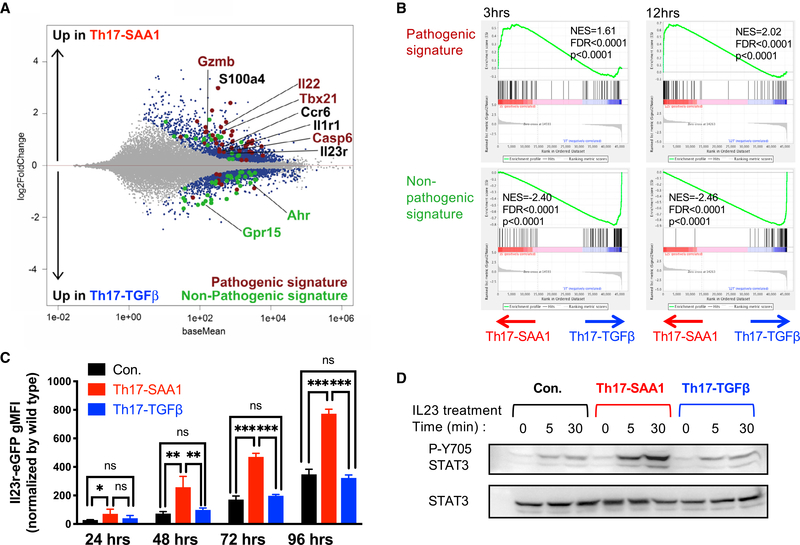
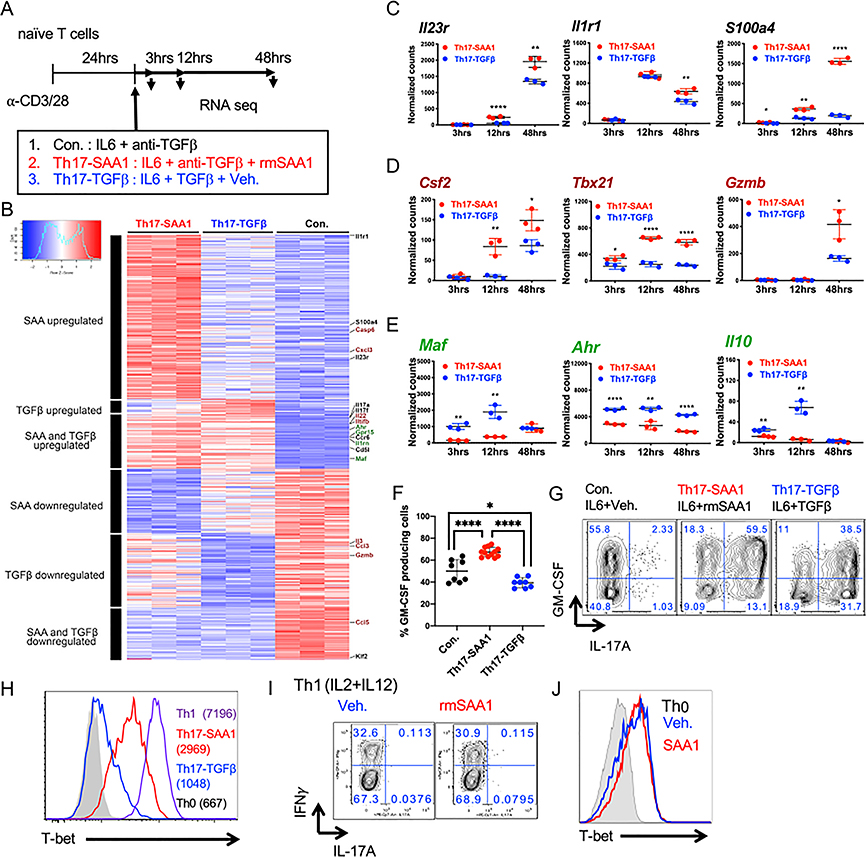
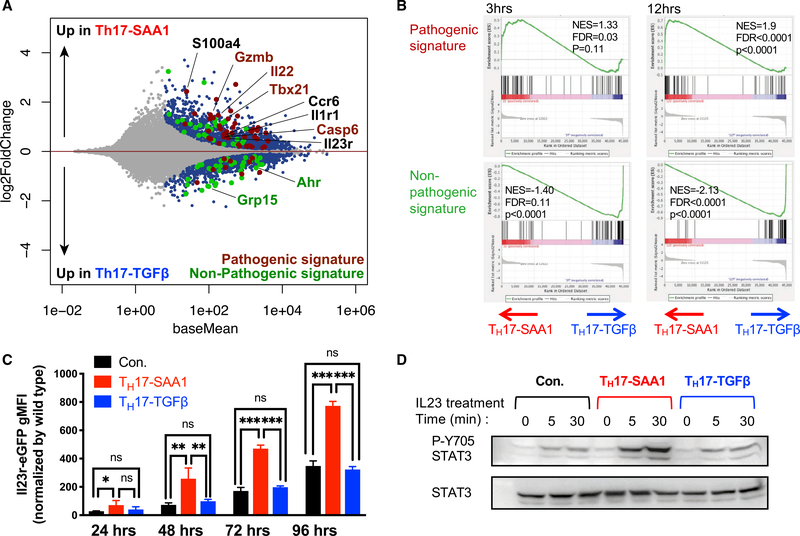
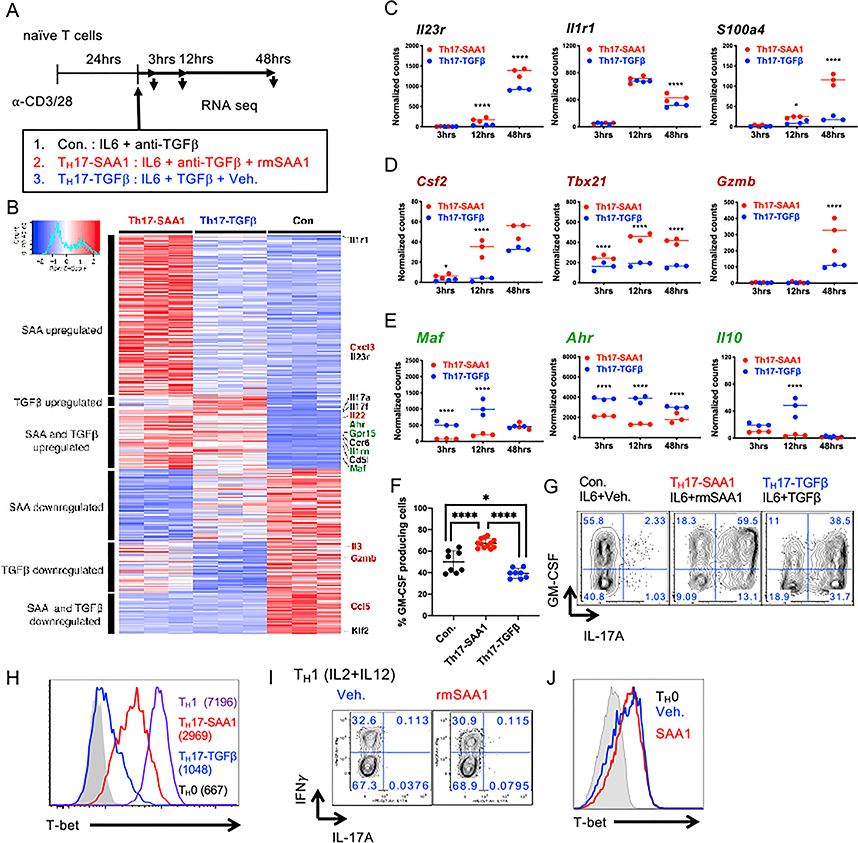
In the published article, there was an error in processing of the RNA-seq data. Multi-mapping and multi-overlapping reads were included in the raw counts, which were counted using the Subread package FeatureCounts with option –M and –O. This led to several errors in Figures 2 and S2. The data were re-analyzed using the STAR aligner with option –quantMode GeneCounts to count reads which overlapped only one gene. The correction led to several minor changes in the fold-change values and significance that resulted in slight alterations in the MA plot (Figure 2A), GSEA (Figure 2B), heatmap (Figure S2B), and normalized counts (Figure S2C–S2E). These figures have been corrected online. The correction does not affect any of the conclusions in the published paper. A more detailed methodology on the RNaseq processing has been added to the section “Quantification and Statistical Analysis” in STAR Methods in the online version. We apologize for this error.
Figure 2.
SAA1 Elicits a Pathogenic Th17 Program In Vitro (original)
Figure S2.
Figure S2. Transcriptional Responses and Validation of Differentially Expressed Genes in Th17 Cells Differentiated with IL6 plus SAA1 or TGF-β (original)
Figure 2.
SAA1 Elicits a Pathogenic Th17 Program In Vitro (corrected)
Figure S2.
Transcriptional Responses and Validation of Differentially Expressed Genes in Th17 Cells Differentiated with IL6 plus SAA1 or TGF-β (corrected)