Abstract
Background
Rett syndrome is a severe neurological disorder with a range of disabling autonomic and respiratory symptoms and resulting predominantly from variants in the methyl-CpG binding protein 2 gene on the long arm of the X-chromosome. As basic research begins to suggest potential treatments, sensitive measures of the dynamic phenotype are needed to evaluate the results of these research efforts. Here we test the hypothesis that the physiological fingerprint of Rett syndrome in a naturalistic environment differs from that of controls, and differs among genotypes within Rett syndrome.
Methods
A comprehensive array of heart rate variability, cardiorespiratory coupling and cardiac repolarisation measures were evaluated from an existing database of overnight and daytime inhome ambulatory recordings in 47 cases and matched controls.
Results
Differences between girls with Rett syndrome and matched controls were apparent in a range of autonomic measures, and suggest a shift towards sympathetic activation and/or parasympathetic inactivation. Daily temporal trends analysed in the context of circadian rhythms reveal alterations in amplitude and phase of diurnal patterns of autonomic balance. Further analysis by genotype class confirms a graded presentation of the Rett syndrome phenotype such that patients with early truncating mutations were most different from controls, while late truncating and missense mutations were least different from controls.
Conclusions
Comprehensive autonomic measures from extensive inhome physiological measurements can detect subtle variations in the phenotype of girls with Rett syndrome, suggesting these techniques are suitable for guiding novel therapies.
INTRODUCTION
Rett syndrome (RTT) is a neurological disorder associated with variants in the methyl-CpG binding protein 2 (MECP2) gene. Recent research in RTT has progressed rapidly,1–6 with increasing classification of MECP2 genotype-Rett phenotype relationships,7–10 advances in understanding cognitive processing,11 and hints at possible therapies. 1–5,12–15 A key feature of RTT is the dysregulation of autonomic circuits that may contribute to sudden death.16–22 Previous research has characterized heart rate variability (HRV),23,24 cardiac repolarisation measures,25,26 and cardiorespiratory coupling as measures of autonomic dysregulation. 18,19,23,27–29 However, many studies were conducted before MECP2 genotype information was available, and they were conducted in hospital settings, where the full range of autonomic behaviour may not be observable.26 Many studies also omitted sleep periods, and failed to analyse autonomic measures in fine temporal detail.
These abovementioned limitations notwithstanding, our research group has contributed two studies that continuously monitored electrocardiogram (ECG), respiratory inductance plethysmography (RIP) of chest and abdomen, and peripheral oxygen saturation (SpO2) in patients with RTT and controls.30,31 However, these characterisations did not include stratification by specific variants. To obtain a deeper understanding of autonomic dysfunction a more comprehensive analysis of autonomic functions was required in relation to the genotype, and with regard to HRV, cardiac repolarisation and cardiorespiratory coupling.
Here we reanalyse these data in the context of underlying MECP2 variants, and provide novel insights into HRV, cardiorespiratory coupling, cardiac repolarisation and circadian modulation.
METHODS
Demographics
The current study is a retrospective analysis of extant data that were the basis for two previous publications describing the respiratory and cardiorespiratory phenotype of girls with RTT during daytime30 and night-time31 ambulatory physiological monitoring, in which methodological details are provided. Here we summarise the methods that are particularly relevant to the current analyses. Girls with clinically identified RTT and genetic testing results indicating MECP2 mutation were identified and recruited for participation (referrals from the Rett Syndrome Research Foundation). Healthy, sex-matched, race-matched and age-matched young (2–7 years of age) female controls without a family history of related disorders were recruited among acquaintances of the other participants or staff. Physiological data from one overnight recording and two daytime recordings (2 hours each) were collected at home using a LifeShirt wearable physiology recording system (VivoMetrics, Ventura, California, USA; no longer in operation) which captured ECG, RIP and SpO2 data. Parents of all patients were encouraged to have their daughters sing along with a favourite video as a way to reproduce a waking respiratory pattern reminiscent of girls with RTT. Although we realise that breath-holds in RTT are behaviorally different from singing, we chose singing, because vocalisations involve a respiratory behaviour that like the breath-holds in RTT is repetitive and occurs during a respiratory phase that is generally referred to as ‘postinspiration’.32,33 All recordings were reviewed using a custom program in MATLAB (MathWorks, Natick, Massachusetts, USA) to develop semiautomated methods in order to identify artefacts, respiratory events and ECG R-peaks. Figure 1 summarises the signal processing and analytical methods used in this study.
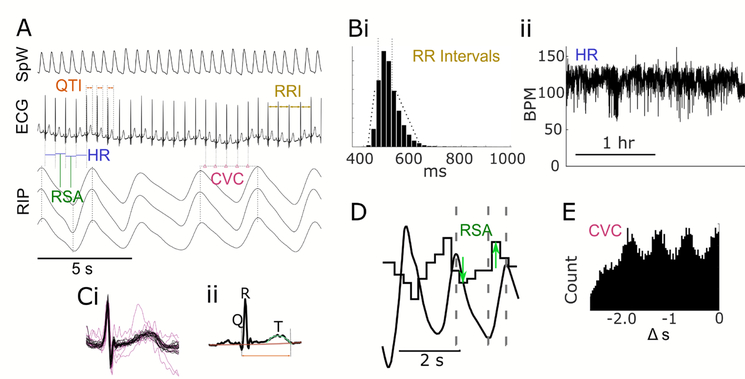
Figure 1.
Illustration of waveforms and analysis techniques. Example waveforms in A include photoplethysmography pulse (SpW), ECG, and respiratory inductance plethysmography (RIP) with schematic representation of derived metrics: QTI (QT interval), R-RI (R-R-interval), HR (heart rate), RSA (respiratory sinus arrhythmia) and CVC (cardioventilatory coupling). HR variability metrics are calculated from distributions of R-RI s (Bi) for time domain, and interpolated heart rate in beats per minute (BPM; Bii), from which frequency domain measures are derived with Fourier analysis. QTI measures are based on automatic artefact rejection (Ci) and averaging, after which waveform morphology metrics are derived (Cii). RSA is calculated as the breath-to-breath difference between the longest R-RI during expiration and the shortest during inspiration (D). CVC is characterised by the Shannon entropy of the distribution of R-wave times preceding inspiratory onset (E).
Patient and public involvement statement
The study was funded by the Rett Syndrome Research Trust, which maintains a close working relationship with patients with RTT and their families.
HRV measures
A range of HRV metrics was calculated from the R-R intervals derived from the ECG. These time domain measures were calculated from 5 min windows for the entirety of the recordings (excluding artefact epochs): heart rate (HR); SD of normal-to-normal intervals (SDNN); the root mean square of successive R-R interval differences (RMSSD); the percentage of successive interval differences greater than 50 ms (pNN50); the triangle index with 8 ms histogram bins (TriInd); and the non-linear sample entropy measure with a two-sample match window and a noise threshold of 0.2 of the sample SD (SampEn). In general SDNN and TriInd are associated with overall HR variability, while RMSSD and pNN50 are associated with rapid variation in HR. SampEn was designed to measure signal complexity from small data sets. In addition, frequency domain measures were calculated from the HR function interpolated from reciprocal R-R intervals at a sample rate of 10 Hz. These were: very low frequency power (VLF; 0.003 to 0.04 Hz); low frequency power (LF; 0.04 to 0.15Hz); high frequency power (HF; 0.15 to 0.6 Hz); broadband power (total; 0.003 to 0.6 Hz); normalised VLF power (VLFN); normalised LF power (LFN); normalised HF power (HFN); and the ratio of LF to HF power (LF:HF). Frequency domain measures are interpreted in the context of slow physiological or circadian regulation (VLF), combined sympathetic and parasympathetic tone (LF) and parasympathetic drive (HF).
Normalised frequency domain measures are designed to isolate sympathetic (LFN and LF:HF) versus parasympathetic components of autonomic regulation (HFN). Data were aggregated in two ways for statistical analysis. Temporal trends over 24 hours were compared between cases and controls in each 1-hour bin (permutation test, n=10 000, α=0.001), and averages of day and night measures were compared in aggregate (permutation, n=10 000, α=0.01). Because 5-min temporal windows are standard for HRV analysis, these metrics could not be calculated separately for regular breathing distinct from breath-holds (as done in our previous studies).30,31
Circadian analysis of HRV
The availability of daytime and night-time data, though not completely sampled across the 24-hour cycle, allowed analysis of circadian variation in the HRV measures. Cosinor analysis was used to find the modulation amplitude and peak phase time for each study participant in which there were more than 12-hour data coverage from all recordings for that child. Case/control comparisons of circadian modulation amplitude were evaluated with paired t-test at α=0.005, and phase differences were examined with a circular mean difference test (p<0.005).
QT interval analysis
QT intervals were calculated from the ECG waveform using peak-detection and automatic methods for rejection of artefact-contaminated waveforms. ECG waveforms were windowed around each detected QRS complex, and an average waveform was calculated from these. This template waveform was then subtracted from each captured QRST complex, and the variance of the residual values was calculated, with waveforms which exceeded a threshold excluded, leaving only those with normal morphology for further analysis. For each of these, the baseline drift was subtracted off using a third-order non-linear fit, and the QT interval was calculated as the difference between the local minimum of the Q wave negative deflection and the time of the T-wave returning to baseline. The beat-to-beat QT series was corrected for R-R interval using the Bazett formula and averaged in 5 min windows. As a check on the assumptions underlying the Bazett correction QRST complexes were averaged with corresponding R-R intervals ranging from 400 ms to 1200 ms in 500 ms bins. QT intervals calculated from these average waveforms were compared with the associated R-R interval bin for RTT and matched controls (CN) in each interval bin. These values were compared with 10 000 permutation resamples at a significance threshold of α=0.001.
Integrative measures
ECG and RIP signals were analysed in a set of beat-to-beat/breath-to-breath integrative measures. Respiratory sinus arrhythmia (RSA) was calculated as the difference of the maximal R-R interval during expiration and minimal R-R interval during inspiration. In this way, RSA was evaluated for every recorded breath in the data set. These were then averaged in 5-min intervals for analysis of 24-hour patterns. Cardiorespiratory coupling was calculated by identifying the temporal relation between inspiratory onsets and R-peaks, which were binned in 20 bins (ranging from 0 s to 3 s offset from the onset), from which Shannon entropy was calculated. Entropy values for 5 min epochs were averaged over time for all participants, again producing time series for further analysis.
Data availability
Reducing barriers to data sharing and reuse is essential to improving the robustness of scientific findings. Nonetheless, specific genotype information in this rare disease could allow reidentification of individual participants who have not given explicit consent for such use. For this reason, data will be shared with any qualified researcher on request only to the extent that it is ethically and legally permissible.
RESULTS
Time domain measures
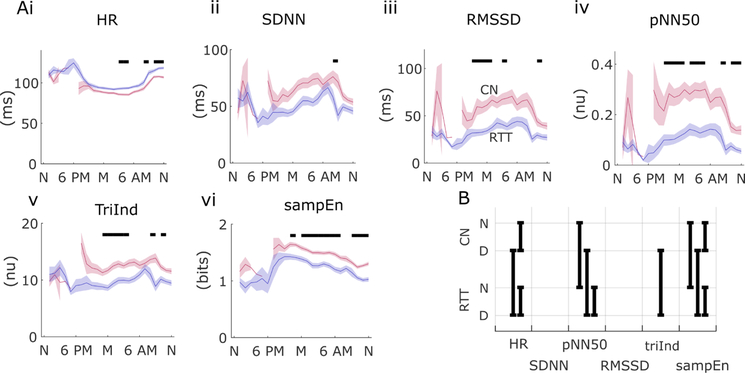
Time domain HRV measures confirmed a pattern of reduced HRV in RTT with respect to controls over a range of several measures. HR itself showed a pattern of significant differences from controls during early morning hours (in overnight recordings) and early hours of daytime recordings (before noon) (figure 2Ai). Aggregate comparisons for day and night recordings were significant for RTT versus CN during the daytime (and for day/night comparisons in both groups) (figure 2B). The SDNN measure was significant in the 24-hour analysis for only 1 hour during the day, whereas RMSSD, pNN50, TriInd and sampEn (figure 2Aii–2Avi) all exhibited a pattern of reduced variability in RTT during evening, overnight and early morning hours (hour-by-hour comparisons, p<0.001). In aggregated comparisons (figure 2B), pNN50 and sampEn were significantly different for day and night recordings, while TriInd differed only between daytime measures. Day versus night differences were evident within groups in RTT for pNN50 and in both cases and controls in sampEn (p<0.01).
Figure 2.
Daytime and night-time variation in time domain heart rate variability in girls with Rett syndrome (RTT) (blue) and matched controls (CN) (red). Twenty-four-hour average values (lines) and standard error of the mean (SEM; shaded regions) are shown for heart rate (HR) (Ai), SD of normal-to-normal intervals (SDNN) (ii), root mean square of successive R-R interval differences (RMSSD) (iii), percentage of successive interval differences greater than 50 ms (pNN50) (iv), ratio of the R-R distribution width and height (TriInd) (v), and sample entropy (sampEn) (vi). Solid bars above indicate statistically significant differences during that 1-hour time bin (p<0.001, permutation test) comparing RTT to CN. (B) Statistically significant comparisons are shown as black bars for aggregate data between RTT and CN for day (D) and night (N) recordings for each of the time domain measures. For example, the left most column indicates day and night differences within case and control cohorts in HR along with between-cohort differences for daytime only.
Frequency domain measures
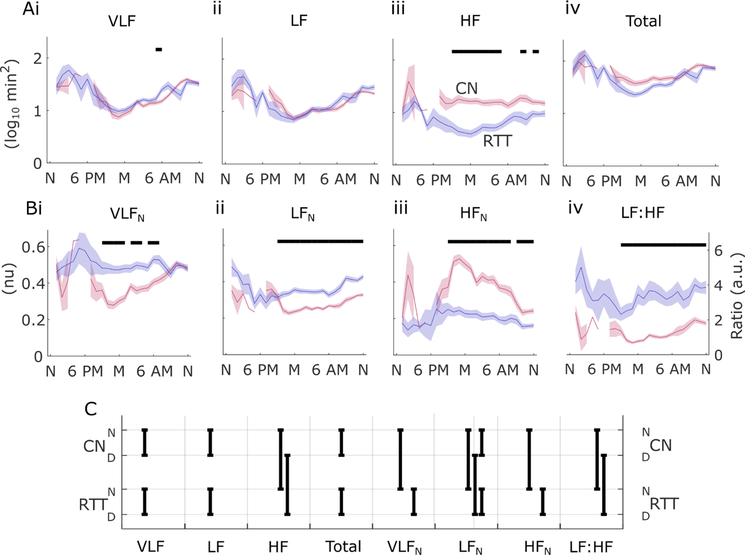
Unnormalised frequency domain measures were generally less sensitive to differences between RTT and controls than normalised versions in the same frequency bands (figure 3). Only HF power showed a consistent pattern of reduction in RTT in the 24-hour analysis, with most significant differences in late evening and overnight hours (figure 3Aiii). Normalised frequency domain analysis showed higher HRV levels for RTT versus controls in the VLFN and LFN bands, but a reversal in HFN range (figure 3Bi–iii). Similarly, the LF:HF ratio showed a consistent elevation in RTT suggesting a shift towards sympathetic drive and/or a withdrawal of parasympathetic influence on HRV (figure 3Biv; hour-by-hour comparisons, p<0.001). These patterns were recapitulated in the aggregate analysis, in which unnormalised HF power was significantly increased in RTT in day and night recordings, while normalised LF power was elevated in RTT during the day and night, normalised VLF and HF differences were significant (and reversed) in night-time recordings, and the LF:HF ratio was elevated in RTT during both day and night epochs (figure 3C; p<0.01).
Figure 3.
Twenty-four-hour temporal trends in frequency domain analysis of heart rate. Solid lines show average and shaded regions show SEM in 1-hour bins for log10 absolute power in very low frequency (VLF) (Ai), low frequency (LF) (ii), high frequency (HF) (iii) bands, as well as total power (iv) in all bands. Normalised power (B) is shown for VLF (Bi), LF (ii) and HF (iii), as well as the ratio of LF to HF power (iv). Significance is shown in black bars above (permutation test; p<0.001) comparing RTT to CN. (C) Statistically significant differences for aggregate data from day (D) and night (N) recordings in CN and RTT (permutation test; n=10000; p<0.01). CN, matched controls; HFN, normalised HF power; LFN, normalised LF power; RTT, Rett syndrome; VLFN, normalized VLF power.
Circadian HRV analysis
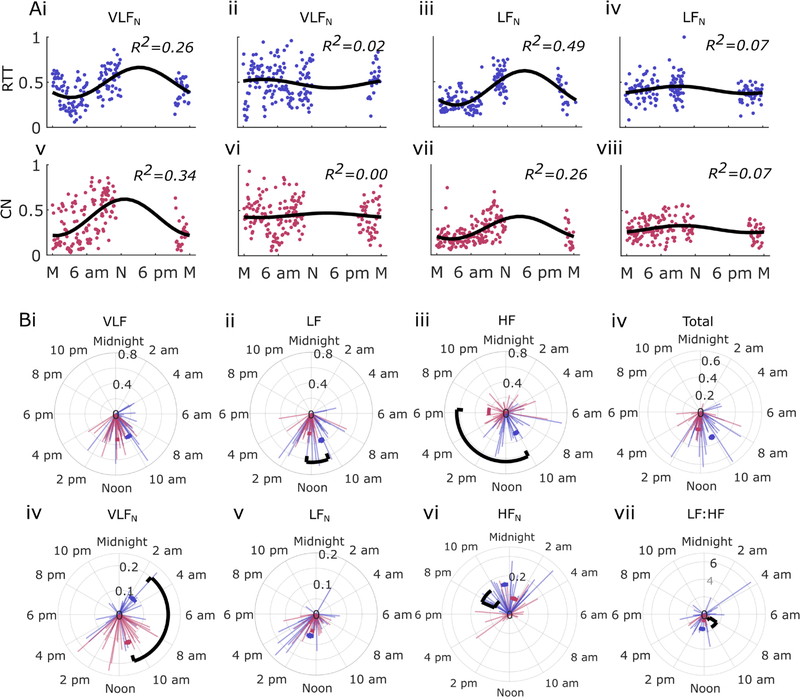
Circadian variation in HRV as assessed with cosinor analysis showed differences in several HRV measures, indicating alteration of both the acrophase (phase of peak amplitude) and the amplitude of 24-hour modulation of HRV between RTT and CN (figure 4). Examples of consinor fits in figure 4Ai–viii illustrate the typical range of goodness-of-fit, as measured by the R2 statistic. Circular statistics for acrophase differences were significant for absolute LF, HF and normalised VLF bands (figure 4B). Differences in the circadian modulation amplitude were noted in normalised HF power and LF:HF ratio. Three cases and three controls had inadequate coverage of the 24-hour period and were excluded from circadian analysis.
Figure 4.
Circadian analysis of HRV. A) Examples of cosinor fits for HRV variables in RTT (blue: Ai–iv) and CN (red: Av–viii). B) Cosinor amplitude and peak phase times shown in polar coordinates (lightly shaded radii are individual results) for frequency domain HRV variables. Mean values are shown as arcs in blue and red (as above), significant differences are indicated with black arcs (p<0.005). CN, matched controls; HF, high frequency; HFN, normalised HF power; HRV, heart rate variability; LF, low frequency; LFN, normalised LF power; RTT, Rett syndrome; VLF, very low frequency; VLFN, normalised VLF power.
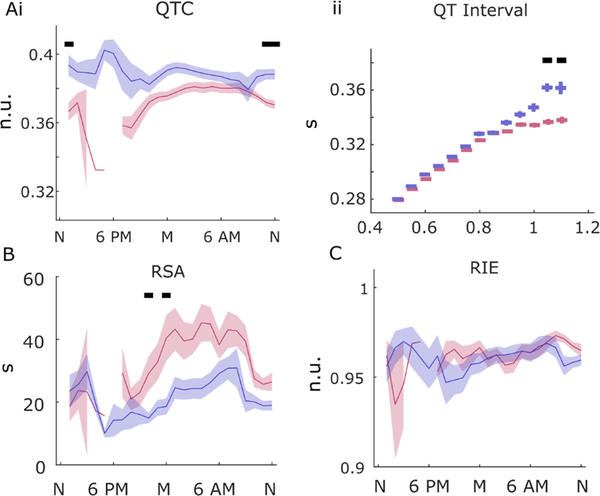
QT interval analysis
QT intervals derived from beat-to-beat analysis of QRS complex waveforms revealed differences between cases and controls both in circadian pattern and in the relationship of QT intervals to R-R intervals. Bazett-corrected QT intervals were longer in RTT than in CN around midday (p<0.001; Figure 5Ai). There was also a notable increase around 18:00 in RTT, but comparison with controls was impossible due to a lack of sampling during that time period. The relationship between R-R intervals and QT intervals was altered for long intervals (>1 s) compared with controls, who showed the expected curvilinear relationship (figure 5Aii).
Figure 5.
QT interval and coupling measures. (A) Corrected QT interval (QTc) (Ai), and QT/R-R (QT interval) (ii) relationship in RTT (blue) and matched controls (CN) (red), significant difference shown in black bars above (p<0.001). Respiratory sinus arrhythmia (RSA) (B) and cardioventilatory coupling (R-wave to inspiration entropy; RIE).
Cardiorespiratory coupling measures
RSA sporadically decreased in RTT in the 24-hour analysis late evening. RSA is thought to be a measure of parasympathetic function as evidenced in the modulation of heart rate by the respiratory cycle (figure 5C). The reverse relationship, as assessed by the cardioventilatory coupling measure RI-entropy (R-wave to inspiration entropy), was largely similar in RTT and CN, with little variation in the 24-hour cycle in either group (figure 5D; p<0.001).
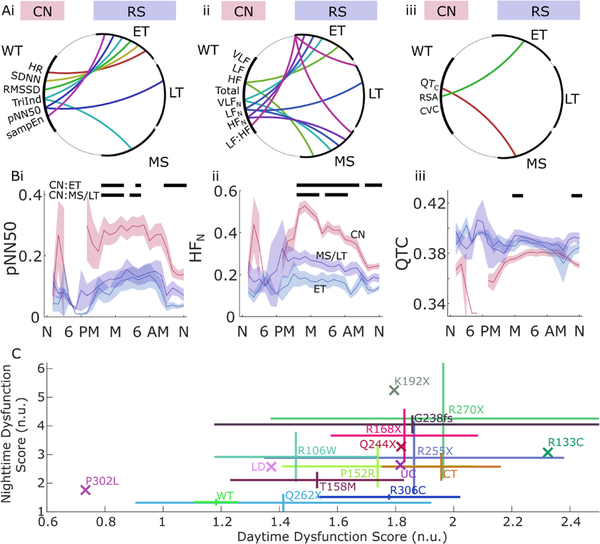
Genotype-phenotype relationships
Genotype-phenotype relationships were assessed for time domain, frequency domain and the supplemental metrics based on categorisation of MECP2 variants as early truncating (ET), late truncating (LT) and missense (MS).34 As expected, patterns of significant differences between groups in these measures (figure 6A) indicate a more robust difference between controls and patients with RTT with ET MECP2 mutations over a range of HRV measures. Differences between controls and RTT with MS mutations were somewhat less common, and only pNN50 and normalised LF power exhibited differences between controls and LT mutations in RTT. Only the LF:HF showed significant differences within RTT MECP2 variant subgroups (figure 6B). Twenty-four-hour temporal trends based on mutation types showed a graded phenotype between ET, combined MS and LT (MS/LT) and controls on some measures (eg, pNN50 and HFN; figure 6Bi–ii), but a less consistent pattern on others (QTC, figure 6Biii) (permutation test n=20 000, α=0.001). Dysfunction across the 17 HRV measures described above was analysed by normalising measures in RTT to those in controls based on the median and IQR of the control distribution for each metric. The absolute value of these normalised scores was averaged for specific mutation types (aside from C-terminal deletions, which were treated as one type, and an intronic mutation that was considered unclassified). For this analysis, the genotypes were: WT (wild type, n=46; one control did not have complete data for both day and night evalautions); C-terminal deletion (n=8); R168X (n=6); T158M (n=5); R106W (n=4); R255X (n=4); R306C (n=4); P152R (n=3); G238fs (n=2); Q262X (n=2); R270X (n=2); K192X, P302L, Q244X, R133C, a large deletion (LD) and and unclassified variant each with n=1 (1 RTT participant was excluded due to imcomplete day/night data). Note that because the absolute value is used on a per-metric basis to reconcile values that in RTT may be either above (as with HF HRV) or below (as with LFN) control values, the population average for controls does not tend towards zero (as it would by definition in a z-score normalisation). For this reason, in figure 6C, where these autonomic dysfunction scores are shown, controls (WT) have a mean score of 1.2 for daytime and 1.3 for night-time recordings. In this figure, each marker shows the average for all participants with that genotype, with the horizontal and vertical extent indicating the interindividual variability within that genotype (±SEM). For example, the R270X variant (which comprised two individuals in this cohort) shows relatively extreme divergence from control values on average along with considerable variability. In contrast, the R306C variant (n=4) was relatively homogeneous in night-time scores, though less so in daytime recordings. Statistical comparison of these mean values was not attempted at this level of detail as the sample size was considered insufficient to power those comparisons.
Figure 6.
Genotype-phenotype differences in several autonomic measures. Time domain HRV (Ai), frequency domain HRV (Aii) and addition measures (Aiii) for wild type (CN, red), early truncating (ET), late truncating (LT) and missense mutations (MS) in RTT (arcs indicate significant differences over day and night recordings tested by permutation tests at n=20 000 for α = 0.001). (B) Examples of genotype subtype analysis in three metrics over 24 hours: pNN50 (Bi), HFN (Bii), QTc (Biii). Black bars above show significant differences for each hour of the day in comparison with controls for ET and combined LT and MS variants (permutation test, n=20 000; α = 0.001). C) RTT-associated mutations plotted in relation to daytime and night-time autonomic dysfunction scores calculated from a composite of metrics (X indicates n=1 subject; others are shown as mean ±SEM; n.u., normalised units). CN, matched controls; CVC, cardioventilatory coupling; HF, high frequency; HFN, normalised HF power; HR, heart rate; HRV, heart rate variability; LF, low frequency; LFN, normalised LF power; pNN50, percentage of successive interval differences greater than 50 ms; RMSSD, root mean square of successive R-R interval differences; RSA, respiratory sinus arrhythmia; RTT, Rett syndrome; SampEN, sample entropy; SDNN, SD of normal-to-normal intervals; TriInd, triangle index; VLF, very low frequency; VLFN, normalised VLF power; WT, wild type.
DISCUSSION
For this study we analysed 1135 hours of inhome ambulatory data for evidence of changes in autonomic function among 47 young girls with MECP2 mutation-confirmed RTT and matched controls. This is the first explicit analysis of HRV over a 24-hour cycle in RTT, as well as the first examination of the results of extensive physiological measures in the context of genotype groupings during inhome ambulatory recordings. These results suggest that most HRV measures are depressed throughout the day/night cycle in RTT as compared with matched controls, with circadian variation largely following a similar pattern in both groups. Nonetheless, normalised HRV measures did indicate differences with controls that suggest that patients with RTT may maintain abnormally high sympathetic tone throughout the day, or that they may fail to exhibit adequate parasympathetic tone during the night (or a combination of both). Interpreted in the context of circadian modulation using cosinor analysis, there may be a phase shift in circadian drive for VLF and HF variability, possibly indicating a dysfunction in circadian autonomic coupling.
QT interval analysis did not reveal a clear circadian component that might help to explain the association between RTT and long QT syndromes. There was a notable increase in QTC during the early evening hours, but there were insufficient data in controls to compare during this time of day. It was notable that in patients with RTT the typical relationship between R-R and QT intervals seemed to be altered for long R-R intervals. The usual curvilinear reduction in QT interval with longer R-R intervals was missing in RTT, possibly suggesting some dysregulation of cardiac repolarisation at low heart rates.
RSA, an indication of respiratory cardiac coupling through the parasympathetic nervous system mediated by brainstem circuits,35 was only slightly reduced in RTT compared with controls. Nominally, the absence of a large reduction in this measure could indicate the shift in autonomic balance in RTT is more attributable to a gain in sympathetic activity with only a mild loss of parasympathetic tone, but frequency domain measures show a much more dramatic loss of presumptively parasympathetic HF variation. It is possible that RSA is less sensitive to autonomic changes present in RTT, but the fact that it is calculated directly from respiratory waveforms and R-R intervals would suggest greater physiological validity compared with HF HRV, which is calculated without respiratory signals within a presumptive respiratory frequency band.
Cardioventilatory coupling as measured through the temporal relationship between R waves and subsequent inspirations has been suggested as a directional estimate of coupling from heart rate to respiration (while RSA is considered to measure the complementary coupling from respiration to heart rate).36 There was little evidence in our data of a difference between RTT and CN on this measure.
Genotype/phenotype studies in RTT have indicated increased severity associated with some subclasses of MECP2 mutations associated with particular functional domains.37–40 For example, the genomic location of neutral and pathological variants has implicated the methyl-CpG binding domain as well as a regulatory co-repressor binding site, with one animal study suggesting that a truncated protein with little more than those domains alone can rescue the phenotype.41 In particular, patients with ET mutations have been shown to exhibit more severe clinical phenotypes.42–45 Our data indicate that HRV metrics are also sensitive to subtle phenotypical differences arising from different MECP2 variant subclasses. In matched analysis by genotype, differences in HRV measures between controls and RTT with ET mutations were the most prevalent, followed by LT and MS mutations. While these differences may be of little clinical utility in themselves—since genotype is readily and reliably determined independent of physiological measurement—the findings do suggest that HRV measures are sensitive enough to detect subtle physiological variation. Detecting such variation offers a biomarker for tracking effectiveness of clinical interventions as we move into an era of being able to treat RTT. In particular, the time domain measures (figure 2) and the normalised frequency domain measures (figure 3B) show the most discriminatory power between RTT and controls. Among the latter, HFN also discriminates mutation subtypes with some precision. Any or all of these could be useful phenotype outcomes for natural history studies or clinical trials, but time domain measures require less meticulous data cleaning, suggesting they are better suited for larger data sets.
Published analyses of variant-specific genotype-phenotype correlations have been somewhat equivocal overall. Such ambiguity is not unexpected given the diverse molecular effects of MECP2 variation, the potential for mosaicism and skewed X-inactivation,46,47 interactions with variants in other genes,48 and the crudeness of many of our phenotype metrics. Still, some genotype-phenotype correlations have been noted. R306C has been reported as connected to a uniquely mild clinical phenotype,49 while our data are suggestive of mild impairment only in the combined night-time autonomic metrics. Jian et al22 indicated that the R270X genotype might be associated with mortality, possibly explaining its lower prevalence among older patients. In our cohort, R270X variant phenotypes were varied, but did tend towards more severe impairment in both daytime and night-time recordings. Neul et al50 found R168X, LDs and R106W were associated with the most severe clinical impairment. Of these, those represented in our data set by more than one individual were R168X (relatively severely impacted in night-time recordings; less so during the day) and R106W (slightly milder for both study types). Neul et al also found milder clinical phenotypes with R306C, C-terminal truncations, R294X and R133C variants, of which in our data R306C was mildly affected during the night only, and C-terminal deletions showed a mixed pattern (more impaired during the day and milder at night).50 In general, our findings (figure 6C) showed a milder phenotype in MS as opposed to nonsense variants, with the exception of Q262X which was comparatively mild. In this sense our findings bear some similarity to transgenic mouse models in which preservation of two key regulatory regions was consistent with phenotype rescue. However, our finding of moderate dysfunction in C-terminal deletions would require some further explanation under this hypothesis.41 Considering the statistical limitations of our sample size, these results should be considered only suggestive, since other studies have found little correlation between specific variants and measures of clinical severity.10,51,52 For this reason, we are hesitant to present clinical recommendations from these results. Our findings should be considered in the context of other sources of information to inform clinical decision-making. Data aggregation across studies could help tease out subtle genotype-phenotype correlations in the future, but data sharing of historical data is hampered by a lack of specific consent for that use. As mentioned above, genotypes in this rare disorder are so unique that data sets containing them cannot be effectively de-identified.
Some additional limitations of the data set used in this study are evident. Clinical severity scoring was not available for this cohort (since data collection predates development of those scoring systems), so physiological and genomic data could not be compared with an objective clinical profile. In addition, though the cohort was relatively large for this rare condition, it may have been inadequate to detect some subtle physiological differences among MECP2 genotypes. It should also be noted that circadian analysis was not envisioned in the study design, so while we believe the analysis is sound under the constraints we imposed, full continuous 24-hour recording is preferred for this kind of analysis. Similarly, ambulatory QT analysis requires strict filtering and artefact rejection protocols, which can be effective but are certainly less accurate than traditional 12-lead ECG assessment for detecting cardiac repolarisation anomalies. In these two areas, as with the specific genotype analysis above, our findings should be considered as providing possible directions for future studies, rather than definitive results in themselves.
CONCLUSIONS
Here we show that comprehensive analysis of extensive physiological monitoring is sensitive enough to detect autonomic dysregulation, circadian variation and genotype-phenotype correlations in RTT. As research into RTT moves into a promising preclinical era, with encouraging results from animal models in which molecular therapies are capable of rescuing at least some deficits even in adult animals, precise physiological measures will be required to pinpoint treatment effects in humans.
Acknowledgments
Funding This work was supported by the Rett Syndrome Research Trust under the project title “Outlining the Autonomic Signature of Rett Syndrome.”
Footnotes
Competing interests None declared.
Patient consent for publication Not required.
Ethics approval The original data were collected under approval from the Rush University Medical CenterCentre Institutional Review Board with written parental consent (Office of Human Research Protections Registration Numbers #00000482; study approval: 02042902). The supplemental analyses presented here were approved by the Institutional Review Board of Ann & Robert H. Lurie Children’s Hospital of Chicago (Office of Human Research Protections Registration Numbers #00000624 and #00009723; study approval: 2019–3120).
Provenance and peer review Not commissioned; externally peer reviewed.
Data availability statement Data are available upon reasonable request. Specific genotype information in this rare disease could allow reidentification of individuals who have not given explicit consent for data sharing. For this reason, data will be shared with any qualified researcher on request only to the extent that it is ethically and legally permissible.
REFERENCES
- 1.Abdala AP, Bissonnette JM, Newman-Tancredi A. Pinpointing brainstem mechanisms responsible for autonomic dysfunction in Rett syndrome: therapeutic perspectives for 5-HT1A agonists. Front Physiol 2014;5:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katz DM, Bird A, Coenraads M, Gray SJ, Menon DU, Philpot BD, Tarquinio DC. Rett syndrome: crossing the threshold to clinical translation. Trends Neurosci 2016;39:100–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khwaja OS, Ho E, Barnes KV, O’Leary HM, Pereira LM, Finkelstein Y, Nelson CA, Vogel-Farley V, DeGregorio G, Holm IA, Khatwa U, Kapur K, Alexander ME, Finnegan DM, Cantwell NG, Walco AC, Rappaport L, Gregas M, Fichorova RN, Shannon MW, Sur M, Kaufmann WE. Safety, pharmacokinetics, and preliminary assessment of efficacy of mecasermin (recombinant human IGF-1) for the treatment of Rett syndrome. Proc Natl Acad Sci U S A 2014;111:4596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krajnc N Severe respiratory dysrhythmia in Rett syndrome treated with topiramate. J Child Neurol 2014;29:NP118–21. [DOI] [PubMed] [Google Scholar]
- 5.Smith-Hicks CL, Gupta S, Ewen JB, Hong M, Kratz L, Kelley R, Tierney E, Vaurio R, Bibat G, Sanyal A, Yenokyan G, Brereton N, Johnston MV, Naidu S. Randomized open-label trial of dextromethorphan in Rett syndrome. Neurology 2017;89:1684–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voituron N, Hilaire G. The benzodiazepine midazolam mitigates the breathing defects of Mecp2-deficient mice. Respir Physiol Neurobiol 2011;177:56–60. [DOI] [PubMed] [Google Scholar]
- 7.Bao X, Downs J, Wong K, Williams S, Leonard H. Using a large international sample to investigate epilepsy in Rett syndrome. Dev Med Child Neurol 2013;55:553–8. [DOI] [PubMed] [Google Scholar]
- 8.Bissonnette JM, Schaevitz LR, Knopp SJ, Zhou Z. Respiratory phenotypes are distinctly affected in mice with common Rett syndrome mutations MECP2 T158A and R168X. Neuroscience 2014;267:166–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah RR, Bird AP. MeCP2 mutations: progress towards understanding and treating Rett syndrome. Genome Med 2017;9:7–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huppke P, Held M, Hanefeld F, Engel W, Laccone F. Influence of mutation type and location on phenotype in 123 patients with Rett syndrome. Neuropediatrics 2002;33:63–8. [DOI] [PubMed] [Google Scholar]
- 11.Rose SA, Djukic A, Jankowski JJ, Feldman JF, Fishman I, Valicenti-Mcdermott M. Rett syndrome: an eye-tracking study of attention and recognition memory. Dev Med Child Neurol 2013;55:364–71. [DOI] [PubMed] [Google Scholar]
- 12.De Filippis B, Nativio P, Fabbri A, Ricceri L, Adriani W, Lacivita E, Leopoldo M, Passarelli F, Fuso A, Laviola G. Pharmacological stimulation of the brain serotonin receptor 7 as a novel therapeutic approach for Rett syndrome. Neuropsychopharmacology 2014;39:2506–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdala APL, Dutschmann M, Bissonnette JM, Paton JFR. Correction of respiratory disorders in a mouse model of Rett syndrome. Proc Natl Acad Sci U S A 2010;107:18208–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gökben S, Ardıç UA, Serdaroğlu G. Use of buspirone and fluoxetine for breathing problems in Rett syndrome. Pediatr Neurol 2012;46:192–4. [DOI] [PubMed] [Google Scholar]
- 15.Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science 2007;315:1143–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guideri F, Acampa M. Sudden death and cardiac arrhythmias in Rett syndrome. Pediatr Cardiol 2005;26:111. [DOI] [PubMed] [Google Scholar]
- 17.Madan N, Levine M, Pourmoghadam K, Sokoloski M. Severe sinus bradycardia in a patient with Rett syndrome: a new cause for a pause? Pediatr Cardiol 2004;25:53–5. [DOI] [PubMed] [Google Scholar]
- 18.Guideri F, Acampa M, Hayek C, Zappella M, Di Perri T. Reduced heart rate variability in patients affected with Rett syndrome. A possible explanation for sudden death. Neuropediatrics 1999;30:146–8. [DOI] [PubMed] [Google Scholar]
- 19.Guideri F, Acampa M, Hayek Y, Zappella M. Effects of acetyl-L-carnitine on cardiac dysautonomia in Rett syndrome: prevention of sudden death? Pediatr Cardiol 2005;26:574–7. [DOI] [PubMed] [Google Scholar]
- 20.Julu POO, Kerr AM, Hansen S, Apartopoulos F, Jamal GA. Immaturity of medullary cardiorespiratory neurones leading to inappropriate autonomic reactions as a likely cause of sudden death in Rett’s syndrome. Arch Dis Child 1997;77:463–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCauley MD, Wang T, Mike E, Herrera J, Beavers DL, Huang T-W, Ward CS, Skinner S, Percy AK, Glaze DG, Wehrens XHT, Neul JL. Pathogenesis of lethal cardiac arrhythmias in Mecp2 mutant mice: implication for therapy in Rett syndrome. Sci Transl Med 2011;3:113ra125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jian L, Archer HL, Ravine D, Kerr A, de Klerk N, Christodoulou J, Bailey MES, Laurvick C, Leonard H. p.R270X MECP2 mutation and mortality in Rett syndrome. Eur J Hum Genet 2005;13:1235–8. [DOI] [PubMed] [Google Scholar]
- 23.Guideri F, Acampa M, DiPerri T, Zappella M, Hayek Y. Progressive cardiac dysautonomia observed in patients affected by classic Rett syndrome and not in the preserved speech variant. J Child Neurol 2001;16:370–3. [DOI] [PubMed] [Google Scholar]
- 24.Glaze DG. Neurophysiology of Rett syndrome. Ment Retard Dev Disabil Res Rev 2002;8:66–71. [DOI] [PubMed] [Google Scholar]
- 25.Crosson J, Srivastava S, Bibat GM, Gupta S, Kantipuly A, Smith-H icks C, Myers SM, Sanyal A, Yenokyan G, Brenner J, Naidu SR. Evaluation of QTc in Rett syndrome: correlation with age, severity, and genotype. Am J Med Genet A 2017;173:1495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellaway CJ, Sholler G, Leonard H, Christodoulou J. Prolonged QT interval in Rett syndrome. Arch Dis Child 1999;80:470–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Julu POO, Witt Engerström I. Assessment of the maturity-related brainstem functions reveals the heterogeneous phenotypes and facilitates clinical management of Rett syndrome. Brain and Development 2005;27:S43–53. [DOI] [PubMed] [Google Scholar]
- 28.Bissonnette JM, Knopp SJ, Maylie J, Thong T. Autonomic cardiovascular control in methyl-C pG-binding protein 2 (Mecp2) deficient mice. Autonomic Neuroscience 2007;136:82–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guideri F, Acampa M, Blardi P, de Lalla A, Zappella M, Hayek Y. Cardiac dysautonomia and serotonin plasma levels in Rett syndrome. Neuropediatrics 2004;35:36–8. [DOI] [PubMed] [Google Scholar]
- 30.Weese-Mayer DE, Lieske SP, Boothby CM, Kenny AS, Bennett HL, Silvestri JM, Ramirez J-M. Autonomic nervous system dysregulation: breathing and heart rate perturbation during wakefulness in young girls with Rett syndrome. Pediatr Res 2006;60:443–9. [DOI] [PubMed] [Google Scholar]
- 31.Weese-Mayer DE, Lieske SP, Boothby CM, Kenny AS, Bennett HL, Ramirez J-M. Autonomic dysregulation in young girls with Rett syndrome during nighttime in-home recordings. Pediatr Pulmonol 2008;43:1045–60. [DOI] [PubMed] [Google Scholar]
- 32.Anderson TM, Garcia AJ, Baertsch NA, Pollak J, Bloom JC, Wei AD, Rai KG, Ramirez J-M. A novel excitatory network for the control of breathing. Nature 2016;536:76–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dutschmann M, Jones SE, Subramanian HH, Stanic D, Bautista TG. The physiological significance of postinspiration in respiratory control. Prog Brain Res 2014; 212:113–30. [DOI] [PubMed] [Google Scholar]
- 34.Charman T, Neilson TCS, Mash V, Archer H, Gardiner MT, Knudsen GPS, McDonnell A, Perry J, Whatley SD, Bunyan DJ, Ravn K, Mount RH, Hastings RP, Hulten M, Ørstavik KH, Reilly S, Cass H, Clarke A, Kerr AM, Bailey MES. Dimensional phenotypic analysis and functional categorisation of mutations reveal novel genotype–phenotype associations in Rett syndrome. Eur J Hum Genet 2005;13:1121–30. [DOI] [PubMed] [Google Scholar]
- 35.Dergacheva O, Griffioen KJ, Neff RA, Mendelowitz D. Respiratory modulation of premotor cardiac vagal neurons in the brainstem. Respir Physiol Neurobiol 2010;174:102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tzeng YC, Larsen PD, Galletly DC. Mechanism of cardioventilatory coupling: insights from cardiac pacing, vagotomy, and sinoaortic denervation in the anesthetized rat. Am J Physiol Heart Circ Physiol 2007;292:H1967–77. [DOI] [PubMed] [Google Scholar]
- 37.Bedogni F, Rossi RL, Galli F, Cobolli Gigli C, Gandaglia A, Kilstrup-N ielsen C, Landsberger N. Rett syndrome and the urge of novel approaches to study MeCP2 functions and mechanisms of action. Neurosci Biobehav Rev 2014;46:187–201. [DOI] [PubMed] [Google Scholar]
- 38.Ogier M, Katz DM. Breathing dysfunction in Rett syndrome: understanding epigenetic regulation of the respiratory network. Respir Physiol Neurobiol 2008;164:55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Halbach N, Smeets EE, Julu P, Witt-E ngerström I, Pini G, Bigoni S, Hansen S, Apartopoulos F, Delamont R, van Roozendaal K, Scusa MF, Borelli P, Candel M, Curfs L. Neurophysiology versus clinical genetics in Rett syndrome: a multicenter study. Am J Med Genet A 2016;170:2301–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoffbuhr K, Devaney JM, LaFleur B, Sirianni N, Scacheri C, Giron J, Schuette J, Innis J, Marino M, Philippart M, Narayanan V, Umansky R, Kronn D, Hoffman EP, Naidu S, Narayanan V. MeCP2 mutations in children with and without the phenotype of Rett syndrome. Neurology 2001;56:1486–95. [DOI] [PubMed] [Google Scholar]
- 41.Tillotson R, Selfridge J, Koerner MV, Gadalla KKE, Guy J, De Sousa D, Hector RD, Cobb SR, Bird A. Radically truncated MeCP2 rescues Rett syndrome-like neurological defects. Nature 2017;550:398–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zappella M, Meloni I, Longo I, Hayek G, Renieri A. Preserved speech variants of the Rett syndrome: molecular and clinical analysis. Am J Med Genet 2001;104:14–22. [DOI] [PubMed] [Google Scholar]
- 43.Mackay J, Downs J, Wong K, Heyworth J, Epstein A, Leonard H. Autonomic breathing abnormalities in Rett syndrome: caregiver perspectives in an international database study. J Neurodev Disord 2017;9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bebbington A, Percy A, Christodoulou J, Ravine D, Ho G, Jacoby P, Anderson A, Pineda M, Ben Zeev B, Bahi-Buisson N, Smeets E, Leonard H. Updating the profile of C-terminal MeCP2 deletions in Rett syndrome. J Med Genet 2010;47:242–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weaving LS, Williamson SL, Bennetts B, Davis M, Ellaway CJ, Leonard H, Thong M-K, Delatycki M, Thompson EM, Laing N, Christodoulou J. Effects ofMECP2 mutation type, location and X-inactivation in modulating Rett syndrome phenotype. Am J Med Genet 2003;118A:103–14. [DOI] [PubMed] [Google Scholar]
- 46.Amir RE, Van den Veyver IB, Schultz R, Malicki DM, Tran CQ, Dahle EJ, Philippi A, Timar L, Percy AK, Motil KJ, Lichtarge O, Smith EO, Glaze DG, Zoghbi HY. Influence of mutation type and X chromosome inactivation on Rett syndrome phenotypes. Ann Neurol 2000;47:670–9. [PubMed] [Google Scholar]
- 47.Huppke P, Maier EM, Warnke A, Brendel C, Laccone F, Gärtner J. Very mild cases of Rett syndrome with skewed X inactivation. J Med Genet 2006;43:814–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grillo E, Lo Rizzo C, Bianciardi L, Bizzarri V, Baldassarri M, Spiga O, Furini S, De Felice C, Signorini C, Leoncini S, Pecorelli A, Ciccoli L, Mencarelli MA, Hayek J, Meloni I, Ariani F, Mari F, Renieri A. Revealing the complexity of a monogenic disease: Rett syndrome exome sequencing. PLoS One 2013;8:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schanen C, Houwink EJF, Dorrani N, Lane J, Everett R, Feng A, Cantor RM, Percy A. Phenotypic manifestations of MECP2 mutations in classical and atypical Rett syndrome. Am J Med Genet 2004;126A:129–40. [DOI] [PubMed] [Google Scholar]
- 50.Neul JL, Fang P, Barrish J, Lane J, Caeg EB, Smith EO, Zoghbi H, Percy A, Glaze DG. Specific mutations in methyl-C pG-binding protein 2 confer different severity in Rett syndrome. Neurology 2008;70:1313–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.G iunti L, Pelagatti S, Lazzerini V, Guarducci S, Lapi E, Coviello S, Cecconi A, Ombroni L, Andreucci E, Sani I, Brusaferri A, Lasagni A, Ricotti G, Giometto B, Nicolao P, Gasparini P, Granatiero M, Giovannucci Uzielli ML. Spectrum and distribution of MeCP2 mutations in 64 Italian Rett syndrome girls: tentative genotype/phenotype correlation. Brain Dev 2001;23:S242–5. [DOI] [PubMed] [Google Scholar]
- 52.Monrós E, Armstrong J, Aibar E, Poo P, Canós I, Pineda M. Rett syndrome in Spain: mutation analysis and clinical correlations. Brain Dev 2001;23:S251–3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Reducing barriers to data sharing and reuse is essential to improving the robustness of scientific findings. Nonetheless, specific genotype information in this rare disease could allow reidentification of individual participants who have not given explicit consent for such use. For this reason, data will be shared with any qualified researcher on request only to the extent that it is ethically and legally permissible.