Abstract
Esophageal squamous cell carcinoma (ESCC) is a common malignant tumor in the human digestive system, which affects the physical and mental health of the patient. Long non-coding (lnc)RNAs have been revealed to play an important role in human malignant tumors. Moreover, long intergenic non-protein coding RNA 491 (LINC00491) is a newly discovered lncRNA that can affect the prognosis of cancer. The present study aimed to explore the expression of LINC00491 in ESCC tissues and cells. The reverse transcription-quantitative PCR results suggested that LINC00491 was upregulated in ESCC tissues and cells. LINC00491 expression in esophageal squamous cell carcinoma cells were knocked down. Cell Counting Kit-8, wound healing, Transwell and apoptosis assays were performed to detect the effects of LINC00491 knockdown on cell biological behavior. The results showed that lower expression of LINC00491 resulted in decreased cell proliferation and migration and increased the apoptosis rate. Therefore, the present results indicated that lncRNA LINC00491 promoted the biological processes of ESCC, and thus LINC00491 may be a potential therapeutic target for ESCC.
Keywords: long intergenic non-protein coding RNA 491, esophageal squamous cell carcinoma, proliferation, apoptosis, migration
Introduction
Esophageal carcinoma (EC) is a common malignant tumor in the upper digestive tract system that occurs worldwide; the incidence of EC ranked seventh and the mortality rate ranked sixth in the world in 2018 according to the GLOBOCAN 2018 estimates of cancer (1). There are two types of esophageal cancer: Esophageal adenocarcinoma and esophageal squamous cell carcinoma (ESCC), whereby the incidence of ESCC accounts for more than esophageal adenocarcinoma in China as of 2015 (2). Based on various factors, such as diet and the environment, China is one of the countries that has a high incidence of EC (3). The overall 5-year survival rate of patients with esophageal cancer is between 15-25% (3). Early diagnosis of the disease can result in better prognosis. Furthermore, the early clinical manifestations of EC are not obvious, and thus most patients are diagnosed at the advanced stage and have a poor prognosis (4,5). Therefore, it is important to understand the molecular mechanisms of EC development and to identify novel biomarkers for the early diagnosis and treatment of EC.
With the rapid development of genome-wide and transcriptome sequencing technologies, the function of long non-coding (lnc)RNAs, which were previously thought to have a 'noise' role in the transcription process, has received increasing attention (6). lncRNAs are a group of RNA molecules with a length of >200 nucleotides, which have no protein coding function due to a lack of complete open reading frame (7). However, previous studies have reported that lncRNAs play an important role in regulating chromatin status, gene activity, gene expression at transcriptional and translational levels, and regulating tumorigenesis (8,9). Moreover, it has been revealed that several lncRNAs, such as metastasis associated lung adenocarcinoma transcript 1 (10), HOX transcript antisense RNA (11) and H19 (12), are abnormally expressed in ESCC and are involved in its development. However, the functional and molecular mechanisms of the majority of lncRNAs remain unknown in ESCC.
Long intergenic non-protein coding RNA 491 (LINC00491) is a recently studied lncRNA, which is transcribed by chromosome 5 (13). Furthermore, the differential expression of LINC00491 is significantly associated with the prognosis of human endometrial cancer (14), colon cancer (15,16) and breast cancer (17). Moreover, LINC00491 acts as a novel molecular biomarker to promote proliferation, migration and invasion of colonic adenocarcinoma cells via sponging microRNA (miR)-145 (13). However, to the best of our knowledge, the expression and function of LINC00491 in ESCC have not been previously reported. The present study identified a large number of differentially expressed lncRNAs in ESCC using high-throughput sequencing, including LINC00491. In addition, the present study examined the expression of LINC00491 in ESCC tissues and cell lines, and then analyzed the effects of LINC00491 on cell proliferation, migration, invasion and apoptosis. It was demonstrated that LINC00491 plays an important role in the development of ESCC and may provide an experimental basis for further clinical research.
Materials and methods
Clinical samples and RNA sequencing assay
In total, two separate samples were collected (20 cases in total). The first group included 4 paired cases of ESCC and normal tissues (53-75 years; 3 males; 1 female), which were collected between May 2018 and July 2018 during resection of cancer via open surgery from the Department of Thoracic Surgery at Zhongda Hospital Affiliated to Southeast University (Nanjing, China). The second group included 16 pairs of ESCC tissues and corresponding normal tissue samples (50-75 years; 10 males; 6 females), which were collected between February 2019 and September 2019 after surgical operation and endoscopic submucosal dissection surgery at the Zhongda Hospital Affiliated to Southeast University (Nanjing, China). Healthy or normal esophageal epithelial tissue was defined as tissues that were ≥5 cm apart from the margin of the cancer tissues. All patients with ESCC had not received local or systemic treatment before operation. Excised ESCC tissue samples and corresponding healthy tissue samples were immediately frozen with liquid nitrogen and stored at 80°C. The study was approved by the Research Ethics Committee of the Southeast University (Nanjing, China; approval no. 2019ZDSYLL022-P01). All patients provided written informed consent.
Next-generation RNA sequencing assay was performed at Kangchen Biotechnology Co., Ltd. using Illumina HiSeq 4000 (Illumina, Inc.) to detect the mRNA and ncRNA expression profiles. Image processing and base recognition were performed using Solexa pipeline version 1.8 software (Off-Line Base Caller software). Then, the reference genome was compared using Hisat2 software (18) (version 2.1.0; http://daehwankimlab.github.io/hisat2/), and a FPKM calculation at the gene and transcript levels was performed using R software 'Ballgown'. The differentially expressed lncRNAs and mRNAs were selected by fold change (FC)/P-value/false discovery rate (FDR) filtration (multiple ≥1.5, P<0.05 and FDR <0.05).
Functional enrichment analyses and coding-non-coding gene co-expression (CNC) network
The Gene Ontology (GO) project (19,20) provides a controlled vocabulary to describe gene and gene product attributes in any organism (http://www.geneontology.org). GO covers three domains: Biological Process, Cellular Component and Molecular Function. Pathway analysis is a functional analysis mapping genes to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The P-value (EASE-score, Fisher-P-value or Hypergeometric-P-value) denotes the significance of the KEGG pathway associated with the conditions. A CNC network of LINC00491 and coding genes (mRNAs) was constructed, and P<0.05 was considered to indicate a significant co-expression association between the two genes. The network was drawn using Cytoscape 2.8.3 (https://cytoscape.org) (21).
Data availability
Gene Expression Omnibus (GEO) is the National Center for Biotechnology Information GEO, from which GSE45670 (22) was downloaded, which contains surgical samples from 28 patients with ESCC and 10 samples from healthy esophageal epithelium. Differential genes were analyzed using R language.
Cell lines and cell culture
The human ESCC cell lines KYSE-410 and KYSE-30 were purchased from Cell Cook Company and the normal esophageal epithelial cell line (HEEC) was purchased from BeNa Culture Collection; Beijing Beina Chunglian Biotechnology Research Institute. Cells were cultured using RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS(Gibco; Thermo Scientific, Inc.) and cells were maintained at 37°C in an incubator containing 5% CO2.
Cell transfection
The short hairpin (sh)RNAs specifically targeting LINC00491 and a negative control shRNA (sh-NC) were designed and synthesized by Shanghai GenePharma Co., Ltd. KYSE30 and KYSE410 cells in the logarithmic growth phase were seeded in a petri dish. Cell transfection with shRNA was performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to manufacturer's protocol; the ratio of shRNA (500 µg/µl): Lipofectamine® 2000 (µl) was 1:3. The sequences of shRNAs were as follows: sh-LINC00491#1, 5′-GGT GTA TTC CAC ATT GTC TCT-3′; sh-LINC00491#2, 5′-GGC CAA AGG TCT GAT AAT TGC-3′; sh-LINC00491#3, 5′-GGA TAT GTG CAG GGA GTC TAG-3′; and sh-NC, 5′-TTC TCC GAA CGT GTC ACG T-3′.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from ESCC tissues, adjacent healthy tissues and HEEC, KYSE-30 and KYSE-410 cells using the Hipure Universal RNA kit (Magen) according to the manufacturer's instructions. Then, the extracted total RNA samples were quantified using a Colibri spectrometer (Titertek Berthold). A total of 1 µg RNA was used for RT, which was performed using the HiScript Q RT SuperMix for qPCR (Including 4X g DNA wiper Mix; Vazyme Biotech Co., Ltd.), following the manufacturer's instructions. qPCR was performed using SYBR Green Master Mixture (Low ROX Premixed; Vazyme Biotech Co., Ltd.) with the Applied Biosystems QuantStudio 3 RT PCR system (Thermo Fisher Scientific, Inc.). The reaction system was as follows: 10 µl qPCR SYBR Green Master mix (Low Rox Premixed), 0.4 µl forward primer (10 µM), 0.4 µl reverse primer (10 µM), 8.2 µl RNase-free water and 1 µl cDNA. The thermocycling conditions were as follows: Initial denaturation at 95°C for 5 min, followed by 40 cycles at 95°C for 10 sec and 60°C for 30 sec, and melting curve stage at 95°C for 15 sec, 60°C for 60 sec and 95°C for 15 sec. GAPDH was used as a standardized internal control. The primers were synthesized by Sangon Biotech Co., Ltd, and the primer sequences were as follows: LINC00491 forward, 5′-CCC CTT AAC CAA CTG GAA-3′ and reverse, 5′-GTG GAC CTT CTC CCA GCA AT-3′; and GAPDH forward, 5′-TGC ACC ACC AAC TGC TTA GC-3′ and reverse, 5′-GGC ATG GAC TGT GGT CAT GAG-3′. The results were quantified by the 2−ΔΔCq method (23). Each test was performed in triplicate.
Cell Counting Kit-8 (CCK-8) assay
KYSE30 and KYSE410 cells in the logarithmic growth phase were adjusted to a cell density of 3×104 cells/ml, and seeded into 96-well cell culture plates, with 100 µl per well and three replicates per group. Until the cells were attached, 20 µl serum-free RPMI-1640 medium was used to dilute shRNA (0.15 µg) and Lipofectamine® 2000 (0.45 µl) for transient transfection. After 48 h of transfection of ESCC cells, 10 µl CCK-8 reagent was added to each well (110 µl/well) of a 96-well plate and incubated for 3 h in a 5% CO2 incubator. Then, the absorbance of 450 nm water soluble tetrazolium salt was measured at 0, 24, 48 and 72 h to quantify cell proliferation.
Wound healing assay
KYSE-30 and KYSE-410 cells in the logarithmic growth phase were seeded into a 6-well plate, and the cell density was adjusted to 1×105 cells/ml, with 3 ml per well and three replicates in each group. Until the cells were attached, transient transfection was performed with 500 µl serum-free RPMI-1640 medium diluted shRNA (2.5 µg) and Lipofectamine® 2000 (7.5 µl). After 48 h of transfection, scratch wounds were created with the tip of a 200-µl pipette to scratch the cells when the cell density reached >90%. The cells were then washed twice with 0.01% PBS and serum-free medium was added. Then, cells were maintained at 37°C in an incubator containing 5% CO2. Random images were taken at 0, 24 and 48 h using a microscope (×50 magnification; Leica Microsystems GmbH) to assess the ability of cells to migrate. The independent experiments were repeated three times.
Cell migration and invasion assay
KYSE-30 and KYSE-410 cells in the logarithmic growth phase were seeded into a 12-well plate. The cell density was adjusted to 1×104 cells/ml, with 3 ml per well and three replicates in each group. After the cells adhered to the wall, shRNA (1.5 µg) and Lipofectamine® 2000 (4.5 µl) were diluted with 200 µl serum-free RPMI-1640 medium for transient transfection. Migration and invasion assays were performed using a Transwell chamber. For the migration assay, 1×104 cells were seeded into the upper chamber of Transwell (Corning, Inc.). For the invasion assay, 1×104 cells were added to the upper chamber (previously coated with Matrigel) and placed in the incubator for 1 h at 37°C. In both assays, cells were maintained in serum-free medium in the upper chamber and medium containing 10% FBS was added as a chemoattractant to the lower chamber. After 24 h of incubation, cells that did not migrate or invade the membrane were removed. The membrane was then fixed with methanol (15 min at room temperature) and stained with 0.1% crystal violet (15 min at room temperature). Each chamber was counted in three random fields using an inverted microscope (×100 magnification; Carl Zeiss AG) and each experiment was repeated three times.
Apoptosis assays
The cell culture and transfection methods for the apoptosis assay were same as that of the wound healing assay, which have been described above. Cells were transfected and incubated for 48 h. The cells were digested with EDTA-free trypsin and collected, and then 300 µl 1X Binding Buffer (Fcmacs Biotech Co., Ltd.) suspended cells were added. Subsequently, 100 µl cell suspension (total number of cells, 1×105) were pipetted into a new tube. Then, 10 µl Annexin V-APC and 5 µl PI (Fcmacs Biotech Co., Ltd.) were added, and after mixing, the cells were incubated for 15 min at room temperature in the dark. Apoptotic cells were detected using flow cytometry (BD FACSVerse; BD Biosciences) and analyzed using FlowJo 7.6.1 (TreeStar, Inc.).
Statistical analysis
All data were analyzed using SPSS 20 (IBM Corp.) and GraphPad Prism 5.0 (GraphPad Software, Inc.). Data are presented as the mean ± standard deviation. Comparisons between the two groups were performed using unpaired Student's t-test, and comparisons between ≥3 groups were performed using one-way ANOVA and post hoc analysis with Tukey's test. Each experiment was repeated three times. P<0.05 was considered to indicate a statistically significant difference.
Results
Differentially expressed lncRNAs in ESCC compared with healthy tissue
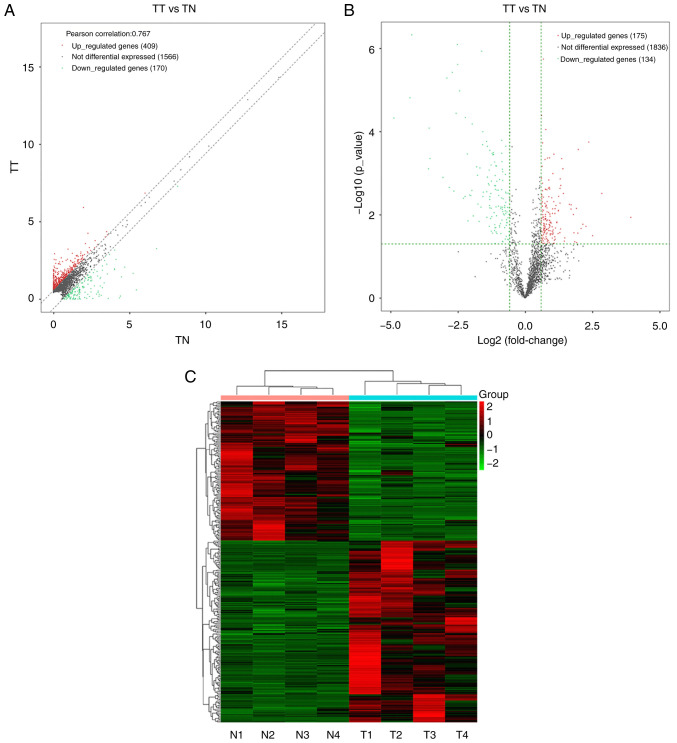
RNA sequences were used to detect 2,145 different lncRNAs from four pairs of human ESCC and healthy esophageal tissue. lncRNAs with FC ≥1.5 and P<0.05 were considered to have significant differences. Among them, 409 were found to be upregulated and 170 were downregulated with FC ≥1.5 (Fig. 1A). Further expressional analysis demonstrated that a series of lncRNAs were differentially expressed in ESCC compared with the control group; of the 309 differentially expressed lncRNAs, 175 were upregulated in ESCC and 134 were downregulated with FC ≥1.5 and P<0.05 (Fig. 1B and C). Moreover, LINC00491 was included in the genes that were significantly differentially expressed.
Figure 1.
Identification of lncRNAs in human ESCC samples. (A) Difference in lncRNA expression in ESCC vs. healthy tissue samples. Data are presented as the mean of lncRNA expression in ESCC (log 2 transformation), and two oblique dashed lines divide the upper and lower genes (1.5-fold difference) and the unmodified gene boundaries. (B) A volcano map representing the differential expression of lncRNA in ESCC. The x-axis represents the fold change (log 2) and the y-axis represents the P-value (−log 10). The green parallel line corresponds to the P-value threshold. The left side of the vertical green line are the downregulated genes, and the opposite are the upregulated genes. (C) Hierarchical cluster heat map of the differentially expressed lncRNAs in RNA-seq analysis of four paired ESCC tumor and adjacent tissues. Each row represents a gene and each column represents a sample. Red represents upregulation of differentially expressed genes, and green represents downregulation of differentially expressed genes. TT, tumor tissue; TN, paired normal tissues; lncRNA, long non-coding RNA; ESCC, esophageal squamous cell carcinoma.
GO and pathway analysis
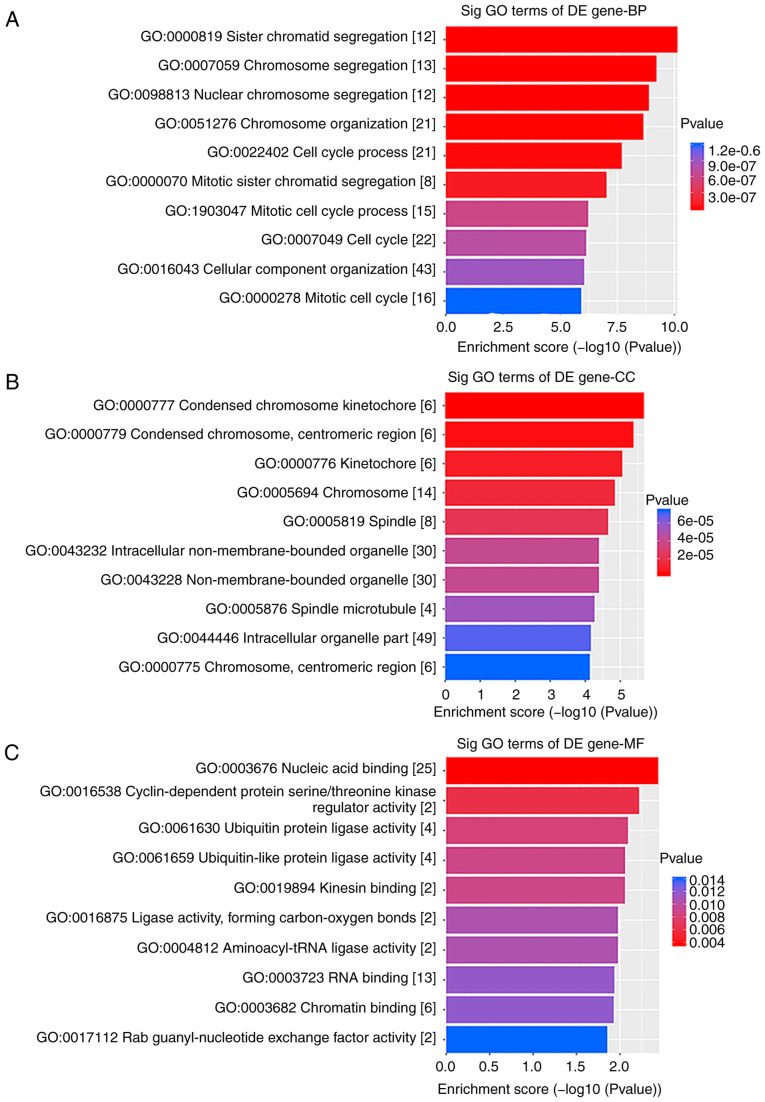
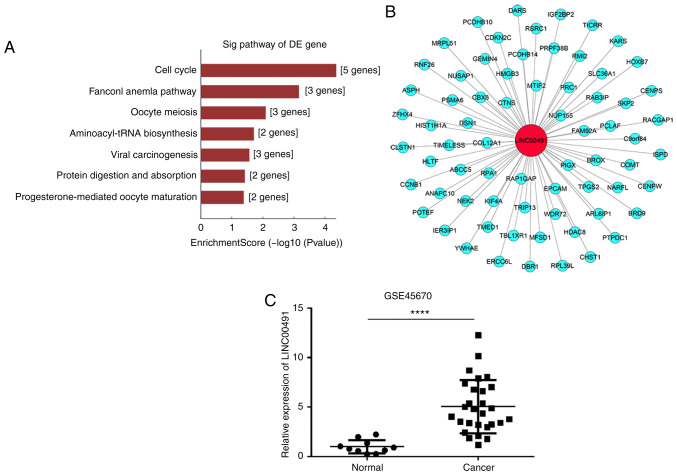
GO analysis results indicated that the differentially expressed genes between ESCC and the matched control group were mainly involved in 'chromosome segregation', 'cell cycle process' and 'mitotic cell cycle process', which affect cell growth and proliferation (Fig. 2). Furthermore, pathway analysis identified that the related genes were also primarily involved in 'cell cycle' and 'oocyte meiosis' (Fig. 3A).
Figure 2.
GO enrichment analysis of LINC00491 in ESCC sample. GO enrichment analysis of (A) BP, (B) MF and (C) CC of LINC00491. DE gene-BP, differentially expressed genes-biological processes; DE gene-CC, differentially expressed genes-cellular components; DE gene-MF, differentially expressed genes-molecular functions; lncRNA, long non-coding RNA; ESCC, esophageal squamous cell carcinoma; GO, Gene Ontology.
Figure 3.
CNC and pathway analysis of LINC00491. (A) Significantly enriched pathways between the ESCC and control group. (B) mRNAs related to LINC00491 were assessed using CNC analysis. Red circle represents lncRNA, blue circle represents mRNA. (C) GSE45670 dataset identified the upregulation of LINC00491 in esophageal squamous cell cancer tissues compared with the control. ****P<0.0001. ESCC, esophageal squamous cell carcinoma; CNC, co-expression network; LINC00491, long intergenic non-protein coding RNA 491; DE, differentially expressed; Sig, significant.
CNC network and GSE45670 analysis
CNC network analysis calculates the correlation co-efficient by comparing the FPKM data of LINC00491 with the differential expression transcripts of all the mRNAs in the transcript, and selecting mRNAs with Pearson's correlation coefficient ≥0.9, P<0.05 and FDR=1. GEO data were obtained from the GEO database. The results of CNC analysis demonstrated that 69 mRNAs were co-expressed with LINC00491, including the oncogenes thyroid hormone receptor interactor 13 (TRIP13) (24) and homeobox B7 (HOXB7) (25), which are associated with ESCC (Fig. 3B). In addition, analysis of the GSE45670 dataset found that compared with the control group, the expression of LINC00491 in EC tissue was significantly higher compared with normal tissue (Fig. 3C).
Increased expression of LINC00491 in ESCC tissues and cells
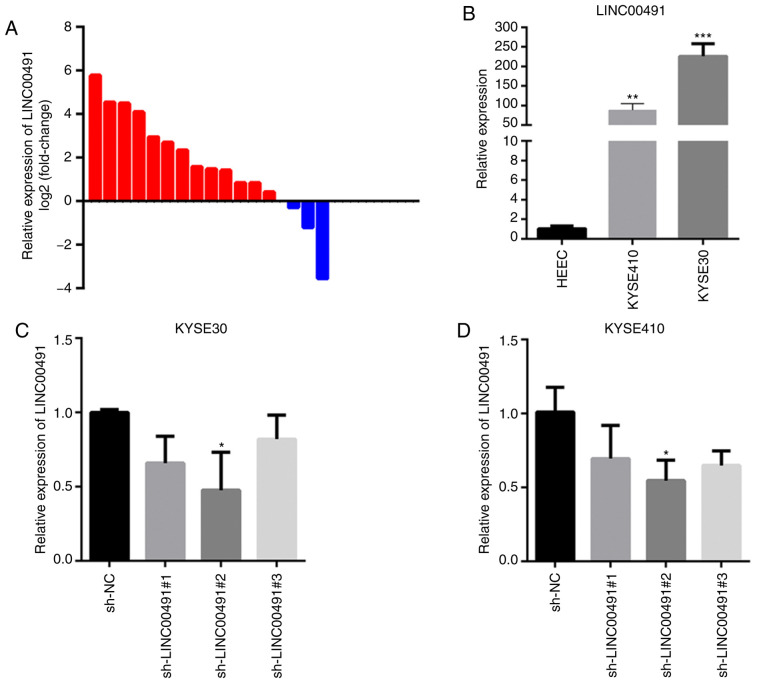
RT-qPCR results demonstrated that LINC00491 expression was significantly higher in 13/16 cases of ESCC compared with the controls (Fig. 4A). Moreover, the expression of LINC00491 was significantly higher in two ESCC cell lines (KYSE-30 and KYSE-410) compared with human normal esophageal epithelial cells (HEEC; Fig. 4B). When sh-LINC00491#1, sh-LINC00491#2, sh-LINC00491#3 or sh-NC were transfected into KYSE-30 and KYSE-410 cells, the expression of LINC00491 was significantly reduced, with the highest reduction in sh-LINC00491#2, compared with the control group (Fig. 4C and D). In summary, the results demonstrated that LINC00491 was highly expressed in ESCC tissues and cell lines, and sh-LINC00491#2 had the highest inhibition efficiency and thus was used for subsequent experiments.
Figure 4.
Expression of LINC00491 in ESCC tissues and cells. (A) RT-qPCR was used to detect the relative expression of LINC00491 in 16 cases of ESCC, among which the expression of LINC00491 was upregulated in 13 cases. Expression of lncRNA was normalized to GAPDH. (B) RT-qPCR was used to detect the relative expression of LINC00491 in ESCC cell lines and human normal esophageal epithelial cells. Data are presented as the mean ± SD, the expressions of LINC00491 in KYSE-410, KYSE-30 and HEEC cells were 88.12±16.59, 225.92±31.79 and 1.03±0.28, respectively. **P<0.01, ***P<0.001 vs. HEEC. (C and D) Three specific sh-LINC00491 were transfected into esophageal squamous cells. After 48 h, RT-qPCR was used to detect the expression of LINC00491 in KYSE-30 cells and KYSE-410 cells. The expressions of LINC00491 in sh-LINC00491#1, sh-LINC00491#2, sh-LINC00491#3 and sh-NC were 0.66±0.18, 0.47±0.25, 0.82±0.16 and 1.00±0.02, respectively. In KYSE-410 cells, the expressions of LINC00491 in sh-LINC00491#1, sh-LINC00491#2, sh-LINC00491#3 and sh-NC were 0.69±0.22, 0.54±0.13, 0.64±0.09 and 1.01±0.16, respectively. *P<0.05 vs. sh-NC. NC, negative control; sh, short hairpin RNA; LINC00491, long intergenic non-protein coding RNA 491; ESCC, esophageal squamous cell carcinoma; RT-qPCR, reverse transcription-quantitative PCR.
Inhibition of the expression of LINC00491 reduces the proliferation of EC cells
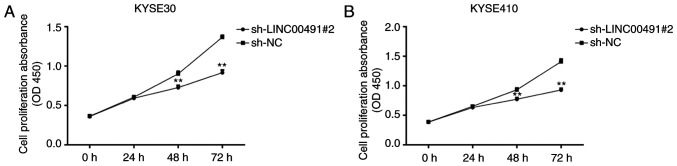
Using the CCK-8 experiment, the effect of knocking down the expression of LINC00491 on cell proliferation was examined in ESCC cells, and the optical density values were measured at 0, 24, 48 and 72 h. It was identified that in KYSE30 and KYSE410 cells, compared with the sh-NC group, the proliferation ability of the sh-LINC00491#2 transfection group was significantly lower after 48 h of culture (P<0.01; Fig. 5A and B).
Figure 5.
CCK8 proliferation assay. Effect of LINC00491 low-expression on proliferation of (A) KYSE30 and (B) KYSE410 cells were detected using Cell Counting Kit-8. Data are presented as the mean ± SD of triplicate samples. **P<0.01 vs. sh-NC. sh, short hairpin RNA; NC, negative control; LINC00491, long intergenic non-protein coding RNA 491; OD, optical density.
Decreased expression of LINC00491 inhibits migration of ESCC cells
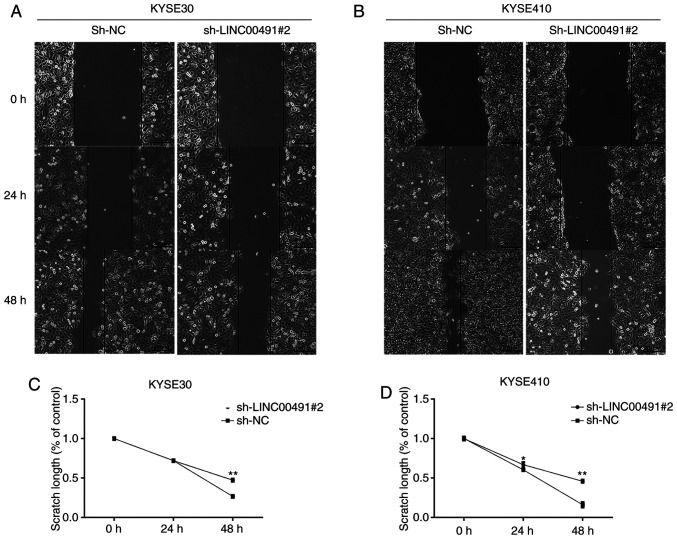
The wound healing assay results indicated that interference of the expression of LINC00491 significantly inhibited the migration of KYSE-30 and KYSE-410 cells (Fig. 6).
Figure 6.
Wound healing assay. After 48 h of transfection, the effect of LINC00491 knockdown on the migration of KYSE30 and KYSE410 cells was detected by wound healing assay. Cell migration at 0, 24 and 48 h of (A) KYSE30 and (B) KYSE 410 cells. Cells transfected with sh-LINC00491#2 showed less migration compared with sh-NC. Graphs illustrate the scratch width based on time intervals in which a larger scratch width was observed in sh-LINC00491#2 compared with sh-NC transfected (C) KYSE30 and (D) KYSE410 cells. sh, short hairpin RNA; NC, negative control; LINC00491, long intergenic non-protein coding RNA 491. The y-axis refers to the width, which is the distance between the margins of area of cell scratch and x-axis is the time interval. Data are shown as the mean ± SD of a representative experiment performed in triplicate. *P<0.05, **P<0.01 vs. sh-NC group. NC, negative control; sh, short hairpin RNA; LINC00491, long intergenic non-protein coding RNA 491.
LINC00491 silencing suppresses ESCC cell migration and invasion
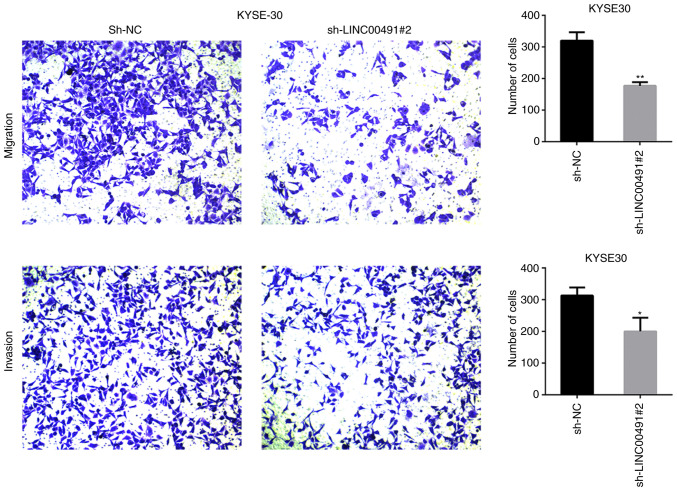
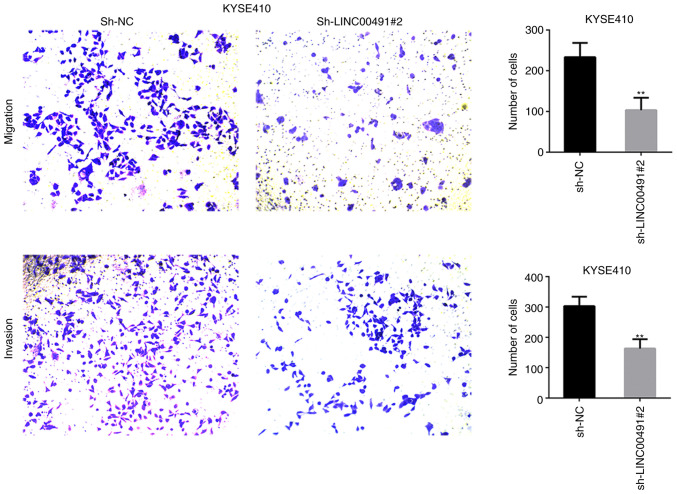
Transwell chamber experiments were performed on ESCC cells to investigate the function of LINC00491 during cell migration and invasion. It was found that silencing LINC00491 expression in KYSE30 and KYSE410 cells significantly inhibited cell invasion and migration compared with the NC group (Figs. 7 and 8). Thus, it was speculated that upregulation of LINC00491 expression in ESCC could accelerate tumor cell migration and invasion.
Figure 7.
Transwell assay was used to assess the invasion and metastasis of KYSE30 cells transfected with LINC00491-specific shRNA or control shRNA after 48 h. Magnification, ×100. Data are presented as the mean ± SD. *P<0.05, **P<0.01 vs. sh-NC. NC, negative control; sh, short hairpin RNA; LINC00491, long intergenic non-protein coding RNA 491.
Figure 8.
Transwell assay was used to assess the invasion and metastasis of KYSE410 cells transfected with LINC00491-specific shRNA or control shRNA after 48 h. Magnification, ×100. Data are presented as the mean ± SD. **P<0.01 vs. sh-NC. NC, negative control; sh, short hairpin RNA; LINC00491, long intergenic non-protein coding RNA 491.
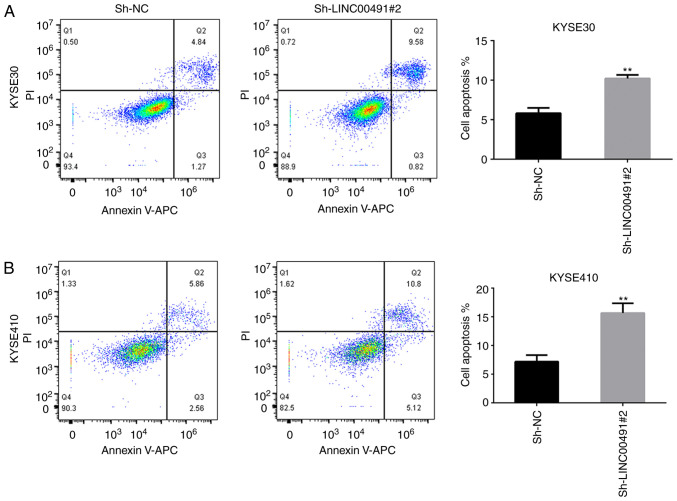
Inhibition of LINC00491 expression promotes apoptosis of ESCC cells
Flow cytometry results demonstrated that the apoptosis rate of KYSE-30 and KYSE-410 ESCC cells transfected with sh-LINC00491#2 was significantly increased compared with the control group (Fig. 9).
Figure 9.
Effects of LINC00491 knockdown on the apoptosis of KYSE30 and KYSE410 cells. Cell apoptosis was determined by flow cytometry using Annexin V/PI staining. (A) Annexin V-APC/PI staining in KYSE30 cells and statistical analysis of apoptotic rates in KYSE30. (B) Annexin V-APC/PI staining in KYSE410 cells and statistical analysis of apoptotic rates in KYSE410. Data are shown as mean ± SD. **P<0.01 vs. sh-NC group. NC, negative control; sh, short hairpin RNA; LINC00491, long intergenic non-protein coding RNA 491; PI, propidium iodide.
Discussion
Cancer is a complex disease associated with a combination of genetic and environmental factors. Currently, although there are various methods for the treatment of malignant tumors, the results of these are not satisfactory, and the main limitations are that the long-term survival rate of patients with cancer have not significantly improved, and relapse is common (26). EC is a common malignant tumor of the digestive system, and is associated with poor prognosis, and high recurrence and invasive rates (27-29). In recent years, although progress has been made in the research of EC, its pathogenesis remains unknown, and in terms of diagnosis and prognosis, there remains a lack of highly specific and sensitive biomarkers.
lncRNA was originally thought to only be a by-product of RNA polymerase II transcription as it does not encode a protein, and thus it was considered to have no biological function (30). However, previous studies have reported that lncRNA is a key regulator of gene transcription and translation (31). Moreover, abnormal expression of lncRNA promotes tumor development by affecting the expression of cancer-related genes at the pre-transcriptional, transcriptional and post-transcriptional levels (32). Furthermore, tumor cell proliferation, invasion, apoptosis, metastasis and drug resistance are all associated with the imbalance of lncRNA expression (33). Therefore, lncRNAs play a key role in tumorigenesis and progression, and can be used as potential biomarkers for various cancer types (34,35). For example, Chen et al (36) revealed that LINC00472 inhibits proliferation, migration and invasion of liver cancer cells via the miR-93-5p/programmed cell death 4 pathway. In addition, Gong et al (37) reported that urothelial cancer associated 1 acts as a competing endogenous lncRNA, which competitively binds to miR-203 and subsequently increases the expression level of the transcription factor zinc finger E-box-binding homeobox 2 to promote the metastasis of gastric cancer. Furthermore, downregulation of lncRNA colon cancer associated transcript 1 enhances the sensitivity of human colon cancer cells to 5-fluorouracil (38). Based on the present RNA sequencing and GEO database analysis results, it was speculated that the expression of LINC00491 in ESCC was upregulated compared with healthy esophageal tissue. The expression of LINC00491 in the healthy tissue is considered as normal expression and the upregulated expression was validated by RT-qPCR. Moreover, LINC00491 CNC network analysis demonstrated that LINC00491 is co-expressed with a variety of oncogenes, such as TRIP13 and HOXB7, and further GO and KEGG enrichment analysis identified that the differential expression of LINC00491 in ESCC may be involved in 'chromosome segregation', 'ubiquitin protein ligase activity' and 'cell cycle process'.
Rapid proliferation is an important characteristic of tumor cells. A normal cell cycle is a key process to ensure the orderly proliferation of cells, and cell cycle disorders may cause normal cells to transform into infinitely increasing tumor cells (39). Thus, LINC00491 may promote the occurrence and development of ESCC by affecting the expression of tumor-related genes.
In the present study, the sample size was expanded to detect the expression of LINC00491 in ESCC and to study the effect of knocking down the expression of LINC00491 on the biological features of ESCC cell lines, such as proliferation, migration, invasion and apoptosis. It was demonstrated that the expression of LINC00491 in 13/16 ESCC tissues was higher compared with healthy tissues. Furthermore, RT-qPCR detection of LINC00491 in KYSE30 and KYSE410 cells revealed it to be highly expressed. Subsequent interference with the expression of LINC00491 in ESCC cell lines revealed that compared with the control group, sh-LINC00491#2 significantly reduced the proliferation, migration and invasion capabilities, and significantly increased the rate of apoptosis. Thus, it was indicated that knockdown of LINC00491 may have an inhibitory effect on ESCC disease progression. Due to time constrains, the main limitation of the present study was that the number of ESCC tissue samples collected was small; however, the experimental tissue sample size will be expanded in future research. In addition, the current study only included in vitro functional experiments, and the potential mechanism of action of LINC00491 and biological functions in vivo require further research. However, the present study provided an experimental basis for future clinical research and may have potentially beneficial effects on the treatment of diseases.
In conclusion, it was demonstrated that the expression of LINC00491 was significantly upregulated in ESCC tissues and cells. Furthermore, in ESCC cells, knockdown of LINC00491 inhibited proliferation and migration, and promoted apoptosis.
Acknowledgments
The authors would like to express their gratitude to Professor Daqing Gao (Immunology Department, School of Medicine, Southeast University, Nanjing, China) for providing laboratory assistance. The authors would also like to thank Dr Qinghua Ji (School of Medicine, Southeast University, Nanjing, China) for his assistance in the laboratory work.
Funding Statement
This study was supported by the National Natural Science Foundation of China (grant no. 6590000161), the Science and Technology Project of Jiangsu Province (grant no. 7790000102) and the Jiangsu Health Commission Fund (grant no. 2017ZXK7QW08).
Availability of data and materials
The datasets used and/or analyzed during the current study can be obtained from the corresponding author on reasonable request.
Authors' contributions
RS designed and supervised the study. HY designed and conducted experiments and performed data analysis. SMS helped in collecting specimens, performed some experiments and guided the experiments. HY and SMS wrote the manuscript. JZ helped in data analysis. YD designed the study, provided technical assistance in operating the equipments and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This research was approved by the Ethics Committee for Clinical Research of Zhongda Hospital affiliated to Southeast University (Nanjing, China; approval no. 2019ZDSYLL022-P01). All patients signed informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400–412. doi: 10.1016/S0140-6736(12)60643-6. [DOI] [PubMed] [Google Scholar]
- 4.Han LC, Chen Y. Small and long non-coding RNAs: Novel targets in perspective cancer therapy. Curr Genomics. 2015;16:319–326. doi: 10.2174/1389202916666150707155851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carninci P, Hayashizaki Y. Noncoding RNA transcription beyond annotated genes. Curr Opin Genet Dev. 2007;17:139–144. doi: 10.1016/j.gde.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: Past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrey G, Duboule D. SnapShot: Hox gene regulation. Cell. 2014;156:856. doi: 10.1016/j.cell.2014.01.060. [DOI] [PubMed] [Google Scholar]
- 8.Gomez-Maldonado L, Tiana M, Roche O, Prado-Cabrero A, Jensen L, Fernandez-Barral A, Guijarro-Muñoz I, Favaro E, Moreno-Bueno G, Sanz L, et al. EFNA3 long noncoding RNAs induced by hypoxia promote metastatic dissemination. Oncogene. 2015;34:2609–2620. doi: 10.1038/onc.2014.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu WD, Wang H, He QF, Xu Y, Wang XC. Long noncoding RNAs in cancer-immunity cycle. J Cell Physiol. 2018;233:6518–6523. doi: 10.1002/jcp.26568. [DOI] [PubMed] [Google Scholar]
- 10.Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y, Yang K. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:7. doi: 10.1186/s13046-015-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Wu Z, Mei Q, Li X, Guo M, Fu X, Han W. Long non-coding RNA HOTAIR, a driver of malignancy, predicts negative prognosis and exhibits oncogenic activity in oesophageal squamous cell carcinoma. Br J Cancer. 2013;109:2266–2278. doi: 10.1038/bjc.2013.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan D, Wu Y, Hu L, He P, Xiong G, Bai Y, Yang K. Long noncoding RNA H19 is up-regulated in esophageal squamous cell carcinoma and promotes cell proliferation and metastasis. Dis Esophagus. 2017;30:1–9. doi: 10.1111/dote.12481. [DOI] [PubMed] [Google Scholar]
- 13.Wan J, Deng D, Wang X, Wang X, Jiang S, Cui R. LINC00491 as a new molecular marker can promote the proliferation, migration and invasion of colon adenocarcinoma cells. Onco Targets Ther. 2019;12:6471–6480. doi: 10.2147/OTT.S201233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia L, Wang Y, Meng Q, Su X, Shen J, Wang J, He H, Wen B, Zhang C, Xu M. Integrated bioinformatic analysis of a competing endogenous RNA network reveals a prognostic signature in endometrial cancer. Front Oncol. 2019;9:448. doi: 10.3389/fonc.2019.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao Z, Fu P, Yu Z, Zhen F, Gu Y. Comprehensive analysis of lncRNA-miRNA-mRNA network ascertains prognostic factors in patients with colon cancer. Technol Cancer Res Treat. 2019;18:1533033819853237. doi: 10.1177/1533033819853237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang WJ, Li HT, Yu JP, Han XP, Xu ZP, Li YM, Jiao ZY, Liu HB. A competing endogenous RNA network reveals novel potential lncRNA, miRNA, and mRNA biomarkers in the prognosis of human colon adenocarcinoma. J Surg Res. 2019;235:22–33. doi: 10.1016/j.jss.2018.09.053. [DOI] [PubMed] [Google Scholar]
- 17.Fan CN, Ma L, Liu N. Systematic analysis of lncRNA-miRNA-mRNA competing endogenous RNA network identifies four-lncRNA signature as a prognostic biomarker for breast cancer. J Transl Med. 2018;16:264. doi: 10.1186/s12967-018-1640-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The Gene Ontology Consortium: The gene ontology resource: 20 Years and still going strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wen J, Yang H, Liu MZ, Luo KJ, Liu H, Hu Y, Zhang X, Lai RC, Lin T, Wang HY, Fu JH. Gene expression analysis of pretreatment biopsies predicts the pathological response of esophageal squamous cell carcinomas to neo-chemoradiotherapy. Ann Oncol. 2014;25:1769–74. doi: 10.1093/annonc/mdu201. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Di S, Li M, Ma Z, Guo K, Li X, Yan X. TRIP13 upregulation is correlated with poor prognosis and tumor progression in esophageal squamous cell carcinoma. Pathol Res Pract. 2019;215:152415. doi: 10.1016/j.prp.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 25.Zhou T, Fu H, Dong B, Dai L, Yang Y, Yan W, Shen L. HOXB7 mediates cisplatin resistance in esophageal squamous cell carcinoma through involvement of DNA damage repair. Thorac Cancer. 2020;11:3071–3085. doi: 10.1111/1759-7714.13142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tiffon C. The impact of nutrition and environmental epigenetics on human health and disease. Int J Mol Sci. 2018;19:3425. doi: 10.3390/ijms19113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhan XH, Jiao JW, Zhang HF, Li CQ, Zhao JM, Liao LD, Wu JY, Wu BL, Wu ZY, Wang SH, et al. A three-gene signature from protein-protein interaction network of LOXL2- and actin-related proteins for esophageal squamous cell carcinoma prognosis. Cancer Med. 2017;6:1707–1719. doi: 10.1002/cam4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu L, Qin YR, Ming XY, Zuo XB, Diao YW, Zhang LY, Ai J, Liu BL, Huang TX, Cao TT, et al. RNA editing of SLC22A3 drives early tumor invasion and metastasis in familial esophageal cancer. Proc Natl Acad Sci USA. 2017;114:E4631–E4640. doi: 10.1073/pnas.1703178114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu SY, Chen W, Chughtai EA, Qiao Z, Jiang JT, Li SM, Zhang W, Zhang J. PIK3CA gene mutations in Northwest Chinese esophageal squamous cell carcinoma. World J Gastroenterol. 2017;23:2585–2591. doi: 10.3748/wjg.v23.i14.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deveson IW, Hardwick SA, Mercer TR, Mattick JS. The dimensions, dynamics, and relevance of the mammalian noncoding transcriptome. Trends Genet. 2017;33:464–478. doi: 10.1016/j.tig.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 31.Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, Zhang Y, Yang L, Shan W, He Q, et al. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell. 2015;28:529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Washietl S, Kellis M, Garber M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. 2014;24:616–628. doi: 10.1101/gr.165035.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ning L, Li Z, Wei D, Chen H, Yang C. LncRNA, NEAT1 is a prognosis biomarker and regulates cancer progression via epithelial-mesenchymal transition in clear cell renal cell carcinoma. Cancer Biomark. 2017;19:75–83. doi: 10.3233/CBM-160376. [DOI] [PubMed] [Google Scholar]
- 34.Sugihara H, Ishimoto T, Miyake K, Izumi D, Baba Y, Yoshida N, Watanabe M, Baba H. Noncoding RNA expression aberration is associated with cancer progression and is a potential biomarker in esophageal squamous cell carcinoma. Int J Mol Sci. 2015;16:27824–27834. doi: 10.3390/ijms161126060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu M, Zou C, Pan L, Liang W, Qian H, Xu W, Jiang P, Zhang X. Long noncoding RNAs in digestive system cancers: Functional roles, molecular mechanisms, and clinical implications (Review) Oncol Rep. 2016;36:1207–1218. doi: 10.3892/or.2016.4929. [DOI] [PubMed] [Google Scholar]
- 36.Chen C, Zheng Q, Kang W, Yu C. Long non-coding RNA LINC00472 suppresses hepatocellular carcinoma cell proliferation, migration and invasion through miR-93-5p/PDCD4 pathway. Clin Res Hepatol Gastroenterol. 2019;43:436–445. doi: 10.1016/j.clinre.2018.11.008. [DOI] [PubMed] [Google Scholar]
- 37.Gong P, Qiao F, Wu H, Cui H, Li Y, Zheng Y, Zhou M, Fan H. LncRNA UCA1 promotes tumor metastasis by inducing miR-203/ZEB2 axis in gastric cancer. Cell Death Dis. 2018;9:1158. doi: 10.1038/s41419-018-1170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang C, Pan Y, Deng SP. Downregulation of lncRNA CCAT1 enhances 5-fluorouracil sensitivity in human colon cancer cells. BMC Mol Cell Biol. 2019;20:9. doi: 10.1186/s12860-019-0188-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Gene Expression Omnibus (GEO) is the National Center for Biotechnology Information GEO, from which GSE45670 (22) was downloaded, which contains surgical samples from 28 patients with ESCC and 10 samples from healthy esophageal epithelium. Differential genes were analyzed using R language.
The datasets used and/or analyzed during the current study can be obtained from the corresponding author on reasonable request.