Abstract
Background:
Familial hypercholesterolemia (FH) is the most common cardiovascular genetic disorder and, if left untreated, is associated with increased risk of premature atherosclerotic cardiovascular disease, the leading cause of preventable death in the United States. Although FH is common, fatal, and treatable, it is underdiagnosed and undertreated due to a lack of systematic methods to identify individuals with FH and limited uptake of cascade testing.
Methods and Results:
This mixed-method, multi-stage study will optimize, test, and implement innovative approaches for both FH identification and cascade testing in 3 aims. To improve identification of individuals with FH, in Aim 1, we will compare and refine automated phenotype-based and genomic approaches to identify individuals likely to have FH. To improve cascade testing uptake for at-risk individuals, in Aim 2, we will use a patient-centered design thinking process to optimize and develop novel, active family communication methods. Using a prospective, observational pragmatic trial, we will assess uptake and effectiveness of each family communication method on cascade testing. Guided by an implementation science framework, in Aim 3, we will develop a comprehensive guide to identify individuals with FH. Using the Conceptual Model for Implementation Research, we will evaluate implementation outcomes including feasibility, acceptability, and perceived sustainability as well as health outcomes related to the optimized methods and tools developed in Aims 1 and 2.
Conclusions:
Data generated from this study will address barriers and gaps in care related to underdiagnosis of FH by developing and optimizing tools to improve FH identification and cascade testing.
Keywords: cardiovascular disease, familial hypercholesterolemia, genetic testing, implementation science, machine learning, phenotype
Familial hypercholesterolemia (FH) is the most common cardiovascular genetic disorder and, if left untreated, is associated with increased risk of premature atherosclerotic cardiovascular disease (ASCVD), the leading cause of preventable death in the United States).1 Because of lifelong exposure to elevated low-density lipoprotein cholesterol (LDL-C) levels, individuals with FH are at very high risk of premature ASCVD without early and aggressive lipid lowering therapy.2 Indeed, 17 500 deaths annually and a significant percentage of heart attacks in people under 45 are attributable to FH.3–5 Individuals with FH have up to a 20-fold increased risk of ASCVD relative to non-FH individuals with normal LDL-C.6 Importantly, when FH is diagnosed and treated early in life, the risk of ASCVD is greatly reduced.3,7,8
Individuals with FH can be identified via clinical criteria and/or genetic testing. Because FH is an autosomal dominant genetic condition, screening at-risk relatives of individuals with FH (cascade testing) is highly effective in identifying additional individuals with FH who require treatment.9,10 However, in the United States, neither FH identification nor cascade testing is systematically performed. When index patient identification and cascade testing are routinely performed, ascertainment is vastly improved, preventive interventions begin at a younger age, and outcomes improve.10 Although FH is common, fatal, and treatable, estimates show 90% or 1.1 million people in the United States with FH are undiagnosed.8
Approximately 1 in 220 individuals has a genetic variant that predisposes them to FH; however, not every individual with clinically diagnosed FH has an identifiable genetic variant, suggesting this approximated prevalence represents an underestimate of total FH prevalence.1,3 The presence of pathogenic variants in FH-associated genes including LDLR, APOB, and PCSK9 increases the risk of ASCVD from 2 to 10 times over age-matched individuals without a variant at equivalent LDL-C levels.1,6,11 The Centers for Disease Control and Prevention has identified FH as a Tier 1 genomic condition, prioritizing approaches to overcome barriers to identification of FH as having significant potential for a positive public health impact.
To improve FH care, the US health care system needs new strategies to find undiagnosed individuals and implement existing evidence-based guidelines.2 Innovative strategies to identify individuals with FH have been developed, including use of DNA sequencing of large populations and use of information technology-based strategies to identify individuals likely to have FH.1,12,13 Although facilitators and barriers to identification, cascade testing, and FH treatment have been studied, to date, no study has unified case-finding and clinical care models to demonstrate the best mechanisms for identifying individuals with FH in the US health care system.
The IMPACT-FH (Identification Methods, Patient Activation, and Cascade Testing for FH) Study is a collaboration among transdisciplinary researchers from Geisinger, an integrated health care system, and expert stakeholders from the FH Foundation, a patient-centered research and advocacy organization dedicated to improving FH care. In IMPACT-FH, we will develop, test, optimize, and implement innovative approaches for both FH identification and cascade testing to address the significant population health problem of FH underdiagnosis. We will compare and optimize machine-learning and genomic approaches to identify individuals likely to have FH. Tools to assist individuals with FH in communicating risk information to their relatives will be developed, optimized, and tested in a prospective trial. Using an implementation science framework, we will examine the outcomes of acceptability and feasibility of these FH identification and FH risk communication methods among patient, family, and provider stakeholders. A comprehensive guide for FH identification will be developed that can be used in other health care settings.
Methods
Overview
IMPACT-FH is a multistage study with 3 aims employing multiple methods. This study was approved by the Geisinger Institutional Review Board and all participants gave informed consent. Figure 1 shows the workflow of IMPACT-FH for all aims. The data that support the findings of this study are available from the corresponding author upon reasonable request.
Figure 1.
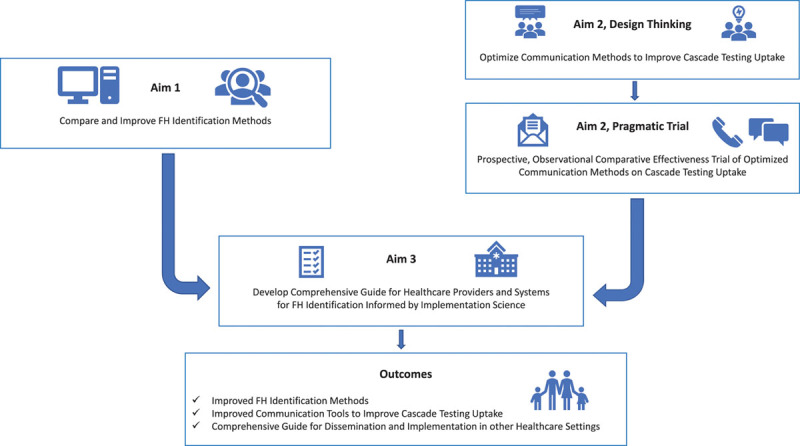
Workflow of aims for IMPACT-FH (Identification Methods, Patient Activation, and Cascade Testing for Familial Hypercholesterolemia). IMPACT-FH is a multi-stage, mixed-methods study composed of 3 aims that will lead to the improvement of FH identification methods, optimized communication tools to improve cascade testing uptake, and a comprehensive guide for improved FH identification.
Setting
IMPACT-FH will leverage Geisinger’s Pennsylvania-based integrated health care delivery system to study identification strategies for individuals with FH. Geisinger’s MyCode Community Health Initiative (MyCode) is a population-based genomics project that includes electronic health records (EHRs) data as well as genomic data generated from exome sequencing.14 Through MyCode, Geisinger collaborates with the Regeneron Genetics Center to combine high-throughput DNA sequencing with longitudinal EHR data for large-scale precision medicine research and genomic medicine implementation, creating the DiscovEHR cohort.15 MyCode also includes a genomic screening initiative, the MyCode Genomic Screening and Counseling Program (GSCP), through which patient-participants receive actionable genetic results, including those for FH.14,16 MyCode currently has over 265 000 patient-participants enrolled with exome sequencing completed on ≈145 000.
Study Aims
Aim 1: Develop and Optimize Methods for FH Identification
Aim 1 will compare and refine 3 approaches to improve identification of individuals likely to have FH. The first 2 approaches will utilize automated natural language processing and machine learning algorithms to screen patients for FH, an approach that can efficiently analyze variables in EHRs to classify and predict outcomes.12,17 Specifically, we will use Flag Identify Network Deliver FH (FIND FH) and Screening Employees and Residents in the Community for Hypercholesterolemia (SEARCH) to analyze structured and unstructured data in EHRs including laboratory results, prescriptions, personal history, family history, clinician notes, and other variables.12,13 In contrast to clinical diagnostic criteria for individual patients, these automated approaches screen large populations to identify individuals who should be evaluated clinically for FH. Finally, a third genomic-based approach will identify individuals with or likely to have FH due to the presence of pathogenic or likely pathogenic variants associated with FH.1,18,19
Data Collection and Analysis
We will evaluate these 3 FH identification methods (FIND FH, SEARCH, and the genomic-based approach) by applying each independently on the DiscovEHR cohort and comparing their performance as described below (Figure 2).
Figure 2.
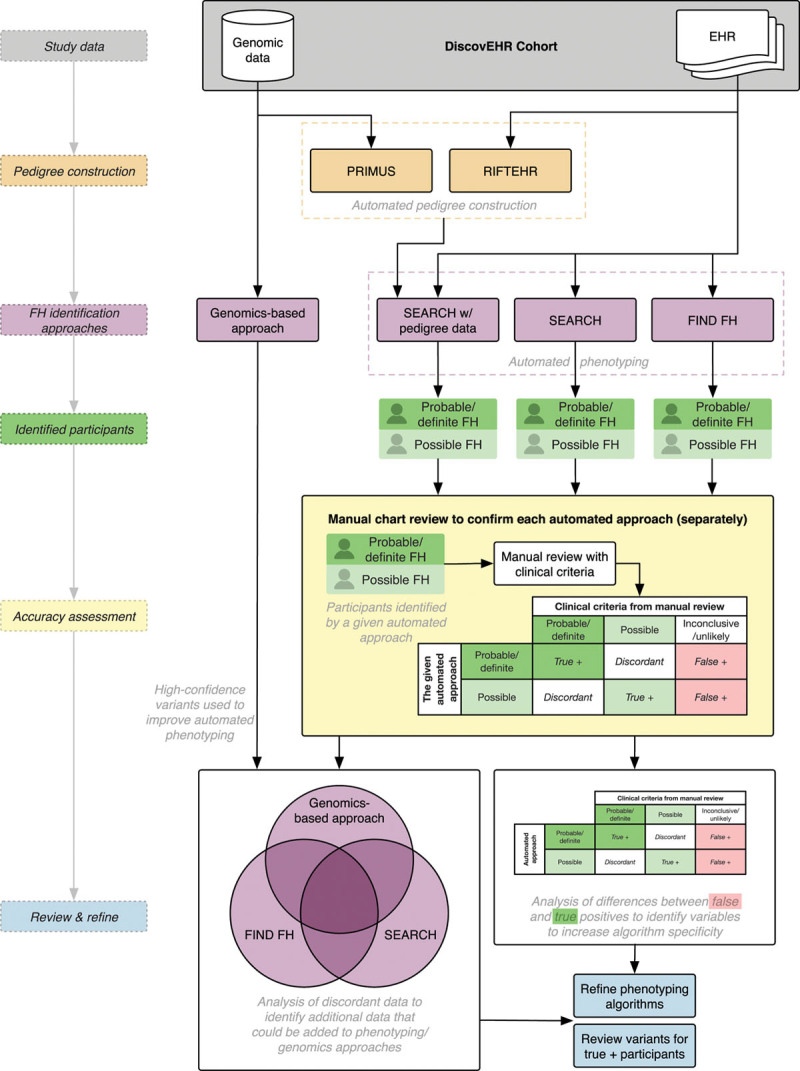
Workflow for evaluation and refinement of familial hypercholesterolemia (FH) identification methods in Aim 1. Aim 1 will reconstruct family histories via automated methods and rigorously compare and evaluate 3 approaches for FH identification to refine these tools. EHR indicates electronic health record; PRIMUS, Pedigree and Identification of the Maximally Unrelated Set; RIFTEHR, The Relationship Inference from the Electronic Health Records; and SEARCH, Screening Employees and Residents in the Community for Hypercholesterolemia.
Incomplete family history information in the EHR is a limitation of identification methods. SEARCH, in particular, can use EHR information from family members to enhance accuracy for the proband (the first individual diagnosed with FH in the family). We will assess 2 methods for providing additional family history information for SEARCH:
The Pedigree Reconstructions and Identification of the Maximally Unrelated Set, a validated software package,20 identifies related individuals using genomic data.
The Relationship Inference from the Electronic Health Records algorithm infers pedigrees from emergency contact information stored in the EHR.21
We will validate the accuracy of pedigrees built via Pedigree Reconstructions and Identification of the Maximally Unrelated Set and Relationship Inference from the Electronic Health Records by manual chart review of a subset of patients who have had a pathogenic or likely pathogenic FH variant disclosed from MyCode and received genetic counseling. We will then use the method(s) with the highest accuracy compared with manual review to reconstruct each individual’s family history. SEARCH will be re-run with the augmented family history information to assess its respective change in accuracy.21
To evaluate the performance of the automated phenotyping approaches (FIND FH and SEARCH), we will conduct manual chart review for a subset of individuals identified as probable/definite, possible, and inconclusive/unlikely FH. A standardized electronic form capturing phenotypic information based on FH clinical diagnostic criteria will be developed. A random sample of participants may be re-contacted to capture additional family history information as needed. Then, each patient will be scored using the Dutch Lipid Clinic Network (DLCN) criteria3,22 and the American Heart Association (AHA) FH diagnostic criteria.3 These scores will be used to compare performance of the automated identification approaches. We chose to use both the DLCN and the AHA criteria, as the DLCN criteria provide higher specificity but may lack sensitivity, while the AHA criteria afford greater sensitivity but are less specific. Both schemas incorporate genetic information; DLCN adds 8 points (confirming a diagnosis) whereas AHA criteria establish a diagnosis with the presence of an LDL-C raising genetic variant.3,22 For the purposes of comparison, the DLCN criteria will be binned into 3 categories; definite and probable criteria will be combined into “Probable FH”, “Possible FH” will be retained, and inconclusive and unlikely will be combined into “Unlikely FH”. These 3 bins are appropriate as the purpose of these automated approaches is not for FH diagnosis but to screen large populations for individuals who should be further evaluated clinically for FH.22
Positive predictive value (PPV) and negative predictive value will be calculated for FIND FH and SEARCH. “Probable FH” and “Possible FH” will be grouped together for the purposes of calculating PPV. PPV will be compared across automated approaches using the Generalized Estimating Equations (GEE) model among those identified as FH cases. Specifically, we will use the DLCN score as the benchmark to which we will compare the automated approaches. The data will be structured such that each patient will have 2 records for their FIND FH and SEARCH classifications. The DLCN score will be the outcome variable, and the automated approach result and approach name (indicator variable that identifies each approach) will be treated as the independent variables.23 We will model the PPVs directly by assuming a binomial distribution and identity link. The GEE model will estimate robust standard errors to account for the correlated data. We will fit a similar model to estimate and compare negative predictive value for those identified as “Unlikely FH” by the automated approaches. The same methodology will also be used to compare the automated approaches against the AHA criteria, which may be a more relevant benchmark in a screening context due to the higher sensitivity of the AHA compared with the DLCN criteria. Additionally, we will re-run SEARCH incorporating pedigree information from the automated pedigree reconstruction methods (Pedigree Reconstructions and Identification of the Maximally Unrelated Set and Relationship Inference from the Electronic Health Records) and compare PPVs and negative predictive values to the pedigree-naïve SEARCH run using the same method described above.
After constructing pedigrees and evaluating identification approaches, individuals with an FH-associated genetic variant who were not detected by FIND FH or SEARCH will be assessed using principal component analysis as a cohort to identify features from the EHR that can be incorporated into these phenotype-based approaches to better identify those at risk for FH. We will compare characteristics of those with an FH-associated genetic variant to those without to identify features that can be used to improve the accuracy of the phenotyping and genomic approaches. We will use these findings to refine these automated approaches and construct improved versions that can be validated on a cohort of ≈45 000 additional patients newly recruited to the DiscovEHR cohort, who will have genomic data available later during the course of this study. We will also use clinical information identified through the phenotypic approaches to better inform interpretation of genetic variants; we will review variants not initially identified through the genomics approach for participants with probable/possible FH based on the phenotyping approaches.
Aim 2: Optimize and Test Strategies to Improve Family Communication and Uptake of FH Cascade Testing
Cascade testing of family members can be highly effective in identifying additional individuals with FH.10,24 Cascade testing relies on identifying an FH proband with subsequent testing of all at-risk relatives, a cycle that is repeated, or cascaded, through the family until all at-risk individuals are tested.16 In the United States, cascade testing for FH is not systematically performed.24 Based on preliminary data, Geisinger’s MyCode GSCP has returned FH results to 114 individuals, who reported 401 living first-degree relatives, but only 3.5% (14/401) of living, at-risk relatives have completed cascade testing. While “Dear Family” letters have been used to support family communication and cascade testing, such passive methods that rely on the proband to transmit genetic risk information to relatives are suboptimal.24,25 A recent systematic review indicated this indirect method resulted in, on average, <1 new relative with FH identified per proband.24 Probands have also reported challenges to sharing genetic information with relatives and have expressed interest in receiving support from health care providers to assist them in communicating with their family about FH.26 While there is support from payers, public health, and clinicians about the importance of cascade testing, how best to inform relatives of risk and implement cascade testing has yet to be determined in the United States.24 Novel, active, acceptable strategies such as digital tools and direct contact have been proposed as solutions.27
One such digital tool is the chatbot, which is a computer-based conversational agent that interacts with people in ways mirroring human dialogue. Use of chatbots can result in activated patients, who have confidence and knowledge to engage in health-promoting behaviors, such as cascade testing.28,29 Chatbots have been designed to support genetic uses cases and deliver standardized medical information designed by clinicians at the user’s pace.30 Geisinger’s MyCode GSCP currently uses 2 types of chatbots to encourage family communication and cascade testing for FH, called the Family Sharing Tool for probands and the Cascade Chatbot for at-risk relatives. Based on preliminary findings, to date, 59% of the 308 probands who have received the Family Sharing Tool opened it and 62% of the 376 relatives who received the Cascade chatbot opened it. These preliminary findings suggest chatbots may be effective communication tools to support family communication and cascade testing for FH. See Figure 3 for an example of a chatbot.
Figure 3.
Example of Chatbot. Chatbots are a form of mobile health technology that can communicate genetic risk information at the user’s pace in a familiar conversational format.
Direct contact of at-risk relatives by providers to share the proband’s result may improve uptake of FH cascade testing by overcoming challenges related to passive approaches to family risk communication.24–27 In the United States, current standard of care puts the burden on probands to notify and encourage at-risk relatives to undergo testing. Programs outside the United States utilizing direct contact have been shown to be more effective in promoting the uptake of cascade testing.10,24,31 In qualitative data from focus groups and interviews with MyCode patient participants, all participants stated that it would be acceptable for Geisinger to contact relatives directly to educate them and facilitate cascade testing. Further, 64 MyCode patient participants completed surveys on direct contact. When participants were asked how helpful it would be if Geisinger offered to contact their relatives to inform them of their genetic risk, 42% indicated very (26.56%) or somewhat (15.6%) helpful, and only 15.6% of participants indicated not helpful at all. Additionally, 42% indicated they would be extremely or somewhat likely to use a direct contact program. Finally, a recent survey study found 85% of participants with FH in the United States and 100% of international participants with FH reported their willingness to provide contact information for certain at-risk relatives to a clinician for the purpose of directly informing relatives.31 Preliminary data and recently published research demonstrate potential acceptability for the development and implementation of a direct contact program in the United States.
This aim will develop, improve, and test strategies of family communication about FH to motivate uptake of cascade testing. It will proceed in 2 phases: a design thinking process followed by a pragmatic trial. The first phase will use a design thinking process in which key stakeholders design and optimize communication methods to improve cascade testing uptake among at-risk relatives. Design thinking is a patient-centered methodology, which re-frames the problem in human-centric ways and begins with empathic engagement with the people most affected by, and knowledgeable about, what needs to be changed.32 In the second phase, optimized communication methods and strategies will be integrated into the MyCode GSCP for patient-participants, and a prospective, observational, comparative-effectiveness pragmatic trial will assess uptake and effectiveness of each novel communication method on cascade testing.
Data Collection
For the design thinking process, we will use purposive sampling to recruit patient stakeholders from Geisinger and the FH Foundation who have a clinical or genetic diagnosis of FH. We will then use snowball sampling, asking patient stakeholders to recruit an at-risk relative to participate in the design thinking process. All participants will be age 18 years or older and speak English. To better represent diverse family relationships and dynamics, we will ask participants to recruit both relationally close (eg, siblings) and distant family members (eg, cousins). Stakeholders will participate in a combination of survey methods and dyadic interviews to optimize the “Dear Family” letter, the chatbot, and the direct contact program for clinicians to share FH results with at-risk relatives. Results from the design thinking process will drive changes to the letter and chatbot and inform the design of a clinical direct contact program for individuals receiving FH results from the GSCP.
For the pragmatic trial phase of Aim 2, individuals receiving results from MyCode for a genetic diagnosis of FH will be offered the improved communication methods to share results with their at-risk relatives. The genetic counselors disclosing the results will utilize a phone script to inform probands of their FH risk variant, recommend sharing with at-risk relatives, and collect data about all living at-risk relatives. These probands will be provided the “Dear Family” letter and the choice of which additional communication method (chatbot and/or direct contact) they prefer to use for each of their at-risk relatives. Genetic counselors will note probands’ choices and collect relatives’ contact information if the proband chooses to use direct contact. Probands who choose to use the chatbot will receive a link to a Family Sharing Tool, a mechanism that allows them to send a cascade chatbot to their relatives via text message, email, or Facebook Messenger. To facilitate seamless logistics for relatives’ cascade testing, each communication method will be offered via the development of a new program called Contact and Support, Counseling, and DNA testing Empowerment. Contact and Support, Counseling, and DNA testing Empowerment will include logistics for cascade testing including at-home sample collection, physician ordering, and no cost testing via Invitae for all relatives within a set period of time from proband result receipt. For those whose testing occurs outside the window, Invitae will coordinate with relatives’ insurance and provide a self-pay option. Cascade testing uptake will be measured at 6 months post-return of the proband’s FH result. If the proband’s at-risk relative(s) did not complete cascade testing within the 6 months, the proband will be offered another communication method for their at-risk relative(s) and cascade testing uptake will be re-assessed at 12 months post-return of result. For example, if the proband chose to use direct contact for a relative and testing was not performed by 6 months, then the proband will be offered a chatbot for that relative at 6 months, and cascade testing uptake will be measured at 12 months.
Data Analysis
Qualitative data from the design thinking process will be audio recorded and transcribed verbatim. Transcripts will be uploaded to www.Atlas.ti for qualitative analysis. The research team will iteratively analyze transcripts and notes from the design thinking process for themes. The study team will start by open coding for emergent (de novo) codes, which will then be categorized for how participants discussed optimizing the letter, chatbot, and designing a direct contact program.33 In the final stage of analysis, axial coding will be used to collapse categorized codes into themes and subthemes to better assess how to optimize and design the communication methods for the prospective, observational comparative effectiveness pragmatic trial.
All quantitative data from the pragmatic trial will be fully described using means, SDs, medians, and interquartile ranges for continuous variables, and frequencies and percentages for categorical variables. If necessary, continuous variables will be transformed via the natural logarithmic or a similar approach to achieve an approximately normal distribution. Analysis at 6 months will be conducted on 3 groups: (1) usual care (letter only), (2) usual care and direct contact, and (3) usual care and chatbot. We will perform comparisons across the 3 groups using GEE for linear and logistic regressions to account for correlation because of the clustering of at-risk relatives within probands. GEE will be used to compare percent cascade testing uptake across the groups. As this is a nonrandomized study design, baseline variables may differ across the 3 groups, possibly confounding unadjusted comparisons. To address this, the GEE models will include any potential confounding variables identified a priori to estimate an unbiased effect of communication method. A similar, secondary analysis will be performed at 12 months on those at-risk relatives that did not complete cascade testing by 6 months. Additionally, we will assess demographics (eg, sex, age, etc) associated with the proband selecting a given method. GEE regression models will be fit with the baseline variables to identify factors associated with choice. Results will be represented as odds ratios and 95% CI.
Aim 3: Develop a Comprehensive Guide for Improved FH Identification Informed by Implementation Science
By using an implementation science framework, this aim will develop a comprehensive guide for other health care settings to identify individuals with FH. We will use the Conceptual Model for Implementation Research to evaluate implementation outcomes including feasibility, acceptability, and perceived sustainability as well as cascade testing uptake related to optimized methods and tools developed in Aims 1 and 2 (See Figure 4).34
Figure 4.
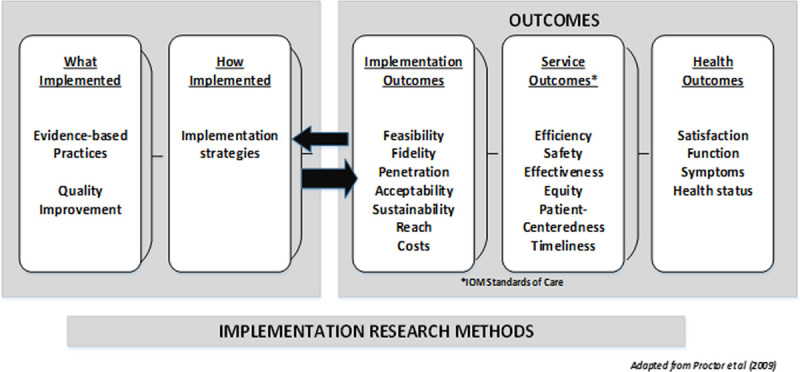
Conceptual model of implementation research. The conceptual model of implementation research provides the analytical framework guiding development of the comprehensive guide to improve familial hypercholesterolemia identification for Aim 3.
Data Collection
We will conduct 2 focus groups for patient stakeholders and 3 focus groups for clinician stakeholders to explore levels of acceptability, feasibility, and perceived need of different FH identification methods. Focus group moderators will explain how machine learning algorithms can be used to analyze EHRs to identify individuals at high risk for FH and elicit feedback from stakeholders. Moderators will also introduce novel communication methods such as chatbots and direct contact to gather feedback on their acceptability and feasibility for improving cascade testing. Demographic data will be collected by a questionnaire administered at each stakeholder focus group. Stakeholder participants, including patients and clinicians, will be recruited from national conferences held by nonprofit organizations such as the FH Foundation, the National Lipid Association, and from Geisinger’s clinics, the MyCode program, and a community hospital in New England not affiliated with a medical school. Patient stakeholders will be 18 years or older and have a clinical or genetic diagnosis of FH. Clinician stakeholders will have been or are currently treating individuals diagnosed with FH.
Then, we will conduct semistructured interviews via telephone with probands from the pragmatic trial of Aim 2, the proband’s at-risk relatives, as well as clinician stakeholders. Approximately 40 probands and 40 at-risk relatives will be selected to participate based on which communication method they used to ensure an equal number of interviews from probands who chose to communicate their results to at-risk relatives via letter, chatbot, direct contact, or a combination. Interviews with probands will gather information on why they chose certain communication methods for each relative, how their at-risk relatives responded, and assess the proband’s experience and satisfaction with each communication method. Interviews with at-risk relatives will gather their experiences and satisfaction with the communication method, their decisions regarding cascade testing, and demographic data. We will conduct ≈10 interviews with clinician stakeholders whose patients participated in the direct contact program.
Finally, we will conduct 2 focus groups with clinician stakeholders to gather perceptions of the acceptability, feasibility, and perceived sustainability of the developed comprehensive guide for FH identification. These focus groups will assess what is needed to implement a comprehensive program to improve identification and cascade testing of FH in health care settings and develop implementation strategies that can be disseminated to other settings.
Data Analysis
All interviews and focus groups will be digitally recorded and transcribed verbatim. Transcripts will be uploaded to www.Atlas.ti for qualitative analysis utilizing the Conceptual Model for Implementation Research.33 This framework approach ensures rigor in the qualitative analytic process and is useful for data that are gathered based on predetermined constructs. The framework method begins with deductive analysis, while also allowing for inductive analysis as new themes emerge from the data. Although focus group and interview transcripts will be analyzed separately, all transcripts will initially be coded using an a priori codebook developed from the guides, summaries, and Conceptual Model for Implementation Research. De novo codes will be added to any other relevant sections of transcript text not fitting the a priori codes. Team members will independently, iteratively code 2-3 transcripts at a time, then discuss their coding to adjust the codebook and create a working analytic framework by grouping codes into categories or themes. This process will continue until the code list is static, all transcripts are coded, and the analytic framework is finalized.
Discussion
Implementation of evidence-based research into practice relies on passive diffusion, is inconsistent, and can take upwards of 17 years.35 This gap between evidence-based guidelines and their implementation into routine clinical practice is one of the most critical issues facing health care and public health today.36 Multiple barriers and facilitators to FH diagnosis and cascade testing have been identified37 and exist at the patient, provider, and health care system levels.38 These barriers include low levels of lipid screening of pediatric individuals,39 diagnostic criteria that are not optimized to facilitate FH identification in EHRs, suboptimal self-reported family history data, proband concerns about having the appropriate communication skills to inform family members about their risks, with resultant low FH identification and recruitment of at-risk relatives to cascade testing.
According to the National Heart, Lung, and Blood Institute, work addressing barriers and gaps in care related to underdiagnosis of FH creates an opportunity to expand translational research agendas in genomics, precision medicine, and implementation science. To address these gaps, IMPACT-FH partners researchers with expertise in genetics, clinical lipidology, health communication, and implementation science with stakeholders from the FH Foundation, including individuals with FH, those with expertise in technology-based tools for FH identification, and health care providers dedicated to improving FH care. Through qualitative and implementation science methodologies, IMPACT-FH will use patient-engaged learnings to build tools for individuals with FH and health care providers that focus on outcomes most important to these individuals, and their families.
Improving identification and diagnosis of individuals with FH is a pivotal first step in the treatment and management of elevated LDL-C levels, preventing ASCVD, and alerting at-risk relatives about the need for cascade testing. Historically, identification of individuals with FH has relied on recognition of clinical and family history features such as lipid levels, premature ASCVD, and physical features such as xanthomas and corneal arcus.22 Genetic testing for FH can identify 60% to 80% of clinically suspected FH cases and population-based genomic screening programs can identify individuals with FH pathogenic or likely pathogenic variants who do not meet clinical diagnostic criteria.1,37,40 Previous research has found FH genetic testing improves diagnosis and is associated with improved adherence to recommended treatment and lower LDL-C and total cholesterol levels.1,7,38,40–42 Evaluating FH identification models and automating pedigree construction can lead to the creation of a novel ensemble model that incorporates elements of both genomic- and phenotype-based approaches to more accurately identify individuals likely to have FH43 who need further clinical evaluation.
Optimizing and applying innovative communication methods to improve family communication and subsequent uptake of cascade testing will provide new tools for application in precision health and genomic medicine at large. IMPACT-FH will pioneer the first clinical direct contact program in the United States, which may be replicated and adapted for other genetic health conditions to improve cascade testing. Results from the pragmatic trial will provide data on uptake of communication methods, including direct contact, as well as compare the effectiveness of each method for improving cascade testing.
By using tools from implementation science, IMPACT-FH rigorously examines how to best disseminate and implement improved methods of identification and cascade testing to other health care settings. Specifically, IMPACT-FH will provide tools for other organizations interested in identifying their FH patient population, and these tools may also be generalizable to other actionable genetic conditions. Developing tools and the comprehensive guide for FH identification informed by implementation science will ensure findings from IMPACT-FH are transferrable to other health care sites. For example, health care sites with biobanks and exome data can use findings from the evaluation of FH identification models to identify individuals in their system likely to have FH. Health care sites without a biobank or exome data can use findings from IMPACT-FH on automated approaches to analyzing their EHR data for FH identification. Further, tools and programs for optimizing communication methods could apply to cholesterol testing uptake when genetic testing is not easily accessible.
The FH Foundation will help export findings from IMPACT-FH to be implemented in communities external to Geisinger. The FH Foundation has the only national FH registry, collecting comprehensive, longitudinal data on individuals with FH. As partners in research on IMPACT-FH, the FH Foundation will communicate findings of this study broadly to 33 major lipid clinics across the country. In the future, the FH Foundation can share chatbots with individuals included in the FH registry to provide additional information about FH and support individuals as they share information about FH with at-risk relatives. Finally, clinicians from the FH Foundation may be involved in a Direct Contact program to help individuals with FH share information about FH with at-risk relatives to improve FH identification and cascade testing. Indeed, the FH Foundation has far-reaching impact to export IMPACT-FH findings.
The IMPACT-FH study design and populations involved have limitations. First, the automated approaches for improved FH identification will be evaluated based on clinical diagnostic criteria from the DLCN and the AHA. These diagnostic criteria may have reduced sensitivity (DLCN) and specificity (AHA) and may also miss individuals with an FH-associated genetic risk variant if they do not have high LDL-C levels or other features of FH noted in their EHR.1,3 The pragmatic trial in Aim 2 is specific to patient-participants in the MyCode GSCP, which may limit the generalizability of findings on effectiveness of the optimized communication methods to improve cascade testing for FH in other patient populations. Although analysis for the pragmatic trial will include the combination of methods, a proband chooses to share FH risk information with their at-risk relatives, it will not include uncontrolled ways probands may communicate and attempt to motivate cascade testing among relatives. Further, cascade testing uptake results from the pragmatic trial may be an underestimation as we may be unable to track at-risk relatives who pursue clinical testing (eg, lipid panel testing) or genetic testing outside of Invitae. The qualitative methods used in Aim 3 to develop the comprehensive guide may limit generalizability as results may not capture all important viewpoints and feedback from stakeholders and we may not reach thematic saturation. Finally, without testing the comprehensive guide for improved FH identification and cascade testing uptake in other health care settings, transferability may be limited.
IMPACT-FH will provide fruitful areas for future research. First, researchers can implement the refined approaches from Aim 1 on EHR and genomic data in health care systems and measure implementation outcomes including acceptability and feasibility, among others, by assessing satisfaction and perception of need by providers and subsequent identification of individuals with FH. Second, future research on family communication of FH risks and cascade testing uptake could examine how optimized communication methods from Aim 2 may need further adaptation for clinical populations outside the GSCP. Finally, future research can extend and improve on the comprehensive guide from Aim 3 by testing and adapting it to other health care settings and programs. Attempts to utilize and replicate findings from IMPACT-FH can further adapt the innovative methods of this research to improve FH identification and care.
In conclusion, IMPACT-FH will develop and optimize innovative, patient-centered solutions through collaborative research between Geisinger and the FH Foundation that can be applied in real-world settings to combat the public health problem of the >1.1 million undiagnosed individuals with FH in the United States.8
Sources of Funding
Research reported in this publication was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health under award number: R01HL148246. This research is 100% supported by Federal money in the amount of $2 837 141.00. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Disclosures
The Regeneron Genetics Center (RGC) provides exome sequencing for the “DiscovEHR” cohort. The RGC is not directly involved in the IMPACT-FH (Identification Methods, Patient Activation, and Cascade Testing for FH) Study. Dr Esplin, A. Morales, M. Snir, and E. Simmons are employees and stockholders of Invitae. Dr Gidding is a consultant for Esperion. HL Kirchner receives funding from RGC. K.D. Myers is the CEO and founder of Atomo, Inc, which has pharmaceutical companies, medical communications agencies, and the FH Foundation as clients.
Footnotes
Nonstandard Abbreviations and Acronyms
- AHA
- American Heart Association
- ASCVD
- atherosclerotic cardiovascular disease
- DLCN
- Dutch Lipid Clinic Network
- EHR
- electronic health record
- FH
- familial hypercholesterolemia
- FIND FH
- Flag Identify Network Deliver FH
- GEE
- generalized estimating equation
- GSCP
- Genomic Screening and Counseling Program
- IMPACT-FH
- Identification Methods, Patient Activation, and Cascade Testing for FH
- LDL-C
- low-density lipoprotein cholesterol
- PPV
- positive predictive value
- SEARCH
- Screening Employees and Residents in the Community for Hypercholesterolemia
For Sources of Funding and Disclosures, see page 120.
Contributor Information
Gemme Campbell-Salome, Email: gemme.campbell@ufl.edu.
Laney K. Jones, Email: ljones14@geisinger.edu.
Max F. Masnick, Email: max@masnick.net.
Nephi A. Walton, Email: Nephi.Walton@imail.org.
Catherine D. Ahmed, Email: cda@thefhfoundation.org.
Adam H. Buchanan, Email: ahbuchanan@geisinger.edu.
Andrew Brangan, Email: ambrangan@geisinger.edu.
Edward D. Esplin, Email: ed.esplin@invitae.com.
David G. Kann, Email: dgkann1@geisinger.edu.
Ilene G. Ladd, Email: igladd@geisinger.edu.
Melissa A. Kelly, Email: makelly2@geisinger.edu.
Iris Kindt, Email: irishommeskindt@hotmail.nl.
H. Lester Kirchner, Email: hlkirchner@geisinger.edu.
Mary P. McGowan, Email: mpm@thefhfoundation.org.
Megan N. McMinn, Email: mnbetts@geisinger.edu.
Ana Morales, Email: ana.morales@invitae.com.
Kelly D. Myers, Email: km@thefhfoundation.org.
Matthew T. Oetjens, Email: mtoetjens@geisigner.edu.
Alanna Kulchak Rahm, Email: akrahm@geisinger.edu.
Tara J. Schmidlen, Email: tjschmidlen@geisinger.edu.
Amanda Sheldon, Email: ams@thefhfoundation.org.
Emilie Simmons, Email: emilie.simmons@invitae.com.
Moran Snir, Email: moran.snir@invitae.com.
Natasha T. Strande, Email: ntstrande@geisinger.edu.
Nicole L. Walters, Email: nlwalters1@geisinger.edu.
Katherine Wilemon, Email: kw@thefhfoundation.org.
Marc S. Williams, Email: mswilliams1@geisinger.edu.
Samuel S. Gidding, Email: samuel.gidding@gmail.com.
References
- 1.Abul-Husn NS, Manickam K, Jones LK, Wright EA, Hartzel DN, Gonzaga-Jauregui C, O’Dushlaine C, Leader JB, Kirchner HL, Lindbuchler DM, et al. Genetic identification of familial hypercholesterolemia within a single US health care system. Science. 2016;354:aaf7000. [DOI] [PubMed] [Google Scholar]
- 2.Grundy SM, Stone NJ, Bailey AL, Beam C, Birtcher KK, Blumenthal RS, Braun LT, de Ferranti S, Faiella-Tommasino J, Forman DE, et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA guideline on the management of blood cholesterol: executive summary: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation. 2019;139:e1046–e1081. doi: 10.1161/CIR.0000000000000624 [DOI] [PubMed] [Google Scholar]
- 3.Gidding SS, Champagne MA, de Ferranti SD, Defesche J, Ito MK, Knowles JW, McCrindle B, Raal F, Rader D, Santos RD, et al. ; American Heart Association Atherosclerosis, Hypertension, and Obesity in Young Committee of Council on Cardiovascular Disease in Young, Council on Cardiovascular and Stroke Nursing, Council on Functional Genomics and Translational Biology, and Council on Lifestyle and Cardiometabolic Health. The agenda for familial hypercholesterolemia: a scientific statement from the American Heart Association. Circulation. 2015;132:2167–2192. doi: 10.1161/CIR.0000000000000297 [DOI] [PubMed] [Google Scholar]
- 4.Cui Y, Li S, Zhang F, Song J, Lee C, Wu M, Chen H. Prevalence of familial hypercholesterolemia in patients with premature myocardial infarction. Clin Cardiol. 2019;42:385–390. doi: 10.1002/clc.23154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Ferranti SD, Rodday AM, Mendelson MM, Wong JB, Leslie LK, Sheldrick RC. Prevalence of familial hypercholesterolemia in the 1999 to 2012 United States National Health and Nutrition Examination Surveys (NHANES). Circulation. 2016;133:1067–1072. doi: 10.1161/CIRCULATIONAHA.115.018791 [DOI] [PubMed] [Google Scholar]
- 6.Khera AV, Won HH, Peloso GM, Lawson KS, Bartz TM, Deng X, van Leeuwen EM, Natarajan P, Emdin CA, Bick AG, et al. Diagnostic yield and clinical utility of sequencing familial hypercholesterolemia genes in patients with severe hypercholesterolemia. J Am Coll Cardiol. 2016;67:2578–2589. doi: 10.1016/j.jacc.2016.03.520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luirink IK, Wiegman A, Kusters DM, Hof MH, Groothoff JW, de Groot E, Kastelein JJP, Hutten BA. 20-year follow-up of statins in children with familial hypercholesterolemia. N Engl J Med. 2019;381:1547–1556. doi: 10.1056/NEJMoa1816454 [DOI] [PubMed] [Google Scholar]
- 8.Nordestgaard BG, Chapman MJ, Humphries SE, Ginsberg HN, Masana L, Descamps OS, Wiklund O, Hegele RA, Raal FJ, Defesche JC, et al. ; European Atherosclerosis Society Consensus Panel. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease: consensus statement of the European Atherosclerosis Society. Eur Heart J. 2013;34:3478–390a. doi: 10.1093/eurheartj/eht273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadfield SG, Horara S, Starr BJ, Yazdgerdi S, Marks D, Bhatnagar D, Cramb R, Egan S, Everdell R, Ferns G, et al. ; Steering Group for the Department of Health Familial Hypercholesterolaemia Cascade Testing Audit Project. Family tracing to identify patients with familial hypercholesterolaemia: the second audit of the Department of Health Familial Hypercholesterolaemia Cascade Testing Project. Ann Clin Biochem. 2009;46pt 124–32. doi: 10.1258/acb.2008.008094 [DOI] [PubMed] [Google Scholar]
- 10.Umans-Eckenhausen MA, Defesche JC, Sijbrands EJ, Scheerder RL, Kastelein JJ. Review of first 5 years of screening for familial hypercholesterolaemia in the Netherlands. Lancet. 2001;357:165–168. doi: 10.1016/S0140-6736(00)03587-X [DOI] [PubMed] [Google Scholar]
- 11.Besseling J, Reitsma JB, Gaudet D, Brisson D, Kastelein JJ, Hovingh GK, Hutten BA. Selection of individuals for genetic testing for familial hypercholesterolaemia: development and external validation of a prediction model for the presence of a mutation causing familial hypercholesterolaemia. Eur Heart J. 2017;38:565–573. doi: 10.1093/eurheartj/ehw135 [DOI] [PubMed] [Google Scholar]
- 12.Myers KD, Knowles JW, Staszak D, Shapiro MD, Howard W, Yadava M, Zuzick D, Williamson L, Shah NH, Banda JM, et al. Precision screening for familial hypercholesterolaemia: a machine learning study applied to electronic health encounter data. Lancet Digit Health. 2019;1:e393–e402. doi: 10.1016/S2589-7500(19)30150-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Safarova MS, Liu H, Kullo IJ. Rapid identification of familial hypercholesterolemia from electronic health records: the SEARCH study. J Clin Lipidol. 2016;10:1230–1239. doi: 10.1016/j.jacl.2016.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carey DJ, Fetterolf SN, Davis FD, Faucett WA, Kirchner HL, Mirshahi U, Murray MF, Smelser DT, Gerhard GS, Ledbetter DH. The Geisinger MyCode community health initiative: an electronic health record-linked biobank for precision medicine research. Genet Med. 2016;18:906–913. doi: 10.1038/gim.2015.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dewey FE, Murray MF, Overton JD, Habegger L, Leader JB, Fetterolf SN, O’Dushlaine C, Van Hout CV, Staples J, Gonzaga-Jauregui C, et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science. 2016;354:aaf6814. [DOI] [PubMed] [Google Scholar]
- 16.Schwartz MLB, McCormick CZ, Lazzeri AL, Lindbuchler DM, Hallquist MLG, Manickam K, Buchanan AH, Rahm AK, Giovanni MA, Frisbie L, et al. A model for genome-first care: returning secondary genomic findings to participants and their healthcare providers in a large research cohort. Am J Hum Genet. 2018;103:328–337. doi: 10.1016/j.ajhg.2018.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banda JM, Sarraju A, Abbasi F, Parizo J, Pariani M, Ison H, Briskin E, Wand H, Dubois S, Jung K, et al. Finding missed cases of familial hypercholesterolemia in health systems using machine learning. NPJ Digit Med. 2019;2:23 doi: 10.1038/s41746-019-0101-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly MA, Caleshu C, Morales A, Buchan J, Wolf Z, Harrison SM, Cook S, Dillon MW, Garcia J, Haverfield E, et al. Adaptation and validation of the ACMG/AMP variant classification framework for MYH7-associated inherited cardiomyopathies: recommendations by ClinGen’s Inherited Cardiomyopathy Expert Panel. Genet Med. 2018;20:351–359. doi: 10.1038/gim.2017.218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42Database issueD980–D985. doi: 10.1093/nar/gkt1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staples J, Qiao D, Cho MH, Silverman EK, Nickerson DA, Below JE; University of Washington Center for Mendelian Genomics. PRIMUS: rapid reconstruction of pedigrees from genome-wide estimates of identity by descent. Am J Hum Genet. 2014;95:553–564. doi: 10.1016/j.ajhg.2014.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polubriaginof FCG, Vanguri R, Quinnies K, Belbin GM, Yahi A, Salmasian H, Lorberbaum T, Nwankwo V, Li L, Shervey MM, et al. Disease heritability inferred from familial relationships reported in medical records. Cell. 2018;173:1692–1704.e11. doi: 10.1016/j.cell.2018.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Defesche JC, Lansberg PJ, Umans-Eckenhausen MA, Kastelein JJ. Advanced method for the identification of patients with inherited hypercholesterolemia. Semin Vasc Med. 2004;4:59–65. doi: 10.1055/s-2004-822987 [DOI] [PubMed] [Google Scholar]
- 23.Leisenring W, Alonzo T, Pepe MS. Comparisons of predictive values of binary medical diagnostic tests for paired designs. Biometrics. 2000;56:345–351. [DOI] [PubMed] [Google Scholar]
- 24.Lee C, Rivera-Valerio M, Bangash H, Prokop L, Kullo IJ. New case detection by cascade testing in familial hypercholesterolemia: a systematic review of the literature. Circ Genom Precis Med. 2019;12:e002723 doi: 10.1161/CIRCGEN.119.002723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dheensa S, Lucassen A, Fenwick A. Limitations and pitfalls of using family letters to communicate genetic risk: a qualitative study with patients and healthcare professionals. J Genet Couns. 2018;27:689–701. doi: 10.1007/s10897-017-0164-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardcastle SJ, Legge E, Laundy CS, Egan SJ, French R, Watts GF, Hagger MS. Patients’ perceptions and experiences of familial hypercholesterolemia, cascade genetic screening and treatment. Int J Behav Med. 2015;22:92–100. doi: 10.1007/s12529-014-9402-x [DOI] [PubMed] [Google Scholar]
- 27.Sturm AC. Cardiovascular cascade genetic testing: exploring the role of direct contact and technology. Front Cardiovasc Med. 2016;3:11 doi: 10.3389/fcvm.2016.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fitzpatrick KK, Darcy A, Vierhile M. Delivering cognitive behavior therapy to young adults with symptoms of depression and anxiety using a fully automated conversational agent (Woebot): a randomized controlled trial. JMIR Ment Health. 2017;4:e19 doi: 10.2196/mental.7785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ledford CJW, Canzona MR, Cafferty LA, Hodge JA. Mobile application as a prenatal education and engagement tool: a randomized controlled pilot. Patient Educ Couns. 2016;99:578–582. doi: 10.1016/j.pec.2015.11.006 [DOI] [PubMed] [Google Scholar]
- 30.Schmidlen T, Schwartz M, DiLoreto K, Kirchner HL, Sturm AC. Patient assessment of chatbots for the scalable delivery of genetic counseling. J Genet Couns. 2019;28:1166–1177. doi: 10.1002/jgc4.1169 [DOI] [PubMed] [Google Scholar]
- 31.Schwiter R, Brown E, Murray B, Kindt I, Van Enkevort E, Pollin TI, Sturm AC. Perspectives from individuals with familial hypercholesterolemia on direct contact in cascade screening. J Genet Couns. 2020;29:1142–1150. doi: 10.1002/jgc4.1266 [DOI] [PubMed] [Google Scholar]
- 32.Roberts JP, Fisher TR, Trowbridge MJ, Bent C. A design thinking framework for healthcare management and innovation. Healthc (Amst). 2016;4:11–14. doi: 10.1016/j.hjdsi.2015.12.002 [DOI] [PubMed] [Google Scholar]
- 33.Saldaña J. The Coding Manual for Qualitative Researchers. 2015. Sage [Google Scholar]
- 34.Proctor EK, Landsverk J, Aarons G, Chambers D, Glisson C, Mittman B. Implementation research in mental health services: an emerging science with conceptual, methodological, and training challenges. Adm Policy Ment Health. 2009;36:24–34. doi: 10.1007/s10488-008-0197-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Green LW. Closing the chasm between research and practice: evidence of and for change. Health Promot J Austr. 2014;25:25–29. doi: 10.1071/HE13101 [DOI] [PubMed] [Google Scholar]
- 36.Mittman B. Brownson RC, Colditz GA, Proctor EK, ed. Implementation science in health care. In: Dissemination and Implementation Research in Health. 2012. Oxford University Press; 400–418. [Google Scholar]
- 37.Hendricks-Sturrup RM, Mazor KM, Sturm AC, Lu CY. Barriers and facilitators to genetic testing for familial hypercholesterolemia in the United States: a review. J Pers Med. 2019;9:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones LK, Kulchak Rahm A, Manickam K, Butry L, Lazzeri A, Corcoran T, Komar D, Josyula NS, Pendergrass SA, Sturm AC, et al. Healthcare utilization and patients’ perspectives after receiving a positive genetic test for familial hypercholesterolemia. Circ Genom Precis Med. 2018;11:e002146 doi: 10.1161/CIRCGEN.118.002146 [DOI] [PubMed] [Google Scholar]
- 39.Wiegman A, Gidding SS, Watts GF, Chapman MJ, Ginsberg HN, Cuchel M, Ose L, Averna M, Boileau C, Borén J, et al. ; European Atherosclerosis Society Consensus Panel. Familial hypercholesterolaemia in children and adolescents: gaining decades of life by optimizing detection and treatment. Eur Heart J. 2015;36:2425–2437. doi: 10.1093/eurheartj/ehv157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sturm AC, Knowles JW, Gidding SS, Ahmad ZS, Ahmed CD, Ballantyne CM, Baum SJ, Bourbon M, Carrié A, Cuchel M, et al. ; Convened by the Familial Hypercholesterolemia Foundation. Clinical genetic testing for familial hypercholesterolemia: JACC Scientific Expert Panel. J Am Coll Cardiol. 2018;72:662–680. doi: 10.1016/j.jacc.2018.05.044 [DOI] [PubMed] [Google Scholar]
- 41.Amor-Salamanca A, Castillo S, Gonzalez-Vioque E, Dominguez F, Quintana L, Lluís-Ganella C, Escudier JM, Ortega J, Lara-Pezzi E, Alonso-Pulpon L, et al. Genetically confirmed familial hypercholesterolemia in patients with acute coronary syndrome. J Am Coll Cardiol. 2017;70:1732–1740. doi: 10.1016/j.jacc.2017.08.009 [DOI] [PubMed] [Google Scholar]
- 42.Minicocci I, Pozzessere S, Prisco C, Montali A, di Costanzo A, Martino E, Martino F, Arca M. Analysis of children and adolescents with familial hypercholesterolemia. J Pediatr. 2017;183:100–107.e3. doi: 10.1016/j.jpeds.2016.12.075 [DOI] [PubMed] [Google Scholar]
- 43.Abbott DW. Combining models to improve classifier accuracy and robustness. 1999;1:In Proceedings of Second International Conference on Information Fusion, Fusion’99; 289–295. [Google Scholar]


