Abstract
Background
Renal multiparametric magnetic resonance imaging (MRI) is a promising tool for diagnosis, prognosis, and treatment monitoring in kidney disease.
Purpose
To determine intrasubject test–retest repeatability of renal MRI measurements.
Study Type
Prospective.
Population
Nineteen healthy subjects aged over 40 years.
Field Strength/Sequences
T1 and T2 mapping, R2* mapping or blood oxygenation level‐dependent (BOLD) MRI, diffusion tensor imaging (DTI), and intravoxel incoherent motion (IVIM) diffusion‐weighted imaging (DWI), 2D phase contrast, arterial spin labelling (ASL), dynamic contrast enhanced (DCE) MRI, and quantitative Dixon for fat quantification at 3T.
Assessment
Subjects were scanned twice with ~1 week between visits. Total scan time was ~1 hour. Postprocessing included motion correction, semiautomated segmentation of cortex and medulla, and fitting of the appropriate signal model.
Statistical Test
To assess the repeatability, a Bland–Altman analysis was performed and coefficients of variation (CoVs), repeatability coefficients, and intraclass correlation coefficients were calculated.
Results
CoVs for relaxometry (T1, T2, R2*/BOLD) were below 6.1%, with the lowest CoVs for T2 maps and highest for R2*/BOLD. CoVs for all diffusion analyses were below 7.2%, except for perfusion fraction (FP), with CoVs ranging from 18–24%. The CoV for renal sinus fat volume and percentage were both around 9%. Perfusion measurements were most repeatable with ASL (cortical perfusion only) and 2D phase contrast with CoVs of 10% and 13%, respectively. DCE perfusion had a CoV of 16%, while single kidney glomerular filtration rate (GFR) had a CoV of 13%. Repeatability coefficients (RCs) ranged from 7.7–87% (lowest/highest values for medullary mean diffusivity and cortical FP, respectively) and intraclass correlation coefficients (ICCs) ranged from −0.01 to 0.98 (lowest/highest values for cortical FP and renal sinus fat volume, respectively).
Data Conclusion
CoVs of most MRI measures of renal function and structure (with the exception of FP and perfusion as measured by DCE) were below 13%, which is comparable to standard clinical tests in nephrology.
Level of Evidence
2
Technical Efficacy
Stage 1
Keywords: repeatability, reproducibility, kidney, MRI, multiparametric, functional
MULTIPARAMETRIC MAGNETIC RESONANCE IMAGING (MRI) of the kidneys is a promising tool for diagnosis, prognosis, and treatment monitoring in kidney disease. Contrary to anatomic imaging, functional imaging allows for quantitative measures of oxygenation, perfusion, tissue microstructure, and water content. Such variables are likely to change in the course of various conditions and are therefore sensitive measures of pathology. 1 Rising interest in renal MRI is driven in Europe by the COST action PARENCHIMA, dedicated to MRI biomarkers in kidney disease (www.renalmri.org). The ultimate goal of this collaboration of kidney MRI researchers is the initiation of large‐scale clinical studies needed to confirm the value of kidney MRI as a clinical biomarker. 1
In various diseases, functional MRI has already been successfully applied to detect pathologic changes. For example, diffusion‐weighted imaging (DWI), an often‐used MRI method, shows a decrease in the apparent diffusion constant (ADC) with increasing fibrosis in chronic kidney disease (CKD). 2 In kidney transplants, the ADC is consistently decreased in patients with acute tubular necrosis, acute rejection, and immunosuppressive toxicity, but the ADC is not able to differentiate between these pathologies. 3 In comparison, tissue T2, for example, is known to increase in response to inflammation, 4 while it tends to decrease in reaction to severe fibrosis, 5 which enables discrimination between those conditions.
Variants of DWI include diffusion tensor imaging, focused on the directionality of diffusion, and intravoxel incoherent motion (IVIM), which corrects for the contribution of microperfusion to the diffusion coefficient and additionally estimates a perfusion fraction. With ASL, cortical perfusion can be mapped, which is known to be impaired in, for example, CKD. 6 Using phase contrast (PC) MRI, renal blood flow can be measured. Blood oxygenation level‐dependent (BOLD) MRI is sensitive oxygenation and can detect renal hypoxia, which is thought to be the driving factor behind the progression of CKD. 7 T1 and T2 are both sensitive to changes in microstructure, water content, and oxygenation and are therefore sensitive, although not very specific markers of pathology. 8 The introduction of other MRI sequences in patient care might increase the specificity of MRI as a diagnostic tool. Ultimately, multiparametric evaluation could allow for recognition of patterns in MRI parameters characteristic of different pathologies.
As a first step towards large diagnostic and prognostic studies, the repeatability of kidney MRI has to be determined. 1 Although for most of those techniques, repeatability studies have been performed, repeatability data of multiparametric MRI in a single group of subjects is scarce, as are data on how repeatability compares between different sequences and methods. Furthermore, as far as we are aware, repeatability data on renal quantitative Dixon and renal T2 mapping are not yet available, apart from a conference proceeding for measurement of T2. 9
In this multiparametric study, we additionally included Dixon‐based fat quantification and dynamic contrast‐enhanced (DCE) imaging. Fat content in both the renal parenchyma and the renal sinus is thought to contribute to the progression of kidney disease. 10 DCE MRI can be used to quantify perfusion and to measure the glomerular filtration rate (GFR), the main clinical measure of kidney function. 11
The aim of this study was to determine the test–retest repeatability of multiparametric kidney MRI in healthy subjects. A secondary aim was to compare the repeatability of different MRI perfusion techniques, ie, PC MRI, arterial spin labeling (ASL) and DCE MRI.
Materials and Methods
Subjects
This study was approved by the local Institutional Review Board and all subjects signed informed consent prior to inclusion. For subjects to be included, they had to be aged 40 years or older. Exclusion criteria were a history of renal or cardiovascular disease and contraindications for MRI, including incompatible implants, claustrophobia, or an allergy to gadolinium‐based contrast agents. Subjects were imaged twice with ~1 week between visits. Data of insufficient image quality on visual assessment were excluded. Both visits of each subject were planned around the same time of the day. Subjects were asked to avoid salt and protein‐rich meals and to drink 2 L per 24 hours of nonalcoholic liquids on the day of the scan to roughly standardize hydration and dietary conditions. Prior to one imaging session, usually the first, blood was sampled to measure creatinine, cystatin C and hematocrit, to allow calculation of estimated glomerular filtration rate (eGFR) using the CKD‐EPI formula 12 (based on creatinine and cystatin C).
Imaging Protocol
All subjects were scanned with a 3T MRI system (Ingenia, Philips Healthcare, Best, The Netherlands; software release 5.3.1) with a 12‐channel posterior and a 16‐channel anterior receive coil array. The imaging protocol started with a localizer scan. B0 and B1 shimming was performed for all acquisitions. All scans were acquired in the coronal plane with a Cartesian readout, except for the DCE, which was acquired in the transverse plane with a radial readout. A detailed overview of the imaging parameters is listed in Table 1.
TABLE 1.
Overview of Scan Parameters in the Same Order as They Were Scanned
| T1w Dixon | Quantitative Dixon | T2* | DTI/IVIM | T2 map | Phase contrast | FAIR‐ASL | T1 map | M0 images | DCE | |
|---|---|---|---|---|---|---|---|---|---|---|
| Sequence | 3D GE | 3D GE | 2D GE | 2D SE | 2D GE | 2D GE | 2D GE | 2D GE | 2D GE | 2D GE |
| Fast imaging | — | — | — | EPI | EPI | TFE | EPI | EPI | EPI | — |
| Echoes | 2 | 6 | 15 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| TE1;ΔTE (ms) | 3.5;1.1 | 0.88;0.6 | 4.6;4.6 | 62 | 21 | 3.3 | 22 | 22 | 22 | 1.42 |
| TR (ms) | 7.5 | 5.1 | 213 | 3247 | 5000 | 5.5 | 6500 | 6500 | 6500 | 3.5 |
| FA (degree) | 8 | 6 | 25 | 90 | 30 | 10 | 90 | 90 | 90 | 10 |
| Halfscan | — | — | — | 0.8 | — | — | — | — | — | 0.7 |
| Orientation | Coronal oblique | Coronal oblique | Coronal oblique | Coronal oblique | Coronal oblique | Sagittal oblique | Coronal oblique | Coronal oblique | Coronal oblique | Transverse |
| Slices | 35 | 51 | 14 | 20 | 7 | 1 | 5 | 11 | 5 | 50 |
| Voxel size (mm) | 1.5 × 1.5 × 3 | 3 × 3 × 6 | 3 × 3 × 3 | 3 × 3 × 3 | 3 × 3 × 6 | 1.5 × 1.5 × 6 | 3 × 3 × 6 | 3 × 3 × 6 | 3 × 3 × 6 | 1.5 × 1.5 × 3 |
| FOV (mm) | 320 × 400 × 70 | 360 × 360 × 178 | 360 × 360 × 48 | 336 × 336 × 60 | 244 × 244 × 48 | 320 × 250 | 244 × 244 × 34 | 244 × 244 × 76 | 244 × 244 × 34 | 280 × 280 × 150 |
| Recon matrix | 400 | 240 | 240 | 112 | 96 | 256 | 96 | 96 | 96 | NA |
| Parallel imaging; SENSEfactor | 2 | 3 | 3 | 2.1 | 1.5 | 3 | 1.5 | 1.5 | 1.5 | None |
| Acquisition time (mm:ss) | 00:19 | 00:08 | 5 × 00:09 | 02:42 | 03:37 | 2 × 00:10 | 4 × 02:23 | 01:12 | 00:33 | 05:27 |
| Respiratory compensation | Breathhold | Breathhold | Breathhold | Free breathing | Free breathing | Breathhold | Synchronized breathing | Synchronized breathing | Synchronized breathing | Free breathing |
| Remarks | Dixon recon of water only images | Dixon recon of fat fraction images | ‐ | b‐values 0,10,20,50, 100,200,500 | MLEV preparation with composite 180° block pulses with T2 weightings 15, 48.75, 82.5, 116.25, 150 ms | 25 phases, venc 150 cm/s, exact acquisition time depends on heart rate | TIs 800, 1400, 2000, 2600 ms; 10 label‐control pairs per TI | Cycled multislice inversion recovery sequence 49 ; TIs: 55–2035 with 198 ms steps | 4 averages | Golden angle stack‐of‐stars radial acquisition, radial percentage 900%. More slices were added if required for full kidney coverage. |
DTI/IVIM: diffusion tensor imaging/intravoxel incoherent motion; FAIR‐ASL: flow attenuated inversion recovery arterial spin labeling; DCE: dynamic contrast enhanced MRI; TE: echo time; GE: spoiled gradient echo; EPI: echo planar imaging; TR: repetition time; FA: flip angle; FOV: field of view; TI: inversion time; recon: reconstruction.
Data were acquired either during breath‐hold (PC, BOLD MRI, the anatomical and quantitative Dixon), synchronized breathing (the subject was asked to breath in between the single‐shot acquisitions, ASL, M0, and T1) or free breathing (T2 mapping, DWI, DCE MRI). The total scan time was ~1 hour. During the last sequence (DCE MRI), 0.05 mmol/kg of gadobutrol (Bayer Healthcare, Berlin, Germany) was infused at a rate of 1 mL/s, followed by a saline flush of 20 mL. DCE MRI had a temporal resolution of 4.1 seconds and images were acquired continuously for 5:27 minutes. Care was taken to acquire T1 maps, ASL images, and M0 images using the same field of view and voxel size, since these images had to be combined for ASL quantification. For timing of the QUIPPS settings in the flow attenuated inversion recovery arterial spin labeling (FAIR‐ASL) acquisition, please refer to the previous study. 13 For the first four patients a slightly different scan protocol for T1 mapping and FAIR‐ASL was used. Those T1 maps and ASL scans were therefore excluded from the current analysis.
Postprocessing
Postprocessing was performed using in‐house‐developed software in MatLab (R2015b, MathWorks Natick, MA), unless stated otherwise. All images were converted to NIfTI format. 14 On the T1‐weighted anatomical Dixon images, coordinates of the center of both kidneys were identified manually. These were used in all series to make a wide crop around each kidney. The remaining processing was performed separately for each kidney.
IMAGE RECONSTRUCTION
All images were reconstructed online using the scanner software except for the radial stack‐of‐stars DCE data, which was reconstructed offline in MatLab using the GRASP compressed sensing algorithm. 15 In short, this algorithm exploits temporal sparsity using a first‐order temporal total variation constraint. DCE data were reconstructed using 21 radial spokes per volume, resulting in a spatial resolution of 1.5 × 1.5 × 3.0 mm3 and a temporal resolution of 4.1 seconds. This was the highest temporal resolution signal‐to‐noise ratio (SNR) would allow as judged by one expert reader (B.S.).
MOTION CORRECTION
Respiratory motion compensation (registration) was performed for all scans except for BOLD, anatomical, and quantitative Dixon scans, since those were acquired in breath‐hold. Image registration was achieved by groupwise deformable registration in Elastix (v. 4.9.0 16 , 17 ). This was performed slicewise for all coronal 2D scans and imagewise for the DCE, which was acquired in 3D. A principal component analysis (PCA)‐based 14 similarity measure was used, since it is insensitive for the sometimes strong contrast changes in the images, especially in the T1 and DCE series. For the phase contrast images, registration was performed on the magnitude images to provide sufficient detail to the registration algorithm.
For ASL quantification, the T1 series and M0 images were required. Since these scans were all acquired with the same geometry settings, these series were registered all at once using the same groupwise approach as described above. For the repeatability analysis of T1, the T1 series was also processed separately.
SEGMENTATION
To allow separate analysis of cortex and medulla segmentation, masks for both regions were defined for all kidneys. Masks were generated using k‐means clustering on the DCE, T1, and BOLD series and copied to the other series as described in the Supplementary Materials. 18 If necessary, they were manually adapted by one expert (A.B., with 5 years of experience in renal imaging) in ITK‐SNAP. 19 Areas affected by artifacts, as identified by visual inspection, were avoided. Detailed information on mask generation, including the approach to copy the masks to the other series, is provided in the Supplementary Materials. For examples of the cortex and medulla mask for different readouts, see Fig. S1 in the Supplemental Materials. Segmentation of the renal artery in the phase contrast data was performed semiautomated (see Supplementary Material). On the quantitative Dixon scans, the renal sinus was manually delineated to enable quantification of renal sinus fat.
QUALITY ASSESSMENT
Quality assessment was performed by three readers (A.B., M.F., and H.H. with 5, 7, and 8 years of experience in renal imaging, respectively) according to the criteria in the Supplementary Material. Data judged to be of insufficient quality by reader A.B were presented to readers M.F. and H.H. If they agreed, the data were discarded. Examples of discarded images are shown in Fig. S1 in the Supplemental Material.
MODELING
Relaxometry data (T1, T2, and R2* mapping) was fitted to monoexponential models in a voxelwise manner, yielding a relaxation time or rate constant map and an S0 map (see Supplementary Material).
For the analysis of the DWI data, two models were used 20 ; a DTI and a biexponential IVIM model. The DTI analysis yields the mean diffusivity (MD) and fractional anisotropy (FA). The IVIM model measures also a diffusion constant (D), excluding the contribution of microperfusion (measured as FP).
For modeling of the ASL data, the Buxton model was used, as previously described. 13 This model yields perfusion per unit of tissue and the arterial transit time (ATT), or the time blood needs to travel from the labeling location (usually the aorta) to tissue.
Modeling of the DCE data was performed on a whole‐parenchyma basis (as opposed to voxel‐by‐voxel fitting). A renal two‐compartment model was used. 11 , 21 , 22 The model was fitted in two steps: 1) tubular flow (GFR per unit of tissue volume) and the tubular transit time were fitted to the second part of the data followed by 2) fitting of the vascular parameters (blood volume, bolus arrival delay, and plasma transit time) to the first part of the data with fixed tubular parameters. A similar approach was proposed by Tofts et al. 11 For details, please refer to the Supplementary Material. For DCE MRI, parenchymal perfusion, mean residence time (MRT, plasma transit time plus bolus arrival delay 11 ), and single kidney GFR (skGFR) were reported. Perfusion was calculated as blood volume divided by mean residence time.
Fat quantification was performed on the scanner using a six‐echo Dixon approach with vendor‐provided software. 23 This yielded a fat fraction (FF), which was multiplied with sinus volume to obtain renal sinus fat volume.
Statistical Analysis
For the MRI measurements, mean values obtained from the first and second scan were separately compared for cortex and medulla on a per‐kidney basis. To assess repeatability, within‐subject coefficients of variations (CoVw) were calculated as follows 24 :
Here, x1 and x2 denote the mean value for the first and second scans. Since the analysis was performed on a per‐kidney basis, the summation is over kidneys, not patients. N and n denote total number of kidneys and kidney number, respectively (or arteries in case of 2DPC). Additionally, intraclass correlation coefficients (ICCs, two‐way mixed effects, absolute agreement, single measurement) and repeatability coefficients (RC) were calculated. The RC gives the expected range of the repeated measurement in 95% of subjects and equals the range between the limits of agreement in the Bland–Altman plot. 25 A Bland–Altman analysis was performed, including scatterplots and Bland–Altman plots with limits of agreement and corresponding confidence intervals. 25 All analyses were performed in R v. 3.4.4. 26 Data are reported as mean (standard deviation) or mean (interquartile range), where appropriate.
Results
Volunteer Characteristics
Twenty healthy subjects were included, of whom one was scanned only once and therefore excluded from the current analysis. Subject characteristics are presented in Table 2. The age distribution was skewed to the left: 10 subjects were aged between 40 and 50. Of the 19 subjects, 14 were scanned with a 7‐day interval, the remaining subjects were scanned at 4, 5, 8, 14, and 16 days. eGFR was above 60 ml/1.73 m2/min in all subjects.
TABLE 2.
Baseline Characteristics of the Study Population
| Median (IQR) | |
|---|---|
| Number of participants (male) | 19 (9) |
| Age, years | 49 (45–57) |
| eGFR, mL/1.73m2/min | 97 (88–101) |
| Creatinine, μmol/L | 73 (69–85) |
| Cystatin C, mg/L | 0.80 (0.77–0.90) |
| Hematocrit, fraction | 0.41 (0.38–0.44) |
IQR: interquartile range.
MRI Measurements
An overview of all calculated repeatability measures is provided in Table 3.
TABLE 3.
Overview of Repeatability Measures for the Multiparametric Scan Protocol
| Baseline | Follow up | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sequence | Parameter | Unit | N | Mean (SD) | Mean (SD) | Bias (%) | CoV (%) | RC (%) | ICC |
| Relaxometry | |||||||||
| T1 map | T1 cortex | ms | 28 | 1526 (54) | 1440 (84) | −5.8 | 5.1 | 14 (9–18) | 0.32 (−0.11–0.71) |
| T1 medulla | ms | 28 | 1870 (61) | 1820 (66) | −2.7 | 2.8 | 7.8 (5.2–10.2) | 0.48 (0.00–0.75) | |
| T2 map | T2 parenchyma | ms | 35 | 113 (8.2) | 114 (7.6) | 0.8 | 2.9 | 8.2 (5.7–10.7) | 0.82 (0.67–0.90) |
| R2* map | R2* cortex | 1/s | 34 | 19.3 (2.1) | 19.0 (1.9) | −1.6 | 6.1 | 18 (13–24) | 0.58 (0.31–0.77) |
| R2* medulla | 1/s | 34 | 26.2 (2.7) | 25.6 (2.2) | −2.0 | 5.8 | 16 (11–22) | 0.62 (0.36–0.79) | |
| DTI and IVIM | |||||||||
| DTI | MD cortex | mm2/s | 36 | 2.3 (0.2) | 2.3 (0.2) | −0.3 | 3.7 | 10 (7–13) | 0.74 (0.55–0.86) |
| MD medulla | mm2/s | 36 | 2.4 (0.1) | 2.4 (0.1) | 0.8 | 2.8 | 7.7 (5.4–10) | 0.76 (0.59–0.87) | |
| FA cortex | — | 36 | 0.31 (0.03) | 0.31 (0.04) | −0.4 | 6.7 | 19 (13–25) | 0.63 (0.38–0.79) | |
| FA medulla | — | 36 | 0.40 (0.04) | 0.40 (0.04) | −0.5 | 5.2 | 15 (10–19) | 0.74 (0.55–0.86) | |
| IVIM | D cortex | mm2/s | 36 | 2.1 (0.1) | 2.1 (0.2) | 1.0 | 6.7 | 18 (13–24) | 0.24 (−0.10–0.52) |
| D medulla | mm2/s | 36 | 1.9 (0.1) | 1.9 (0.2) | −0.5 | 7.2 | 20 (14–26) | 0.23 (−0.11–0.52) | |
| FP cortex | % | 36 | 10 (3) | 10 (3) | −4.0 | 24 | 87 (61–113) | −0.01 (−0.34–0.32) | |
| FP medulla | % | 36 | 13 (3) | 12 (3) | −5.2 | 18 | 56 (39–73) | 0.35 (0.03–0.61) | |
| Perfusion and GFR | |||||||||
| FAIR | Cortical perfusion | mL/100 mL/min | 28 | 340 (51) | 350 (49) | 2.8 | 10 | 29 (19–39) | 0.47 (0.13–0.72) |
| ATT | s | 28 | 0.50 (0.13) | 0.50 (0.14) | −1.3 | 12 | 34 (22–46) | 0.80 (0.62–0.90) | |
| DCE | Parenchymal perfusion | mL/100 mL/min | 30 | 443 (110) | 435 (132) | −1.8 | 16 | 44 (30–59) | 0.67 (0.41–0.83) |
| MRT | s | 30 | 5.1 (1.0) | 5.2 (0.9) | 3.1 | 12 | 32 (21–43) | 0.50 (0.30–0.78) | |
| skGFR | mL/min | 30 | 45 (10) | 44 (9) | −3.5 | 13 | 36 (24–47) | 0.66 (0.40–0.82) | |
| PC | RBF per artery | mL/min | 37* | 388 (147) | 381 (141) | −2.0 | 13 | 37 (26–47) | 0.88 (0.77–0.93) |
| Fat quantification | |||||||||
| Dixon | Fat fraction sinus | — | 38 | 0.41 (0.12) | 0.41 (0.12) | −0.2 | 9.2 | 19 (23–24) | 0.95 (0.90–0.97) |
| Fat volume sinus | mL | 38 | 16.3 (8.4) | 15.8 (8.3) | −3.2 | 9.5 | 20 (24–25) | 0.98 (0.96–0.99) | |
In Table S1 in the Supplemental Material, CoV values are compared to literature values.
N: number of kidneys; SD: standard deviation; CoV: within‐subject coefficient of variation; ICC: intraclass correlation coefficient (two‐way mixed effects, absolute agreement, single measurement); DTI: diffusion tensor imaging; MD: mean diffusivity; FA: fractional anisotropy; IVIM: intravoxel incoherent motion; D: diffusion coefficient; Fp: perfusion fraction; FAIR: flow alternating inversion recovery; ATT: arterial transit time; DCE: dynamic contrast enhanced MRI; skGFR: single kidney glomerular filtration rate; MRT: mean residence time; PC: phase contrast; RBF: renal blood flow; *number of arteries.
RELAXOMETRY
All acquired T1 maps were included except for one examination which had to be excluded because of severe motion obscuring the kidneys (see Fig. S1 in the Supplementary Material). Furthermore, the T1 maps of the first four subjects were excluded since they were obtained with a slightly different imaging protocol.
Some R2* source images were affected by macroscopic susceptibility artifacts where air bubbles were present in stomach or intestines adjacent to the kidneys. Two R2* examinations had to be excluded because of severe (respiratory) motion artifacts (see Fig. S1 in the Supplementary Material).
The T2 images sometimes suffered from banding artifacts due to sensitivity of the T2 preparation to B0 inhomogeneities and insufficient fat suppression (for examples, see Figs. S1 and S2 in the Supplementary Material), which in one kidney led to exclusion of the examination. Representative acquired and processed relaxometry images from a single subject are shown in Fig. 1.
FIGURE 1.
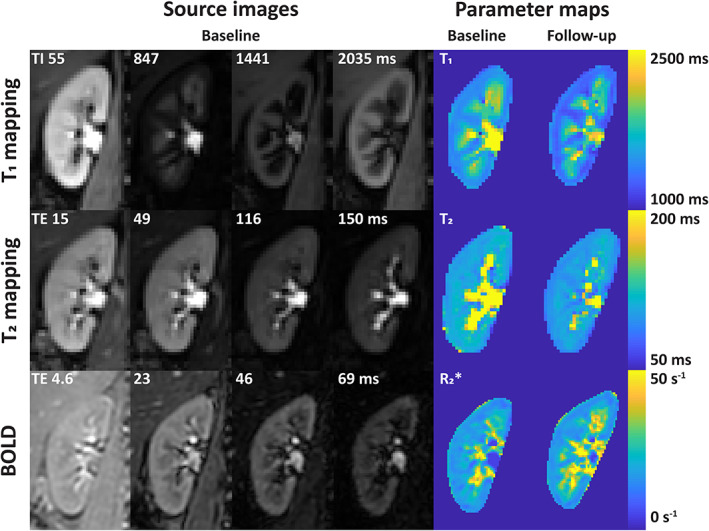
Source images and parameter maps of the relaxometry measures. BOLD: blood oxygen level‐dependent.
Bland–Altman plots and scatterplots for all relaxometry sequences are presented in Fig. 2. Repeatability was best for T2 mapping, with a CoV of 2.9%. For R2* mapping, the CoV was around 6% for cortex and medulla. For T1 mapping, despite low CoVs (5.1% and 2.8% for cortex and medulla, respectively), a small but systematic bias was present, which was more pronounced in the cortex compared to the medulla. This is evident in Fig. 2a–c.
FIGURE 2.
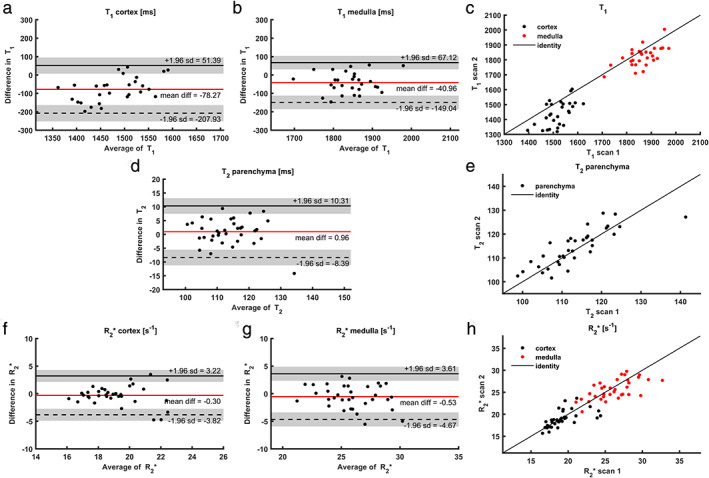
Bland–Altman plots and scatterplots for all relaxometry measures; (a) mean cortical T1; (b) mean medullary T1; (c) scatterplot for both cortical and medullary T1; (d) parenchymal T2; (e) scatterplot of parenchymal T2; (f) mean cortical R2*; (g) mean medullary R2*; (h) scatterplot for both cortical and medullary R2*. For larger image, see digital version.
DIFFUSION
Diffusion images were available in all but one subject, in whom DWI was not performed due to time constraints. In Fig. 3, source images and parameters maps of the diffusion images are shown for the same subject as in Fig. 1. Sometimes on the diffusion images the top of the kidneys were obscured by foldover artifacts which could not be sufficiently suppressed by saturation slabs.
FIGURE 3.
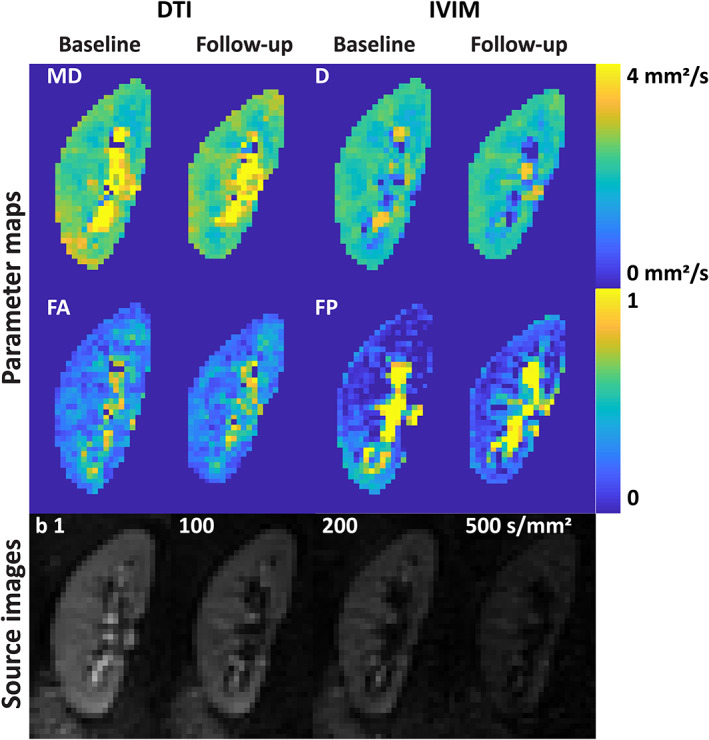
DTI and IVIM images. DTI: diffusion tensor imaging; IVIM: intravoxel incoherent motion; MD: mean diffusivity; D: diffusion coefficient; FA: focal anisotropy; FP: perfusion fraction.
Bland–Altman plots and corresponding scatterplots are depicted in Fig. 4. Regarding the diffusion constants, the CoV of mean diffusivity (MD, DTI analysis) was slightly lower than the apparent diffusion constant (IVIM analysis): 3.7% vs. 6.7% in the cortex and 2.8% vs. 5.2% in the medulla. FA had CoVs of 6.7% and 5.2% in cortex and medulla, respectively. Repeatability of the perfusion fraction (FP, IVIM analysis) was markedly less, with CoVs of 24% and 18% in cortex and medulla, respectively (Table 3).
FIGURE 4.
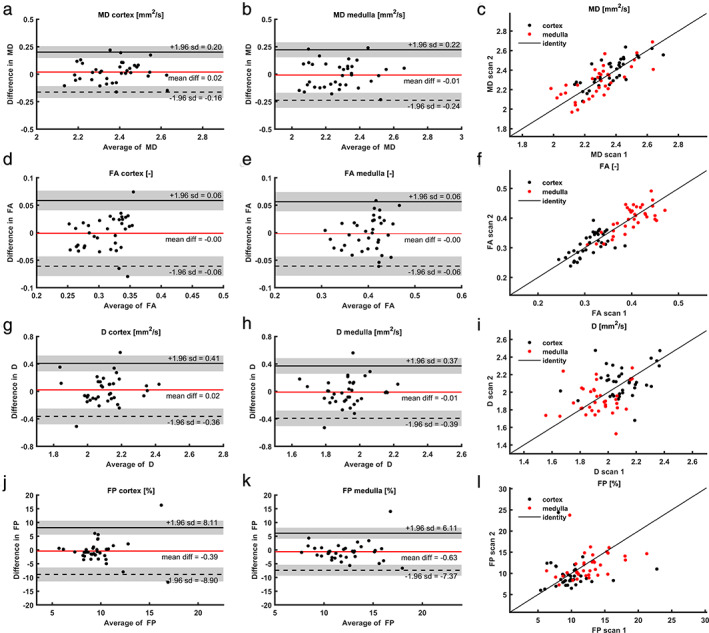
Bland–Altman plots and scatterplots for all DWI measures; (a) mean cortical MD (DTI); (b) mean medullary MD (DTI); (c) scatterplot for both cortical and medullary MD (DTI); (d) mean cortical FA (DTI); (e) mean medullary FA (DTI); (f) scatterplot for both cortical and medullary FA (DTI); (g) mean cortical D (IVIM); (h) mean medullary D (IVIM); (i) scatterplot for both cortical and medullary D (IVIM); (j) mean cortical FP (IVIM); (k) mean medullary FP (IVIM); (l) scatterplot for both cortical and medullary FP (IVIM) For larger image, see digital version. DTI: diffusion tensor imaging; IVIM: intravoxel incoherent motion; MD: mean diffusivity; D: diffusion coefficient; FA: focal anisotropy; FP: perfusion fraction.
PERFUSION AND GFR
For FAIR‐ASL and M0, which used the same readout, no datasets had to be excluded because of insufficient image quality. Artifacts (Fig. 5) were limited to geometrical distortion due to B0 inhomogeneities and susceptibility effects due to air in the lungs and digestive tract, which did not affect the perfusion quantification. In one subject, FAIR‐ASL could not be planned because the kidneys were located in the same coronal plane as the aorta. In the first four subjects FAIR‐ASL was performed with slightly different settings, and therefore they were excluded from the current analysis. This resulted in 13 complete datasets for FAIR ASL.
FIGURE 5.
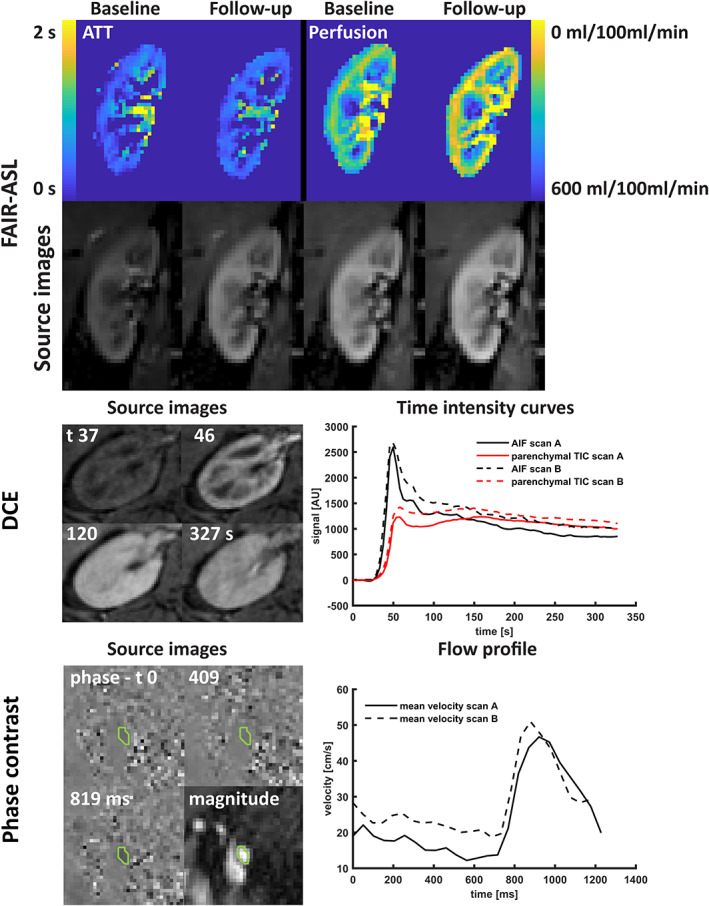
Top: Source images and parameter maps of arterial spin labeling; middle: transverse source images of DCE imaging at four timepoints (precontrast, cortical phase, medullary phase, and late phase) and the AIF and parenchymal TIC measured at both scan sessions; bottom: source images (phase and magnitude) of the phase contrast scan, including the region‐of‐interest and blood flow velocity over the cardiac cycle as measured during the first and second scan session. FAIR‐ASL: flow‐attenuated alternating inversion recovery arterial spin labeling; ATT: arterial transit time; DCE: dynamic contrast enhanced MRI; AIF: arterial input function; TIC: time intensity curve; AU: arbitrary units.
In some DCE images (Fig. 5) streaking artifacts originating from the radial acquisition were visible. In two subjects DCE MRI could not be performed due to an unavailable software license, in one subject the image SNR was too low for analysis, and in one subject there were problems with intravenous access, resulting in 15 complete datasets.
Although respiratory motion during the acquisition of 2DPC sometimes resulted in misalignment of the images, it did not affect image quality (Fig. 5). One subject had two renal arteries bilaterally and one subject had two left renal arteries. One subject was excluded due to failed cardiac synchronization, and two arteries were excluded due to erroneous planning (see Fig. S1 in the Supplemental Material). This resulted in 37 examined renal arteries from 18 subjects.
CoVs of all perfusion measures were generally lower compared to both relaxometry and diffusion analysis. Repeatability of perfusion obtained by ASL was better than DCE (10% vs. 17%), but the measures of transit time (ATT in ASL and MRT in DCE) were equally repeatable for both sequences (12%). Repeatability of renal blood flow, as measured as the total blood flow through the main renal artery by 2DPC, was comparable to that of the perfusion imaging methods with a CoV of 13%. For Bland–Altman plots and corresponding scatterplots, see Fig. 6.
FIGURE 6.
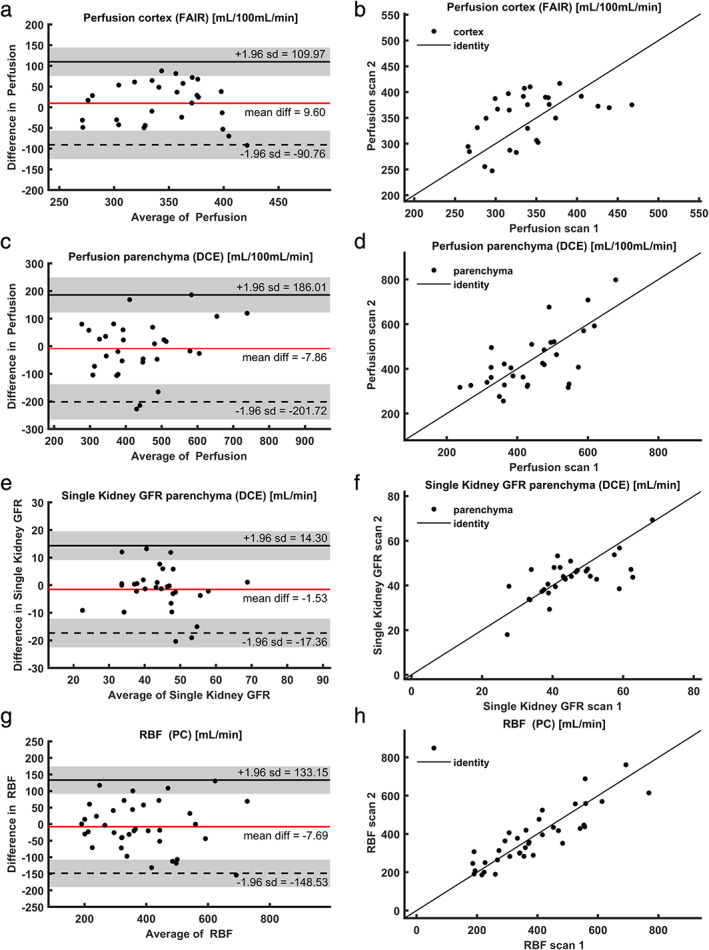
(a) Mean cortical perfusion (FAIR‐ASL); (b) scatterplot for cortical perfusion (FAIR‐ASL); (c) mean parenchymal perfusion (DCE); (d) scatterplot for parenchymal perfusion (DCE); (e) single kidney GFR (DCE); (f) scatterplot for single kidney GFR (DCE); (g) mean renal artery flow (phase contrast); (h) scatterplot for renal artery flow (phase contrast). FAIR: flow‐attenuated alternating inversion recovery; ASL: arterial spin labeling; DCE: dynamic contrast enhanced MRI; GFR: glomerular filtration rate; RBF: renal blood flow; PC: phase contrast.
FAT QUANTIFICATION
Acquisition of Dixon images succeeded in all subjects (Fig. 7a). The main artifacts included motion artifacts and water‐fat interference artifacts. Fat quantification in the renal parenchyma did not yield realistic values, ranging from below zero to more than 10% (for additional images, see Fig. S3 in the Supplementary Material). Repeatability of both the fat fraction and the total fat volume in the renal sinus was around 9% as assessed by the CoV. For Bland–Altman plots, see Fig. 7b–e.
FIGURE 7.
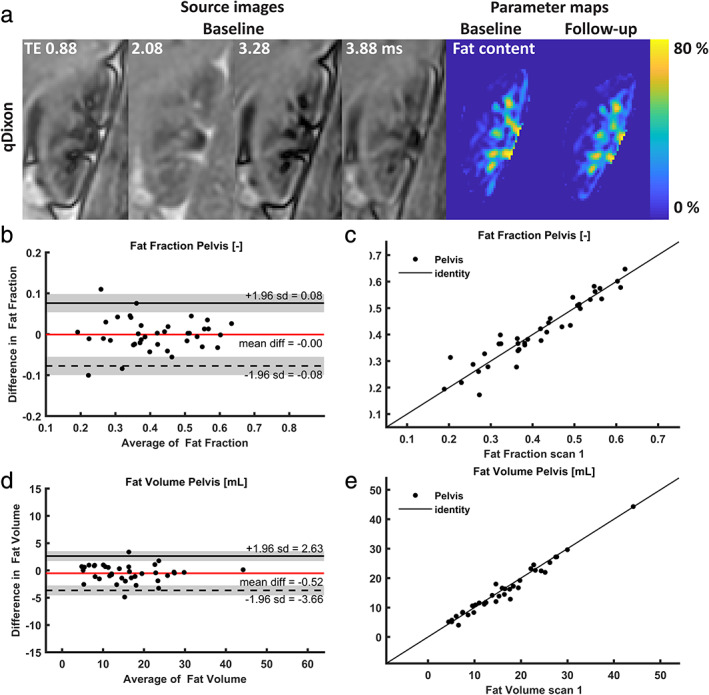
(a)Source images and parameter maps of the quantitative Dixon; (b–e) Bland–Altman plots and scatterplots for (b): pelvic fat fraction; (c) scatterplot for pelvic fat fraction; (d) pelvic fat volume; (e) scatterplot for pelvic fat volume. qDixon: quantitative Dixon.
Robustness
In 13 out of 19 subjects, all sequences could be acquired successfully when only considering patient‐related reasons for exclusion. Out of the six subjects where this was not the case, two had multiple scans affected by motion artifacts, in one patient FAIR‐ASL could not be acquired due to anatomy, in two subjects 2DPC acquisition failed due to erroneous planning, and in one patient 2DPC failed due to erroneous cardiac triggering. Missing data due to time constraints, absence of software keys, or acquisition with (slightly) different protocols were not considered patient‐related and were thus excluded in the evaluation of exam robustness.
Discussion
In this study the repeatability of various functional and structural MRI measurements of the kidney was assessed. Repeatability of most functional MRI measurements were in the range of other commonly used kidney function tests. For example, for a serum creatinine blood test a CoV of 5.8% has been reported (healthy subjects, month‐to‐month variability). 27 Inulin clearance by constant infusion, which is considered the gold standard for GFR measurement, has been reported to have a CoV of 11.3% under strictly controlled dietary and hydration conditions. 28
CoVs of more structural measures like T1, T2, and diffusion constants were generally lower (indicating better repeatability) than those of functional measures like R2* and perfusion. T1 and T2 both had an excellent repeatability, with CoVs below 5.1% for both cortex and medulla. Note that the CoV of 5.1% for cortical T1 was slightly higher than reported previously. 29 , 30 , 31 , 32 This can likely be attributed to the remaining contrast agent from the administration during the previous scan session, about a week before. 33 This caused a significant negative bias in the follow‐up measurement. For repeatability of renal T2 values, we could only find one preliminary report, which reported comparable repeatability. 9 R2* mapping or BOLD MRI is sensitive to renal oxygenation and therefore a more functional measure compared to the other relaxometry measurements. This is reflected by the slightly higher CoV, in line with previous reports. 29 , 30 , 31 , 34 , 35 , 36
Considering repeatability of the diffusion data, the DTI analysis yielded lower CoVs compared to the IVIM analysis. Repeatability of FP was poor, especially in the cortex, in line with previous reports. 29 , 37 , 38 Calculation of FP is based on a double exponential model with four free parameters and is therefore prone to fitting errors.
Perfusion measurements were the least reproducible, which likely reflects physiological fluctuations in renal perfusion. 39 Although ASL, DCE, and PC MRI all measure slightly different perfusion indices, they can all be used to monitor changes in perfusion over time, or, in research settings, between groups. Therefore, it is relevant to compare their performance. Overall, the perfusion values obtained agreed reasonably well. Cortical perfusion as measured with ASL (on average, 345 mL/100 mL/min) was slightly lower compared to perfusion as measured with DCE (439 mL/100 mL/min). Considering that renal parenchymal volume is around 150 mL, 40 the flow through the renal arteries (385 mL/min) seems relatively low. However, six of those arteries were one of a pair of renal arteries and small accessory renal arteries might have been present that were not detected on the vascular survey. In terms of repeatability, renal blood flow as measured with 2DPC MRI was relatively reliable, with a CoV of 13%. It must be noted that 2DPC is challenging to plan on the scanner, since the imaging slice has to be positioned exactly perpendicular to the artery.
Cortical perfusion measurement by FAIR‐ASL had the lowest CoV of all perfusion measurements: 10%, outperforming both phase contrast and DCE. Vascular transit times were equally repeatable when measured using FAIR‐ASL or DCE MRI. This confirms Cutajar et al, 41 who also reported lower CoVs for FAIR‐ASL compared to DCE (16% vs. 30%). A disadvantage of FAIR‐ASL, however, is that it cannot be performed on both kidneys simultaneously in a minority of subjects whose kidneys lie in the same plane as the aorta, since the aorta should not be included in the imaging plane. 42 In accordance with a recent recommendation article, medullary perfusion was not reported for FAIR‐ASL since it was deemed unreliable due to low SNR. 42
DCE perfusion measurements are heavily influenced by the first‐pass peak of the arterial input function (AIF), which probably explains the higher CoV compared to FAIR‐ASL. Accurate measurement of this peak is challenging due to the high contrast agent concentration resulting in saturation of the signal. Also, when the temporal resolution is too low, the peak can be missed. Considering this, it is surprising that with a temporal resolution of only 4.1 seconds we managed to obtain a CoV of only 16% for the perfusion (while reported CoVs range from 14–29.5% 11 , 37 , 41 , 43 , 44 ). This is comparable to Tofts et al 11 and Cutajar et al, 44 who reported CoVs of 14% and 17%, respectively, both with acquisitions with a temporal resolution of 2.5 seconds. The relatively good repeatability found in this study might be explained by the radial acquisition, which constantly samples the center of k‐space and therefore is less likely to miss the AIF peak. In Cartesian acquisitions, which are more commonly used, this is not the case. To avoid saturation of the signal, we injected the contrast agent (1 mL/s) relatively slowly, probably leading to a broader and lower AIF peak. Furthermore, we gave a half dose of contrast agent, but in future studies this will be decreased to a quarter dose. Also, the two‐step fitting approach, where tubular parameters were fitted to the second part of the time–intensity curve and vascular parameters like perfusion to the first part, might have contributed to the better CoV compared to other reports.
Single kidney GFR as measured by DCE had a CoV of 13%, which is slightly higher than the CoV of inulin clearance (11.3% 28 ) and lower than that of other reports on DCE MRI. 37 , 43 Also for tubular flow, which is multiplied by cortical volume to obtain (sk)GFR, higher CoVs are generally reported (15–18%). 11 , 43 , 44 Again, the two‐step fitting approach might explain the relatively good repeatability. This approach was already suggested by Tofts et al, 11 but has not yet been generally adopted. Its effectiveness should be confirmed in further analyses.
Although repeatability of the DCE measures was relatively good compared to other studies, we noted that the temporal regularization applied in the compressed sensing reconstruction affected the shape of the time–intensity curves, leading to flattening of both the AIF and the parenchymal first‐pass peak. In principle, if the temporal regularization on both uptake curves can be described by the same fixed temporal kernel that is weighted equally, it would cancel out. However, in the compressed sensing reconstruction the relative weight of temporal regularization can vary over the uptake curve, depending on the magnitude of the data mismatch term. Thus, it likely will affect the perfusion and filtration quantification. When we compared perfusion and GFR values obtained with and without compressed sensing, we found ~10% lower perfusion values and no effect on GFR with compressed sensing. Indeed, the temporal smoothing in compressed sensing mainly affects the AIF and the first pass in tissue which determines perfusion.
Measurement of renal sinus fat fraction and fat volume yielded CoVs of around 9%. We could not find reports of repeatability of quantitative Dixon for renal fat quantification. Fat quantification of the renal parenchyma using this approach proved to be impossible, yielding values ranging from negative to >10%, which is unrealistic. The unrealistic values seemed to be caused by a combination of low fat content and the water–fat interference on the border of the kidney (see Fig. S3 in the Supplemental Material). Furthermore, the quantitative Dixon acquisition that was used in the current study has been developed for liver fat quantification, and is therefore based on the liver fat spectrum. Ideally, a renal fat spectrum should be used for the quantification, although its influence on the fat quantification is expected to be limited.
Recently, recommendation articles have been published for renal BOLD, ASL, DWI, and relaxometry. 42 , 45 , 46 , 47 Despite the current study being finished before publication of these articles, acquisition and processing was largely performed in accordance with those recommendations. The main deviation from the recommendations is the T1 mapping, which was performed using inversion recovery with a cycled multislice 2D readout, in contrast to the recommended modified Look–Locker inversion (MOLLI) scheme. 47 However, in the recommendation article it was noted that a MOLLI scheme is not developed for renal imaging and has limited spatial resolution. Furthermore, with a MOLLI scheme an apparent T1 is measured, which is not the case for our approach. For FAIR‐ASL, instead of a spin‐echo EPI readout, a gradient echo EPI readout was used, which was in our experience is less affected by artifacts. 42 For BOLD MRI, fasting was recommended, which was not performed in this study. 45
Most subjects were capable of paced breathing during the ASL and T1 acquisitions. One subject fell asleep during the scan and could not easily be awakened, resulting in severe motion artifacts on the breath‐hold scans. While motion artifacts were absent on all free‐breathing acquisitions, breath‐hold acquisitions were regularly affected by either motion or inconsistent breath‐holds, resulting in incomplete multiparametric datasets in two subjects. This was even the case while breath‐hold duration was less than 10 seconds. In patients, breath‐holding can be even more problematic. Therefore, robust, free‐breathing acquisitions are preferred. Alternatively, radial acquisitions can be employed as we did for the DCE MRI. Motion artifacts in the DCE datasets, which were acquired in free breathing with radial sampling, were virtually absent. 2DPC was susceptible to acquisition errors, which led to incomplete multiparametric datasets in three subjects. This was partly due to the learning curve of the operators, illustrated by the fact that 2DPC acquisition in one of both renal arteries failed in the first two subjects.
Limitations
A limitation of the study is that blood samples were only taken during one visit. Therefore, we could not correct for physiological variation in kidney function. Diet and water intake were loosely controlled to be within normal ranges, but exercise and smoking were not. Therefore, the physiological state of subjects might have differed between the scans. However, the influence of those factors on measurement of MRI parameters is uncertain and normal hydration status was specifically recommended for BOLD, DWI, ASL, and relaxometry. 42 , 45 , 46 , 47 Another limitation is the relatively short time between the scan sessions (4–14 days, median 7). Based on the pharmacokinetics reported in the summary of product characteristics 48 we expected the contrast agent to have already been eliminated during the second scan session. Surprisingly, the second T1 measurement both in cortex and medulla was significantly lower, presumably due to remaining contrast agent. 33 Based on the measured T1 difference we can conclude that the contrast agent concentration during the second scan session (~4 nmol/g–8 nmol/g tissue) was too low to affect other measurements. Other measurements indeed did not show significant bias between the first and the second imaging session. ASL and DCE perfusion measurements, which directly depend on T1, were corrected for the T1 measured during the same session. Therefore, we expect the influence of the remaining contrast agent to be limited to the T1 measurements.
Conclusion
Various measures of renal structure and function were obtained within an acceptable acquisition time of 1 hour. Repeatability of all measures except for the perfusion fraction of the IVIM analysis was comparable with other tests of renal function. Furthermore, we compared various perfusion measurements in terms of repeatability and can conclude that either FAIR‐ASL or 2DPC can be used for renal perfusion measurements, while DCE is less reliable for perfusion analysis.
Supporting information
Appendix S1 Supporting Information.
Acknowledgment
The authors thank Tobias T. Pieters for his assistence with the data acquisition.
Contract grant sponsor: Anneloes de Boer was supported by an Alexandre Suerman stipend granted to MD‐PhD students by the University Medical Center Utrecht, the Netherlands. Clemens Bos, Suzanne L. Franklin, and Anita A. Harteveld acknowledge funding from the Netherlands Organization for Scientific Research (14951).
REFERENCES
- 1. Selby NM, Blankestijn PJ, Boor P, et al. Magnetic resonance imaging biomarkers for chronic kidney disease: A position paper from the European Cooperation in Science and Technology Action PARENCHIMA. Nephrol Dial Transplant 2018;33:ii4‐ii14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Friedli I, Crowe LA, Berchtold L, et al. New magnetic resonance imaging index for renal fibrosis assessment: A comparison between diffusion‐weighted imaging and T1 mapping with histological validation. Sci Rep 2016;6:30088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Caroli A, Schneider M, Friedli I, et al. Diffusion‐weighted magnetic resonance imaging to assess diffuse renal pathology: A systematic review and statement paper. Nephrol Dial Transplant 2018;33:ii29‐ii40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Spieker M, Katsianos E, Gastl M, et al. T2 mapping cardiovascular magnetic resonance identifies the presence of myocardial inflammation in patients with dilated cardiomyopathy as compared to endomyocardial biopsy. Eur Heart J Cardiovasc Imaging 2018;19:574‐582. [DOI] [PubMed] [Google Scholar]
- 5. Di Cesare E. MRI of the cardiomyopathies. Eur J Radiol 2001;38:179‐184. [DOI] [PubMed] [Google Scholar]
- 6. Odudu A, Nery F, Harteveld AA, et al. Arterial spin labelling MRI to measure renal perfusion: A systematic review and statement paper. Nephrol Dial Transplant 2018;33:ii15‐ii21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pruijm M, Mendichovszky IA, Liss P, et al. Renal blood oxygenation level‐dependent magnetic resonance imaging to measure renal tissue oxygenation: A statement paper and systematic review. Nephrol Dial Transplant 2018;33:ii22‐ii28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wolf M, de Boer A, Sharma K, et al. Magnetic resonance imaging T1‐ and T2‐mapping to assess renal structure and function: A systematic review and statement paper. Nephrol Dial Transplant 2018;33:ii41‐ii50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang JL, Storey P, Rusinek H, et al. Reproducibility of R2* and R2 measurements in human kidneys. In: Proceedings of the 19th Annual Meeting of ISMRM, Montréal; 2011 (abstract 2954).
- 10. Spit KA, Muskiet MHA, Tonneijck L, et al. Renal sinus fat and renal hemodynamics: A cross‐sectional analysis. Magn Reson Mater Phy 2020;33:73‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tofts PS, Cutajar M, Mendichovszky IA, Peters AM, Gordon I. Precise measurement of renal filtration and vascular parameters using a two‐compartment model for dynamic contrast‐enhanced MRI of the kidney gives realistic normal values. Eur Radiol 2012;22:1320‐1330. [DOI] [PubMed] [Google Scholar]
- 12. Inker LA, Eckfeldt J, Levey AS, et al. Expressing the CKD‐EPI (chronic kidney disease epidemiology collaboration) cystatin C equations for estimating GFR with standardized serum cystatin C values. Am J Kidney Dis 2011;58:682‐684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harteveld AA, de Boer A, Franklin SL, Leiner T, van Stralen M, Bos C. Comparison of multi‐delay FAIR and pCASL labeling approaches for renal perfusion quantification at 3T MRI. Magn Reson Mater Phy 2020;33:81‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cox RW, Ashburner J, Breman H, et al. A (sort of) new image data format standard: NiFTI‐1. In: Proceedings of the 10th Annual Meeting Of Organisation of Human Brain Mapping, Budapest; 2004.
- 15. Stemkens B, Prins FM, Bruijnen T, et al. A dual‐purpose MRI acquisition to combine 4D‐MRI and dynamic contrast‐enhanced imaging for abdominal radiotherapy planning. Phys Med Biol 2019;64:06nt02. [DOI] [PubMed] [Google Scholar]
- 16. Klein S, Staring M, Murphy K, Viergever MA, Pluim JP. Elastix: A toolbox for intensity‐based medical image registration. IEEE Trans Med Imaging 2010;29:196‐205. [DOI] [PubMed] [Google Scholar]
- 17. Shamonin DP, Bron EE, Lelieveldt BP, Smits M, Klein S, Staring M. Fast parallel image registration on CPU and GPU for diagnostic classification of Alzheimer's disease. Front Neuroinform 2013;7:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Boer A, Leiner T, Vink EE, Blankestijn PJ, van den Berg CAT. Modified Dixon‐based renal dynamic contrast‐enhanced MRI facilitates automated registration and perfusion analysis. Magn Reson Med 2018;80:66‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yushkevich PA, Piven J, Hazlett HC, et al. User‐guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 2006;31:1116‐1128. [DOI] [PubMed] [Google Scholar]
- 20. van Baalen S, Leemans A, Dik P, Lilien MR, Ten Haken B, Froeling M. Intravoxel incoherent motion modeling in the kidneys: Comparison of mono‐, bi‐, and triexponential fit. J Magn Reson Imaging 2017;46:228‐239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Annet L, Hermoye L, Peeters F, Jamar F, Dehoux JP, Van Beers BE. Glomerular filtration rate: Assessment with dynamic contrast‐enhanced MRI and a cortical‐compartment model in the rabbit kidney. J Magn Reson Imaging 2004;20:843‐849. [DOI] [PubMed] [Google Scholar]
- 22. Sourbron SP, Michaely HJ, Reiser MF, Schoenberg SO. MRI‐measurement of perfusion and glomerular filtration in the human kidney with a separable compartment model. Invest Radiol 2008;43:40‐48. [DOI] [PubMed] [Google Scholar]
- 23. Kukuk GM, Hittatiya K, Sprinkart AM, et al. Comparison between modified Dixon MRI techniques, MR spectroscopic relaxometry, and different histologic quantification methods in the assessment of hepatic steatosis. Eur Radiol 2015;25:2869‐2879. [DOI] [PubMed] [Google Scholar]
- 24. Hyslop NP, White WH. Estimating precision using duplicate measurements. J Air Waste Manag Assoc 2009;59:1032‐1039. [DOI] [PubMed] [Google Scholar]
- 25. Bland MJ , Altman D. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986;327:307–310. [PubMed] [Google Scholar]
- 26. R Development Core Team . R: A language and environment for statistical computing. R foundation for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2018. [Google Scholar]
- 27. Toffaletti JG, McDonnell EH. Variation of serum creatinine, cystatin C, and creatinine clearance tests in persons with normal renal function. Clin Chim Acta 2008;395:115‐119. [DOI] [PubMed] [Google Scholar]
- 28. Florijn KW, Barendregt JN, Lentjes EG, et al. Glomerular filtration rate measurement by "single‐shot" injection of inulin. Kidney Int 1994;46:252‐259. [DOI] [PubMed] [Google Scholar]
- 29. Cox EF, Buchanan CE, Bradley CR, et al. Multiparametric renal magnetic resonance imaging: Validation, interventions, and alterations in chronic kidney disease. Front Physiol 2017;8:696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Buchanan CE, Mahmoud H, Cox EF, et al. Quantitative assessment of renal structural and functional changes in chronic kidney disease using multi‐parametric magnetic resonance imaging. Nephrol Dial Transplant 2019; in press. 10.1093/ndt/gfz129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bane O, Hectors SJ, Gordic S, et al. Multiparametric magnetic resonance imaging shows promising results to assess renal transplant dysfunction with fibrosis. Kidney Int 2020;97:414‐420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dekkers IA, Paiman EHM, de Vries APJ, Lamb HJ. Reproducibility of native T1 mapping for renal tissue characterization at 3T. J Magn Reson Imaging 2019;49:588‐596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. de Boer A, Harteveld AA, Pieters TT, et al. Decreased native renal T1 up to one week after gadobutrol administration in healthy volunteers. J Magn Reson Imaging 2019; in press. 10.1002/jmri.27014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Simon‐Zoula SC, Hofmann L, Giger A, et al. Non‐invasive monitoring of renal oxygenation using BOLD‐MRI: A reproducibility study. NMR Biomed 2006;19:84‐89. [DOI] [PubMed] [Google Scholar]
- 35. Khatir DS, Pedersen M, Jespersen B, Buus NH. Reproducibility of MRI renal artery blood flow and BOLD measurements in patients with chronic kidney disease and healthy controls. J Magn Reson Imaging 2014;40:1091‐1098. [DOI] [PubMed] [Google Scholar]
- 36. Kline TL, Edwards ME, Garg I, et al. Quantitative MRI of kidneys in renal disease. Abdom Radiol (NY) 2018;43:629‐638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bane O, Wagner M, Zhang JL, et al. Assessment of renal function using intravoxel incoherent motion diffusion‐weighted imaging and dynamic contrast‐enhanced MRI. J Magn Reson Imaging 2016;44:317‐326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pan J, Zhang H, Man F, et al. Measurement and scan reproducibility of parameters of intravoxel incoherent motion in renal tumor and normal renal parenchyma: A preliminary research at 3.0 T MR. Abdom Radiol (NY) 2018;43:1739‐1748. [DOI] [PubMed] [Google Scholar]
- 39. Koopman MG, Koomen GC, Krediet RT, de Moor EA, Hoek FJ, Arisz L. Circadian rhythm of glomerular filtration rate in normal individuals. Clin Sci (Lond) 1989;77:105‐111. [DOI] [PubMed] [Google Scholar]
- 40. Johnson S, Rishi R, Andone A, et al. Determinants and functional significance of renal parenchymal volume in adults. Clin J Am Soc Nephrol 2011;6:70‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cutajar M, Thomas DL, Hales PW, Banks T, Clark CA, Gordon I. Comparison of ASL and DCE MRI for the non‐invasive measurement of renal blood flow: Quantification and reproducibility. Eur Radiol 2014;24:1300‐1308. [DOI] [PubMed] [Google Scholar]
- 42. Nery F, Buchanan CE, Harteveld AA, et al. Consensus‐based technical recommendations for clinical translation of renal ASL MRI. Magn Reson Mater Phy 2020;33:141‐161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eikefjord E, Andersen E, Hodneland E, et al. Dynamic contrast‐enhanced MRI measurement of renal function in healthy participants. Acta Radiol 2017;58:748‐757. [DOI] [PubMed] [Google Scholar]
- 44. Cutajar M, Mendichovszky IA, Tofts PS, Gordon I. The importance of AIF ROI selection in DCE‐MRI renography: Reproducibility and variability of renal perfusion and filtration. Eur J Radiol 2010;74:e154‐e160. [DOI] [PubMed] [Google Scholar]
- 45. Bane O, Mendichovszky IA, Milani B, et al. Consensus‐based technical recommendations for clinical translation of renal BOLD MRI. Magn Reson Mater Phy 2020;33:199‐215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ljimani A, Caroli A, Laustsen C, et al. Consensus‐based technical recommendations for clinical translation of renal diffusion‐weighted MRI. Magn Reson Mater Phy 2020;33:177‐195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dekkers IA, de Boer A, Sharma K, et al. Consensus‐based technical recommendations for clinical translation of renal T1 and T2 mapping MRI. Magn Reson Mater Phy 2020;33:163‐176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bayer Healthcare Pharmaceuticals . Highlights of prescribing information ‐ GADAVIST (gadobutrol) injection, for intravenous use. US Food and Drug Administration, 2011.
- 49. Ordidge RJ, Gibbs P, Chapman B, Stehling MK, Mansfield P. High‐speed multislice T1 mapping using inversion‐recovery echo‐planar imaging. Magn Reson Med 1990;16:238‐245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Supporting Information.


