Abstract
The dysfunctions of human umbilical vein endothelial cells (HUVECs) are important features of atherosclerosis (AS). Circular RNAs (circRNAs) are regulators of a wide range of human diseases, including AS. The present study aimed to investigate the role of circUSP36 in the ectopic phenotype of HUVECs and to provide evidence of the involvement of circUSP36 in the pathogenesis of AS. AS cell models in vitro were established using HUVECs exposed to oxidized low-density lipoprotein (ox-LDL). Cell viability, cell cycle progression and apoptosis, and cell migration and invasion were assessed by cell counting kit-8 (CCK-8) assay, flow cytometric assay and Transwell assay, respectively. The expression levels or releases of pro-inflammatory factors were detected by western blot analysis or enzyme-linked immunosorbent assay (ELISA). The mRNA expression of circUSP36, miR-20a-5p and Rho-associated coiled-coil kinase 2 (ROCK2) was detected by reverse transcription-quantitative polymerase chain reaction (RT-qPCR), and the protein level of ROCK2 was detected by western blot analysis. The targeted association between miR-20a-5p and circUSP36 or ROCK2 was verified by dual-luciferase reporter assay and RNA immunoprecipitation (RIP) assay. The results revealed that circUSP36 was highly expressed in ox-LDL-exposed HUVECs. CircUSP36 knockdown attenuated ox-LDL-induced cell cycle arrest, cell apoptosis and inflammatory responses, and promoted cell migration and invasion which had been blocked by ox-LDL. miR-20a-5p was found to be a target of circUSP36, and miR-20a-5p inhibition reversed the effects of circUSP36 knockdown. Moreover, miR-20a-5p directly bound to ROCK2, and miR-20a-5p inhibition aggravated ox-LDL-induced injury by increasing the ROCK2 level. More importantly, circUSP36 targeted miR-20a-5p to regulate the expression of ROCK2. On the whole, the present study demonstrates that, circUSP36 regulates ox-LDL-induced HUVEC injury and inflammation by modulating ROCK2 via competitively targeting miR-20a-5p.
Keywords: circUSP36, miR-20a-5p, ROCK2, atherosclerosis, human umbilical vein endothelial cell
Introduction
Atherosclerosis (AS) is a common type of cardiovascular disease, characterized by progressive inflammation and lesions on the arterial wall (1). Lipid deposition in the arterial wall is the main cause of AS, and lipoproteins and lipid-related factors are implicated in the pathogenesis of AS (2). Furthermore, the dysfunctions of endothelial cells substantially contribute to the development of AS, as endothelial cells are important components that protect the histological layers of the arterial wall from damage (3,4). Thus, it is a feasible strategy to study the induction factors and molecular mechanisms of endothelial cell injury and dysfunction for the treatment of AS.
Oxidized low-density lipoprotein (ox-LDL) evokes cellular oxidative stress, inflammatory responses and cell apoptosis, leading to endothelial cell dysfunction, and ox-LDL is thus regarded as a crucial element in the pathogenesis of AS (5,6). Human umbilical vein endothelial cells (HUVECs) exposed to ox-LDL are widely used to as an in vitro cell model of AS (7,8). However, the mechanisms through which ox-LDL induces HUVEC damage and dysfunction are complex and variable, and have not yet been fully elucidated. RNA sequencing technology provides a precise approach for the identification of circRNAs that are differently regulated in human diseases (9). CircRNAs derive from precursor mRNAs in a 'back-splicing' manner (10). With the increase in circRNA research, circRNAs as indicators or tools of disease diagnosis, treatment and prognosis have been widely recognized (11). Pan et al obtained circRNA expression profiles using RNA sequencing and identified 66 specific circRNAs in coronary artery AS (12). Li et al utilized circRNA expression profiles to analyze significantly dysregulated circRNAs in ox-LDL-induced HUVECs (13), and circ_0003204 was found as one of the differentially expressed circRNAs with an upregulated expression in ox-LDL-induced HUVECs (13). Circbase (http://www.circbase.org/) indicates that circ_0003204 is derived from precursor mRNA USP36, thus terming it as circUSP36. However, the function of circUSP36 in ox-LDL-induced HUVECs is largely unclear.
MicroRNAs (miRNAs or miRs) are characterized by RNAs with 18-24 nucleotides in length that govern extensive biological functions (14). Accumulating evidence has suggested that miRNAs can be used as diagnostic and therapeutic markers for AS and that they regulate the pathologies of vascular diseases (15,16). Previous studies have demonstrated that miR-20a-5p is involved in the development of cardiovascular diseases (17,18). Research into the role of miR-20a-5p in ox-LDL-induced HUVECs is not limited. Rho-associated coiled-coil kinase 2 (ROCK2) is one of the isoforms of the ROCK family (19). It has been demonstrated that ROCKs are substantially implicated in cardiovascular diseases, including AS, and the dysregulation of ROCKS regulates various cardiovascular pathological processes, such as abnormal apoptosis, cardiac hypertrophy and cardiac fibrosis (19,20). The functional role of ROCK2 in AS is largely clear, and the mechanisms of ROCK2 action need to be further explored, particularly in association with circUSP36 and miR-20a-5p.
In the present study, the expression of circUSP36 was verified in ox-LDL-exposed HUVECs, and a loss-of-function assay was performed to explore the function of circUSP36 in HUVECs. Moreover, the interaction between miR-20a-5p and circUSP36 or ROCK2 was validated. The present study provides a novel mechanism (at least to the best of our knowledge) to explain the functional mode of circUSP36 in ox-LDL-induced HUVEC dysfunction.
Materials and methods
HUVEC treatment
HUVECs were obtained from Procell Life Science & Technology Co., Ltd. (no. CL-0122) and maintained in HUVEC specific medium [Ham's F-12K+10% fetal bovine serum (FBS); Procell Life Science & Technology] at 37°C conditions with 5% CO2. HUVECs were exposed to various concentrations of ox-LDL (40, 60, 80 and 100 µg/ml) and incubated for different periods of time (6, 12, 24 and 48 h) at 37°C conditions with 5% CO2. The cell model of AS was constructed using HUVECs exposed to 60 µg/ml ox-LDL with a 24-h incubation at 37°C conditions with 5% CO2.
Cell viability detection
Cell counting kit-8 (CCK-8) assay was implemented to detect cell viability. HUVECs exposed to various concentrations of ox-LDL were supplemented with 10 µl CCK-8 reagent (Beyotime Institute of Biotechnology) at 24 h post-treatment and mixed for a further 2 h. Moreover, HUVECs exposed to 60 µg/ml ox-LDL were reacted with 10 µl CCK-8 reagent at 6, 12, 24 and 48 h post-treatment and mixed for a further 2 h. The absorbance at 450 nm was then monitored using a microplate reader (Thermo Fisher Scientific, Inc.) to observe cell viability.
Cell cycle analysis
HUVECs were collected and resuspended in fresh culture medium containing 10% FBS. Cells were collected and gently resuspended in 70% ice-cold ethanol for fixing at -20°C overnight. Subsequently, the cells were collected, washed and resuspended in fresh culture medium containing 10% FBS. The cells were then collected and resuspended in 200 µl Cell Cycle Solution [containing propidium iodide (PI), RNase A, and Triton X-100; Invitrogen; Thermo Fisher Scientific, Inc.] at a density of 2×105 cells. Cells were then incubated at room temperature for 30 min without light and finally analyzed using an EPICS XL-MCL FACScan flow cytometer (Beckman Coulter, Inc.).
Cell apoptosis analysis
Cell apoptosis was monitored using the Annexin V-FITC Apoptosis Detection kit (Beyotime Institute of Biotechnology). Briefly, a total of 5×104 HUVECs were collected at 48 h post-transfection and resuspended in cold phosphate-buffered saline (PBS). Cells were then collected and resuspended in 195 µl Annexin V-FITC binding buffer followed by the addition of 5 µl Annexin V-FITC and 10 µl PI. Cells were incubated at room temperature for 20 min in the dark and subsequently analyzed using an EPICS XL-MCL FACScan flow cytometer (Beckman Coulter, Inc.).
Cell migration and invasion assay
Transwell migration and invasion assays were implemented using Transwell chambers (Corning, Inc.). Chambers were pre-coated with or without Matrigel (Corning, Inc.) at 4°C overnight. HUVECs were collected at 24 h post-transfection, resuspended in fresh culture medium with serum depletion, and transferred into the top of chambers (5×104 cells) coated with or without Matrigel for invasion or migration analysis, respectively. The bottom chambers were filled with fresh culture medium containing 10% FBS to induce cell migration and invasion. Allowing for migration and invasion for 24 h at 37°C, the cells that had migrated or invaded into the lower surface were fixed with formaldehyde, stained with 0.1% crystal violet (Beyotime Institute of Biotechnology) for 20 min at 37°C and then observed under a CKX41 light microscope (Olympus Corporation) at a magnification of ×100.
Western blot analysis
Total proteins were extracted from the HUVECs and quantified using the Enhanced BCA Protein Assay kit (Beyotime Institute of Biotechnology). Equal proteins (20 µg per lane) were then separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and loaded onto polyvinylidene fluoride (PVDF) membranes (Beyotime Institute of Biotechnology). The membranes were subjected to blocking buffer (5% skim milk) at room temperature for 1 h and probed with the primary antibodies, including anti-interleukin (IL)-6 (ab6672; 1/1,000), anti-IL-1β (ab226918; 1/1,000), anti-β-actin (ab8227; 1/2,000) and anti-ROCK2 (ab71598; 1/1,000) (all from Abcam) at 4°C overnight. The membranes were then probed with the secondary antibodies (ab205718; 1/5,000; Abcam) at room temperature for 2 h. Finally, the enhanced chemiluminescence kit (Beyotime Institute of Biotechnology) was adopted to visualize the protein blots, and the blot bands were then quantified using ImageJ software (version 1.46; National Institutes of Health).
Enzyme-linked immunosorbent assay (ELISA)
The release of IL-6 and IL-1β in the culture medium was detected using ELISA, with the application of the Human IL-6 ELISA kit (cat. no. PI330; Beyotime Institute of Biotechnology) and the human IL-1β ELISA kit (cat. no. PI305; Beyotime Institute of Biotechnology). The assay was performed in line with the guidelines from the manufacturer's protocols.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated using a Total RNA Extraction kit (Invitrogen; Thermo Fisher Scientific, Inc.) according to the product manual. Complementary DNA (cDNA) for circUSP36 and ROCK2 was synthesized using a Universal RT-PCR kit (cat. no. RP1100; Beijing Solarbio Science & Technology Co., Ltd.), and cDNA for miR-20a-5p was synthesized using a miRNA cDNA Synthesis kit (cat. no. 638315; Takara Biotechnology, Inc.). Subsequently, SYBR Master Mix (cat. no. SR1110; Beijing Solarbio Science & Technology Co., Ltd.) was used for qPCR analysis on a CFX96 Touch real-time PCR system (Bio-Rad Laboratories, Inc.). All experimental procedures were conducted in line with the manufacturers' protocols. β-actin or U6 was set as the housekeeping gene for circUSP36 and ROCK2 or miR-20a-5p, respectively. Relative expression was calculated using the 2−ΔΔCq method (21). The primer sequences are listed as follows: circUSP36 forward, 5′-GGC AGT AGA AGA GGA TGG GC-3′ and reverse, 5′-GGA GCA GGT GAC ACA GCC-3′; miR-20a-5p forward, 5′-GCC CGC TAA AGT GCT TAT AGT G-3′ and reverse, 5′-GCT GTC AAC GAT ACG CTA CGT-3′; ROCK2 forward, 5′-GAG AGC TTG CTG GAT GGC TT-3′ and reverse, 5′-CGA ACC AAC TGC ACT TCA CC-3′; U6 forward, 5′-ATT GGA ACG ATA CAG AGA AGA TT-3′ and reverse, 5′-GGA ACG CTT CAC GAA TTT G-3′; and β-actin forward, 5′-AGA TCA AGA TCA TTG CTC CTC CTG A-3′ and reverse, 5′-ATA CTC CTG CTT GCT GAT CCA CAT C-3′.
Cell transfection
Small interfering RNA (siRNA) targeting circUSP36 (si-circUSP36: 5′-GGG GAC CGC ATG GGG CTG TGT-3′) or ROCK2 (si-ROCK2: 5′-CAG AAG TGC AAG TCT ATT A-3′) for circUSP36 knockdown or ROCK2 knockdown and their corresponding controls (si-NC; scramble) were synthesized by Guangzhou RiboBio Co., Ltd. The mimic for miR-20a-5p for the restoration of miR-20a-5p expression (miR-20a-5p mimic), the inhibitor of miR-20a-5p for the inhibition of miR-20a-5p expression (miR-20a-5p inhibitor), and their corresponding controls (miR-NC mimic or miR-NC inhibitor; scramble) were purchased from Guangzhou RiboBio Co., Ltd. CircUSP36 was assembled into the pCD-ciR vector for circUSP36 overexpression (oe-circUSP36) (GenePharma, Inc.), and pCD-ciR empty vector (Vector) was used as its control. Lipofectamine 3000 kit (Invitrogen; Thermo Fisher Scientific, Inc.) was applied to perform cell transfection (siRNAs, 30 nM; miRNA mimic or inhibitor, 40 nM; vector, 1.5 µg) according to the manufacturer's instructions.
Bioinformatics analysis
For the prediction of putative targets of circUSP36, the online tool starBase (http://starbase.sysu.edu.cn/) was adopted. For the prediction of putative targets of miR-20a-5p, the online tool Targetscan (http://www.targetscan.org/vert_72/) was utilized.
Dual-luciferase reporter assay
According to the wild-type sequence of circUSP36 or ROCK2 3′UTR containing miR-20a-5p binding sites, the mutant sequence of circUSP36 or ROCK2 3′UTR (binding sites mutation) was designed. These sequences (wild-type and mutant-type) were amplified and separately inserted into pGL4 reporter plasmid (Promega Corporation) to generate luciferase reporter plasmids, which were named as WT-circUSP36, MUT-circUSP36, WT-ROCK2 and MUT-ROCK2, respectively. WT-circUSP36 (2 µg), MUT-circUSP36 (2 µg), WT-ROCK2 (2 µg) or MUT-ROCK2 (2 µg) together with miR-20a-5p mimic (40 nM) or miR-NC mimic (40 nM) were co-transfected into the HUVECs using Lipofectamine 3000 kit (Invitrogen; Thermo Fisher Scientific, Inc.). Transfected cells were then incubated for 48 h at 37°C, and the luciferase activities in these cells were investigated using the dual-luciferase reporter system (Promega Corporation) according to the manufacturer's protocol. Relative luciferase activity was expressed as normalization of Renilla luciferase activity to Firefly luciferase activity.
RNA binding protein immunoprecipitation (RIP) assay
HUVECs were transfected with miR-20a-5p mimic (60 nM) or miR-NC mimic (60 nM) using Lipofectamine 3000 kit (Invitrogen; Thermo Fisher Scientific, Inc.) and lysed in RIP lysis buffer from a Magna RIP kit (cat. no. 17-700; EMD Millipore). Cell lysates were reacted with magnetic beads conjugated with Argonaute 2 (Ago2) antibody (cat. no. ab186733; 1/1,000; Abcam) or immunoglobulin G (IgG; control) antibody (cat. no. ab190475; 1/1,000; Abcam) at 4°C overnight. The unbound materials were washed off, and complexes were eluted to extract RNAs. RT-qPCR analysis was then carried out to examine the expression of circUSP36 and ROCK2 as described above.
Statistical analysis
All experiments contained 3 independent repeats. Statistical analysis was processed using GraphPad Prism 5.0 (GraphPad Software, Inc.). The comparison of the differences between 2 groups was evaluated by unpaired Student's t-test, and the comparison of differences among ≥3 groups was distinguished by one- and two-way analyses of variance (ANOVA) with Tukey's post hoc test. Data are presented as the means ± standard deviation. A P-value <0.05 was considered to indicate a statistically significant difference.
Results
CircUSP36 is highly expressed in ox-LDL-exposed HUVECs
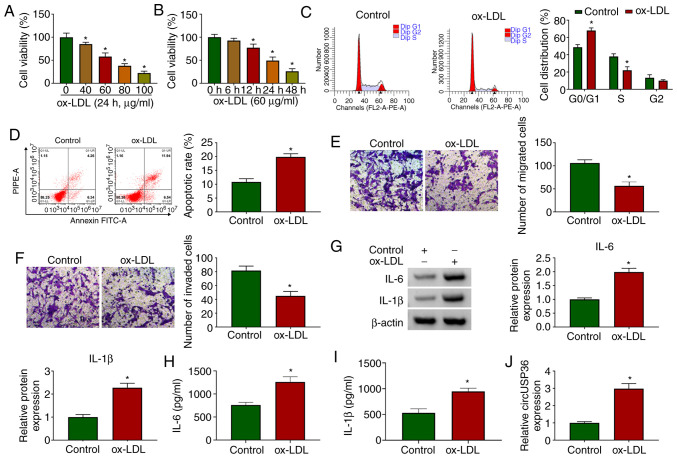
HUVECs were exposed to ox-LDL to examine the effects of ox-LDL on HUVECs. As shown by CCK-8 assay, the viability of the HUVECs was significantly decreased with the addition of ox-LDL (24 h) in a dose-dependent manner (Fig. 1A). Moreover, the viability of the HUVECs was also significantly decreased with the addition of ox-LDL (60 µg/ml) in a time-dependent manner (Fig. 1B). For the HUVECs exposed to 60 µg/ml ox-LDL for 24 h, flow cytometric assay revealed that the cell cycle was notably arrested (Fig. 1C), and cell apoptosis was notably promoted compared to the control (Fig. 1D). In addition, the addition of ox-LDL visibly blocked HUVEC migration and invasion, as shown by Transwell assay (Fig. 1E and F). The protein levels of pro-inflammatory factors (IL-6 and IL-1β) were significantly elevated in the HUVECs exposed to ox-LDL (Fig. 1G). The release of IL-6 and IL-1β was significantly enhanced in the culture medium of HUVECs exposed to ox-LDL (Fig. 1H and I). All these data suggested that the in vitro model of AS was successfully established, and that ox-LDL impaired HUVEC survival and metastasis, but induced apoptosis and inflammatory responses. Furthermore, it was observed that the expression of circUSP36 was significantly increased in the ox-LDL-exposed HUVECs (Fig. 1J), in a dose- and time-dependent manner (Fig. S1), suggesting that circUSP36 plays a role in ox-LDL-induced HUVEC injury.
Figure 1.
CircUSP36 is upregulated in a model of AS in vitro. (A) HUVECs were exposed to various concentrations of ox-LDL (40, 60, 80 and 100 µg/ml) for 24 h, and cell viability was detected by CCK-8 assay (n=3). (B) HUVECs were exposed to 60 µg/ml ox-LDL and for different periods of time (6, 12, 24 and 48 h), and cell viability was detected by CCK-8 assay (n=3). In HUVECs exposed to 60 µg/ml ox-LDL for 24 h, (C and D) cell cycle distribution and cell apoptosis were monitored by flow cytometric assay (n=3). (E and F) Cell migration and invasion were assessed by Transwell assay (n=3). (G) The expression of IL-6 and IL-1β was detected by western blot analysis (n=3). (H and I) The release of IL-6 and IL-1β in medium was measured by ELISA (n=3). (J) The expression of circUSP36 in ox-LDL-treated HUVECs was measured by RT-qPCR (n=3). *P<0.05 vs. control. HUVECs, human umbilical vascular endothelial cells; ox-LDL, oxidized low-density lipoprotein.
CircUSP36 inhibition attenuates ox-LDL-induced cell cycle arrest, apoptosis and inflammatory responses, but enhances the impaired migration and invasion of HUVECs induced by ox-LDL
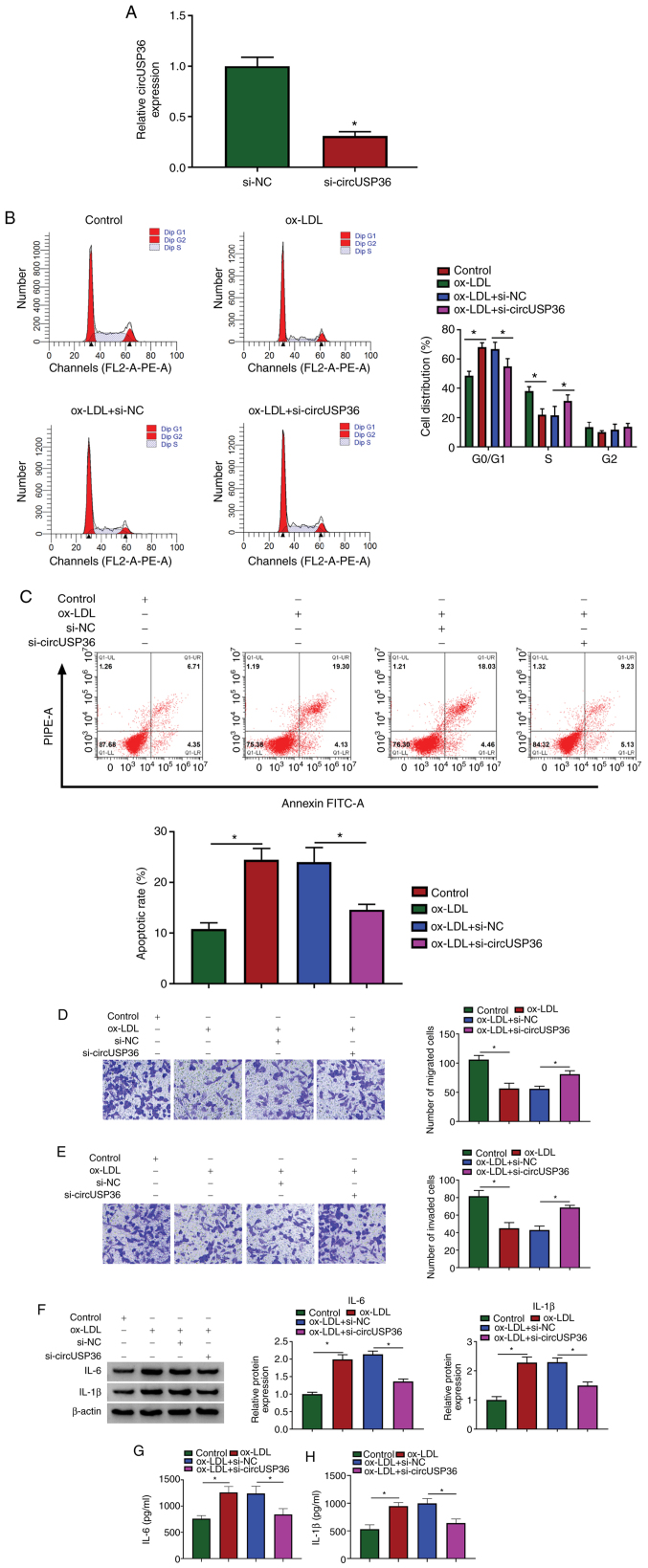
Given that the expression of circUSP36 was elevated in the ox-LDL-exposed HUVECs, the endogenous level of circUSP36 was decreased to explore its potential function. The expression of circUSP36 was significantly decreased in the HUVECs following transfection with si-circUSP36 (Fig. 2A). Flow cytometric assay revealed that si-circUSP36 transfection substantially diminished the cell cycle arrest and apoptosis which were induced by ox-LDL (Fig. 2B and C). Transwell assay revealed that the number of migrated and invaded cells which were blocked by ox-LDL exposure was largely recovered with the knockdown of circUSP36 (Fig. 2D and E). In addition, the protein levels of IL-6 and IL-1β in the HUVECs stimulated by ox-LDL were blocked by the knockdown of circUSP36 (Fig. 2F), and the release of IL-6 and IL-1β in culture medium was increased by ox-LDL, but weakened by transfection with si-circUSP36 (Fig. 2G and H). These data indicated that ox-LDL-induced HUVEC dysfunctions and inflammatory responses were alleviated by circUSP36 inhibition.
Figure 2.
CircUSP36 knockdown inhibits ox-LDL-induced cycle arrest, apoptosis and inflammatory responses, but promotes migration and invasion of HUVECs which had been blocked by ox-LDL. (A) Silencing efficiency of circUSP36 was examined by RT-qPCR. In ox-LDL-exposed HUVECs transfected with si-circUSP36 or si-NC, (B and C) cell cycle distribution and apoptosis were investigated using flow cytometric assay (n=3). (D and E) Cell migration and invasion were monitored by Transwell assay (n=3). (F) The protein levels of IL-6 and IL-1β were detected by western blot analysis (n=3). (G and H) The release of IL-6 and IL-1β in the culture medium was detected by ELISA (n=3). *P<0.05 vs. respective control. HUVECs, human umbilical vascular endothelial cells; ox-LDL, oxidized low-density lipoprotein.
CircUSP36 targets miR-20a-5p and negatively regulates miR-20a-5p expression
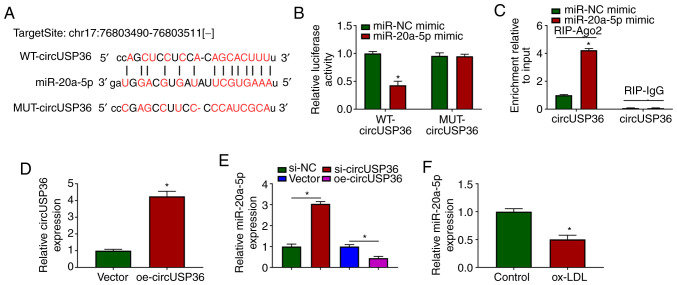
To ascertain whether circUSP36 serves as a molecular sponge to mediate the expression of downstream miRNAs, the potential target miRNAs of circUSP36 were identified. The targeting sites between circUSP36 and miR-20a-5p were analyzed using the bioinformatics tool, starBase, which revealed that miR-20a-5p may be a target of circUSP36 (Fig. 3A). Subsequently, dual-luciferase reporter assay demonstrated that transfection with miR-20a-5p mimic significantly suppressed the luciferase activities in HUVECs transfected with WT-circUSP36 reporter plasmid, but not with the MUT-circUSP36 reporter plasmid (Fig. 3B). In addition, RIP assay revealed that transfection with miR-20a-5p mimic notably enriched circUSP36 in the RIP-Ago group compared with miR-NC mimic, while circUSP36 in the RIP-IgG group was hardly enriched (Fig. 3C). A circUSP36 overexpression plasmid was constructed, and transfection with oe-circUSP36 markedly enhanced the expression of circUSP36 (Fig. 3D). It was noted that the expression of miR-20a-5p was markedly increased in the HUVECs in which circUSP36 was knocked down, while it was decreased in the HUVECs in which circUSP36 was overexpressed (Fig. 3E). Notably, the expression of miR-20a-5p was markedly decreased in the ox-LDL-exposed HUVECs (Fig. 3F). On the whole, these findings demonstrated that miR-20a-5p was a target of circUSP36, and that circUSP36 negatively regulated the expression of miR-20a-5p.
Figure 3.
miR-20a-5p is a target of circUSP36. (A) The potential association between miR-20a-5p and circUSP36 was predicted using the online tool, starBase. (B and C) The association between miR-20a-5p and circUSP36 was verified by dual-luciferase reporter assay and RIP assay (n=3). (D) The overexpression efficiency of circUSP36 was detected by RT-qPCR. (E) The effects of circUSP36 knockdown or overexpression on the expression of miR-20a-5p were detected by RT-qPCR (n=3). (F) The expression of miR-20a-5p in ox-LDL-treated HUVECs was measured by RT-qPCR (n=3). *P<0.05 vs. respective control. HUVECs, human umbilical vascular endothelial cells; ox-LDL, oxidized low-density lipoprotein.
CircUSP36 downregulation mediates ox-LDL-induced HUVEC dysfunction and inflammation by upregulating miR-20a-5p
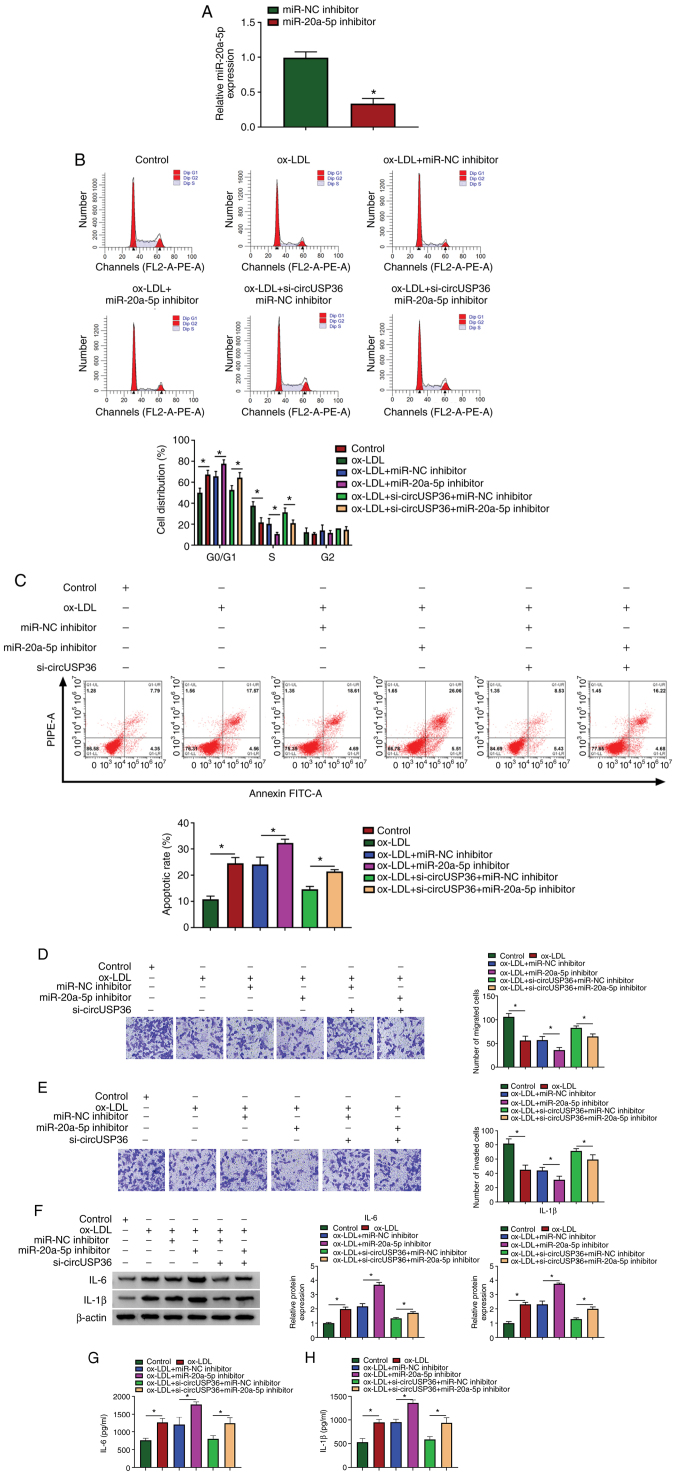
To determine whether circUSP36 targets miR-20a-5p to regulate ox-LDL-induced apoptosis and inflammatory responses, circUSP36 and miR-20a-5p were knocked down alone or in combination in the ox-LDL-exposed HUVECs. The expression of miR-20a-5p was predominantly decreased in the HUVECs following transfection with miR-20a-5p inhibitor compared to miR-NC inhibitor (Fig. 4A). Functionally, miR-20a-5p inhibition aggravated ox-LDL-induced cell cycle arrest and apoptosis, and the effects of circUSP36 knockdown were partly abolished by miR-20a-5p inhibition, leading to the promotion of cell cycle arrest and apoptosis (Fig. 4B and C). Moreover, miR-20a-5p inhibition further suppressed HUVEC migration and invasion which were blocked by ox-LDL exposure, and cell migration/invasion which were partly attenuated by circUSP36 knockdown was suppressed by miR-20a-5p inhibition (Fig. 4D and E). Moreover, miR-20a-5p inhibition increased the protein levels of IL-6 and IL-1β in the HUVECs, which were stimulated by ox-LDL exposure, and miR-20a-5p inhibition also impaired the effects of circUSP36 knockdown, increasing the levels of IL-6 and IL-1β (Fig. 4F). The release of IL-6 and IL-1β in the culture medium stimulated by ox-LDL was further promoted by transfection with miR-20a-5p inhibitor, and the release of IL-6 and IL-1β, which was suppressed by si-circUSP36 transfection was partly enhanced by the reintroduction of miR-20a-5p inhibitor (Fig. 4G and H). Combined with the above-mentioned data, it was thus concluded that circUSP36 exerts its functions in ox-LDL-exposed HUVECs by targeting miR-20a-5p.
Figure 4.
CircUSP36 regulates ox-LDL-mediated HUVEC injury and inflammation by targeting miR-20a-5p. (A) The efficiency of miR-20a-5p inhibition was examined by RT-qPCR. HUVECs exposed to ox-LDL were transfected with miR-20a-5p inhibitor, miR-NC inhibitor, si-circUSP36 + miR-20a-5p inhibitor or si-circUSP36 + miR-NC inhibitor. (B and C) Cell cycle distribution and cell apoptosis were investigated by flow cytometric assay (n=3). (D and E) Cell migration and invasion were assessed by Transwell assay (n=3). (F) The expression of IL-6 and IL-1β in cells was examined by western blot analysis (n=3). (G and H) The release of IL-6 and IL-1β in the culture medium was detected by ELISA (n=3). *P<0.05 vs. respective control. HUVECs, human umbilical vascular endothelial cells; ox-LDL, oxidized low-density lipoprotein.
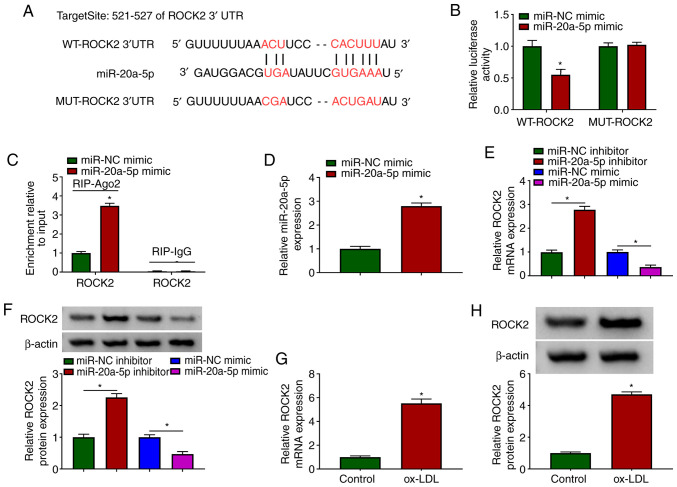
miR-20a-5p binds to ROCK2 3′UTR and negatively regulates ROCK2 expression
To further address the functional mechanism of circUSP36, mRNAs targeted by miR-20a-5p were identified. The bioinformatics tool, TargetScan revealed that miR-20a-5p bound to ROCK2 through several targeting sites (Fig. 5A). Likewise, their interaction was validated by dual-luciferase reporter assay and RIP assay. The data indicated that the reintroduction of miR-20a-5p mimic significantly diminished the luciferase activity in HUVECs transfected with WT-ROCK2, but did not affect the luciferase activity in the HUVECs transfected with MUT-ROCK2 (Fig. 5B). In addition, transfection with miR-20a-5p mimic markedly enriched ROCK2 in the RIP-Ago2 group, but not in the RIP-IgG group (Fig. 5C). In the HUVECs transfected with miR-20a-5p mimic, it was found that the expression of miR-20a-5p was notably increased (Fig. 5D). It was then found that the expression of ROCK2 in the HUVECs transfected with miR-20a-5p inhibitor was substantially enhanced, while it was markedly decreased in the HUVECs transfected with miR-20a-5p mimic (Fig. 5E and F), suggesting that miR-20a-5p negatively regulated ROCK2. Moreover, the expression of ROCK2 was aberrantly elevated in the ox-LDL-exposed HUVECs (Fig. 5G and H). Collectively, ROCK2 was found to be a target of miR-20a-5p, and its expression was negatively regulated by miR-20a-5p.
Figure 5.
ROCK2 is targeted by miR-20a-5p. (A) The potential association between miR-20a-5p and ROCK2 was predicted using the online tool, TargetScan. (B and C) The association between miR-20a-5p and ROCK2 was validated by dual-luciferase reporter assay and RIP assay (n=3). (D) The overexpression efficiency of miR-20a-5p was detected by RT-qPCR. (E and F) The effects of miR-20a-5p knockdown or overexpression on the expression of ROCK2 were examined by RT-qPCR and western blot analysis (n=3). (G and H) The expression of ROCK2 in ox-LDL-exposed HUVECs was measured by RT-qPCR and western blot analysis (n=3). *P<0.05 vs. respective control. HUVECs, human umbilical vascular endothelial cells; ox-LDL, oxidized low-density lipoprotein.
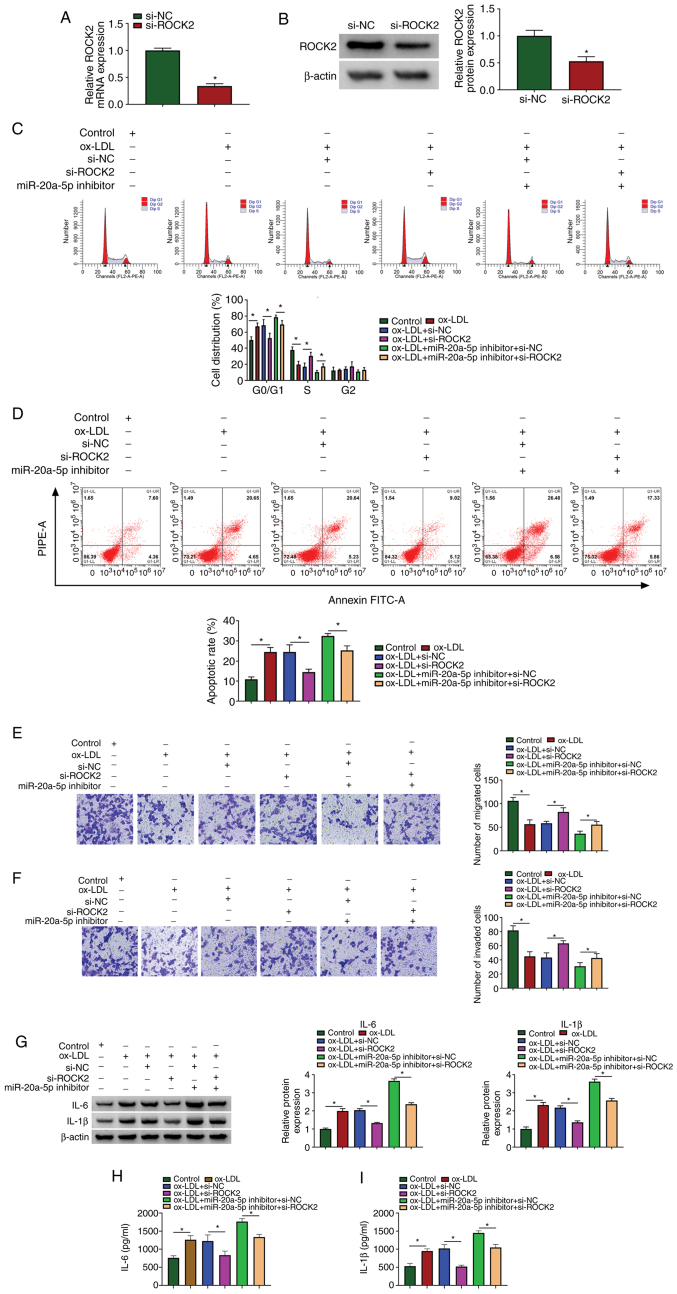
miR-20a-5p inhibition aggravates ox-LDL-induced HIVEC injury and inflammation by increasing the expression of ROCK2
The roles of miR-20a-5p and ROCK2 in ox-LDL-induced HUVEC injury and were then investigated. siRNA targeting ROCK2 was used to downregulate the expression of ROCK2, and the data from RT-qPCR and western blot analysis revealed that the expression of ROCK2 was notably decreased in the HUVECs following transfection with si-ROCK2 (Fig. 6A and B). Cell cycle arrest and cell apoptosis induced by ox-LDL exposure were attenuated by transfection with si-ROCK2. In addition, cell cycle arrest and cell apoptosis, which were aggravated in the ox-LDL-exposed HUVECs transfected with miR-20a-5p inhibitor were partly suppressed by the reintroduction of si-ROCK2 (Fig. 6C and D). Cell migration and cell invasion, which were blocked by ox-LDL exposure, were recovered by ROCK2 downregulation, and ROCK2 downregulation also partly reversed the effects of miR-20a-5p inhibition, promoting cell migration and invasion (Fig. 6E and F). The protein levels of IL-6 and IL-1β stimulated in the ox-LDL-exposed HUVECs were blocked by transfection with si-ROCK2, and transfection with si-ROCK2 also reversed the effects of miR-20a-5p inhibitor, and partly reduced the levels of IL-6 and IL-1β (Fig. 6G). The release of IL-6 and IL-1β in the culture medium was also impaired by ROCK2 knockdown, and the release of IL-6 and IL-1β induced by miR-20a-5p inhibition was largely suppressed by ROCK2 knockdown (Fig. 6H and I). These data indicated that ROCK2 knockdown attenuated ox-LDL-induced HUVEC injury and inflammation, and ROCK2 knockdown also reversed the effects of miR-20a-5p inhibition.
Figure 6.
miR-20a-5p regulates ox-LDL-induced HUVEC injurys and inflammation by mediating ROCK2. (A and B) The inference efficiency of ROCK2 was examined by RT-qPCR and western blot analysis. HUVECs exposed to ox-LDL were transfected with si-ROCK2, si-NC, miR-20a-5p inhibitor + si-ROCK2 or miR-20a-5p inhibitor + si-NC. (C and D) Cell cycle distribution and cell apoptosis were monitored by flow cytometric assay (n=3). (E and F) Transwell assay was performed to observe cell migration and invasion (n=3). (G) The protein levels of IL-6 and IL-1β in cells were examined by western blot analysis (n=3). (H and I) The release of IL-6 and IL-1β in the culture medium was detected by ELISA (n=3). *P<0.05 vs. respective control. HUVECs, human umbilical vascular endothelial cells; ox-LDL, oxidized low-density lipoprotein.
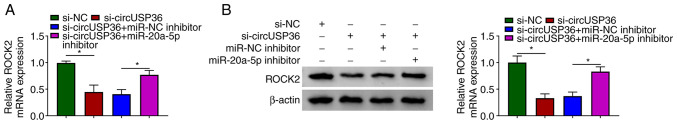
CircUSP36 indirectly regulates ROCK2 expression by targeting miR-20a-5p
Notably, it was found that the expression of ROCK2 was markedly decreased in the HUVECs following transfection with si-circUSP36 compared to si-NC, but was partly recovered in the HUVECs following transfection with si-circUSP36 + miR-20a-5p inhibitor compared to si-circUSP36 + miR-NC inhibitor at both the mRNA and protein level (Fig. 7). These data illustrate that circUSP36 functions as miR-20a-5p sponge to modulate the expression of ROCK2.
Figure 7.
CircUSP36 regulates ROCK2 expression by targeting miR-20a-5p. HUVECs were transfected with si-circUSP36, si-NC, si-circUSP36 + miR-20a-5p inhibitor or si-circUSP36 + miR-NC inhibitor. (A and B) The expression of ROCK2 in these transfected cells was measured by RT-qPCR and western blot analysis (n=3). *P<0.05 vs. respective control. HUVECs, human umbilical vascular endothelial cells; ox-LDL, oxidized low-density lipoprotein.
Discussion
The pathogenesis of AS is complex, and the characteristics of AS include the uncontrolled proliferation of smooth muscle cells (SMCs), necrosis and fibrosis, local inflammatory responses and endothelial damage (22). The present study focused on the molecular mechanisms of endothelial cell injury. The findings mainly demonstrated that circUSP36 was dysregulated in ox-LDL-exposed HUVECs, exhibiting a high expression. The silencing of circUSP36 attenuated ox-LDL-induced HUVEC injury and dysfunction, resulting in the inhibition of cell cycle arrest, cell apoptosis and inflammatory responses, and in the activation of cell migration and invasion. Mechanistically, circUSP36 functioned in the HUVECs by targeting the miR-20a-5p/ROCK2 axis.
Several circRNAs have been functionally identified in ox-LDL-exposed cell models (13,23). For example, circ_0003575 has been found to be highly expressed in ox-LDL-stimulated HUVECs, and its downregulation restores the ability of HUVEC proliferation and angiogenesis (13). The reduced expression of circANRIL has been shown to block mouse vascular endothelial cell apoptosis and inflammatory factor expression, thus preventing the progression of coronary AS (24). As for circUSP36, it was identified to be highly expressed in ox-LDL-exposed HUVECs in a previous study (13), and Zhang et al and Liu et al reported that circUSP36 (circ_0003204) reintroduction suppressed the proliferation, migration, angiogenesis and tube formation of human aortic endothelial cells (HAECs) or HUVECs exposed to ox-LDL (25,26). Consistent with these findings, the results of the present study demonstrated that circUSP36 knockdown attenuated ox-LDL-induced cell cycle arrest and apoptosis, recovered ox-LDL-blocked cell migration and invasion, and reduced ox-LDL-induced inflammatory responses. All these functional effects suggested that circUSP36 aggravates the progression of AS.
Cytoplasmic circRNAs harbor binding sites with miRNAs and serve as competing endogenous RNAs (ceRNAs) to sponge miRNAs (27). CircUSP36 has been demonstrated to be mainly distributed in the cytoplasm (25). The present study thus screened and validated the target miRNAs of circUSP36. Through the current analyses, miR-20a-5p was identified as a target of circUSP36 by dual-luciferase reporter assay and RIP assay. Moreover, a previous study documented the role and mechanisms of miR-20a-5p in AS in vitro, and the data indicated that miR-20a-5p expression was suppressed by the exposure of HAECs to ox-LDL, and miR-20a-5p restoration attenuated ox-LDL-induced inflammatory injury (28). Another study reported that miR-20a-5p was weakly expressed in coronary artery disease, and that miR-20a-5p agomir suppressed hydrogen peroxide-induced apoptosis of HUVECs (29). Consistently, miR-20a-5p in the present study was downregulated in ox-LDL-exposed HUVECs, and its inhibition aggravated ox-LDL-triggered HUVEC dysfunction and inflammatory injury.
The circRNA-miRNA-mRNA network has been found to be an important mode involving in the pathogenesis of human diseases (11). In the presen study, to construct the same network of circUSP36, the potential target mRNAs of miR-20a-5p were obtained. The current analysis indicated that miR-20a-5p directly bound to ROCK2 3′UTR to degrade the expression of ROCK2. Reports pf the role of ROCK2 in AS are abundant. Zhou et al demonstrated that ROCK2 regulated the formation of foam cells and contributed to the development of AS (30). Mattaliano et al indicated that ox-LDL exposure catalyzed the dynamic combination of ROCK2 and ox-LDL receptor-1 (LOX-1) and stimulated the activity of ROCK2, and ROCK2 suppression impaired ox-LDL-induced inflammation in endothelial cells (31). Huang et al demonstrated that ROCK2 knockdown blocked apoptosis and increased cytoskeleton stability of HUVECs, thus ameliorating HUVEC dysfunction (32). In brief, ROCK2 contributes to HUVEC dysfunction and AS progression through multiple mechanisms. The data of the present study demonstrated that ROCK2 silencing attenuated ox-LDL-induced HUVEC cycle arrest, apoptosis and inflammation, but promoted HUVEC migration and invasion which had been blocked by ox-LDL. Moreover, ROCK2 expression was activated by circUSP36 through miR-20a-5p, thereby participating in ox-LDL-induced HUVEC dysfunction.
However, the present study is only a preliminary study of the role of circUSP36 in ox-LDL-induced HUVEC dysfunction. Some effects of circUSP36, such as oxidative stress, were not investigated in the present study. Moreover, the similar role of circUSP36 in the progression of AS was not examined in vivo. Therefore, further extensive in-depth studies are required, including oxidative stress detection and animal experiments.
In conclusion, the present study identified that circUSP36 regulated ox-LDL-induced HUVEC dysfunction and provided the circUSP36/miR-20a-5p/ROCK2 network as a novel mechanism for circUSP36 involvement in these processes. The present study provides a molecular basis for the involvement of circUSP36 in the pathogenesis of AS, and the targeting of the circUSP36/miR-20a-5p/ROCK2 axis may prove to be a novel strategy against AS.
Supplementary Data
Acknowledgments
Not applicable.
Availability of data and materials
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request.
Authors' contributions
JM and BW designed the study. RS and YW conducted the reviewing and editing of the manuscript and were involved in the design of the study. All the authors had full access to all the data in the study and took responsibility for the integrity of the data and the accuracy of the data analysis. JM supervised the study. JM, BW and YW wrote the manuscript. RS revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Libby P, Hansson GK. Inflammation and immunity in diseases of the arterial tree: Players and layers. Circ Res. 2015;116:307–311. doi: 10.1161/CIRCRESAHA.116.301313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li B, Li W, Li X, Zhou H. Inflammation: A novel therapeutic target/direction in atherosclerosis. Curr Pharm Des. 2017;23:1216–1227. doi: 10.2174/1381612822666161230142931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gimbrone MA, Jr, García-Cardeña G. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ Res. 2016;118:620–636. doi: 10.1161/CIRCRESAHA.115.306301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Wang Y, Li Y, Wang S, Zhu X. Platelet-rich plasma protects HUVECs against oX-LDL-induced injury. Open Med (Wars) 2018;13:41–52. doi: 10.1515/med-2018-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kattoor AJ, Goel A, Mehta JL. LOX-1: Regulation, signaling and its role in atherosclerosis. Antioxidants (Basel) 2019;8:218. doi: 10.3390/antiox8070218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitra S, Deshmukh A, Sachdeva R, Lu J, Mehta JL. Oxidized low-density lipoprotein and atherosclerosis implications in anti-oxidant therapy. Am J Med Sci. 2011;342:135–142. doi: 10.1097/MAJ.0b013e318224a147. [DOI] [PubMed] [Google Scholar]
- 7.Yin J, Hou X, Yang S. microRNA-338-3p promotes ox-LDL-induced endothelial cell injury through targeting BAMBI and activating TGF-β/Smad pathway. J Cell Physiol. 2019;234:11577–11586. doi: 10.1002/jcp.27814. [DOI] [PubMed] [Google Scholar]
- 8.Peng J, Tang ZH, Ren Z, He B, Zeng Y, Liu LS, Wang Z, Wei DH, Zheng XL, Jiang ZS. TET2 protects against oxLDL-Induced HUVEC dysfunction by upregulating the CSE/H2S system. Front Pharmacol. 2017;8:486. doi: 10.3389/fphar.2017.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng X, Lin W, Guo M, Zou Q. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput Biol. 2017;13:e1005420. doi: 10.1371/journal.pcbi.1005420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA. 2015;6:563–579. doi: 10.1002/wrna.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bayoumi AS, Aonuma T, Teoh JP, Tang YL, Kim IM. Circular noncoding RNAs as potential therapies and circulating biomarkers for cardiovascular diseases. Acta Pharmacol Sin. 2018;39:1100–1109. doi: 10.1038/aps.2017.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan RY, Zhao CH, Yuan JX, Zhang YJ, Jin JL, Gu MF, Mao ZY, Sun HJ, Jia QW, Ji MY, et al. Circular RNA profile in coronary artery disease. Am J Transl Res. 2019;11:7115–7125. [PMC free article] [PubMed] [Google Scholar]
- 13.Li CY, Ma L, Yu B. Circular RNA hsa_circ_0003575 regulates oxLDL induced vascular endothelial cells proliferation and angiogenesis. Biomed Pharmacother. 2017;95:1514–1519. doi: 10.1016/j.biopha.2017.09.064. [DOI] [PubMed] [Google Scholar]
- 14.Feinberg MW, Moore KJ. MicroRNA regulation of atherosclerosis. Circ Res. 2016;118:703–720. doi: 10.1161/CIRCRESAHA.115.306300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dlouhá D, Hubáček JA. Regulatory RNAs and cardiovascular disease- with a special focus on circulating microRNAs. Physiol Res. 2017;66(Suppl 1):S21–S38. doi: 10.33549/physiolres.933588. [DOI] [PubMed] [Google Scholar]
- 16.Wojciechowska A, Braniewska A, Kozar-Kamińska K. MicroRNA in cardiovascular biology and disease. Adv Clin Exp Med. 2017;26:865–874. doi: 10.17219/acem/62915. [DOI] [PubMed] [Google Scholar]
- 17.Gao G, Chen W, Liu M, Yan X, Yang P. Circulating MicroRNAs as novel potential biomarkers for left ventricular remodeling in postinfarction heart failure. Dis Markers. 2019;2019:5093803. doi: 10.1155/2019/5093803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muendlein A, Geiger K, Leiherer A, Saely CH, Fraunberger P, Drexel H. Evaluation of the associations between circulating microRNAs and kidney function in coronary angiography patients. Am J Physiol Renal Physiol. 2020;318:F315–F321. doi: 10.1152/ajprenal.00429.2019. [DOI] [PubMed] [Google Scholar]
- 19.Shahbazi R, Baradaran B, Khordadmehr M, Safaei S, Baghbanzadeh A, Jigari F, Ezzati H. Targeting ROCK signaling in health, malignant and non-malignant diseases. Immunol Lett. 2020;219:15–26. doi: 10.1016/j.imlet.2019.12.012. [DOI] [PubMed] [Google Scholar]
- 20.Okamoto R, Li Y, Noma K, Hiroi Y, Liu PY, Taniguchi M, Ito M, Liao JK. FHL2 prevents cardiac hypertrophy in mice with cardiac-specific deletion of ROCK2. FASEB J. 2013;27:1439–1449. doi: 10.1096/fj.12-217018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Singh RB, Mengi SA, Xu YJ, Arneja AS, Dhalla NS. Pathogenesis of atherosclerosis: A multifactorial process. Exp Clin Cardiol. 2002;7:40–53. [PMC free article] [PubMed] [Google Scholar]
- 23.Yang L, Yang F, Zhao H, Wang M, Zhang Y. Circular RNA circCHFR facilitates the proliferation and migration of vascular smooth muscle via miR-370/FOXO1/Cyclin D1 pathway. Mol Ther Nucleic Acids. 2019;16:434–441. doi: 10.1016/j.omtn.2019.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song CL, Wang JP, Xue X, Liu N, Zhang XH, Zhao Z, Liu JG, Zhang CP, Piao ZH, Liu Y, Yang YB. Effect of circular ANRIL on the inflammatory response of vascular endothelial cells in a rat model of coronary atherosclerosis. Cell Physiol Biochem. 2017;42:1202–1212. doi: 10.1159/000478918. [DOI] [PubMed] [Google Scholar]
- 25.Zhang S, Song G, Yuan J, Qiao S, Xu S, Si Z, Yang Y, Xu X, Wang A. Circular RNA circ_0003204 inhibits proliferation, migration and tube formation of endothelial cell in atherosclerosis via miR-370-3p/TGFbetaR2/phosph-SMAD3 axis. J Biomed Sci. 2020;27:11. doi: 10.1186/s12929-019-0595-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Liu H, Ma X, Mao Z, Shen M, Zhu J, Chen F. Circular RNA has_circ_0003204 inhibits oxLDL-induced vascular endothelial cell proliferation and angiogenesis. Cell Signal. 2020;70:109595. doi: 10.1016/j.cellsig.2020.109595. [DOI] [PubMed] [Google Scholar]
- 27.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 28.Chen M, Li W, Zhang Y, Yang J. MicroRNA-20a protects human aortic endothelial cells from Ox-LDL-induced inflammation through targeting TLR4 and TXNIP signaling. Biomed Pharmacother. 2018;103:191–197. doi: 10.1016/j.biopha.2018.03.129. [DOI] [PubMed] [Google Scholar]
- 29.Wang D, Wang Y, Ma J, Wang W, Sun B, Zheng T, Wei M, Sun Y. MicroRNA-20a participates in the aerobic exercise-based prevention of coronary artery disease by targeting PTEN. Biomed Pharmacother. 2017;95:756–763. doi: 10.1016/j.biopha.2017.08.086. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Q, Mei Y, Shoji T, Han X, Kaminski K, Oh GT, Ongusaha PP, Zhang K, Schmitt H, Moser M, et al. Rho-associated coiled-coil-containing kinase 2 deficiency in bone marrow-derived cells leads to increased cholesterol efflux and decreased atherosclerosis. Circulation. 2012;126:2236–2247. doi: 10.1161/CIRCULATIONAHA.111.086041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mattaliano MD, Wooters J, Shih HH, Paulsen JE. ROCK2 associates with lectin-like oxidized LDL receptor-1 and mediates oxidized LDL-induced IL-8 production. Am J Physiol Cell Physiol. 2010;298:C1180–1187. doi: 10.1152/ajpcell.00483.2009. [DOI] [PubMed] [Google Scholar]
- 32.Huang L, Dai F, Tang L, Bao X, Liu Z, Huang C, Zhang T, Yao W. Distinct roles for ROCK1 and ROCK2 in the regulation of oxldl-mediated endothelial dysfunction. Cell Physiol Biochem. 2018;49:565–577. doi: 10.1159/000492994. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request.