SUMMARY
Antibody memory protects humans from many diseases. Protective antibody memory responses require activation of transcriptional programs, cell proliferation, and production of antigen-specific antibodies, but how these aspects of the response are coordinated is poorly understood. We profile the molecular and cellular features of the antibody response to influenza vaccination by integrating single-cell transcriptomics, longitudinal antibody repertoire sequencing, and antibody binding measurements. Single-cell transcriptional profiling reveals a program of memory B cell activation characterized by CD11c and T-bet expression associated with clonal expansion and differentiation toward effector function. Vaccination elicits an antibody clone, which rapidly acquired broad high-affinity hemagglutinin binding during affinity maturation. Unexpectedly, many antibody clones elicited by vaccination do not bind vaccine, demonstrating non-specific activation of bystander antibodies by influenza vaccination. These results offer insight into how molecular recognition, transcriptional programs, and clonal proliferation are coordinated in the human B cell repertoire during memory recall.
Graphical Abstract
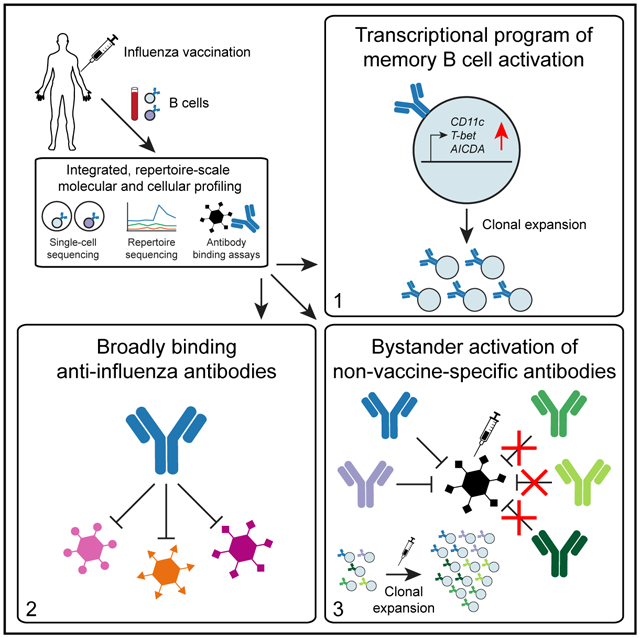
In Brief
Antibody memory requires coordination of molecular recognition, gene expression programs, and clonal dynamics. Horns et al. study the human antibody memory response using single-cell and repertoire sequencing, revealing a transcriptional program of memory B cell activation, broadly binding anti-influenza antibodies, and widespread bystander activation of non-vaccine-binding antibodies after influenza vaccination.
INTRODUCTION
Antibody memory is a hallmark of adaptive immunity and confers life-saving protection against many pathogens. During an initial encounter with a pathogen, clonal selection and affinity maturation focus the antibody repertoire onto variants that bind specifically to pathogen-derived antigens with high affinity, and these antibodies are preserved in memory B cells. In subsequent encounters, memory B cells are rapidly activated, leading to clonal expansion and differentiation to antibody-secreting cells. This robust immune response can prevent reinfection or reduce severity of disease.
Although a protective memory response requires the coordination of antigen recognition, gene expression, and clonal expansion, studies linking these facets of the response have been lacking. In particular, deep-sequencing-based measurements of the population dynamics and clonal structure of the B cell repertoire have shown that vaccination typically induces rapid expansion of a small set of B cell clones within 7 days (Horns et al., 2019; Jiang et al., 2013; Vollmers et al., 2013). However, the transcriptional programs of these expanded clones and the antigen specificity of their antibodies have not been characterized.
Analogously, antigen-resolved measurements, such as serum binding assays and antigen-specific cell sorting, have demonstrated that antigen-specific serum antibody (Belshe et al., 2004; Treanor et al., 2002), memory B cells (Crotty et al., 2004), and antibody-secreting cells (Wrammert et al., 2008) become more abundant after vaccination. However, these approaches have not been able to resolve clonal relationships among antigen-specific cells, the population dynamics of these clones, or their gene expression programs.
Finally, bulk transcriptome measurements have detected transient expression signatures associated with memory recall after vaccination in blood (Gaucher et al., 2008; Henn et al., 2013; Li et al., 2014), but it is not known how these transcriptional programs are related to clonal dynamics and antigen specificity within the B cell repertoire. Thus, an integrated portrait of how the memory response unfolds with cellular and molecular detail at the scale of the entire organism’s antibody repertoire remains lacking, despite its importance for protective immunity and vaccine design.
To address these questions, we developed an integrative approach that combines information from single-cell transcriptomics, longitudinal antibody repertoire sequencing, and antibody binding measurements and applied it to study the human antibody response to influenza vaccination. We tracked the population dynamics of B cell clones in a time course after vaccination and profiled transcriptomes of single B cells within those clones, revealing an activated memory B cell state associated with vaccine-elicited clonal expansion. We then assessed the relationship between clonal expansion and antigen specificity by expressing native human antibodies isolated from single B cells and characterizing their binding properties.
RESULTS
Integrating Single B Cell Phenotypes with Clonal Population Dynamics after Vaccination
We studied the antibody repertoire response of one healthy young adult (age 18) to seasonal influenza vaccination in 2012. A deep multimodal study of a single individual’s vaccine response enabled us to extensively investigate the relationships between global repertoire structure and molecular function using a diverse suite of experimental techniques. To measure B cell population dynamics during the vaccine response, we sequenced the peripheral blood antibody repertoire (Rep-seq) at the time of vaccination (D0) and 1, 4, 7, 9, and 11 days afterward (D1, D4, D7, D9, and D11), as well as 3 and 5 days before vaccination (D-3 and D-5) (Figures 1A and 1B), as we previously reported (Horns et al., 2019). We detected ~625,000 unique antibody heavy-chain sequences belonging to ~55,000 clones, each of which originated by expansion from a single naive B cell. Vaccination elicited rapid recall of 16 vaccine-responsive clones, which we defined as those having >50-fold expansion in unique sequences detected between D0 and D7. These clones bear the hallmarks of memory B cells, including extensive somatic mutation, class-switched isotypes, and population genetic signatures of positive selection (Horns et al., 2019).
Figure 1. Experimental Design and Workflow for Studying B Cell Response to Influenza Vaccination Using Integrated Single-Cell and Antibody Repertoire Sequencing.
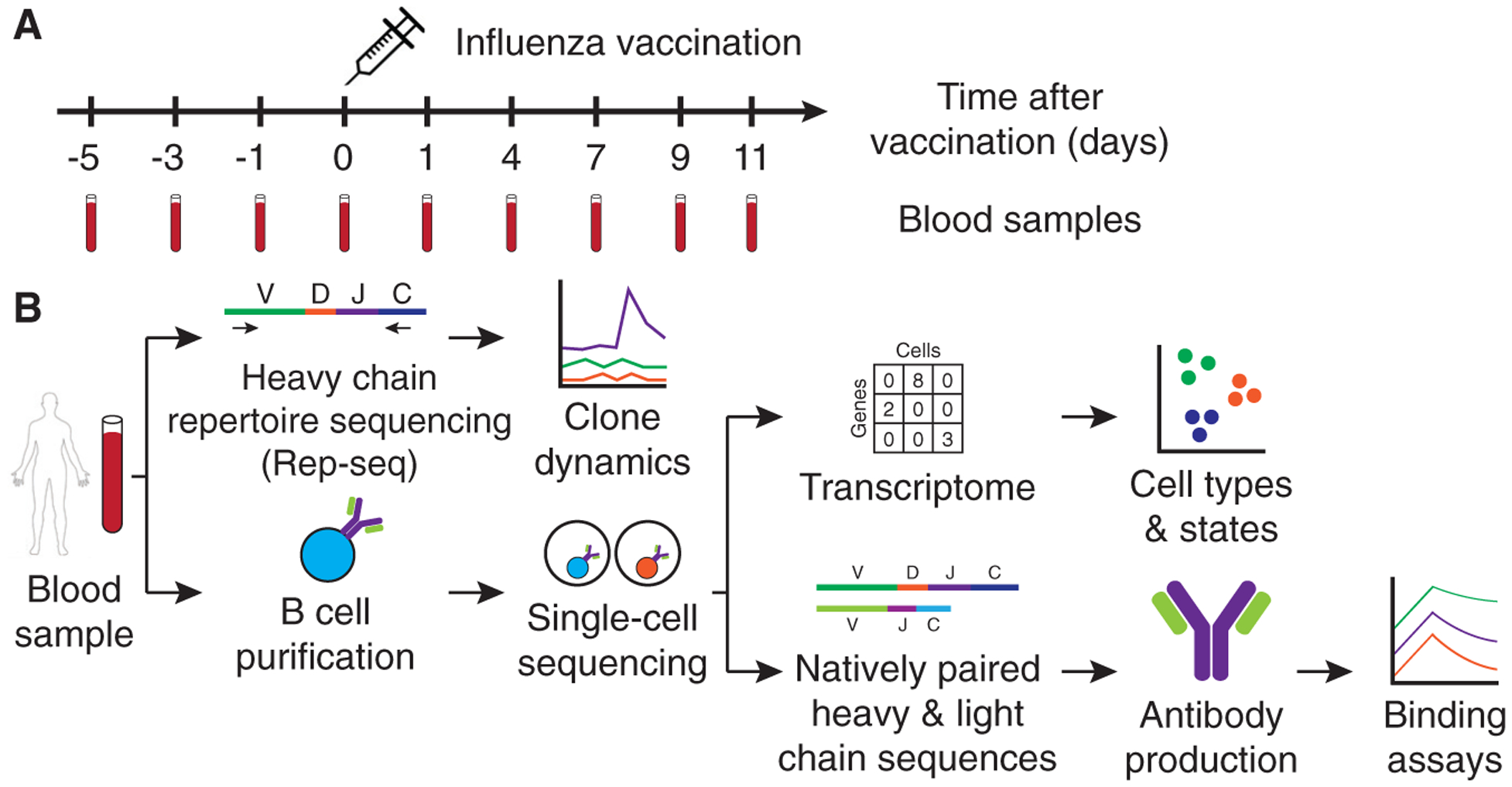
(A) Study design. 2011–2012 seasonal trivalent influenza vaccine was administered, and peripheral blood samples were collected for analysis at the indicated days before and after vaccination.
(B) Experiment workflow. Antibody heavy-chain repertoire sequencing (Rep-seq) was performed on samples from all time points. Single-cell transcriptional profiling and antibody heavy- and light-chain sequencing were performed on samples from d7 and d9 after vaccination.
We also sequenced antibody heavy- and light-chain transcripts in single B cells purified from peripheral blood samples of the same subject at D7 and D9, which correspond to the peak of the memory response (Figure 1B). After quality filtering and computational removal of doublets, we obtained 94,259 single B cells having exactly one productive heavy-chain transcript and one productive light-chain transcript (Figures S1A and S1B). We detected cells producing antibodies of every class and the majority of cells produced immunoglobulin (Ig)M antibodies, as expected from pan-B cell purification, which includes naive B cells (Figure S1C).
To connect single-cell phenotypes with clonal population dynamics, we mapped these single B cells to clones detected by Rep-seq using an established approach for identifying clones via single-linkage clustering (Figure S1D; Gupta et al., 2017; Horns et al., 2016). Clones were identified in the Rep-seq repertoire for 8% of cells, with the nearest heavy-chain complementarity determining region 3 (HCDR3) exhibiting high identity (97% ± 3% [mean ± SD]) for these matches (Figure S1E). Matches were strongly enriched for class-switched isotypes and depleted for IgD, as expected for memory B cells (Figure S1F). The majority of cells did not match a clone in Rep-seq data, because most cells are naive B cells, as confirmed later by transcriptome profiling. Additionally, the resampling probability of low-abundance memory B cell clones across replicate samples is low (Vollmers et al., 2013). Nevertheless, for clones detected in both measurements, quantification of clone size was highly consistent across the two methods (Figure 2A; Spearman’s rho = 0.57, p < 10−91).
Figure 2. Clone Dynamics during B Cell Response to Influenza Vaccination.
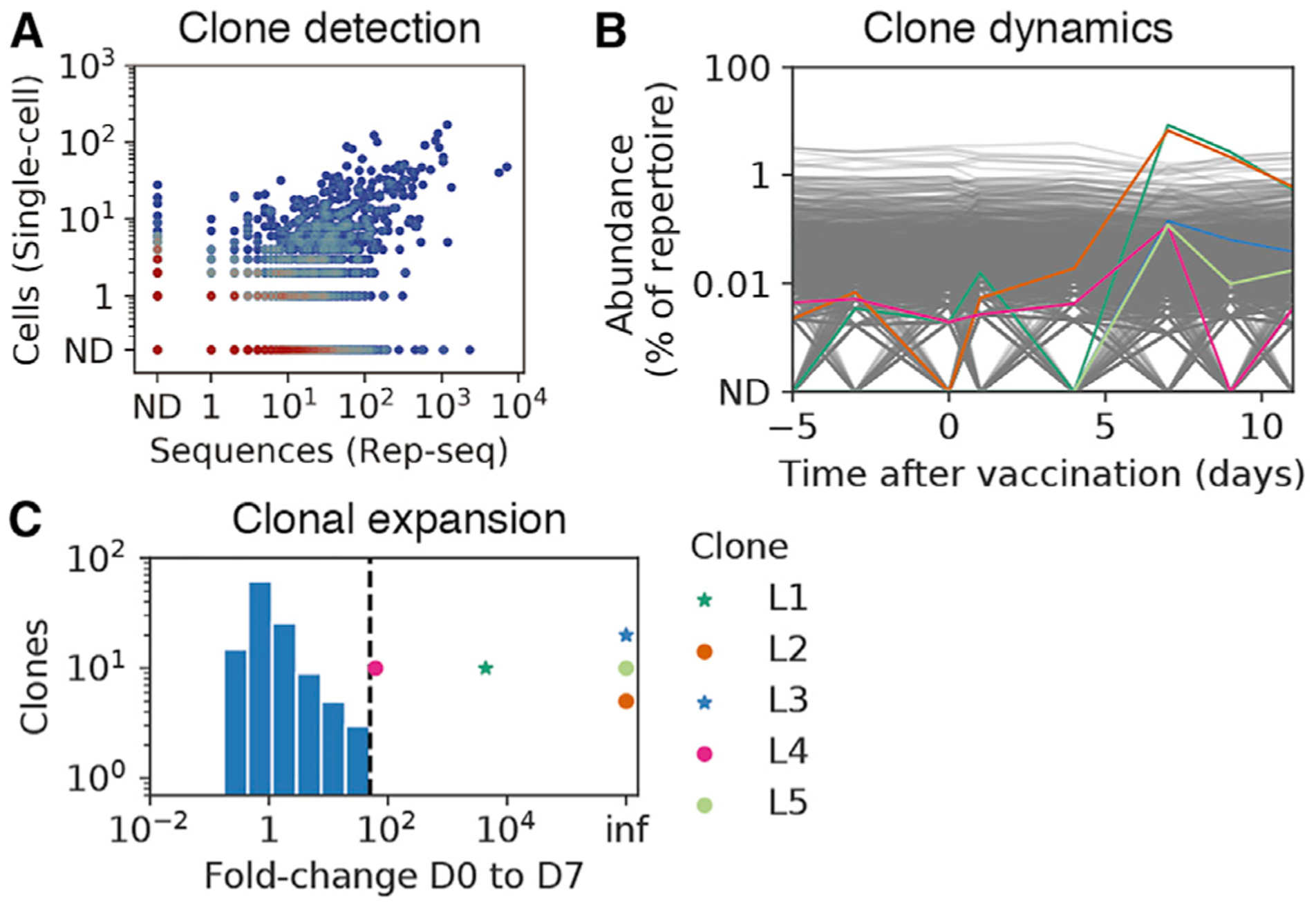
(A) Comparison of clonal abundance measurements across platforms, showing cells detected by single-cell sequencing and sequences detected by Rep-seq within each clone. Color indicates density of clones. ND, not detected.
(B) Population dynamics of B cell clones. Each line represents a clone. Colored lines indicate vaccine-responsive clones (>50-fold expansion from D0 to D7 and >0.1% of repertoire at D7, as in Horns et al., 2019), according to the key in (C). Gray lines indicate non-vaccine-responsive clones.
(C) Repertoire-wide distribution of the extent of clonal expansion after vaccination. Histogram shows the fold change from D0 to D7 of clones annotated as not vaccine responsive, which composed a substantial fraction of the repertoire at D7 (>0.1%) and had ≥1 cell detected by single-cell sequencing. Clones identified as vaccine responsive are indicated by markers (stars indicate vaccine binding, and circles indicate not vaccine binding). Dashed line shows the extent of expansion used as cutoff for identifying vaccine-responsive clones. Inf indicates undefined fold change arising because zero cells in the clone were detected at D0.
D0, day of vaccination; D7, 7 days after vaccination; ND, not detected. See also Figure S1.
Based on the Rep-seq measurement of clonal population dynamics, we identified five vaccine-responsive clones that both expanded dramatically after vaccination (>50-fold change from D0 to D7) (Figures 2B and 2C) and contained sequenced single cells. Although we detected 16 vaccine-responsive clones by Rep-seq, only 5 of these clones were also detected by single-cell sequencing, likely due to limited sampling of the repertoire. Nevertheless, this included the two globally most abundant clones at the peak of recall at D7, and each of these five vaccine-responsive clones comprised >0.1% of the repertoire at D7 (range = 0.1%–8%) (Figure 2B). Antibodies in these vaccine-responsive clones were mostly IgG (Figure S1G; 94% ± 5% IgG [mean ± SD]) and had extensive somatic hypermutation (mutation density in V gene; 3.8% ± 1.4% [mean ± SD]). Assembly of full-length heavy-chain sequences from single cells revealed the presence of diverse IgG subclasses in these clones (48% IgG1, 22% IgG2, and 9% IgG3 among cells belonging to vaccine-responsive clones). These results establish that the combination of longitudinal Rep-seq and single-cell sequencing captures a rich portrait of B cell population dynamics at the scale of the entire circulating antibody repertoire and links single-cell phenotypes, such as paired heavy-light chain antibody sequences, with clonal population dynamics.
Because single-cell sequencing preserves the native pairing between heavy- and light-chain sequences, we were able to assess the fidelity of the widely used strategy of clone identification based on the heavy-chain sequence alone by using the light chain as an independent marker of clonal identity. Light-chain genes were highly concordant within the vast majority of clones, as evidenced by the majority light-chain gene representing a very high proportion of cells within each clone (Figure S1H; median = 100%; 90% ± 18% [mean ± SD] for light-chain constant region genes; similar results were found for light-chain V and J genes). We observed that a minority of clones (16%) had substantial impurity based on the presence of cells containing a plurality of different light-chain genes. We determined that these impure clones were strongly enriched for short HCDR3 sequences (Figure S1I; p = 3.8 × 10−91, Mann-Whitney U test; median HCDR3 length, 14 aa (amino acids) in impure clones and 16 aa in pure clones) and usage of the IGHJ4 gene, which contributes a longer templated insert to the HCDR3 and, thus, tends to reduce sequence diversity (Figure S1I; p = 5.1 × 10−225, Fisher’s exact test; 64% IGHJ4 usage in impure clones and 28% in pure clones). We conclude that the fidelity of clone identification based on clustering of heavy-chain sequences is high for most clones. Clone assignment errors predominantly arise from low diversity compartments of the repertoire, and assignment can be improved by using light-chain sequences when pairing information is available. By examining the natively paired heavy- and light-chain genes from individual cells, we also found that the extent of somatic mutation in the heavy and light chains is highly correlated (Figure S1J; Pearson’s r = 0.68, p < 10−307, two-tailed).
Transcriptional Program of Vaccine-Induced Memory B Cell Activation
We performed single-cell transcriptome profiling on 35,631 cells, comprising a subset of the cells for which we sequenced antibody transcripts. We detected a median of 2,015 unique molecular identifiers (UMIs) and 766 genes per cell (Figure S2A), as typical for microfluidic droplet-based single-cell sequencing (The Tabula Muris Consortium, 2018). Similar transcriptional profiles were obtained across 4 technical replicates (Figure S2B) and these data were pooled for analysis. Using t-distributed stochastic neighbor embedding (tSNE) visualization and DBSCAN clustering, we identified distinct immune cell types, which we manually annotated based on established type-specific genes (Figure 3A; STAR Methods). Three clusters corresponded to CD4+ and CD8+ T cells and macrophages, which displayed specific expression of markers such as CD3E for T cells and LYS for macrophages (Figure S2G) and lacked antibody expression (Figure 3B). These cell types were present at low abundance due to the imperfect purity of B cell isolation and were not analyzed further. B cells formed two distinct clusters, which we annotated as memory B cells and naive B cells based on established markers and antibody features (Figure 3A). Memory B cells expressed CD27 (Figure S2H) and made predominantly class-switched antibodies (Figure 3B; Figure S2D) that were somatically mutated (Figures S2E and S2F). Naive B cells expressed TCL1A (Figure S2H) and made exclusively IgM and IgD antibodies (Figure 3B; Figure S2D) that were not somatically mutated (Figures S2E and S2F). In total, we analyzed 16,653 memory and 18,953 naive B cells.
Figure 3. Characterization of Gene Expression in Single B Cells Isolated from Peripheral Blood after Influenza Vaccination.
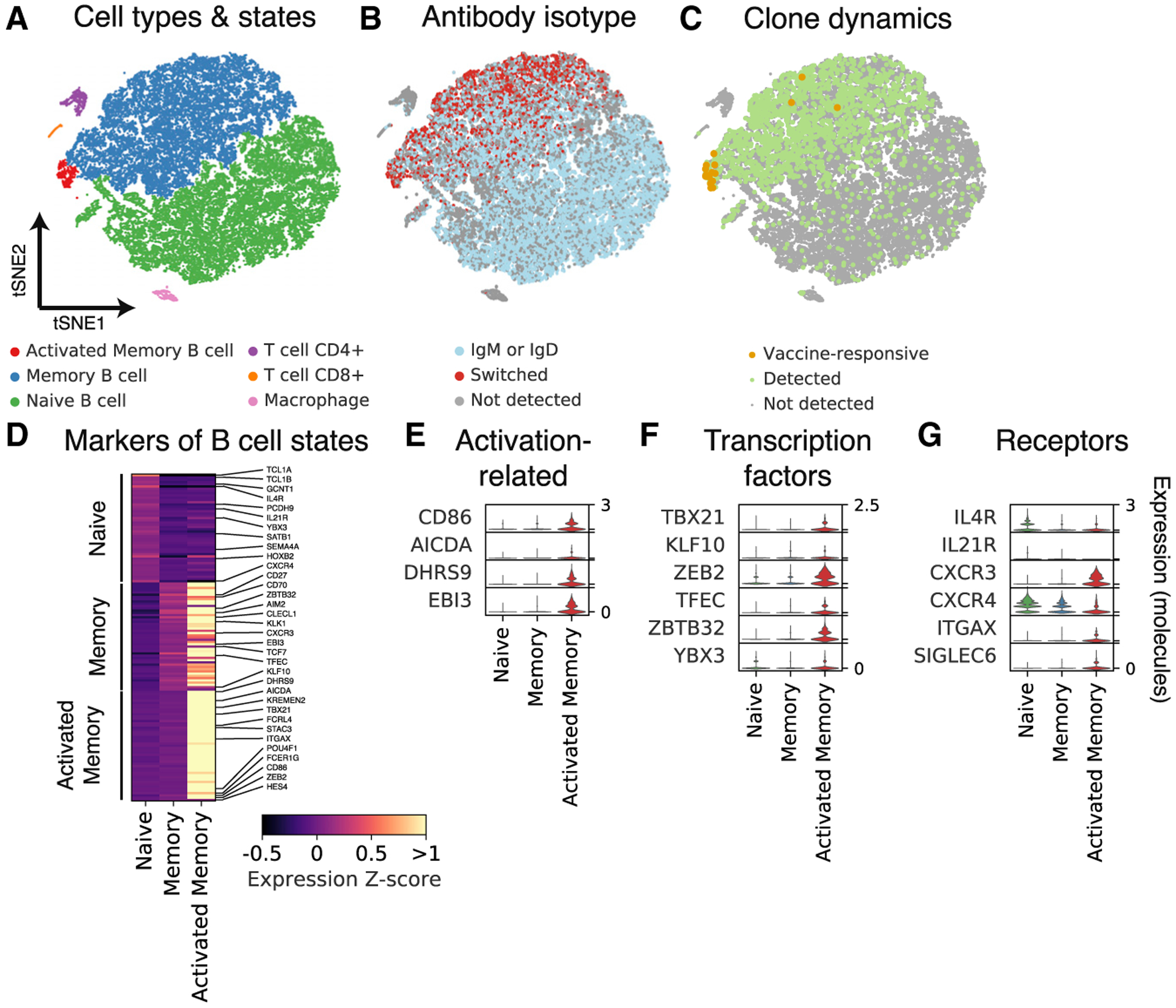
(A–C) Principal-component analysis and t-distributed stochastic neighbor embedding (tSNE) separates cells into distinct clusters. Each dot is a cell, colored by type or state revealed by gene expression profile (A), antibody isotype as revealed by single-cell antibody heavy-chain sequencing (B), or clonal population dynamics as revealed by Rep-seq (C).
(D) Differential expression analysis identifies markers of distinct B cell states. Genes of immunological interest are labeled.
(E–G) Gene expression distributions in distinct B cell states of established immune activation-related genes (E), transcription factors (F), and signaling receptors (G).
To address how clonal population dynamics are related to transcriptome state, we mapped the single B cell transcriptomes to the clones identified using Rep-seq based on heavy-chain sequence, as described earlier. Matches to clones were obtained almost exclusively for memory B cells as expected (Figure 3C). Remarkably, we found that cells belonging to vaccine-responsive clones had a distinct transcriptional profile characteristic of a small neighborhood of transcriptional state space within the memory B cell cluster (Figure 3C; Figure S2C). Cells in this neighborhood expressed established genes related to B cell activation, including the activation marker CD86 and the somatic hypermutation gene AICDA, also known as AID (Figures 3D and 3E; Figure S2H). Thus, on the basis of these markers, we annotated cells in this neighborhood as activated memory B cells, comprising 421 cells in total (1% of all B cells; 2.5% of memory B cells).
To define the transcriptional programs of B cell states, we identified genes exhibiting differential expression across naive, memory, and activated memory B cells. We found 755 differentially expressed genes between naive and memory B cells (false discovery rate [FDR] = 0.1%, Mann-Whitney U test with Benjamini-Hochberg correction), including established markers such as CD27 and IGHD (Figure 3D). About half of these genes were upregulated in naive B cells, while the other half were upregulated in memory B cells (Figure S2J). By contrast, we found 172 differentially expressed genes between memory and activated memory B cells, all of which were upregulated in activated memory B cells (Figure S2J). Dominant upregulation of genes in the activated memory state was consistently observed across a range of significance thresholds defining differential expression (Figure S2J). We also detected more genes (median = 1,786) and more UMIs (median = 5,517) in activated memory B cells than in memory B cells (Figure S2I; median gene count in memory B cells = 849; UMI count = 2,406), possibly reflecting greater mRNA content due to elevated transcription. Together, these results suggest that the transcriptional program of memory B cell activation predominantly involves the activation rather than deactivation of gene expression.
To characterize the activated memory B cell state, we sought to identify transcription factors (TFs), which may be central regulators of the program of activation. We identified 6 TFs specifically expressed in activated memory B cells (Figure 3F). These TFs include T-bet, also known as TBX21 (Figure S2H), which is required for IgG2a class switching (Wang et al., 2012) and clearing chronic viral infections (Barnett et al., 2016), and ZBTB32, which modulates the duration of memory B cell recall responses in mice (Jash et al., 2016).
Several cytokine receptors are downregulated in activated memory B cells (Figure 3G). IL4R and IL21R are highly expressed in naive B cells but downregulated in memory and activated memory B cells (Figure 3G; Figure S2H), suggesting that naive B cells are more responsive than memory or activated memory B cells to interleukin (IL)-4 and IL-21, which regulate class switching to IgG4 or IgE (Stavnezer, 1996) and to IgG1 or IgG3 (Pène et al., 2004), respectively. The chemokine receptor CXCR4, which controls entry to anatomical locations of B cell maturation such as lymph nodes and Peyer’s patches (Okada et al., 2002), is also progressively downregulated from naive to memory and activated memory B cells (Figure 3G).
Other genes related to humoral activation are upregulated in activated memory B cells. The chemokine receptor CXCR3, which is required for cell migration to sites of inflammation (Lacotte et al., 2009), is specifically expressed in activated memory B cells (Figure 3G; Figure S2H). Interestingly, CD11c, also known as ITGAX, is specifically expressed in activated memory B cells (Figure 3G), suggesting that this state overlaps with the recently described age/autoimmune-associated B cells (ABCs) (Hao et al., 2011; Rubtsov et al., 2011). Finally, EBI3, which is known to be expressed in germinal-center B cells (Larousserie et al., 2006), is found exclusively in activated memory B cells (Figure 3E). Complete lists of differentially expressed genes across naive, memory, and activated memory B cells are shown in Table S1 and Table S2. Together, these results define a transcriptional program of memory B cell activation associated with vaccine-induced clonal expansion, which bears hallmarks of an effector B cell response.
Many Vaccine-Responsive Antibodies Do Not Bind Vaccine
To study how clone dynamics and antigen specificity are related, we expressed and functionally characterized 21 antibodies obtained from single B cells within 5 vaccine-responsive clones (Figure S3A). We first measured binding of these antibodies to the vaccine (trivalent influenza vaccine from the 2011–2012 flu season) by ELISA. Surprisingly, only 57% of the vaccine-responsive antibodies (12 of 21) and 40% of the vaccine-responsive antibody clones (2 of 5) exhibited binding to vaccine (Figure 4; Figure S3B). For the non-vaccine-binding antibodies, we further screened for binding by ELISA against a panel of purified influenza proteins, including hemagglutinins, neuraminidases, nucleoprotein, matrix protein, and non-structural proteins, but found no binding (Figure S3C). Notably, despite not binding vaccine or influenza proteins, these three clones expanded dramatically after vaccination (>62-fold) and were highly abundant at D7, including one clone that was the second most globally abundant clone, representing 6.7% of the repertoire at D7 (Figure S3A). These results indicate that many vaccine-responsive antibodies do not bind vaccine or purified components of the vaccine. This suggests that vaccination induced activation of some antibody clones in an antigen-independent manner. We found no binding of these non-vaccine-binding antibodies to a panel of common viral and bacterial antigens, such as herpes simplex, measles, and varicella zoster virus (Figure S3C), and we were unable to determine the specificities of these antibodies. We also found no apparent relationship between vaccine-binding specificity and transcriptional profile: cells from vaccine-binding and non-vaccine-binding clones alike exhibited the activated memory B cell transcriptional program (Figure S2C).
Figure 4. Binding of Influenza Vaccine-Responsive Antibodies to Vaccine.

Binding of 21 monoclonal antibodies from 5 clones to the trivalent inactivated influenza vaccine from the 2011–2012 season was measured using enzyme-linked immunosorbent assay (ELISA), revealing that many vaccine-responsive antibodies do not bind vaccine. Ab, antibody; OD, optical density; hIgG1, human IgG1. See also Figure S3.
A Broadly Binding High-Affinity Anti-influenza Antibody Clone Elicited by Vaccination
To determine the specificity of the vaccine-binding antibodies, we screened them for binding by ELISA against purified influenza proteins, including the major antigenic determinants of influenza virus, hemagglutinin (HA) and neuraminidase (NA). One vaccine-responsive clone, which we refer to as L3, displayed strong binding to diverse HA proteins, including the influenza A variants contained in the vaccine, H1 A/California/7/2009 and H3A/Perth/16/2009, as well as H5 and H9 variants (Figure 5A). These antibodies had similar binding strength and breadth as established broadly neutralizing antibodies MEDI8852 (Kallewaard et al., 2016) and CR9114 (Dreyfus et al., 2012) (Figure 5A). L3 antibodies use the IGHV4-34*01 and IGHJ3*01 genes, have a 19 amino acid HCDR3, and are heavily mutated (28 ± 5 [mean ± SD] mutations from inferred germline heavy chain) (Figure S4A).
Figure 5. Reconstructing Evolution of a Broadly Binding High-Affinity Anti-influenza Antibody Clone.
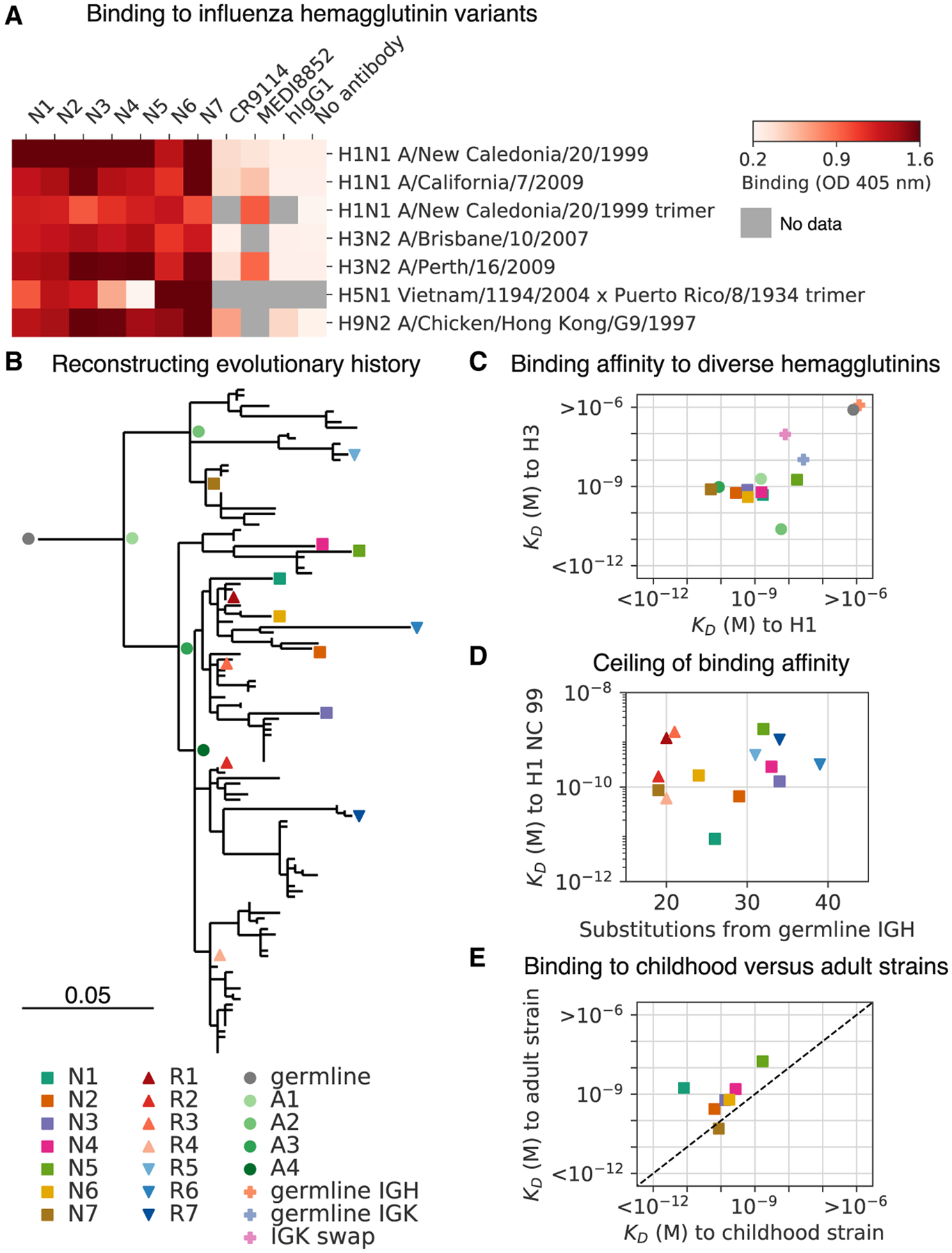
(A) Binding of antibodies from the L3 clone to a panel of influenza hemagglutinin (HA) variants was measured using ELISA. OD, optical density; hIgG1, human IgG1.
(B) Evolutionary history of L3 depicted as a maximum-likelihood phylogeny based on heavy-chain sequence. Markers indicate antibodies detected by single-cell sequencing (N1–N7) or repertoire sequencing (R1–R7), or reconstructed ancestral sequences (germline and A1–A4).
(C) Dissociation constants (KDs) of binding between L3 antibody variants and H1 (A/California/7/2009) and H3 (A/Perth/16/2009) hemagglutinin variants, as determined by biolayer interferometry. L3 antibodies include extant sequences (N1–N7), reconstructed ancestral sequences (germline and A1–A4), and engineered variants having the L3N6 sequences, but with heavy chain reverted to the inferred germline sequence (germline immunoglobulin heavy chain [IGH]), light chain reverted to the inferred germline sequence (germline immunoglobulin kappa chain [IGK]), or a light chain sequence substituted from a different clone (IGK swap). Jitter was added to germline and germline IGH to improve visualization of the data points.
(D) Dissociation constants of binding between L3 antibodies compared with extent of somatic hypermutation.
(E) Dissociation constants of binding between L3 antibodies and H1 variants from childhood (A/New Caledonia/20/1999) and adulthood (A/California/7/2009). Dashed line indicates equal KD for binding to both variants. Uncertainty of fitted parameters was smaller than the size of the markers used for plotting; therefore, error bars are not shown.
We measured the binding affinity of L3 antibodies to diverse H1 and H3 variants using biolayer interferometry. Most L3 antibodies bound with sub-nanomolar affinity to both H1 and H3, which are highly divergent HA variants drawn from the two major groups of influenza A virus and share only 44% amino acid identity (Figure 5C; Figure S4C; dissociation constants (KD) from 18 nM to 50 pM). Thus, L3 broadly binds diverse HA variants with high affinity. A second vaccine-binding clone, which we call L1, displayed strong but narrow specificity binding to HA B (Figure S3C). Given that the isolation of broadly neutralizing antibodies against influenza has been a major goal for therapeutics and vaccine design, we chose to focus on L3 and did not analyze L1 further.
Evolution of a Broadly Binding Anti-influenza Antibody Clone
To shed light on the evolutionary trajectories leading to broad high-affinity anti-influenza binding, we reconstructed the clonal evolution of L3 (Figure 5B). Using maximum-likelihood phylogenetic models, we reconstructed the ancestral sequences of the unmutated germline precursor and four intermediate ancestors (Figures S4A and S4B) and then expressed these antibodies and measured their binding affinities to diverse HAs. While the germline precursor bound weakly to H1 and H3 (KD > 1 μM) (Figure S4E), the first intermediate ancestor A1 bound to both H1 and H3 with nanomolar affinity (KD = 1.5 nM and 2 nM, respectively) (Figure 5C), despite having acquired only 11 amino acid substitutions (6 in the heavy chain and 5 in the light chain) (Figures S4A and S4B).
To dissect the contributions of heavy- and light-chain mutations to binding affinity, we engineered variants of the high-affinity L3N6 antibody in which the heavy- and light-chain sequences were separately reverted to the respective germline precursor sequence (Figures S4A and S4B). We found that germline reversion of the heavy chain greatly reduced binding affinity to both H1 and H3 (KD > 1 μM) (Figure 5C; Figure S4E). In contrast, germline reversion of the light chain minimally affected binding to H1 and H3 (KD = 27 nM and 10 nM, respectively) (Figure 5C; Figure S4E). To further test the contribution of light-chain mutations, we created a variant of L3N6 in which the light chain was swapped for a different immunoglobulin kappa (IGK) sequence originating from a distinct clone having a different light chain complementarity determining region 3 (LCDR3) (Figure S4B). This alteration of the light chain also minimally affected binding to H1 and H3 (KD = 8 nM and 94 nM, respectively) (Figures 5C and S4E). These findings show that heavy-chain mutations were predominantly responsible for affinity maturation, indicating that broad nanomolar-affinity binding was achieved via ≰6 amino acid substitutions in the heavy chain.
L3 antibodies, therefore, rapidly evolved broad high-affinity binding to diverse HA variants through a small number of somatic mutations. Affinity improvements were predominantly driven by decreasing the dissociation rate, which varied ~10,000-fold across the clone, rather than increasing the association rate, which varied only ~10-fold (Figure S4D). We found evidence for an affinity ceiling: acquisition of mutations beyond the intermediate ancestor A1 did not substantially affect affinity, and there was no trend toward enhanced affinity with additional mutations (across the range of 18–38 mutations from the inferred germline heavy-chain sequence) (Figure 5D; Spearman’s rho = 0.25, p = 0.37). Instead, L3 antibody affinity evidently drifted neutrally after rapid acquisition of high-affinity binding.
To determine how L3 antibodies bind HA, we performed cross-competition binding experiments using biolayer interferometry. We compared L3N1 and L3N6 with a panel of broadly binding antibodies consisting of stem-binding antibodies CR9114 (Dreyfus et al., 2012) and MEDI8852 (Kallewaard et al., 2016), receptor-binding site antibodies CH65 (Whittle et al., 2011) and H2897 (Liu et al., 2017), and lateral patch antibody 6649 (Raymond et al., 2018). We found that L3N1 and L3N6 did not compete with any of these antibodies (Figure S5). In contrast, CR9114 and MEDI8852 are known to have overlapping epitopes based on crystal structures (Dreyfus et al., 2012; Kallewaard et al., 2016), and we confirmed that they compete with each other, serving as a positive control for our measurement (Figure S5E). This result indicates that the epitopes recognized by L3 antibodies do not overlap with any antibodies in this panel, suggesting that L3 achieves broad specificity by a distinct structural mechanism. Furthermore, the L3 epitope may be conserved across HA variants belonging to groups 1 and 2.
It has been proposed that the antibody memory response is biased toward antigens seen early in an individual’s life, and this priming influences subsequent responses (Schmidt et al., 2015). To test this hypothesis using L3, we compared binding affinity to H1 variants that circulated during the subject’s childhood and adulthood. We found that extant antibodies of the L3 clone nearly all bound with higher affinity to the childhood strain (H1 New Caledonia/20/1999) than the adult strain (H1 California/07/2009) (Figure 5E; fold change in KD between childhood and adult strains = 35 ± 74 [mean ± SD]). This indicates that the affinity of a broad binding anti-HA antibody clone is biased toward antigenic variants associated with childhood exposure, supporting the idea that affinity maturation most efficiently focuses the antibody repertoire on antigens encountered in early life, leaving a lasting imprint on subsequent responses.
DISCUSSION
Mobilization of an effective antibody memory response requires coordination across widely varying length and time scales, from antibody-antigen recognition and transcriptional activation in single cells to clonal population dynamics that globally remodel an organism’s antibody repertoire. This multi-scale nature of the immune system creates challenges for understanding its function. To address these challenges, we have developed an experimental approach that integrates single B cell sequencing, longitudinal antibody repertoire sequencing, and biophysical measurements of antibody function. Our results show that this strategy yields a unified portrait of the molecular and cellular features of the memory B cell response to vaccination, giving insights into mechanisms of immune memory. Building upon these proof-of-principle results, future studies with more subjects should reveal how the molecular and cellular underpinnings of immunity and their coordination vary across human populations.
Much recent interest has focused on a functionally specialized B cell subset marked by CD11c and T-bet expression named age- or autoimmune-associated B cells (ABCs). B cells with these features are associated with viral infections, autoimmunity, and aging in mouse and human (Hao et al., 2011; Moir et al., 2008; Rubtsov et al., 2011; Rubtsova et al., 2017, 2013), but, to our knowledge, the phenotype has not been described as a transcriptional state at single-cell resolution. Using single-cell transcriptomics and longitudinal clone tracking, we have defined an activated memory B cell state, which displays hallmarks of an effector B cell response and shares many features with previously described ABCs, including high expression of CD11c (Hao et al., 2011; Rubtsov et al., 2011), T-bet (Rubtsova et al., 2013), FCRL4, and CXCR3 (Moir et al., 2008). Several genes that define this activated memory B cell state are directly involved in germinal center migration (EBI3) (Larousserie et al., 2006), somatic hypermutation (AICDA) (Muramatsu et al., 2000), and class switching (AICDA and T-bet) (Muramatsu et al., 2000; Wang et al., 2012), suggesting that these cells are poised for secondary affinity maturation. Our results indicate that these CD11c+ T-bet+ B cells are associated with vaccine-elicited clonal expansion in a healthy young adult human. These findings support the view that CD11c+ T-bet+ B cells are essential to health, but aberrant regulation of them can lead to autoimmunity. Defining the transcriptional program of these cells opens avenues to understanding their origin, function, and regulation, which might, in turn, reveal therapeutic targets in both pathogen immunity and autoimmunity.
Unexpectedly, several antibody clones elicited by vaccination did not bind vaccine. Formally, we cannot exclude that the lack of binding between recombinant vaccine-responsive antibodies and the vaccine in our in vitro measurements is due to conformational changes occurring under physiological conditions. Notwithstanding this alternative explanation, our results suggest that bystander activation of memory B cells bearing non-vaccine specificities is common after influenza vaccination. Polyclonal activation of memory B cells bearing non-vaccine specificities after vaccination has previously been described at the level of serum antibody (Bernasconi et al., 2002) and antibody-secreting (Wrammert et al., 2008) cells. Similarly, infection with both measles and varicella induces non-specific B cell activation (Arneborn et al., 1983). Our results show that many, perhaps even the majority of, memory B cells elicited by influenza vaccination produce antibodies that do not bind the vaccine, revealing an unanticipated extent of this phenomenon. This extent comports with some previous studies based on single-cell cloning of antibody-secreting cells (Wrammert et al., 2008) but may have been underestimated in other studies that tested binding against limited panels of antigens (Bernasconi et al., 2002; Lee et al., 2011). Non-specific polyclonal activation has been proposed as a mechanism for maintenance of long-term immune memory, enabling memory cell proliferation in the absence of antigen encounter (Moticka and Streilein, 1978). We were not able to identify antigens for the non-vaccine-binding antibodies by screening against a panel of common viral and bacterial antigens; conclusive identification of non-vaccine specificities will require high-throughput screening methods. Nevertheless, our integrated strategy of single-cell sequencing and Rep-seq offers a direct route to characterization of these non-vaccine-specific yet vaccine-elicited antibodies. Our results also suggest that bystander activation is confined to a small number of clones by an unknown mechanism, perhaps related to the localization of activated T cells (Jasiulewicz et al., 2015; Juy et al., 1987; Lanzavecchia et al., 1983).
We discovered a broadly binding anti-HA antibody clone in which fewer than six somatic mutations in the heavy chain alone were sufficient to confer broad high-affinity binding, offering a striking example of rapid affinity maturation. Together with prior examples of influenza antibodies that emerged via a small number of mutations (Lingwood et al., 2012; Pappas et al., 2014), this suggests that a single-dose vaccine could be sufficient to confer lasting protection against influenza. Unlike prior examples which use the heavy-chain variable region VH1-69 gene (Lingwood et al., 2012; Pappas et al., 2014), L3 uses the VH4-34 gene, which may be a target for germline-targeting immunogens. L3 antibodies appear to bind a distinct epitope compared with many previously identified classes of broadly binding anti-HA antibodies (Dreyfus et al., 2012; Kallewaard et al., 2016; Liu et al., 2017; Raymond et al., 2018; Whittle et al., 2011), suggesting that structural characterization of the interaction may reveal a new site of vulnerability on HA.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to the Lead Contact, Stephen R. Quake (steve@quake-lab.org). All unique and stable reagents generated in this study are available from the Lead Contact by request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Study subject gave informed consent and protocols were approved by the Stanford Institutional Review Board. Subject was a female human aged 18 who was recruited in 2011. The subject was apparently healthy and showed no signs of disease.
METHOD DETAILS
Sample collection
As previously described (Horns et al., 2019), blood was drawn by venipuncture, then peripheral blood mononuclear cells (PBMCs) were isolated using a Ficoll gradient and frozen in 10% (vol/vol) DMSO and 40% fetal bovine serum (FBS) according to Stanford Human Immune Monitoring Center/CTRU protocol. Subject was vaccinated with the 2011–2012 seasonal trivalent inactivated influenza vaccine. Blood was collected 3 and 5 days before vaccination (D-3 and D-5); immediately before vaccination (D0); and 1, 4, 7, 9, and 11 days afterward (D1, D4, D7, D9, D11).
Antibody repertoire sequencing
Antibody repertoire sequencing was previously performed on samples from all time points and preprocessed data was downloaded (Horns et al., 2019). Briefly, PBMCs were thawed and RNA was extracted. This RNA was reverse transcribed using immunoglobulin heavy chain constant region-specific primers and cDNA was amplified by PCR. UMIs were incorporated during reverse transcription and PCR. These libraries were sequenced using the Illumina HiSeq 2500 and MiSeq platforms using paired-end 101 or 300 bp reads, respectively. Consensus-based error correction was performed using UMIs. UMIs also enable the computational removal of PCR bias. Sequences were annotated with V and J germline gene usage at the resolution of alleles using IgBlast (Ye et al., 2013) and isotype using BLASTN (Altschul et al., 1990). Clones were identified based on V and J gene usage, HCDR3 length, and HCDR3 sequence composition. Dynamics of clones were determined by comparing fractional abundance across study time points. As in our previous study (Horns et al., 2019), vaccine-responsive clones were identified as those having > 50-fold expansion from D0 to D7 and composing >0.1% of the repertoire at D7.
Single-cell isolation and sequencing
PBMCs from D7 and D9, which correspond to the peak of the B cell memory recall response, were thawed. B cells were magnetically enriched using the B Cell Isolation Kit II (Miltenyi). Single cells were encapsulated in droplets using 16 lanes of the Chromium device (10X Genomics) with target loading of 14,000 cells per lane. Reverse transcription and complementary DNA (cDNA) amplification were performed using the Single Cell V(D)J kit (10X Genomics). In 12 lanes, direct enrichment of VDJ was performed. In the remaining 4 lanes, VDJ and gene expression measurement was performed; these 4 lanes were considered technical replicates. All steps were done according to manufacturer’s instructions, except with additional cycles of polymerase chain reaction (PCR) (19 total cycles for direct enrichment of VDJ; 22 total cycles for VDJ and gene expression). 50 ng of cDNA was used as input for library preparation. Libraries were sequenced using the Illumina NextSeq 500 platform with paired-end reads for VDJ of 150 bp each and for gene expression of 26 bp and 98 bp.
Preprocessing of single-cell sequence data
Sequences were preprocessed to map reads to the human reference genome (GRCh38) using STAR 2.5.1b (Dobin et al., 2013), count molecules aligning to each gene, and assemble antibody heavy and light chain transcripts within cellranger 2.1.0. To distinguish bona fide single cells from multiplets, we examined the number of productive heavy and light chain contigs assembled for each cell barcode. Single B cells were identified by the presence of a single productive heavy chain and a single productive light chain, yielding a total of 94,259 single B cells for analysis. All other cells were excluded from further analysis.
Mapping single B cells into clones
Single B cells were mapped to clones using a custom algorithm similar to that used for identification of clones previously (Horns et al., 2019, 2016). Sequences detected by repertoire sequencing (n = 625,750) were annotated for V and J gene usage, HCDR3 length, and HCDR3 sequence and formed the database of subject sequences. The heavy chain variable region sequence from each single B cell was used as a query to search this database. For each query, the set of subjects sharing the query’s V and J genes and CDR3 length was identified. Within this set, the identity between the query and subject sequences within the HCDR3 and outside the HCDR3 were calculated based on Hamming distance, and hits were defined as having >90% nucleotide identity in both regions. Previous studies have demonstrated that this cutoff of sequence identity enables identification of clonally related sequences with high sensitivity and specificity (Gupta et al., 2017; Horns et al., 2016). This yielded 8,377 single B cells that had matching clones detected by repertoire sequencing.
Fidelity of clonal clustering was assessed using the light chain as an independent marker of clonal identity. In clones having multiple B cells detected by single-cell sequencing, the percentage of cells possessing the dominant light chain was determined. Impure clones were identified as those having <80% of cells within the clone sharing the dominant light chain. All of the vaccine-responsive clones were pure.
Analysis of gene expression in single cells
Gene expression profiles were log-transformed and normalized to counts per million molecules. Dimensionality reduction using principal components analysis (PCA) retaining the top 10 principal components followed by t-distributed Stochastic Neighbor Embedding (tSNE; perplexity = 30, theta = 0.5, max_iter = 1,000) (van der Maaten and Hinton, 2008) were performed using cellranger 2.1.0. Clusters were identified in an automated, unbiased manner using Density-Based Spatial Clustering of Applications with Noise (DBSCAN; eps = 0.66, min_samples = 10) (Ester et al., 1996), yielding 77 clusters. These clusters were manually annotated (i.e., labels were assigned) based on expression of marker genes for each cell type: CD19 and CD20, also known as MS4A1, for B cells; TCL1A, IGHM, and IGHD for naive B cells; CD27 for memory B cells; CD3E for T cells; and LYZ for macrophages. Activated memory B cell clusters were annotated based on the expression of the established activation markers AICDA and CD86. Differentially expressed genes were identified using the negative binomial exact test adjusted for multiple testing using the Benjamini-Hochberg procedure as implemented in Loupe 2.0.0 (10X Genomics). For visualization of differential expression, the Z-score of expression of each group of cells was computed in comparison with the mean and standard deviation of expression in all other cells. Data visualization and analysis were performed using Scanpy (Wolf et al., 2018) within JupyterLab (Kluyver et al., 2016).
Reconstructing the evolutionary history of antibody clone L3
Evolutionary analysis was conducted sequences in clone L3 obtained by repertoire sequencing using paired-end 300 bp reads (n = 125) and single-cell sequencing (n = 7). Sequences were initially aligned in an ungapped manner using the start and end positions of the HCDR3 as anchor points. This alignment was refined using MUSCLE 3.8.31 with “-refine -maxiters 1 -diags -gapopen −5000” (Edgar, 2004), then trimmed to remove positions which were only covered by single-cell sequencing contigs (which are substantially longer than repertoire sequencing assemblies). We added an inferred germline sequence consisting of the reference heavy chain V and J genes and the consensus of the alignment for the untemplated regions of the HCDR3. Phylogenetic reconstruction was performed using FastTree 2.1.7 with “-nt -gtr” (Price et al., 2010). We concatenated light chain sequences to this alignment, then performed reconstruction by maximum-likelihood assuming equal rates for all mutations.
To assess the contribution of heavy and light chain mutations to binding, we engineered variants of the high-affinity antibody L3N6 by substituting either the inferred germline heavy (germline IGH) or light (germline IGK) chain sequence. We also substituted the light chain with a randomly chosen sequence from a different clone that used the same VK gene, but had a distinct LCDR3 (IGK swap). For cloning and expression of antibodies derived from repertoire sequencing (R1-7), we used the light chain sequence originating from the single cell nearest the selected antibody, using the metric of heavy chain nucleotide sequence identity. These antibodies were chosen to span a wide range of somatic mutation levels.
Recombinant antibody expression
Recombinant antibodies were cloned and expressed by Genscript. Variants were selected to represent a broad range of sequence diversity within clones L1 and L2 (five antibodies each). Within clones L3, L4, and L5, all antibodies for which single-cell information was obtained were cloned and expressed (seven, three, and one antibody respectively). Selected antibodies were codon-optimized for human expression. These DNA sequences were synthesized and cloned into heavy and light chain pcDNA3.4 expression vectors. Heavy chains were expressed as human IgG1 and light chains were expressed as either human IGK or IGL as appropriate. Vectors were transiently transfected in HEK293-6E cells and antibodies were purified from supernatant using Robocolumn Eshmuno A (EMD Millipore) or Monofinity A Resin prepacked columns (Genscript). Purity generally >95% was confirmed using SDS-PAGE and immunoblots under reducing and non-reducing conditions.
Antigens for binding measurements
Fluzone trivalent inactivated influenza vaccine from the 2011–2012 flu season (Sanofi Pasteur) containing H1N1 A/California/7/2009, H3N2 A/Perth/16/2009, and B/Brisbane/60/2008 was obtained as a gift from Dr. Harry Greenberg. Purified influenza proteins expressed in human cells (typically HEK293) where possible, otherwise baculovirus or E. coli, were purchased from Sino Biological (11683-V08H, 11085-V08H, 11056-V08H, 40043-V08H, 11048-V08H1, 40104-V08H, 40036-V08H, 11053-V08H, 40197-V07H, 40017-V07H, 40569-V07H, 40502-V07B, 40205-V08B, 40499-V08B, 40010-V07E, 40107-V08E, 40011-V07E, 40012-VNAE). Viruses inactivated by irradiation or formaldehyde treatment were purchased from Biorad (PIP005, PIP009, PIP010, PIP013, PIP014, PIP023, PIP008, PIP015, PIP016). Tetanus toxin was purchased from Sigma Aldrich (T3194).
Binding measurements using ELISA
Semiquantitative measurements of binding were carried out using enzyme-linked immunosorbent assay (ELISA). Antigen was immobilized on clear polystyrene 96- or 384-well MaxiSorp plates (ThermoFisher) by overnight incubation at 4 C at 2 ng/uL diluted in phosphate-buffered saline (PBS) pH 7.4, then three washes were performed. When vaccine was used as antigen, vaccine was immobilized at a 50-fold dilution in PBS pH 7.4. The plate was incubated for 2 hours at room temperature with blocking buffer (PBS pH 7.4 with 0.05% Tween-20 and 2% bovine serum albumin [BSA]), then washed twice. The plate was incubated with primary antibody diluted to 2 ng/uL unless otherwise noted in blocking buffer for 2 hours at room temperature, then washed four times. The plate was incubated with detection antibody (mouse anti-human IgG1 Fc conjugated to horseradish peroxidase clone HP6069; ThermoFisher) for 2 hours at room temperature, then washed five times. All washes consisted of 5 minute incubation with PBS pH 7.4 with 0.05% Tween-20. Detection was performed by adding 1-Step ABTS Substrate (ThermoFisher), then measuring absorbance at 405 nm at 1 or 3 min intervals for 45 min. Time point used for analysis was determined based on the dynamic range of the data (increasing signal, but no saturation). Positive controls included the broadly binding anti-influenza antibodies MEDI8852 (Kallewaard et al., 2016) and CR9114 (Dreyfus et al., 2012) obtained as a gift from Dr. Peter Kim. As negative controls, we used natural human IgG1 prepared from myeloma plasma (Abcam), or incubated wells with PBS alone instead of antigen (referred to as “no antigen”) or blocking buffer alone instead of antibody (referred to as “no antibody”).
Binding measurements using biolayer interferometry
Kinetic measurements of antibody-antigen interactions were performed using biolayer interferometry on a ForteBio Octet 96 instrument with anti-human IgG Fc capture (AHC) biosensors. All assays were carried out in PBS with 1% BSA and 0.05% Tween-20 with a total volume of 250 uL per well using the following protocol: 60 s baseline, 300 s loading of antibody, 60 s baseline, 300 s association of antigen, and dissociation of variable duration up to 600 s for high affinity interactions. Antibody was loaded at 1.5 ng/uL and antigen concentrations ranged from 2.5 to 100 nM. Between assays, sensors were regenerated by cycling between assay buffer and 10 mM glycine pH 1.5 for 30 s, then quenched for 30 s in assay buffer. Data were processed using ForteBio software and custom Python scripts to perform global fitting of a 1:1 binding model across 2–5 antigen concentrations after double reference subtraction (using buffer only and analyte only conditions).
To determine whether antibodies bind similar or overlapping epitopes, competitive binding of antibody pairs to a specific antigen was characterized using anti-penta-HIS (HIS1K) biosensors. We used the following protocol: 60 s baseline, 300 s loading of antigen, 60 s baseline, 900 s association of blocking antibody, 60 s baseline, 600 s association of test antibody. Antigen was HA H1N1 A/New Caledonia/20/1999 with an isoleucine zipper trimerization domain and polyhistidine tag obtained as a gift from Dr. Peter Kim and used at 25 nM. Blocking antibodies were used at 400 nM and included MEDI8852 (Kallewaard et al., 2016), CR9114 (Dreyfus et al., 2012), CH65 (Whittle et al., 2011), H2897 (Liu et al., 2017), and 6649 (Raymond et al., 2018) obtained as gifts from Dr. Peter Kim. Test antibodies were used at 100 nM and included L3N1 and L3N6. As a control, self-blocking assays were performed using the same antibody for blocking and test steps, except with test antibody at 100 nM. Data were processed using ForteBio software and custom Python scripts. We note that complete blocking was observed between MEDI8852 and CR9114, which have overlapping epitopes. Partial blocking was observed between 6649 and H2897, which have partially overlapping epitopes.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis (calculation of Pearson correlations; Spearman correlations; Mann-Whitney U test, two-sided; Fisher’s exact test, two-sided, and associated P values) was performed using Scipy 1.1.0 in Python 2.7. Differential expression analysis was performed using Loupe Cell Browser 2.0.0 (10X Genomics) to perform the negative exact binomial test with the Benjamini-Hochberg correction for multiple testing. Uncertainty of fitted parameters based on biolayer interferometry measurements was calculated using ForteBio Data Analysis Software 7.1. Statistical details of experiments (number of sequences or cells; dispersion and precision measures; statistical tests used) can be found in Results and Star Methods.
DATA AND CODE AVAILABILITY
Data Availability Statement
The accession number for the sequence data reported in this paper (10X Genomics single-cell transcriptional profiling and paired heavy-light chain sequencing) is Sequence Read Archive: PRJNA512111. Preprocessed data are available via Google Drive at http://bit.ly/2LuR4Bw.
Code Availability Statement
Code is available at https://github.com/felixhorns/SingleBCell-FluVaccine.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal antibody L1N1 | This study | L1N1 |
| Monoclonal antibody L1N2 | This study | L1N2 |
| Monoclonal antibody L1N3 | This study | L1N3 |
| Monoclonal antibody L1N4 | This study | L1N4 |
| Monoclonal antibody L1N5 | This study | L1N5 |
| Monoclonal antibody L2N1 | This study | L2N1 |
| Monoclonal antibody L2N2 | This study | L2N2 |
| Monoclonal antibody L2N3 | This study | L2N3 |
| Monoclonal antibody L2N4 | This study | L2N4 |
| Monoclonal antibody L2N5 | This study | L2N5 |
| Monoclonal antibody L3N1 | This study | L3N1 |
| Monoclonal antibody L3N2 | This study | L3N2 |
| Monoclonal antibody L3N3 | This study | L3N3 |
| Monoclonal antibody L3N4 | This study | L3N4 |
| Monoclonal antibody L3N5 | This study | L3N5 |
| Monoclonal antibody L3N6 | This study | L3N6 |
| Monoclonal antibody L3N7 | This study | L3N7 |
| Monoclonal antibody L4N1 | This study | L4N1 |
| Monoclonal antibody L4N2 | This study | L4N2 |
| Monoclonal antibody L4N3 | This study | L4N3 |
| Monoclonal antibody L5N1 | This study | L5N1 |
| Monoclonal antibody L3_germline | This study | L3_germline |
| Monoclonal antibody L3N6_germlineIGH | This study | L3N6_germlineIGH |
| Monoclonal antibody L3_germlineIGK | This study | L3N6_germlineIGK |
| Monoclonal antibody L3N6_IGKswap | This study | L3N6_IGKswap |
| Monoclonal antibody L3R1 | This study | L3R1 |
| Monoclonal antibody L3R2 | This study | L3R2 |
| Monoclonal antibody L3R3 | This study | L3R3 |
| Monoclonal antibody L3R4 | This study | L3R4 |
| Monoclonal antibody L3R5 | This study | L3R5 |
| Monoclonal antibody L3R6 | This study | L3R6 |
| Monoclonal antibody L3R7 | This study | L3R7 |
| Anti-influenza antibody MEDI8852 | Gift from Peter Kim | MEDI8852 |
| Anti-influenza antibody CR9114 | Gift from Peter Kim | CR9114 |
| Anti-influenza antibody CH65 | Gift from Peter Kim | CH65 |
| Anti-influenza antibody H2897 | Gift from Peter Kim | H2897 |
| Anti-influenza antibody 6649 | Gift from Peter Kim | 6649 |
| Natural human IgG1 prepared from myeloma plasma | Abcam | ab90283 |
| Mouse anti-human IgG1 Fc conjugated to horseradish peroxidase clone | ThermoFisher | HP6069 |
| Biological Samples | ||
| Human peripheral blood mononuclear cells (PBMCs) | This study | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Ficoll/Hypaque | GE Healthcare | 45001750 |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | D8418 |
| Fetal bovine serum (FBS) | Atlanta Biologicals | S11150 |
| Trivalent inactivated influenza vaccine (2011 – 2012 season) | Sanofi Pasteur | 271 |
| Influenza A H1N1 (A/New Caledonia/20/1999) Hemagglutinin / HA Protein (His Tag) | Sino Biological | 11683-V08H |
| Influenza A H1N1 (A/California/07/2009) Hemagglutinin / HA Protein (His Tag) | Sino Biological | 11085-V08H |
| Influenza A H3N2 (A/Brisbane/10/2007) Hemagglutinin / HA Protein (His Tag) | Sino Biological | 11056-V08H |
| Influenza A H3N2 (A/Perth/16/2009) Hemagglutinin / HA Protein (His Tag) | Sino Biological | 40043-V08H |
| Influenza A H5N1 (A/Anhui/1/2005) Hemagglutinin / HA Protein (His Tag) | Sino Biological | 11048-V08H1 |
| Influenza A H7N9 (A/Shanghai/1/2013) Hemagglutinin / HA Protein (His Tag) | Sino Biological | 40104-V08H |
| Influenza A H9N2 (A/Chicken/Hong Kong/G9/97) Hemagglutinin / HA Protein (His Tag) | Sino Biological | 40036-V08H |
| Influenza B virus (B/Florida/4/2006) Hemagglutinin / HA Protein (His Tag) | Sino Biological | 11053-V08H |
| Influenza A H1N1 (A/USSR/90/77) Neuraminidase / NA (His Tag) | Sino Biological | 40197-V07H |
| Influenza A H3N2 (A/Babol/36/2005) Neuraminidase / NA (His Tag) | Sino Biological | 40017-V07H |
| Influenza A H3N2 (A/Hong Kong/4801/2014) Neuraminidase / NA (His Tag) | Sino Biological | 40569-V07H |
| Influenza virus B (B/PHUKET/3073/2013) Neuraminidase / NA Protein (His Tag) | Sino Biological | 40502-V07B |
| Influenza A H3N2 (A/Switzerland/9715293/2013) Nucleoprotein / NP Protein (His Tag) | Sino Biological | 40499-V08B |
| Influenza A H1N1 (A/Puerto Rico/8/34/Mount Sinai) Matrix protein 1 / M1 Protein (His Tag) | Sino Biological | 40010-V07E |
| Influenza A H7N9 (A/Anhui/1/2013) Matrix protein 1 / M1 Protein (His Tag) | Sino Biological | 40107-V08E |
| Influenza A H1N1 (A/Puerto Rico/8/34/Mount Sinai) Non-structural / NS1 Protein (His Tag) | Sino Biological | 40011-V07E |
| Influenza A H1N1 (A/Puerto Rico/8/34/Mount Sinai) Non-structural Protein 2 / NS2 | Sino Biological | 40012-VNAE |
| Native Cytomegalovirus | Biorad | PIP005 |
| Native Herpes simplex Virus 1 | Biorad | PIP009 |
| Native Herpes simplex Virus 2 | Biorad | PIP010 |
| Native Measles Virus | Biorad | PIP013 |
| Native Mumps Virus | Biorad | PIP014 |
| Native Varicella Zoster Virus | Biorad | PIP023 |
| Native Hepatitis A Virus | Biorad | PIP008 |
| Native Parainfluenza Virus 1 | Biorad | PIP015 |
| Native Parainfluenza Virus 2 | Biorad | PIP016 |
| Tetanus toxin from Clostridium tetani | Sigma Aldrich | T3194 |
| Clear Flat-Bottom Immuno Nonsterile 384-Well Plates | ThermoFisher | 464718 |
| Nunc MaxiSorp flat-bottom 96-Well Plates | ThermoFisher | 44-2404-21 |
| 1-Step ABTS Substrate | ThermoFisher | 37615 |
| Anti-hIgG Fc Capture (AHC) Biosensors | ForteBio | 18-5063 |
| Anti-Penta-HIS (HIS1K) Biosensors | ForteBio | 18-5120 |
| 10 mM Glycine pH 1.5 | GE Healthcare | BR100354 |
| Critical Commercial Assays | ||
| Chromium Single Cell 5′ Library & Gel Bead Kit | 10X Genomics | 1000006 |
| Chromium Single Cell V(D)J Enrichment Kit, Human B Cell | 10X Genomics | 1000016 |
| Chromium Single Cell A Chip Kit | 10X Genomics | 1000009 |
| Chromium i7 Multiplex Kit | 10X Genomics | 120262 |
| NextSeq 500/550 High Output Kit v2.5 (300 Cycles) | Illumina | 20024908 |
| B Cell Isolation Kit II, human | Milteyi | 130-091-151 |
| Deposited Data | ||
| Antibody repertoire sequencing data | Horns et al., 2019 | https://bit.ly/2BL83JV |
| Single-cell transcriptional profiling data | This paper | http://bit.ly/2LuR4Bw (preprocessed) and Sequence Read Archive PRJNA512111 (raw) |
| Single-cell paired heavy-light chain antibody sequencing data | This paper | http://bit.ly/2LuR4Bw (preprocessed) and Sequence Read Archive PRJNA512111 (raw) |
| Software and Algorithms | ||
| IgBlast | Ye et al., 2013 | 1.3.0 |
| blastn | Altschul et al., 1990 | 2.2.29+ |
| cellranger | 10X Genomics | 2.1.0 |
| STAR | Dobin et al., 2013 | 2.5.1b |
| JupyterLab | Kluyver et al., 2016 | 4.4.0 |
| Scanpy | Wolf et al., 2018 | 1.3.6 |
| Loupe Cell Browser | 10X Genomics | 2.0.0 |
| MUSCLE | Edgar, 2004 | 3.8.31 |
| FastTree | Price et al., 2010 | 2.1.7 |
| ForteBio Data Analysis Software | ForteBio | 7.1 |
| Custom analysis code | This study | https://github.com/felixhorns/SingleBCell-FluVaccine/ |
Highlights.
Human antibody memory response studied using single-cell and repertoire sequencing
Single-cell transcriptomics reveals a program of memory B cell activation
Previously unknown broadly binding anti-influenza antibodies are identified
Bystander activation: many vaccine-responsive antibodies do not bind vaccine
ACKNOWLEDGMENTS
We thank Peter Kim, Derek Croote, and Yael Rosenberg-Hasson for helpful discussions; 10X Genomics for providing reagents for single-cell sequencing; Krista McCutcheon for expressing antibodies; Payton Weidenbacher and Peter Kim for discussions and providing antibodies and influenza proteins; and Harry Greenberg and Caiqiu Zhang for providing trivalent inactivated influenza vaccine. This work was supported by United States of America National Institutes of Health U19A1057229 (to S.R.Q.) and the National Science Foundation Graduate Research Fellowship Program (to F.H.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.12.063.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, and Lipman DJ (1990). Basic local alignment search tool. J. Mol. Biol 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Arneborn P, Biberfeld G, Forsgren M, and von Stedingk LV (1983). Specific and non-specific B cell activation in measles and varicella. Clin. Exp. Immunol 51, 165–172. [PMC free article] [PubMed] [Google Scholar]
- Barnett BE, Staupe RP, Odorizzi PM, Palko O, Tomov VT, Mahan AE, Gunn B, Chen D, Paley MA, Alter G, et al. (2016). Cutting ddge: B cell-intrinsic T-bet expression is required to control chronic viral infection. J. Immunol 197, 1017–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshe RB, Newman FK, Cannon J, Duane C, Treanor J, Van Hoecke C, Howe BJ, and Dubin G (2004). Serum antibody responses after intradermal vaccination against influenza. N. Engl. J. Med 351, 2286–2294. [DOI] [PubMed] [Google Scholar]
- Bernasconi NL, Traggiai E, and Lanzavecchia A (2002). Maintenance of serological memory by polyclonal activation of human memory B cells. Science 298, 2199–2202. [DOI] [PubMed] [Google Scholar]
- Crotty S, Aubert RD, Glidewell J, and Ahmed R (2004). Tracking human antigen-specific memory B cells: a sensitive and generalized ELISPOT system. J. Immunol. Methods 286, 111–122. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, et al. (2012). Highly conserved protective epitopes on influenza B viruses. Science 337, 1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ester M, Kriegel H-P, Sander J, and Xu X (1996). A density-based algorithm for discovering clusters in large spatial databases with noise In KDD’96: Proceedings of the Second International Conference on Knowledge Discovery and Data Mining (AAAI Press; ), pp. 226–231. [Google Scholar]
- Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, Moser JM, Mehta RS, Drake DR 3rd, Castro E, et al. (2008). Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J. Exp. Med 205, 3119–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta NT, Adams KD, Briggs AW, Timberlake SC, Vigneault F, and Kleinstein SH (2017). Hierarchical clustering can identify B cell clones with high confidence in Ig repertoire sequencing data. J. Immunol 198, 2489–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, O’Neill P, Naradikian MS, Scholz JL, and Cancro MP (2011). A B-cell subset uniquely responsive to innate stimuli accumulates in aged mice. Blood 118, 1294–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn AD, Wu S, Qiu X, Ruda M, Stover M, Yang H, Liu Z, Welle SL, Holden-Wiltse J, Wu H, and Zand MS (2013). High-resolution temporal response patterns to influenza vaccine reveal a distinct human plasma cell gene signature. Sci. Rep 3, 2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horns F, Vollmers C, Croote D, Mackey SF, Swan GE, Dekker CL, Davis MM, and Quake SR (2016). Lineage tracing of human B cells reveals the in vivo landscape of human antibody class switching. eLife 5, e16578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horns F, Vollmers C, Dekker CL, and Quake SR (2019). Signatures of selection in the human antibody repertoire: Selective sweeps, competing subclones, and neutral drift. Proc. Natl. Acad. Sci. USA 116, 1261–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jash A, Wang Y, Weisel FJ, Scharer CD, Boss JM, Shlomchik MJ, and Bhattacharya D (2016). ZBTB32 restricts the duration of memory B cell recall responses. J. Immunol 197, 1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasiulewicz A, Lisowska KA, Pietruczuk K, Frąckowiak J, Fulop T, and Witkowski JM (2015). Homeostatic ‘bystander’ proliferation of human peripheral blood B cells in response to polyclonal T-cell stimulation in vitro. Int. Immunol 27, 579–588. [DOI] [PubMed] [Google Scholar]
- Jiang N, He J, Weinstein JA, Penland L, Sasaki S, He X-S, Dekker CL, Zheng N-Y, Huang M, Sullivan M, et al. (2013). Lineage structure of the human antibody repertoire in response to influenza vaccination. Sci. Transl. Med 5, 171ra19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juy D, Sterkers G, Gomez A, Zelizewski D, and Lévy J-P (1987). Polyclonal B-cell activation by influenza A/Texas virus-specific human T-cell clones. Ann. Inst. Pasteur Immunol 138, 371–382. [DOI] [PubMed] [Google Scholar]
- Kallewaard NL, Corti D, Collins PJ, Neu U, McAuliffe JM, Benjamin E, Wachter-Rosati L, Palmer-Hill FJ, Yuan AQ, Walker PA, et al. (2016). Structure and function analysis of an antibody recognizing all influenza A sub-types. Cell 166, 596–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluyver T, Ragan-Kelley B, Pérez F, Granger B, Bussonnier M, Frederic J, Kelley K, Hamrick J, Grout J, Corlay S, et al. (2016). Jupyter Notebooks—a publishing format for reproducible computational workflows In Positioning and Power in Academic Publishing: Players, Agents and Agendas. In Proceedings of the 20th International Conference on Electronic Publishing, Loizides F and Schmidt B, eds. (IOS Press; ), pp. 87–90. [Google Scholar]
- Lacotte S, Brun S, Muller S, and Dumortier H (2009). CXCR3, inflammation, and autoimmune diseases. Ann. N Y Acad. Sci 1173, 310–317. [DOI] [PubMed] [Google Scholar]
- Lanzavecchia A, Parodi B, and Celada F (1983). Activation of human B lymphocytes: frequency of antigen-specific B cells triggered by alloreactive or by antigen-specific T cell clones. Eur. J. Immunol 13, 733–738. [DOI] [PubMed] [Google Scholar]
- Larousserie F, Bardel E, Coulomb L’Herminé A, Canioni D, Brousse N, Kastelein RA, and Devergne O (2006). Variable expression of Epstein-Barr virus-induced gene 3 during normal B-cell differentiation and among B-cell lymphomas. J. Pathol 209, 360–368. [DOI] [PubMed] [Google Scholar]
- Lee FE-H, Halliley JL, Walsh EE, Moscatiello AP, Kmush BL, Falsey AR, Randall TD, Kaminiski DA, Miller RK, and Sanz I (2011). Circulating human antibody-secreting cells during vaccinations and respiratory viral infections are characterized by high specificity and lack of bystander effect. J. Immunol 186, 5514–5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Rouphael N, Duraisingham S, Romero-Steiner S, Presnell S, Davis C, Schmidt DS, Johnson SE, Milton A, Rajam G, et al. (2014). Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat. Immunol 15, 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D, McTamney PM, Yassine HM, Whittle JRR, Guo X, Boyington JC, Wei C-J, and Nabel GJ (2012). Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature 489, 566–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Pan J, Jenni S, Raymond DD, Caradonna T, Do KT, Schmidt AG, Harrison SC, and Grigorieff N (2017). CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface. J. Mol. Biol 429, 1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir S, Ho J, Malaspina A, Wang W, DiPoto AC, O’Shea MA, Roby G, Kottilil S, Arthos J, Proschan MA, et al. (2008). Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J. Exp. Med 205, 1797–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moticka EJ, and Streilein JW (1978). Hypothesis: nonspecific polyclonal activation of memory B cells by antigen as a mechanism for the preservation of long term immunologic anamnesis. Cell. Immunol 41, 406–413. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, and Honjo T (2000). Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102, 553–563. [DOI] [PubMed] [Google Scholar]
- Okada T, Ngo VN, Ekland EH, Förster R, Lipp M, Littman DR, and Cyster JG (2002). Chemokine requirements for B cell entry to lymph nodes and Peyer’s patches. J. Exp. Med 196, 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas L, Foglierini M, Piccoli L, Kallewaard NL, Turrini F, Silacci C, Fernandez-Rodriguez B, Agatic G, Giacchetto-Sasselli I, Pellicciotta G, et al. (2014). Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature 516, 418–422. [DOI] [PubMed] [Google Scholar]
- Pène J, Gauchat J-F, Lécart S, Drouet E, Guglielmi P, Boulay V, Del-wail A, Foster D, Lecron J-C, and Yssel H (2004). Cutting edge: IL-21 is a switch factor for the production of IgG1 and IgG3 by human B cells. J. Immunol 172, 5154–5157. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, and Arkin AP (2010). FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond DD, Bajic G, Ferdman J, Suphaphiphat P, Settembre EC, Moody MA, Schmidt AG, and Harrison SC (2018). Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc. Natl. Acad. Sci. USA 115, 168–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov AV, Rubtsova K, Fischer A, Meehan RT, Gillis JZ, Kappler JW, and Marrack P (2011). Toll-like receptor 7 (TLR7)-driven accumulation of a novel CD11c+ B-cell population is important for the development of autoimmunity. Blood 118, 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsova K, Rubtsov AV, van Dyk LF, Kappler JW, and Marrack P (2013). T-box transcription factor T-bet, a key player in a unique type of B-cell activation essential for effective viral clearance. Proc. Natl. Acad. Sci. USA 110, E3216–E3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsova K, Rubtsov AV, Thurman JM, Mennona JM, Kappler JW, and Marrack P (2017). B cells expressing the transcription factor T-bet drive lupus-like autoimmunity. J. Clin. Invest 127, 1392–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AG, Do KT, McCarthy KR, Kepler TB, Liao H-X, Moody MA, Haynes BF, and Harrison SC (2015). Immunogenic stimulus for germline precursors of antibodies that engage the influenza hemagglutinin receptor-binding site. Cell Rep. 13, 2842–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J (1996). Immunoglobulin class switching. Curr. Opin. Immunol 8, 199–205. [DOI] [PubMed] [Google Scholar]
- The Tabula Muris Consortium (2018). Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treanor J, Keitel W, Belshe R, Campbell J, Schiff G, Zangwill K, Wolff M, Klimov A, Levandowski R, and Lambert L (2002). Evaluation of a single dose of half strength inactivated influenza vaccine in healthy adults. Vaccine 20, 1099–1105. [DOI] [PubMed] [Google Scholar]
- van der Maaten L, and Hinton G (2008). Visualizing data using t-SNE. J. Mach. Learn. Res 9, 2579–2605. [Google Scholar]
- Vollmers C, Sit RV, Weinstein JA, Dekker CL, and Quake SR (2013). Genetic measurement of memory B-cell recall using antibody repertoire sequencing. Proc. Natl. Acad. Sci. USA 110, 13463–13468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang NS, McHeyzer-Williams LJ, Okitsu SL, Burris TP, Reiner SL, and McHeyzer-Williams MG (2012). Divergent transcriptional programming of class-specific B cell memory by T-bet and RORα. Nat. Immunol 13, 604–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle JRR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, et al. (2011). Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. USA 108, 14216–14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng N-Y, Mays I, Garman L, Helms C, et al. (2008). Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 453, 667–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Ma N, Madden TL, and Ostell JM (2013). IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 41, W34–W40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data Availability Statement
The accession number for the sequence data reported in this paper (10X Genomics single-cell transcriptional profiling and paired heavy-light chain sequencing) is Sequence Read Archive: PRJNA512111. Preprocessed data are available via Google Drive at http://bit.ly/2LuR4Bw.
Code Availability Statement
Code is available at https://github.com/felixhorns/SingleBCell-FluVaccine.
The accession number for the sequence data reported in this paper (10X Genomics single-cell transcriptional profiling and paired heavy-light chain sequencing) is Sequence Read Archive: PRJNA512111. Preprocessed data are available via Google Drive at http://bit.ly/2LuR4Bw.


