Abstract
Perfused three-dimensional (3D) cultures enable long-term in situ growth and monitoring of 3D organoids making them well-suited for investigating organoid development, growth, and function. One of the limitations of this long-term on-chip perfused 3D culture is unintended and disruptive air bubbles. To overcome this obstacle, we invented an imaging platform that integrates an innovative microfluidic bubble pocket for long-term perfused 3D culture of gastrointestinal (GI) organoids. We successfully applied 3D printing technology to create polymer molds that cast polydimethylsiloxane (PDMS) culture chambers in addition to bubble pockets. Our developed platform traps unintended, or induced, air bubbles in an integrated PDMS pocket chamber, where the bubbles diffuse out across the gas permeable PDMS or an outlet tube. We demonstrated that our robust platform integrated with the novel bubble pocket effectively circumvents the development of bubbles into human and mouse GI organoid cultures during long-term perfused time-course imaging. Our platform with the innovative integrated bubble pocket is ideally suited for studies requiring long-term perfusion monitoring of organ growth and morphogenesis as well as function.
INTRODUCTION
Human “organs-on-chips” are a new platform in which advanced microfluidic technologies are combined with tissue engineering approaches to recapitulate the biological and physiological microenvironment of human organs.1–4 These organ-like microdevices enable more accurate prediction of human physiology and, more importantly, offer the potential to develop specialized in vitro human disease models that could revolutionize drug development.2,3,5–9 In particular, there has been a rapid development in new in vitro models that mimic gastrointestinal (GI) physiology and disease. Besides the human “gut-on-a-chip” platform,9 advances in the areas of tissue engineering have led to the development of intestinal organoids, a physiologically relevant in vitro model of the intestine with the potential for regenerative and personalized medicine.10–13 Moreover, the advances in microfluidic systems allow the fabrication of microbioreactors for growing these engineered intestinal organoids under continuous perfusion and provide cells with various physiological perturbations.14–16 Consequently, the integration of microfluidic devices with 3D human tissue models, “organoids-on-chips,” or “microphysiological platforms,” has emerged as superior predictive platforms for forecasting safety and efficacy of drugs as well as studying the effects of environmental toxins on the human tissues in preclinical studies.15–17
Compared to static cell culture with their associated periodic media changes, perfused 3D culture provides controlled delivery and removal of soluble biochemical factors in the extracellular microenvironment in addition to controlled application of mechanical cues exerted via fluid flow due to the continuous unidirectional nutrient supply and waste elimination. Long-term perfused 3D culture of cells or organoids with real-time imaging technique is particularly useful for exploring organ development and morphogenesis as well as function. Despite the several advantages listed above, long-term microfluidic 3D culture platforms still face various problems, including contamination caused by repeated manual interventions, fluctuation of the cell microenvironment due to the routine medium replacement, and undesired disruptive air bubble accumulation. Bubble accumulation is the most challenging critical hurdle to avoid in most polydimethylsiloxane (PDMS)-based long-term on-chip culture systems. Undesired accumulation of air bubbles in the microfluidic perfusion culture device abruptly changes the microenvironment of adherent cells and can even rupture cell membranes, negatively affecting the experimental outcomes.18–20 Existing approaches to avoid air bubbles have various drawbacks even though many groups have developed various bubble traps integrated with microfluidic systems.20–24 The manufacturing process for an existing report18 remained relatively complex, so it is not simply flexible to all types of microfluidic applications. Another approach20 required relatively longer degassing time due to the low gas removal rates in the system. Some methods22 need an external pressure or vacuum source that is especially inconvenient for portable, point-of-care, integrated microfluidic devices. Several bubble trap chambers23 launch large dead volumes, which is not appropriate for microfluidic devices involving multiple reaction steps as well as costly reagents. Additionally, bubble traps containing hydrophobic membrane do not provide acceptably fast degassing.24
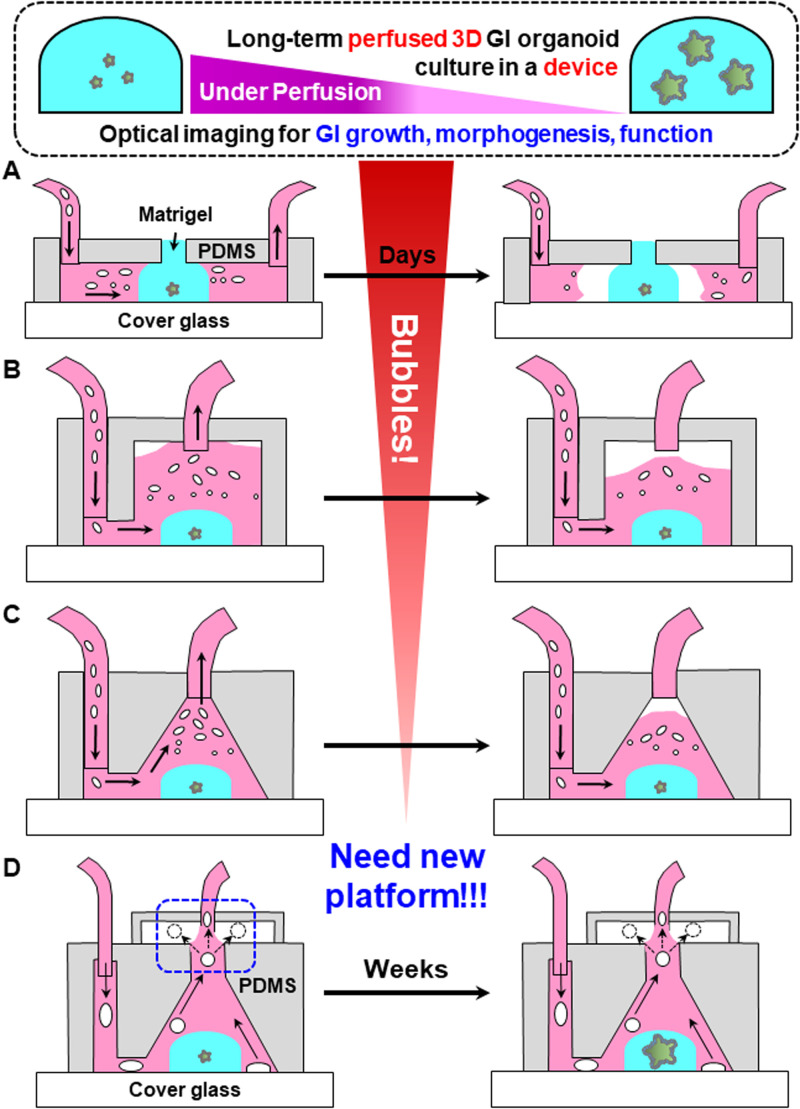
Due to the current lack of existing technologies that adequately eliminate air bubbles, we sought to create a robust, long-term perfused 3D “intestine-on-a-chip” platform that is directly integrated with a novel bubble pocket (Fig. 1). We effectively characterized the trapping and de-bubbling capability of the incorporated bubble pocket on the perfused 3D platform in a normal device operation, including a continuous flow at atmospheric pressure. Our developed bubble pocket captures bubbles by utilizing the buoyancy of air bubbles in the liquid media, providing a space for the bubbles that have floated to the top of the chamber, and serves to pull the air bubbles out through the gas-permeable PDMS and the connected outlet tube [Fig. 1(d)]. Ultimately, we demonstrated that our robust platform integrated with the novel bubble pocket effectively evades the adverse effects of bubbles onto human and mouse GI organoids during a long-term (over 30 days) perfused time-course imaging.
FIG. 1.
Three different types of microfluidic perfusion devices [(a)–(c)] for long-term 3D culture of organoids show problems generated from introduced air bubbles. Air bubbles through the inlet tube were accumulated in the culture chambers over time, causing disruptive effects on normal device operation. (d) The de-bubbling principle of an integrated bubble pocket model for long-term perfusion culture in the 3D intestine-on-a-chip platform. Device operation, including perfusion flow at atmospheric pressure, is normally sustained during the bubble trapping and de-bubbling processes.
EXPERIMENTAL PROCEDURES
Device fabrication
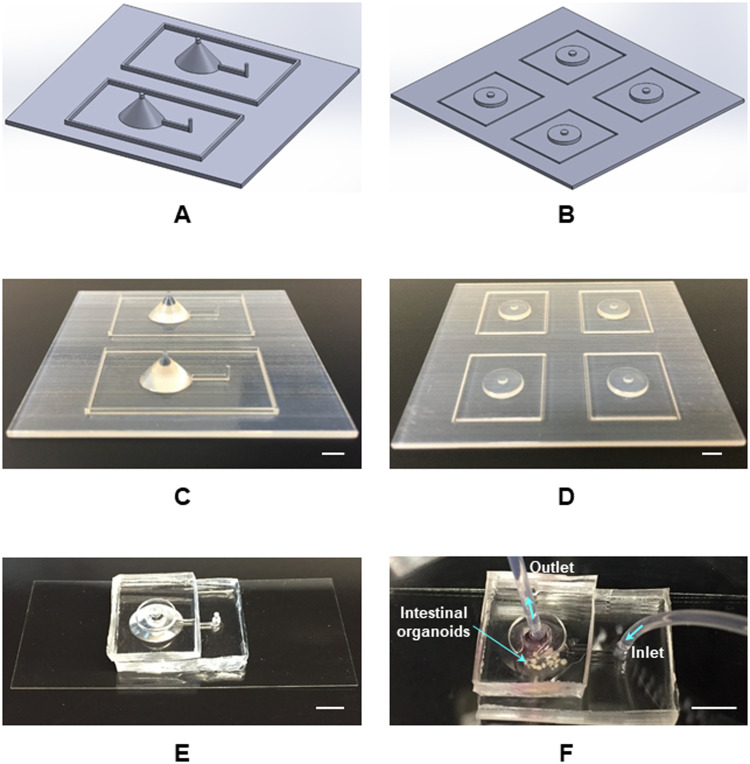
Figure 2 presents the fabrication procedure for long-term perfused 3D intestine-on-a-chip platform. 3D structures for culture chambers and bubble pockets were designed and modeled using SolidWorks 2017 (Dassault Systems SolidWorks Corp.) CAD package [Figs. 2(a) and 2(b)]. To produce the perfusion culture and imaging platform, polymer master molds were effectively printed via a fused deposition modeling (FDM) process in a 3D printing system (uPrint SE, Stratasys) [Figs. 2(c) and 2(d)]. The material for this process was acrylonitrile butadiene styrene (ABS) polymer that is a terpolymer resin made by polymerizing styrene and acrylonitrile in the presence of polybutadiene. The perfused 3D intestine-on-a-chip platform was constructed with PDMS (10:1 wt./wt.; Sylgard 184, Dow Corning) by a standard PDMS replica molding technique. After cleaning the surface using a 1:5 ratio of (HCl:H2O) (v/v) solution for 10 min, the PDMS replica and cover glasses were activated using the oxygen plasma treatment (CS-1701 RIE, MARCH Instruments) to generate surface hydroxyl groups. The platform with bubble pocket was assembled by spontaneous bonding between the patterned PDMS replica and cover glass [Fig. 2(e)]. Since the production of these fabricated PDMS devices does not require cleanroom facilities, this technology reduced the overall cost and time required for fabrication. Moreover, our 3D intestine-on-a-chip platform [Fig. 2(e)] maintained the biocompatibility and oxygen permeability advantages of PDMS. We embedded several GI organoids in Matrigel™ into the culture chamber in the platform and delivered culture media via a mechanical pump for the perfused 3D culture of the organoids [Fig. 2(f)].
FIG. 2.
Fabrication procedure for long-term perfused 3D intestine-on-a-chip platform. (a) SolidWorks design for culture chambers. (b) SolidWorks design for bubble pockets. (c) 3D printed mold for culture chambers. (d) 3D printed mold for bubble pockets. (e) Fabricated 3D intestine-on-a-chip system with a novel bubble pocket model. (f) Long-term perfused 3D intestine-on-a-chip device for embedded GI organoids. Scale bars, 5 mm.
Preparation of human and mouse intestinal organoids
Human intestinal organoids (hIOs) were prepared as previously described;11,13 however, ethics approval is not required in our study. Briefly, human pluripotent stem cells (hPSCs), including both human embryonic stem cells (hESCs) and human induced pluripotent stem cells (iPSCs), were maintained on hESC-qualified Matrigel (BD Biosciences) in a mTesR1 medium without feeders in a standard tissue culture incubator set to 37 °C and 5% CO2. Prior to initiating the differentiation protocol, hPSCs were dissociated into single cells using Accutase (Thermo Fisher Scientific) and passaged into a 24-well plate in mTeSR1 supplemented with ROCK inhibitor Y-27632 (10 μM; Tocris). Approximately 24 h later, the media was exchanged with fresh mTeSR1 media alone. The following day, cells were treated with Activin A (100 ng/ml, R&D Systems) for three consecutive days in RPMI 1640 media (Invitrogen) supplemented with NEAA (1×; Gibco) and increasing concentrations of 0%, 0.2%, 2% HyClone defined fetal bovine serum (dFBS) (Thermo Scientific) to induce differentiation into a definitive endoderm (DE) monolayer. After DE formation, cells were cultured in RPMI 1640 media supplemented with NEAA, 2.0% dFBS, 500 ng/ml FGF4 (R&D Systems), and 3 μM CHIR99021 (Stemgent) for 4 days to generate mid-hindgut intestinal spheroids. On the 4th day of spheroid generation media, 3D free floating spheroids were then transferred into 3D culture, previously shown to promote intestinal growth and differentiation.10,12 Spheroids were collected and embedded in Matrigel™ (Corning) in ∼50 μl 3D Matrigel™ droplets with ∼100 spheroids per droplet. After the Matrigel™ droplets were allowed to solidify for 10–15 min in a tissue culture incubator, spheroids were overlaid with “Gut media:” Advanced DMEM/F12 (Gibco) supplemented with N2 (1×; Invitrogen), B27 (without Vitamin A) (1×; Invitrogen), 2 mM l-glutamine (1×; Life Technologies), HEPES (15 mM; Gibco), 100 units/ml penicillin/streptomycin (1×; Life Technologies), and EGF (100 ng/ml; R&D Systems).
Mouse small intestinal organoids or enteroids were prepared from fluorescent ubiquitination-based cell-cycle indicator (FUCCI) transgenic mice25 utilizing a previously published protocol.26,27 FUCCI transgenic mice possess two distinct fluorescent reporters, mCherry-hCDT1 and mVenus-hGeminin, which are expressed during G0/G1 and S/G2/M phases, respectively. Briefly, mouse intestinal crypts were isolated from fresh mid-jejunum. Intestinal crypts were suspended with Matrigel™ (Corning) and then plated onto tissue culture dish. The Matrigel™ suspension was allowed to polymerize at 37 °C for between 15 min and 1 h before the fresh Minigut medium26,27 was supplied. The Minigut medium containing growth factors was replaced every 3–4 days. Mouse enteroids were maintained in culture for ∼6–9 days before being propagated into additional wells or used for experimental procedures.
Imaging human and mouse intestinal organoids in a perfused 3D intestine-on-a-chip
Approximately 12 days after embedding human intestinal spheroids or mouse intestinal crypts in Matrigel™ for 3D culture, several developing intestinal organoids were collected and re-embedded into the culture chamber of a 3D intestine-on-a-chip platform with the fresh Matrigel™. The Matrigel™ droplet was allowed to solidify for 10–15 min in a tissue culture incubator prior to media reintroduction. Two polymer tubes (1/16 in. outside diameter, IDEX Health & Science) were inserted into the inlet and outlet sides of the platform, and the inlet tube was connected to a syringe pre-loaded with gut media for the hIOs. Media was perfused through the entire system manually by using a syringe plunger. Following full immersion, the syringe was directly connected to a mechanical syringe pump for long-term automated system perfusion (New Era Pump Systems). The perfused 3D intestine-on-a-chip system immediately returned to the tissue culture incubator and was allowed to equilibrate for several days to over one month, demonstrating the feasibility of our developed bubble pocket for a long-term, perfused time-course imaging system. We continuously provided the culture media into the device via a mechanical pump (0.5 μl/min) and repeatedly observed the morphologies of growing hIOs using a Nikon TE2000-U inverted microscope (10× magnification).
FUCCI2 enteroids were collected and similarly re-embedded into the culture chamber of a 3D intestine-on-a-chip platform with the fresh Matrigel™ before imaging experiments. Under continuous media flow for 30 days, the enteroids had equilibrated within the perfused system in the incubator. Then, the platform was securely transferred on a confocal microscope to image the fluorescent FUCCI2 signal within enteroids. Image acquisition was performed through LSM710 LIVE Duo Confocal Microscope (Zeiss) with a 20×, 0.85 NA, dry objective. We integrated the platform, tubes, and the mechanical pump on the stage of the confocal microscope to acquire live bright-field and fluorescent images. For the fluorescent images of FUCCI2 enteroids, sequential excitation of mVenus and mCherry was performed by using a 514 nm and 560 nm lasers, respectively. We used QCapture Pro and Zen software to acquire bright field and two-photon images, respectively. ImageJ software was used to analyze images and to create fluorescence movies and images.
RESULTS AND DISCUSSION
Figures 1(a)–1(c) depict the flow disruption issues caused by air bubbles in three different types of microfluidic perfusion devices for long-term 3D culture of organoids. Air bubbles brought into the device through the inlet tube were accumulated in the 3D perfusion culture chambers over time, causing disruptive effects on normal device operation. The liquid–gas interface generated from the induced air bubbles could interrupt steady and continuous fluid flow, reduce cell viability and adhesion, which disrupts experiments. We proposed a novel solution to alleviate bubble-induced device failure in Fig. 1(d) that describes the de-bubbling principle of an integrated bubble pocket for long-term perfused 3D culture. Air bubbles generated under perfusion quickly floated to the upper portion of the culture chamber, due to the cone-shaped design of the 3D printed mold. Upon reaching the upper portion of the culture chamber, the bubbles diffused into the free space in a pocket chamber directly connected to the culture chamber [Fig. 1(d)]. The permeable PDMS pocket chamber then allowed the bubble to dissipate out of the device. An equilibrium interface exists between the liquid flow via outlet tube and the confined space of the top chamber. Additionally, the PDMS itself is highly hydrophobic, so the flowing liquid through the chamber and the outlet tube will not fill up the top chamber over time [Fig. 1(d)]. Device operation, including perfusion flow at atmospheric pressure, was sustained during the bubble trapping and de-bubbling processes. Therefore, the novel integrated bubble pocket enables the prevention of air bubbles entering culturing areas in the device and disrupting consistent flow while imaging in a long-term manner.
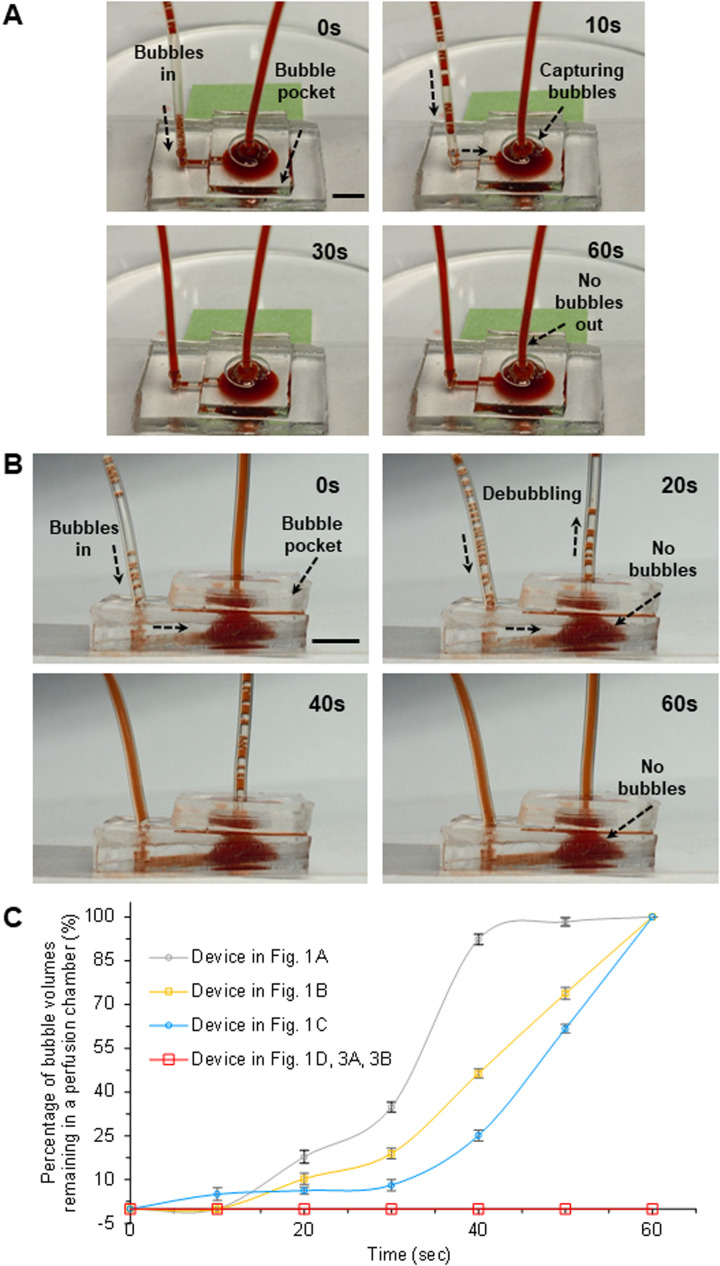
Figure 3 clearly describes the de-bubbling capabilities through a PDMS pocket chamber [Fig. 3(a)] and outlet tube [Fig. 3(b)] depending on the flow rates in the 3D perfused system. The flow rate in Fig. 3(a) was relatively low (<10 μl/min) in comparison with the (25 μl/min) present in Fig. 3(b). Before the injection through an inlet tube by the mechanical pump, the syringe containing the dyed water has been simply shaken to intentionally generate certain amounts of air bubbles in it. The generated bubbles in the syringe were easily introduced into the device through an inlet tube depending on the flow rates. Figure 3(a) shows that the small air bubbles introduced through an inlet tube were moving to the microchannel, followed by the culture chamber. Through our design of the culture chamber, the introduced air bubbles were floated upward in the principle of buoyancy of the bubbles in the liquid. The floated bubbles were pulled out through the bubble pocket directly attached onto the culture chamber followed by the gas permeable PDMS. Our bubble pocket successfully showed the ability to trap bubbles while maintaining device operation as well as to then remove these bubbles from the system, still under normal device operation. We intentionally introduced air bubbles into the device for 30 s, which were successfully removed by the device demonstrating no bubbles within the outlet tube [Fig. 3(a)]. Figure 3(b) also depicts that the introduced air bubbles were successfully diffused out through the outlet tube. It is noted that the imaging region of interest in the 3D perfused platform would be safely protected by our bubble pocket model, regardless of the flow rate.
FIG. 3.
(a) Characterization of de-bubbling capability through the PDMS pocket chamber for a long-term perfused 3D intestine-on-a-chip system. Introduced air bubbles were successfully trapped in the bubble pocket in the flow rate of <10 μm/min. The air bubbles were introduced into the device for 30 s, but bubbles were absent in the outlet tube. (b) Characterization of de-bubbling capability through the outlet tube for a long-term perfused 3D intestine-on-a-chip device. Introduced air bubbles were successfully removed in the culture chamber and bubble pocket with the flow rate of 25 μm/min. The introduced air bubbles were successfully diffused out through both bubble pocket and outlet tubing. (c) Quantified de-bubbling capability of an integrated bubble pocket for a long-term perfused 3D intestine-on-a-chip platform (N = 3). The bubble volumes were assessed in the various perfusion chambers [Figs. 1, 3(a), and 3(b)]. There were no remaining air bubbles in our developed device [Fig. 1(d)], showing the high efficiency of the novel bubble pocket. The bubble pocket we generated successfully trapped the introduced bubbles and removed them through the pocket or the outlet tube. Scale bars, 5 mm.
Figure 3(c) (N = 3) quantifies the de-bubbling capabilities of an integrated bubble pocket for a long-term perfused 3D intestine-on-a-chip platform. The air bubbles in dyed water were brought into the devices via the mechanical pump. The bubble volumes residing in the devices could be evaluated in various perfusion chambers [Figs. 1, 3(a), 3(b), and S2 in the supplementary material] over time because we already had the information about the internal diameter of the inlet and outlet tubes. It is noted that there were no remaining air bubbles in our developed device [Fig. 1(d)], confirming the high efficiency of the integrated bubble pocket model. However, all air bubbles continuously accumulated in the perfusion chambers [Figs. 1(a)–1(c)] within 60 s, so we could simply determine volume of the accumulated bubbles every 10 s. Then, we technically made the volume fraction of bubbles while placing the bubble volumes every 10 s over those at 60 s as depicted in Fig. 3(c). The most severe accumulation of air bubbles was obtained from the device in Fig. 1(a) (Fig. S1 in the supplementary material).
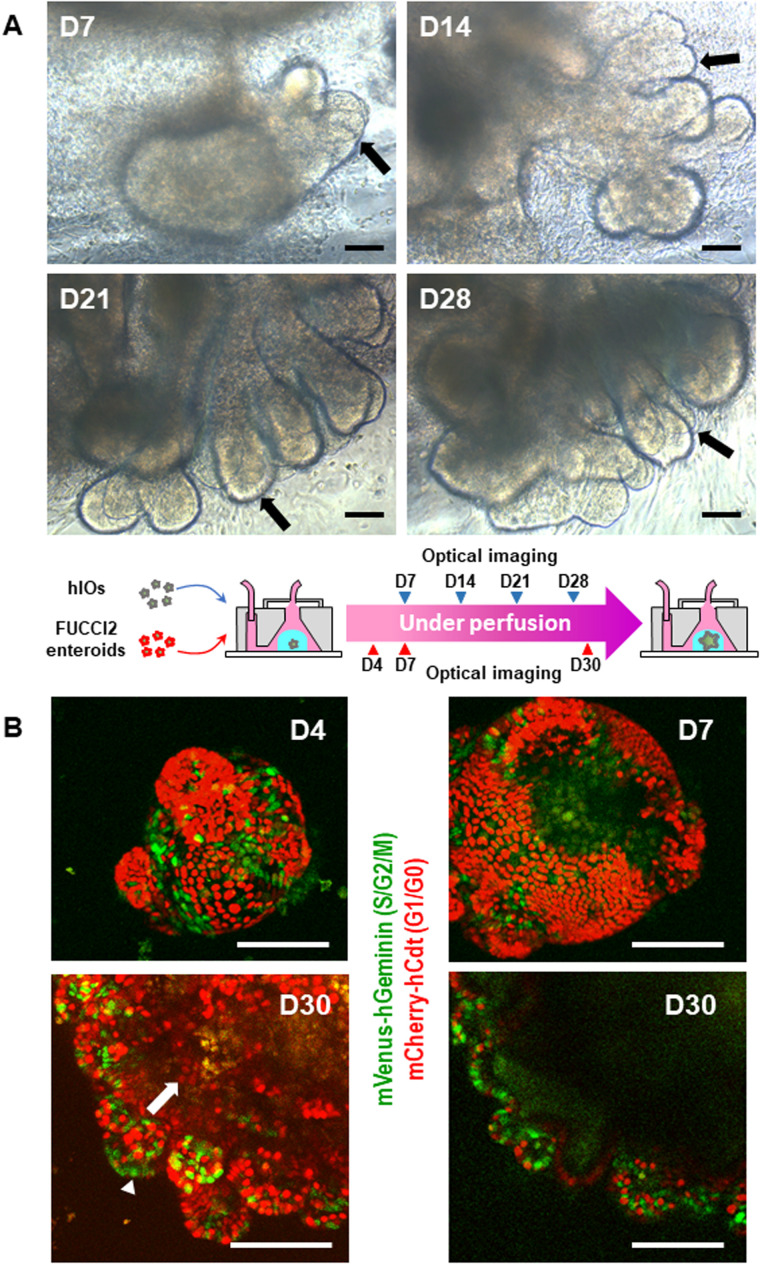
To validate the feasibility of our developed bubble pocket for a long-term perfused 3D intestine-on-a-chip platform, we embedded hIOs or FUCCI2 mouse enteroids in Matrigel™ into the 3D perfused system. We introduced culture media into the device using a mechanical pump, cultured the GI organoids over 4 weeks, and regularly monitored the organoid morphologies using a microscope. Figure 4(a) shows bright-field images of hIOs cultured in a long-term 3D perfusion device (10× magnification). When placed into the 3D perfusion culture device, hIOs (12 days of 3D growth) embedded in the Matrigel™ continued to develop and matured into intestinal tissue in vitro under the continuous perfusion of media (0.5 μl/min). The epithelial morphology of hIOs matured from villus-like involutions over time [arrows in Fig. 4(a)]. By 28 days in culture, the epithelium of hIOs in the 3D perfusion device clearly showed the villus-like protrusions similar to the intervillus epithelium11,13 of fetal mouse intestines. Thus, under continuous perfusion iPSC-derived hIOs continued to grow and undergo morphogenesis in vitro.
FIG. 4.
(a) Optical images of cultured hIO under a long-term perfused (0.5 μl/min) 3D intestine-on-a-chip device (10× magnification). The hIO epithelium matured, expanded, and developed villus-like involutions over time in a perfused intestine-on-a-chip system (arrows). (b) Confocal images of cultured FUCCI2 mouse enteroid in a long-term perfused 3D intestine-on-a-chip platform (20× magnification). Green fluorescent signal from a S/G2/M phase reporter, mVenus-hGeminin, was observed mainly at the crypts, where intestinal stem cells and progenitor cells reside (an arrow head). Red fluorescent signal from a G0/G1 phase reporter, mCherry-hCdt1, was primarily observed within villus-like domains, where terminally differentiated cells reside (an arrow). Long-term, perfused time-course imaging (over 30 days) of mouse enteroids was feasible in our robust, perfused 3D intestine-on-a-chip system with a novel bubble pocket. Top left, top right, and bottom left images are maximum intensity projection of a FUCCI2 enteroid at days 4, 7, and 30, respectively. The bottom right image shows a single confocal section of a FUCCI2 enteroid. Scale bars, 100 μm.
Figure 4(b) shows the confocal images of a fluorescent cell cycle sensor, FUCCI2, expressing mouse enteroids embedded in our 3D perfusion platform over 30 days (20× magnification). Among genetic expressions in FUCCI2 transgenic mice, we obtained red fluorescent signal from a G0/G1 cell cycle phase reporter, mCherry-hCdt1, primarily within villus-like domains, where terminally differentiated cells reside [an arrow in Fig. 4(b)]. We also identified green fluorescent signal from a S/G2/M cell cycle phase reporter, mVenus-hGeminin, mainly at the crypt, where intestinal stem cells and progenitor cells reside [an arrow head in Fig. 4(b)]. These results suggested that mouse enteroids continuously maintain the niche, proliferation, and differentiation ability of intestinal stem cells in our 3D intestine-on-a-chip device. Overall, long-term, perfused time-course imaging (over 30 days) of mouse enteroids was successfully demonstrated in our robust, 3D microfluidic platform with a novel integrated bubble pocket [Fig. 4(b)].
CONCLUSION
In conclusion, we demonstrated an on-chip, time-course imaging platform that enables the long-term perfused 3D culture for an in vitro GI organoid model. A combination of an on-chip 3D culture chamber with a novel microfluidic bubble pocket allowed us to effectively avoid undesirable effects of air bubbles to GI organoids in addition to circumvent culture medium evaporation. The de-bubbling capability of the integrated bubble pocket in our 3D platform was successfully characterized in terms of a range of flow rates and induced bubble volumes. The embedded hIOs or mouse enteroids were grown well over 4 weeks in our robust, long-term perfused system. Therefore, our 3D intestine-on-a-chip platform with an innovative bubble pocket is a milestone that promotes studies for in situ growth and morphogenesis as well as function of various human organoids during long-term, perfused time-course imaging. Since our new platform was successful for the GI organoids, we propose to extend our application for other types of organoids. Additionally, this investigation will be studied using specific cell types (i.e., endothelial cell monolayers). We suppose that the platform with our integrated bubble pocket is expected to work for other culture systems that are more sensitive to bubble exposure.
SUPPLEMENTARY MATERIAL
See the supplementary material for Fig. S1 which displays several device images of the accumulated air bubbles in Fig. 1(a). Figure S2 shows several images of the accumulated bubbles in the devices without a bubble trap (control).
ACKNOWLEDGMENTS
We acknowledge Dr. Atsushi Miyawaki (RIKEN BSI) and Dr. Hiroyuki Miyoshi (RIKEN BRC) for donating plasmid vectors to express mVenus-hGeminin. We acknowledge Dr. Hiroshi Kiyonari, Dr. Takaya Abe, and Dr. Yasuhide Furuta (RIKEN CDB) for donating FUCCI2 (CDB0264K) mice. This work was partially supported by the Defense Advanced Research Projects Agency (DARPA) (No. D12AP00005) (C.I.H.), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (No. R01 DK117005) (C.I.H.), National Cancer Institute (NCI) (No. R21 CA227379) (C.I.H.), National Institute of Allergy and Infectious Diseases (NIAID) (No. U19AI116491) (J.M.W. and C.I.H.), Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) (No. P01HD093363) (J.M.W.), (No. NIDDK UG3 DK119982) (J.M.W.), the Shipley Foundation (J.M.W.), and the Allen Foundation (J.M.W.). This work was also supported by the funds from the Office of Research, University of Cincinnati. We also acknowledge core support from the Pluripotent Stem Cell Facility of Cincinnati Children's Hospital Medical Center and the Live Microscopy Core at the University of Cincinnati College of Medicine. We acknowledge core support from the Cincinnati Digestive Disease Center award (No. P30DK0789392).
Contributor Information
Kang Kug (Paul) Lee, Email: .
Christian I. Hong, Email: .
DATA AVAILABILITY
The data that support the findings of this study are available within the article and its supplementary material.
REFERENCES
- 1.Paul S. M., Mytelka D. S., Dunwiddie C. T., Persinger C. C., Munos B. H., Lindborg S. R., and Schacht A. L., Nat. Rev. Drug Discov. 9, 203–214 (2010). 10.1038/nrd3078 [DOI] [PubMed] [Google Scholar]
- 2.Esch E. W., Bahinski A., and Huh D., Nat. Rev. Drug Discov. 14, 248–260 (2015). 10.1038/nrd4539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benam K. H., Dauth S., Hassell B., Herland A., Jain A., Jang K. J., Karalis K., Kim H. J., MacQueen L., Mahmoodian R., Musah S., Torisawa Y. S., van der Meer A. D., Villenave R., Yadid M., Parker K. K., and Ingber D. E., Annu. Rev. Pathol. 10, 195–262 (2015). 10.1146/annurev-pathol-012414-040418 [DOI] [PubMed] [Google Scholar]
- 4.Bhatia S. N. and Ingber D. E., Nat. Biotechnol. 32, 760–772 (2014). 10.1038/nbt.2989 [DOI] [PubMed] [Google Scholar]
- 5.Knowlton S., Yenilmez B., and Tasoglu S., Trends Biotechnol. 34, 685–688 (2016). 10.1016/j.tibtech.2016.06.005 [DOI] [PubMed] [Google Scholar]
- 6.Lee H. and Cho D. W., Lab Chip 16, 2618–2625 (2016). 10.1039/C6LC00450D [DOI] [PubMed] [Google Scholar]
- 7.Benam K. H., Villenave R., Lucchesi C., Varone A., Hubeau C., Lee H. H., Alves S. E., Salmon M., Ferrante T. C., Weaver J. C., Bahinski A., Hamilton G. A., and Ingber D. E., Nat. Methods 13, 151–157 (2016). 10.1038/nmeth.3697 [DOI] [PubMed] [Google Scholar]
- 8.Kim H. J., Li H., Collins J. J., and Ingber D. E., Proc. Natl. Acad. Sci. U.S.A. 113, E7–E15 (2016). 10.1073/pnas.1522193112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esch M. B., Ueno H., Applegate D. R., and Shuler M. L., Lab Chip 16, 2719–2729 (2016). 10.1039/C6LC00461J [DOI] [PubMed] [Google Scholar]
- 10.Sato T., Vries R. G., Snippert H. J., van de Wetering M., Barker N., Stange D. E., van Es J. H., Abo A., Kujala P., Peters P. J., and Clevers H., Nature 459, 262–265 (2009). 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- 11.Spence J. R., Mayhew C. N., Rankin S. A., Kuhar M. F., Vallance J. E., Tolle K., Hoskins E. E., Kalinichenko V. V., Wells S. I., Zorn A. M., Shroyer N. F., and Wells J. M., Nature 470, 105–109 (2011). 10.1038/nature09691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Amour K. A., Agulnick A. D., Eliazer S., Kelly O. G., Kroon E., and Baetge E. E., Nat. Biotechnol. 23, 1534–1541 (2005). 10.1038/nbt1163 [DOI] [PubMed] [Google Scholar]
- 13.McCracken K. W., Howell J. C., Wells J. M., and Spence J. R., Nat. Protoc. 6(12), 1920–1928 (2011). 10.1038/nprot.2011.410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin X., Mead B. E., Safaee H., Langer R., Karp J. M., and Levy O., Cell Stem Cell 18, 25–38 (2016). 10.1016/j.stem.2015.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee K. K., McCauley H. A., Broda T. R., Kofron M. J., Wells J. M., and Hong C. I., Lab Chip 18, 3079–3085 (2018). 10.1039/C8LC00910D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skardal A., Shupe T., and Atala A., Drug Discov. Today 9, 1399–1411 (2016). 10.1016/j.drudis.2016.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Low L. A. and Tagle D. A., Lab Chip 17, 3026–3036 (2017). 10.1039/C7LC00462A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu C., Thompson J. A., and Bau H. H., Lab Chip 11, 1688–1693 (2011). 10.1039/c1lc20089e [DOI] [PubMed] [Google Scholar]
- 19.Karlsson J. M., Gazin M., Laakso S., Haraldsson T., Malhotra-Kumar S., Mäki M., Goossens H., and van der Wijngaart W., Lab Chip 13, 4366–4373 (2013). 10.1039/c3lc50778e [DOI] [PubMed] [Google Scholar]
- 20.Skelley A. M. and Voldman J., Lab Chip 8, 1733–1737 (2008). 10.1039/b807037g [DOI] [PubMed] [Google Scholar]
- 21.Hibara A., Iwayama S., Matsuoka S., Ueno M., Kikutani Y., Tokeshi M., and Kitamori T., Anal. Chem. 77, 943–947 (2005). 10.1021/ac0490088 [DOI] [PubMed] [Google Scholar]
- 22.Zheng W., Wang Z., Zhang W., and Jiang X., Lab Chip 10, 2906–2910 (2010). 10.1039/c005274d [DOI] [PubMed] [Google Scholar]
- 23.Yang Z., Matsumoto S., and Maeda R., Sens. Actuator A Phys. 95, 274–280 (2002). 10.1016/S0924-4247(01)00741-5 [DOI] [Google Scholar]
- 24.Kang J. H., Kim Y. C., and Park J.-K., Lab Chip 8, 176–178 (2008). 10.1039/B712672G [DOI] [PubMed] [Google Scholar]
- 25.Abe T., Sakaue-Sawano A., Kiyonari H., Shioi G., Inoue K., Horiuchi T., Nakao K., Miyawaki A., Aizawa S., and Fujimori T., Development 140, 237–246 (2013). 10.1242/dev.084111 [DOI] [PubMed] [Google Scholar]
- 26.Moore S. R., Pruszka J., Vallance J., Aihara E., Matsu-Ura T., Montrose M. H., Shroyer N. F., and Hong C. I., Dis. Models Mech. 7, 1123–1130 (2014). 10.1242/dmm.014399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsu-Ura T., Dovzhenok A., Aihara E., Rood J., Le H., Ren Y., Rosselot A. E., Zhang T., Lee C., Obrietan K., Montrose M. H., Lim S., Moore S. R., and Hong C. I., Mol. Cell 64, 900–912 (2016). 10.1016/j.molcel.2016.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See the supplementary material for Fig. S1 which displays several device images of the accumulated air bubbles in Fig. 1(a). Figure S2 shows several images of the accumulated bubbles in the devices without a bubble trap (control).
Data Availability Statement
The data that support the findings of this study are available within the article and its supplementary material.