map (A), negative correlations lncRNA LINC02323 and miR‐1343‐3p in patients with ovarian cancer (B), miR‐1343‐3p expression by over‐expression of LINC02323 (C) and down‐regulation of LINC02323 (D), lncRNA LINC02323 targeted miR‐1343‐3p (E), luciferase assay activity (F). All data are expressed as the mean ± SD and were analyzed by the SPSS 19.0 statistical software. Negative, negative group; LINC02323, over‐expression of LINC02323 group; siLINC02323, down‐regulation of LINC02323 group. **p < 0.01 compared with negative group
Next, we analyzed the mechanism of miR‐1343‐3p in the function of lncRNA LINC02323 on ovarian cancer cell growth. Heat map and volcano map showed that Over‐expression of miR‐1343‐3p reduced the expression of TGF‐β in vitro model (Figure 4A‐C). TGF‐β targeted miR‐1343‐3p and luciferase assay activity were reduced in over‐expression of miR‐1343‐3p group (Figure 4D,E). Over‐expression of miR‐1343‐3p suppressed TGF‐β1, vimentin, and N‐cadherin protein expressions, and induced E‐cadherin protein expression levels in vitro model (Figure 4F‐J). Down‐regulation of miR‐1343‐3p induced TGF‐β1, vimentin, and N‐cadherin protein expressions and suppressed E‐cadherin protein expression levels in vitro model (Figure 4J‐N).
Figure 4.
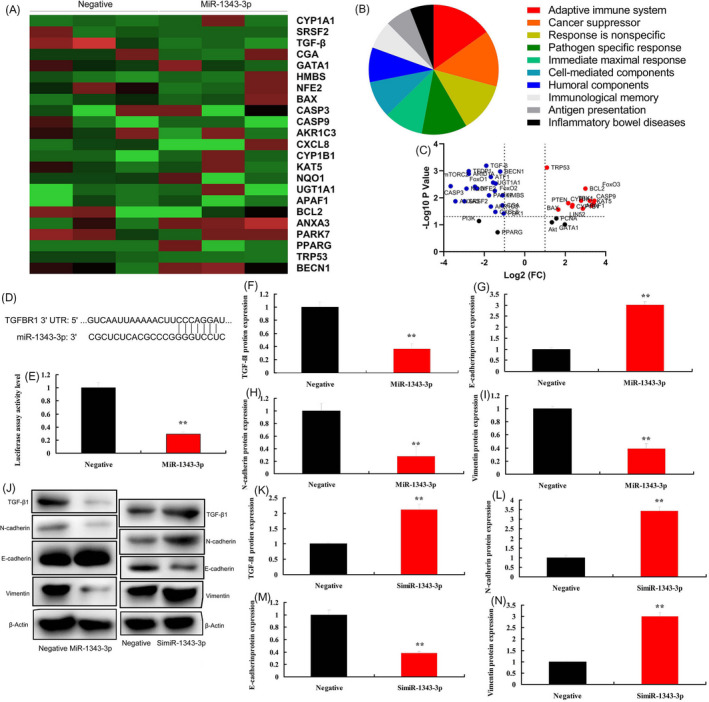
MiR‐1343‐3p regulated TGF‐β in vitro model. Heat map, results, and volcano figure (A, B and C), lncRNA LINC02323 targeted miR‐1343‐3p (D), luciferase assay activity (E), TGF‐β1, N‐cadherin, E‐cadherin, and vimentin protein expressions (F, G, H, I, and J) by over‐expression of miR‐1343‐3p; TGF‐β1, N‐cadherin, E‐cadherin, and vimentin protein expressions (J, K, L, M, and N) by down‐regulation of miR‐1343‐3p. All data are expressed as the mean ± SD and were analyzed by the SPSS 19.0 statistical software. Negative, negative group; MiR‐1343‐3p, over‐expression of miR‐1343‐3p group; simiR‐1343‐3p, down‐regulation of miR‐1343‐3p group. **p < 0.01 compared with negative group
Lastly, we confirmed the function of TGF‐β1 in the effects of lncRNA LINC02323 on ovarian cancer cell growth. Over‐expression of lncRNA LINC02323 induced TGF‐β1, vimentin and N‐cadherin protein expressions and suppressed E‐cadherin protein expression levels in vitro model (Figure 5A‐D,I). Down‐regulation of suppressed TGF‐β1, vimentin, and N‐cadherin protein expressions and induced E‐cadherin protein expression levels in vitro model (Figure 5E‐I).
Figure 5.
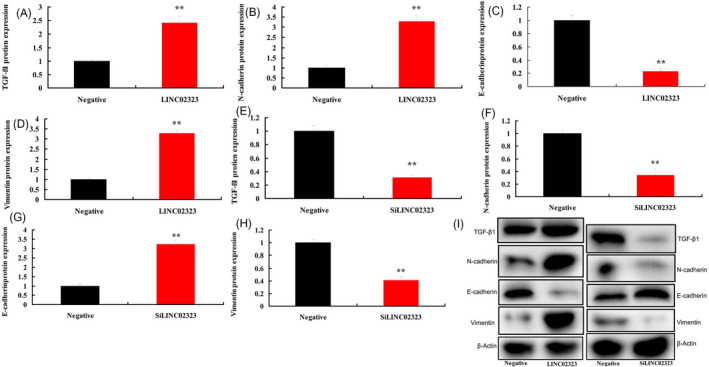
LncRNA LINC02323 plays a crucial role in TGF‐β of ovarian cancer by targeting miR‐1343‐3p. TGF‐β1, N‐cadherin, E‐cadherin, and vimentin protein expressions (A, B, C, D, and I) by over‐expression of LINC02323; TGF‐β1, N‐cadherin, E‐cadherin, and vimentin protein expressions (E, F, G, H, and I) by down‐regulation of LINC02323. All data are expressed as the mean ± SD and were analyzed by the SPSS 19.0 statistical software. Negative, negative group; LINC02323, over‐expression of LINC02323 group; siLINC02323, down‐regulation of LINC02323 group. **p < 0.01 compared with negative group
3.4. MiR‐1343‐3p regulated ovarian cancer cell proliferation and migration
To explore the underlying roles of miR‐1343‐3p in ovarian cancer cell proliferation and migration, we used miR‐1343‐3p plasmid and simiR‐1343‐3p plasmid to up‐regulated or down‐regulated the expression of miR‐1343‐3p in vitro model (Figure 6A,B). Over‐expression of lncRNA LINC02323 reduced cell proliferation and migration rate and increased caspase‐3/caspase‐9 activity levels and LDH activity levels in vitro model (Figure 6C‐H). Down‐regulation of lncRNA LINC02323 promoted cell proliferation and migration rate, and reduced caspase‐3/caspase‐9 activity levels and LDH activity levels in vitro model (Figure 6I‐N).
Figure 6.
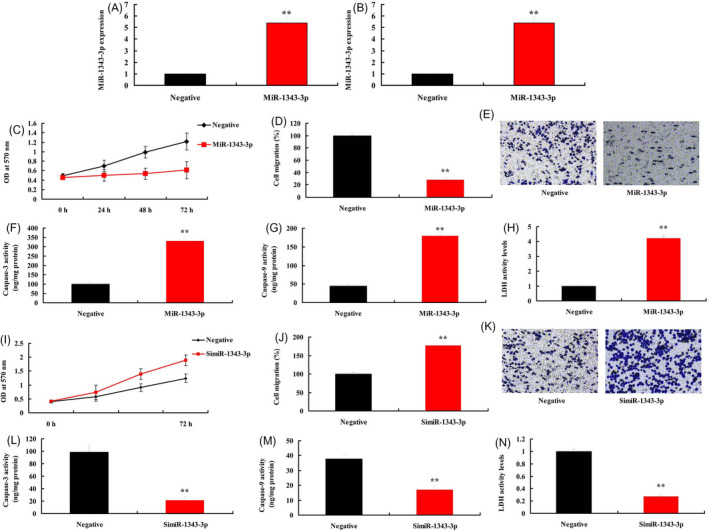
MiR‐1343‐3p regulated ovarian cancer cell proliferation and migration. MiR‐1343‐3p (A and B); cell proliferation (C), cell migration (D and E), caspase‐3/caspase‐9 activity levels (F and G), LDH activity levels (H) in vitro model by over‐expression of miR‐1343‐3p; cell proliferation (I), cell migration (J and K), caspase‐3/caspase‐9 activity levels (L and M), LDH activity levels (N) in vitro model by down‐regulation of miR‐1343‐3p. All data are expressed as the mean ± SD and were analyzed by the SPSS 19.0 statistical software. Negative, negative group; MiR‐1343‐3p, over‐expression of miR‐1343‐3p group; simiR‐1343‐3p, down‐regulation of miR‐1343‐3p group. **p < 0.01 compared with negative group
3.5. MiR‐1343‐3p reversed the effects of lncRNA LINC02323 in ovarian cancer via TGF‐β
To determine whether miR‐1343‐3p affected the pro‐cancer effects of lncRNA LINC02323 in cell proliferation and migration of ovarian cancer, miR‐1343‐3p and lncRNA LINC02323 plasmid or simiR‐1343‐3p and silncRNA LINC02323 plasmid were co‐transfected into vitro model, respectively. MiR‐1343‐3p plasmid increased the expression of miR‐1343‐3p in vitro model of lncRNA LINC02323 over‐expression, compared with LINC02323 over‐expression group (Figure 7A). Compared with LINC02323 over‐expression group, over‐expression of miR‐1343‐3p reduced cell proliferation and migration, suppressed TGF‐β1, vimentin and N‐cadherin protein expressions, induced E‐cadherin protein expression levels, and increased caspase‐3/caspase‐9 activity levels and LDH activity levels in vitro model (Figure 7B‐K).
Figure 7.
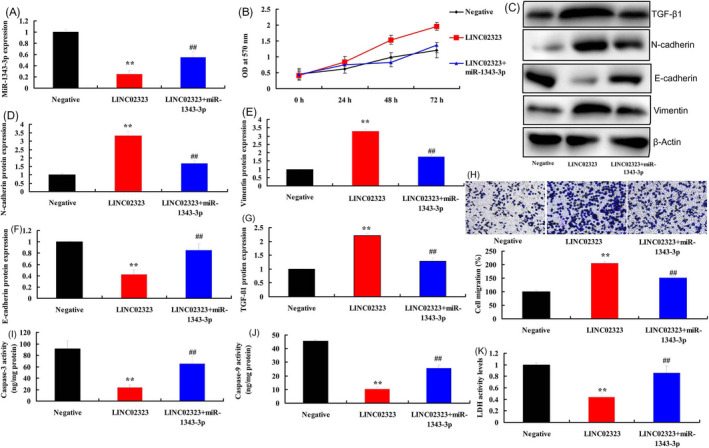
MiR‐1343‐3p reversed the effects of lncRNA LINC02323 in ovarian cancer via TGF‐β. MiR‐1343‐3p (A), cell proliferation (B), TGF‐β1, N‐cadherin, E‐cadherin, and vimentin protein expressions (C, D, E, F, and G), cell migration (H), caspase‐3/caspase‐9 activity levels (I and J), LDH activity levels (K). All data are expressed as the mean ± SD and were analyzed by the SPSS 19.0 statistical software. Negative, negative group; LINC02323, over‐expression of LINC02323 group; LINC02323 + miR‐1343‐3p, over‐expression of LINC02323 and miR‐1343‐3p group. **p < 0.01 compared with negative group, ## p < 0.01 compared with over‐expression of LINC02323 group
SiMiR‐1343‐3p plasmid decreased the expression of miR‐1343‐3p in vitro model of lncRNA LINC02323 down‐regulation, compared with LINC02323 down‐regulation group (Figure 8A). Compared with LINC02323 down‐regulation group, down‐regulation of miR‐1343‐3p promoted cell proliferation and migration, induced TGF‐β1, vimentin and N‐cadherin protein expressions, suppressed E‐cadherin protein expression levels, and decreased caspase‐3/caspase‐9 activity levels and LDH activity levels in vitro model (Figure 8B,K).
Figure 8.
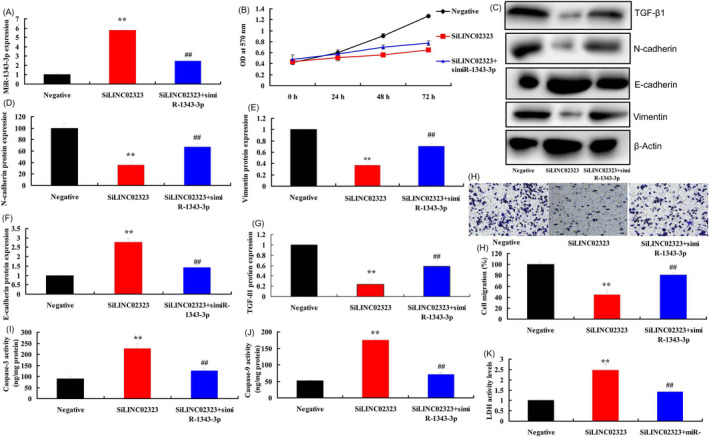
SimiR‐1343‐3p reversed the effects of silncRNA LINC02323 in ovarian cancer via TGF‐β. MiR‐1343‐3p (A), cell proliferation (B), TGF‐β1, N‐cadherin, E‐cadherin, and vimentin protein expressions (C, D, E, F, and G), cell migration (H), caspase‐3/caspase‐9 activity levels (I and J), LDH activity levels (K). All data are expressed as the mean ± SD and were analyzed by the SPSS 19.0 statistical software. Negative, negative group; siLINC02323, down‐regulation of LINC02323 group; siLINC02323, down‐regulation of LINC02323 and miR‐1343‐3p group. **p < 0.01 compared with negative group, ## p < 0.01 compared with down‐regulation of LINC02323 group
4. DISCUSSION
Ovarian cancer is the second most common female reproductive malignancy, with the highest mortality rate among all gynecological tumors. 13 Approximately 200,000 people are diagnosed with ovarian cancer each year globally, and 125,000 people die from ovarian cancer. 14 Due to the relatively insidious onset, a lack of effective screening, and early diagnosis methods, 70% to 80% of patients are diagnosed in advanced stage, with extremely poor therapeutic effect and prognosis, and the 5‐year survival rate less than 20%. 15 Herein, our results demonstrated that serum levels of lncRNA LINC02323 in patients with ovarian cancer were up‐regulated. OS and DFS of lncRNA LINC02323 high expressions were lower than those of lncRNA LINC02323 low expression, which is consistent with the LINC02323‐mediated metastasis of ovarian cancer. Zhang et al showed that LINC01819 signature had a regulatory role in the lung adenocarcinom cell mobility. 16
According to the previous description, different lncRNAs can interact with specific miRNAs, and lncRNAs and miRNAs synergistically regulate various aspects of gene expression, thereby maintaining the balance of internal environment. 17 , 18 , 19 miRNA sponge function has the unique characteristics as an ideal ceRNA. 20 Relevant research and construction not only provide important bases to study the functions of lncNAs and miRNAs, but also provide a powerful tool for the antitumor treatment, simultaneously shedding novel light on the treatment of other diseases. 21 , 22 In the present study, our data showed that LncRNA LINC02323 plays a crucial role in TGF‐β of ovarian cancer by targeting miR‐1343‐3p. Zhang et al 23 reported that INC02323 promotes cell metastasis via sponging miR‐1343‐3p in lung adenocarcinoma. These findings suggest that LncRNA LINC02323/ miR‐1343‐3p/ TGF‐β axis was involved in the molecular mechanism of ovarian cancer cell growth, and it might play a leading role in ovarian cancer metastasis development.
The disorder of TGF‐β signal transduction pathway can protect cells from the growth inhibitory effect mediated by TGF‐β, leading to the occurrence of various types of tumors. 24 Because TGF‐β signal transduction pathway can mediate the inhibitory effect of cell growth, especially the inhibitory effect on epithelial cell growth, thereby promoting the tumorigenesis and tumor progression. 25 These results indicated that miR‐1343‐3p reversed the effects of lncRNA LINC02323 in ovarian cancer via TGF‐β.
Our study provided, for the first time, serum levels of lncRNA LINC02323 in patients with ovarian cancer was up‐regulated, and over‐expression of lncRNA LINC02323 promoted cell proliferation and migration rate in vitro model via TGF‐β receptor 1 by miR‐1343‐3p, which indicated that it was a key molecule in the process of invasion and metastasis of ovarian cancer and might be used as a potential target in ovarian cancer.
CONFLICT OF INTEREST
There are no potential conflicts of interest to disclose.
AUTHOR CONTRIBUTIONS
Yingchun Li is responsible for the guarantor of integrity of the entire study, study concepts, and data analysis; Zheng Zhao is responsible for the study design, definition of intellectual content, and data acquisition. Dan Sun is responsible for the literature research and statistical analysis; Yanfei Li is responsible for the clinical studies and manuscript preparation. All authors read and approved the final manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Serum samples were obtained from patients with ovarian cancer patients or normal volunteer in Cangzhou Center Hospital. Written informed consent was obtained from all participants, and the research protocols were approved by the Ethics Committee of Cangzhou Center Hospital.
Li Y, Zhao Z, Sun D, Li Y. Novel long noncoding RNA LINC02323 promotes cell growth and migration of ovarian cancer via TGF‐β receptor 1 by miR‐1343‐3p. J Clin Lab Anal.2021;35:e23651 10.1002/jcla.23651
REFERENCES
- 1. Burger RA, Brady MF, Bookman MA, et al. Incorporation of bevacizumab in the primary treatment of ovarian cancer. N Engl J Med. 2011;365:2473‐2483. [DOI] [PubMed] [Google Scholar]
- 2. Jacobs IJ, Menon U, Ryan A, et al. Ovarian cancer screening and mortality in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): a randomised controlled trial. Lancet (London, England). 2016;387:945‐956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ledermann J, Harter P, Gourley C, et al. Olaparib maintenance therapy in platinum‐sensitive relapsed ovarian cancer. N Engl J Med. 2012;366:1382‐1392. [DOI] [PubMed] [Google Scholar]
- 4. Chang H, Zhang X, Li B, Meng X. MAGI2‐AS3 suppresses MYC signaling to inhibit cell proliferation and migration in ovarian cancer through targeting miR‐525‐5p/MXD1 axis. Cancer Med. 2020;9(17):6377‐6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hao T, Huang S, Han F. LINC‐PINT suppresses tumour cell proliferation, migration and invasion through targeting miR‐374a‐5p in ovarian cancer. Cell Biochem Funct. 2020, Jul 7. 10.1002/cbf.3565 [DOI] [PubMed] [Google Scholar]
- 6. Kong S, Xue H, Li Y, et al. The long noncoding RNA OTUD6B‐AS1 enhances cell proliferation and the invasion of hepatocellular carcinoma cells through modulating GSKIP/Wnt/β‐catenin signalling via the sequestration of miR‐664b‐3p. Exp Cell Res. 2020;395(1):e112180. [DOI] [PubMed] [Google Scholar]
- 7. Shen W, Xie X, Liu M, Wang L. Diagnostic value of lncRNA ROR in differentiating ovarian cancer patients. Clin Lab. 2020;66(7):e191035. [DOI] [PubMed] [Google Scholar]
- 8. Du W, Feng Z, Sun Q. LncRNA LINC00319 accelerates ovarian cancer progression through miR‐423‐5p/NACC1 pathway. Biochem Biophys Res Commun. 2018;507:198‐202. [DOI] [PubMed] [Google Scholar]
- 9. Li N, Zhan X, Zhan X. The lncRNA SNHG3 regulates energy metabolism of ovarian cancer by an analysis of mitochondrial proteomes. Gynecol Oncol. 2018;150:343‐354. [DOI] [PubMed] [Google Scholar]
- 10. Ahn JH, Lee HS, Lee JS, et al. nc886 is induced by TGF‐β and suppresses the microRNA pathway in ovarian cancer. Nat Commun. 2018;9:1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guo Y, Cui W, Pei Y, Xu D. Platelets promote invasion and induce epithelial to mesenchymal transition in ovarian cancer cells by TGF‐β signaling pathway. Gynecol Oncol. 2019;153:639‐650. [DOI] [PubMed] [Google Scholar]
- 12. Al Ameri W, Ahmed I, Al‐Dasim FM, et al. Cell type‐specific TGF‐β mediated EMT in 3D and 2D models and its reversal by TGF‐β receptor kinase inhibitor in ovarian cancer cell lines. Int J Mol Sci. 2019;20(14):3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pujade‐Lauraine E, Fujiwara K, Dychter SS, Devgan G, Monk BJ. Avelumab (anti‐PD‐L1) in platinum‐resistant/refractory ovarian cancer: JAVELIN Ovarian 200 Phase III study design. Future Oncol. 2018;14:2103‐2113. [DOI] [PubMed] [Google Scholar]
- 14. Pujade‐Lauraine E, Ledermann JA, Selle F, et al. SOLO2/ENGOT‐Ov21 investigators. Olaparib tablets as maintenance therapy in patients with platinum‐sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT‐Ov21): a double‐blind, randomised, placebo‐controlled, phase 3 trial. Lancet Oncol. 2017;18:1274‐1284. [DOI] [PubMed] [Google Scholar]
- 15. van Driel WJ, Koole SN, Sikorska K, et al. Hyperthermic intraperitoneal chemotherapy in ovarian cancer. N Engl J Med. 2018;378(3):230‐240. [DOI] [PubMed] [Google Scholar]
- 16. Zhang X, Han J, Du L, et al. Unique metastasis‐associated lncRNA signature optimizes prediction of tumor relapse in lung adenocarcinoma. Thorac Cancer. 2020;11:728‐737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin X, Yang F, Qi X, et al. LncRNA DANCR promotes tumor growth and angiogenesis in ovarian cancer through direct targeting of miR‐145. Mol Carcinog. 2019;58:2286‐2296. [DOI] [PubMed] [Google Scholar]
- 18. Zhao X, Tang DY, Zuo X, Zhang TD, Wang C. Identification of lncRNA‐miRNA‐mRNA regulatory network associated with epithelial ovarian cancer cisplatin‐resistant. J Cell Physiol. 2019;234:19886‐19894. [DOI] [PubMed] [Google Scholar]
- 19. Vallino L, Ferraresi A, Vidoni C, et al. Modulation of non‐coding RNAs by resveratrol in ovarian cancer cells: In silico analysis and literature review of the anti‐cancer pathways involved. J Tradit Complement Med. 2020;10:217‐229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yi K, Hou M, Yuan J, et al. LncRNA PVT1 epigenetically stabilizes and post‐transcriptionally regulates FOXM1 by acting as a microRNA sponge and thus promotes malignant behaviors of ovarian cancer cells. Am J Transl Res. 2020;12:2860‐2874. [PMC free article] [PubMed] [Google Scholar]
- 21. Zhao L, Li JF, Tong XJ. Long noncoding RNA PROX1‐AS1 promoted ovarian cancer cell proliferation and metastasis by suppressing KLF6. Eur Rev Med Pharmacol Sci. 2020;24:6561‐6568. [DOI] [PubMed] [Google Scholar]
- 22. Liang H, Yu T, Han Y, et al. LncRNA PTAR promotes EMT and invasion‐metastasis in serous ovarian cancer by competitively binding miR‐101‐3p to regulate ZEB1 expression. Mol cancer. 2018;17:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang X, Du L, Han J, et al. Novel long non‐coding RNA LINC02323 promotes epithelial‐mesenchymal transition and metastasis via sponging miR‐1343‐3p in lung adenocarcinoma. Thorac cancer. 2020;11(9):2506‐2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matsumoto T, Yokoi A, Hashimura M, Oguri Y, Akiya M, Saegusa M. TGF‐β‐mediated LEFTY/Akt/GSK‐3β/Snail axis modulates epithelial‐mesenchymal transition and cancer stem cell properties in ovarian clear cell carcinomas. Mol carcinog. 2018;57:957‐967. [DOI] [PubMed] [Google Scholar]
- 25. Yeung TL, Leung CS, Wong KK, et al. TGF‐β modulates ovarian cancer invasion by upregulating CAF‐derived versican in the tumor microenvironment. Cancer Res. 2013;73:5016‐5028. [DOI] [PMC free article] [PubMed] [Google Scholar]


