Abstract
The SARS-CoV-2 pandemic, an ongoing global health crisis, has revealed the need for new technologies that integrate the sensitivity and specificity of RT-PCR tests with a faster time-to-detection. Here, an emulsion loop-mediated isothermal amplification (eLAMP) platform was developed to allow for the compartmentalization of LAMP reactions, leading to faster changes in emulsion characteristics, and thus lowering time-to-detection. Within these droplets, ongoing LAMP reactions lead to adsorption of amplicons to the water-oil interface, causing a decrease in interfacial tension, resulting in smaller emulsion diameters. Changes in emulsion diameter allow for the monitoring of the reaction by use of angle-dependent light scatter (based off Mie scatter theory). Mie scatter simulations confirmed that light scatter intensity is diameter-dependent and smaller colloids have lower intensity values compared to larger colloids. Via spectrophotometers and fiber optic cables placed at 30° and 60°, light scatter intensity was monitored. Scatter intensities collected at 5 min, 30° could statistically differentiate 10, 103, and 105 copies/μL initial concentrations compared to NTC. Similarly, 5 min scatter intensities collected at 60° could statistically differentiate 105 copies/μL initial concentrations in comparison to NTC. The use of both angles during the eLAMP assay allows for distinction between high and low initial target concentrations. The efficacy of a smartphone-based platform was also tested and had a similar limit of detection and assay time of less than 10 min. Furthermore, fluorescence-labeled primers were used to validate target nucleic acid amplification. Compared to existing LAMP assays for SARS-CoV-2 detection, these times-to-detections are very rapid.
Keywords: SARS-CoV-2, COVID-19, Emulsion, Mie scatter, Loop-mediated isothermal amplification, Interfacial tension
1. Introduction
In late 2019, a novel coronavirus called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was identified (Xu et al., 2020) and a COVID-19 pandemic followed. As of December 2020, the virus has infected 64 million people worldwide, and tragically killed 1.5 million individuals according to the WHO Coronavirus Disease Dashboard. Due to its rapid escalation in new cases and potentially fatal disease progression, the SARS-CoV-2 pandemic has become a serious public health concern (Wang et al., 2020). One of the most effective methods to curtail the spread of the disease is to rapidly test local populations for the virus in order to properly quarantine them from non-infected individuals. One of the primary test types used during the pandemic is reverse transcription polymerase chain reaction (RT-PCR), which is used primarily for its high sensitivity and specificity (Corman et al., 2020). However, the long run time for the assay can lead to delays in results on the scale of hours, if not days. This, combined with the fact that RT-PCR assays require trained personnel and complex laboratory equipment, limits the effectiveness of this technology in combatting the outbreak, particularly in low-resource areas (Cui and Zhou, 2020; Ravi et al., 2020). Because of these limitations, there is a great need for a more rapid testing technology that can aid in the detection of SARS-CoV-2.
Isothermal nucleic acid amplification techniques have gained interest in previous years due to the attractiveness of utilizing a single temperature. A constant temperature for nucleic acid amplification mitigates the need for specialized equipment such as thermocyclers to finely and rapidly adjust temperatures in an intricate manner like in PCR reactions. This aspect has been especially appealing for field-deployable, point-of-care platforms, where simplicity is necessary. However, it is notoriously known that isothermal nucleic acid amplification techniques are susceptible to nonspecific amplification, rendering in less specific than PCR methods, particularly when utilizing dirty samples such as blood, saliva, and tissue samples (Deguo et al., 2008).
Emulsion platforms can be utilized to address the problem of background signals due to non-specific amplification. Water-in-oil emulsion protocols are advantageous in nucleic acid amplification techniques due to the ability to compartmentalize target-gene-of-interest into individual containment units. This allows for an intrinsic separation mechanism of components that may inhibit amplification or induce non-specific amplification (Nakano et al., 2003). Emulsions can be especially advantageous due to the intrinsic presence of surfactants and agitation during their formation that could be utilized as an alternative method of “extracting” DNA or RNA.
Nucleic acid amplification by PCR can be measured by gel electrophoresis at the end point of thermal cycling or by fluorescence quantification during thermal cycling (i.e., quantitative PCR or qPCR). Similar methods can be used for LAMP. Alternative methods have been suggested for measuring nucleic acid amplification that can provide extreme sensitivity and/or detect amplification in early thermal cycles to significantly shorten the assay time. One such method was the measurement of the interfacial tension changes of the aqueous nucleic acid amplification reaction in a droplet. Harshman et al. (2015) utilized a moving droplet-on-a-thermocouple PCR instrumentation to amplify targets-of-interest and monitor its progression in real-time by measuring the droplet size. The phenomenon attributing such change in droplet size was due to a decrease in the water-oil interface, rendering the droplet unstable as amplicon amount increased within. Similarly, a droplet loop-mediated isothermal amplification (LAMP) nucleic acid method was utilized for monitoring amplification progression via interfacial-effect detection. The platform was comprised of a static, aqueous LAMP droplet immersed in mineral oil, in which the change in contact angle was monitored over time and related to interfacial tension (Ulep et al., 2019). In both platforms, time-to-results were reduced immensely in comparison to conventional nucleic acid amplification techniques and showed significant amplification in complex sample matrices due to inhibition relief at the water-oil interface.
In this paper, we focus specifically on LAMP in a water-in-oil emulsion platform, reducing the droplet volume of 7.5 μL or 10 μL (Harshman et al., 2015; Ulep et al., 2019) to around 524 fL (diameter of 10 μm), to address non-specific signals and slow time-to-detection. These emulsions compartmentalize the LAMP reaction, reducing the amount of potentially amplifiable targets. In addition, this compartmentalization into small droplets causes protein adsorption to the oil-water interface to occur rapidly, attenuating amplification inhibition due to protein presence. The emulsion platform also leads to faster detection times due to faster saturation of the droplets with LAMP amplicons. Only a small number of amplicons is necessary to saturate the water-oil interface that can significantly change the interfacial tension and subsequently emulsion droplet size. All these factors result in a more specific LAMP assay. We also utilize interfacial effect-based real-time monitoring through Mie scatter detection to achieve rapid time-to-results. Adsorption of LAMP amplicons to the water-oil interface will change the interfacial tension and subsequently emulsion size, leading to emulsion destabilization (Nicolini et al., 2017). Based on Mie scatter theories, which relate angle-dependent light scatter intensity in relation to particle size, we can monitor LAMP reactions progression. Specifically, the N (nucleoprotein) protein gene in SARS-CoV-2 was amplified via emulsion LAMP at varying concentrations of 105, 103, 10, and 0 copies/μL. Firstly, the proposed detection mechanism was validated with a miniature spectrophotometer and fiber optic cables from a cylindrical glass tube containing the emulsion, placed on a magnetic stirrer with temperature control function. Secondly, a smartphone camera with blinking LED set up was used and resulted in similar light scatter intensity measurements and correlation to LAMP reaction progression as the spectrophotometer set up. Light scatter intensity showed to decrease in relation to amount of initial target concentration within 3 min of emulsion LAMP reaction for both detection set ups. Furthermore, experiments utilizing fluorescently labeled primers were conducted to validate target nucleic acid detection. Overall, we have demonstrated the use of angle-dependent light scatter intensity as a means of rapid real-time monitoring of isothermal nucleic acid amplification in a simple and user-friendly platform.
2. Materials and methods
2.1. Preparation of specimens
Escherichia coli O157:H7 (part number 0801622; ZeptoMetrix Corporation, Buffalo, NY, USA) was used as a proof-of-concept target. The stock bacterial solution was diluted to different concentrations of 106, 103, 1, 0.1, and 0 CFU/μL in nuclease free water. For SARS-CoV-2 detection, 2019-nCoV_N_Positive Control (catalog number 10006625; Integrated DNA Technologies, Coralville, IA, USA) stock solution was diluted to different concentrations of 105, 103, 101, and 0 copies/μL in nuclease free water.
2.2. LAMP reaction
Escherichia coli O157:H7 LAMP primers were selected from literature (Zhao et al., 2010) and purchased from Sigma-Aldrich (St. Louis, MO, USA). SARS-CoV-2 LAMP primers were also selected from literature (Zhang et al., 2020) and purchased from Integrated DNA Technologies. Sequences for the primers used in this study can be found in Supplementary Table S1. 10X target-specific primer sets were formulated to contain 16 μM each of FIP and BIP primers, 8 μM each of Loop-F and Loop-B primers, and 2 μM of F3 and B3 primers. LAMP reactions were prepared on ice and utilized the WarmStart® LAMP Kit DNA & RNA (E1700; New England Biolabs Inc, Ipswich, MA, USA). Final mixtures were comprised of Bst 2.0 WarmStart DNA Polymerase and WarmStart RTx Reverse Transcriptase in manufacturer's optimized LAMP buffer solution. The LAMP final mixture contained 5:1:0.4:1:2.6 ratio of Warm Start LAMP 2X master mix, 10X primer mix, target solution (or nuclease-free water as no target control, NTC), 20 mg/mL bovine serum albumin (catalog number B8667; Sigma-Aldrich), and nuclease-free water. For SARS-CoV-2 detection, target solution was contained within a 1% v/v saliva-in-nuclease-free water solution (catalog number 991-05-P; Lee Biosolutions, Maryland Heights, MO). Conventionally amplified samples were conducted in a thermocycler (MJ Research, Waltham, MA, USA) programmed for amplification to occur at 65 °C for 30 min, followed by refrigeration at 4 °C.
2.3. Emulsion LAMP assay
Water-in-oil colloidal emulsions were prepared similarly as the published method (Nicolini et al., 2017). The bulk oil phase was made daily by mixing 25 mL of light mineral oil, 1.25 mL of Span 80, 100 μL of Tween 80, and 12.5 μL of Triton-X 100 (catalog numbers M5904-500 ML, S6760-250 ML, P5188-100 ML, and T8787-50 ML, respectively, all from Sigma-Aldrich, St. Louis, MO) and then allowing the mixture to rock for at least 30 min in room temperature conditions. A single emulsion reaction consisted of the addition of a 10 μL aqueous LAMP droplet from a blunt end needle tip suspended and dropped uniformly into 2 mL of preheated 65 °C oil phase. Emulsions were formed and agitated by a micro stir bar (8 × 1.5 mm, catalog number 1179Z30, Thomas Scientific, Swedesboro, NJ) set to 1500 rotations per minute (RPM) for 30 min. Post-reaction, emulsions were collected and byproducts were extracted via 3 iterations of water saturated diethyl ether (Williams et al., 2006).
2.4. Light scatter detection
An incident light of 650 nm (LS-450 LED; Ocean Optics, Dunedin, FL, USA) illuminated emulsion samples via fiber optical cable, while another fiber optical cable connected to a miniature spectrophotometer (USB4000, Ocean Optics) placed at 30° and 60° collected light scatter intensity, which was saved to a laptop computer via a CSV file. Light scatter intensity measurements were collected every 3 s. A 3D printed holder was used to secure these fiber optic cables and a vial with emulsion throughout assays (Fig. 1 A). This data was normalized by subtracting each datapoint by the initial light scatter intensity value so that the final data indicates change in intensity over time.
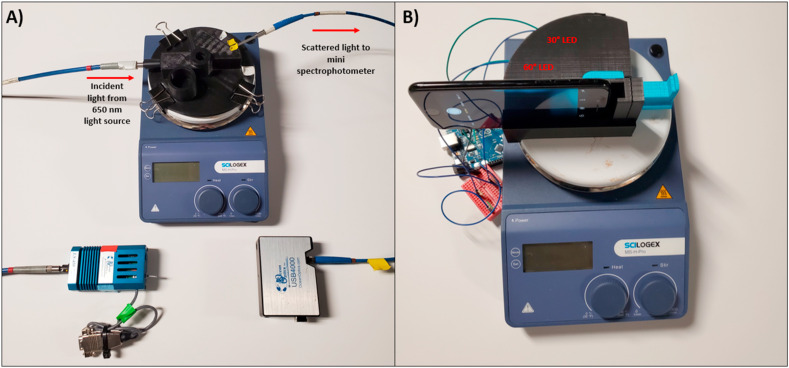
Fig. 1.
A) Spectrophotometer-based emulsion LAMP platform. Bottom-left light source feeds 650 nm light into the 3D printed platform via the left optical fiber, while the right optical fiber feeds the scattered light to the miniature spectrophotometer, which is on the bottom-right. B) Smartphone-based emulsion LAMP platform. A 3D printed housing holds the smartphone, reaction vial, and two red LEDs in place while an Arduino Uno-controlled circuit alternates LEDs every 3 s. The iPhone's built-in timelapse feature is used to capture videos. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.5. Interfacial tension (IFT) measurements
Interfacial tension (mN m-1) measurements were made with an FTÅ200 contact angle analyzer (First Ten Ångstroms, Portsmouth, VA, USA) using the pendant droplet method. 10 μL aqueous LAMP samples were pre-amplified (not in emulsion) on a conventional thermocycler and stopped at several time points prior to interfacial tension measurements.
2.6. Light scatter measurement of emulsion from pre-amplified samples
10 μL aqueous LAMP reactions containing Escherichia coli O157:H7 were first amplified in a conventional thermocycler for several different reaction times. These samples were then emulsified in 2 mL of the pre-mixed oil phase, where light scatter was then monitored for 1 min.
2.7. Emulsion diameter measurement of emulsion LAMP
An emulsion LAMP reaction was conducted (see section 2.3) using the 105 copies/μL SARS-CoV-2. 10 μL aqueous phase of this reaction was taken out at several time points (0, 5, 10, 20, and 30 min), where the samples were placed on a glass slide, and five random images were taken throughout the sample. The emulsified aqueous droplet diameters were then measured using ImageJ software (U.S. National Institutes of Health; Bethesda, MD, USA).
2.8. Mie scatter simulations
Mie scattering simulations were performed using MiePlot v4.6 (Phlip Laven, www.philiplaven.com/mieplot.htm). The simulation assumed a refractive index of sample medium (mineral oil) = 1.47, refractive index of emulsions (water) = 1.33, a 650 nm incident light, and emulsion sizes varying from 9.42 μm to 0.42 μm (taken from the microscopic images). Light intensity at 30° and 60° angles were investigated.
2.9. End-point spectroscopic confirmation of emulsion amplification
After emulsion amplification was performed then broken, the amplicon precipitates dissolved in an aqueous solution were analyzed. Presence of nucleic acid was determined by measurement of absorption at 260 nm using a miniature spectrophotometer (USB4000, Ocean Optics) and cuvette holder and UV/visible light source (CHEMUSB4, Ocean Optics).
2.10. End-point gel electrophoresis confirmation of emulsion amplification
The same amplicon precipitates after breaking the emulsion were analyzed using gel electrophoresis. 3% w/v agarose gel (A0169; Sigma-Aldrich) in 1X Tris-acetate-EDTA (TAE) buffer (35100131; Quality Biological Inc, Gaithersburg, MD, USA) was prepared and placed at 120 V for 50 min with an electrophoresis power supply (Fischer Scientific; FB200). TrackIt™ 100 bp DNA ladder was used as a standard for fragment sizing. Gels were stained with ethidium bromide (E1510; Sigma) and imaged under UV light. Gel images were analyzed using ImageJ software (U.S. National Institutes of Health).
2.11. Fluorescent probe detection of SARS-CoV-2 amplicons
To detect amplification of SARS-CoV-2 target, fluorescent probes (FLOS-LAMP) were utilized according to literature (Gadkar et al., 2018)]. In particular, a fluorescein molecule was attached to the thymine residue at the 3′ end of the FIP primer for SARS-CoV-2 detection, as this was the only primer that fulfilled the three requirements proposed by Gadkar et al. (2018). The three requirements are that the primer chosen to be a fluorescent probe must have 1) the presence of cytosine or guanine residue at the 3′-terminal end, 2) thymine at the second or third position from the 3′-end, and (optionally) 3) may have at least one guanine nucleotides neighboring said thymine nucleotide. This fluorescent probe took the place of the standard primer in these experiments. The fluorescent-probe-attached FIP primer is included in the sample and the neighboring guanine residues self-quench the fluorescein fluorophore. When the primer attaches to the target sequence that it is specific to, the self-quenching effect is removed, thus allowing for fluorescent emission when in the presence of target sequences. Once amplification was completed on the eLAMP platform, the aqueous byproduct was collected via water-saturated diethyl ether extraction, and the fluorescence of the final product was measured via a miniature spectrophotometer (USB4000; Ocean Optics) at 520 nm and UV excitation using LS-450 (Ocean Optics) with 350 nm UV LED. These experiments were done over a range of concentrations (NTC, 101, 103, and 105 copies per μL).
2.12. Smartphone-based light scatter detection
A 3D printed enclosure was fabricated to house the ongoing LAMP reaction on top of the existing hotplate while red LEDs set at 30° and 60° were utilized to illuminate the emulsion sample (Fig. 1B). The LEDs, which were powered by an Arduino Uno-based circuit, would alternate being on/off every 3 s, with one LED always being on. The light scatter was then monitored using an iPhone 9 smartphone camera using the “time lapse” feature, which captured an image every second and stitched together all the images in a single experiment into a MOV movie file. A Python-based algorithm (using the OpenCV library) was then utilized to separate out the red channel in each image, where a region of interest was then selected and average red intensity in this region was read over the entire movie file.
3. Results and discussion
3.1. Standard thermocycler amplification of SARS-CoV-2 samples
10 μL LAMP reactions containing various concentrations of SARS-CoV-2 (NTC, 101, 103, and 105 copies/μL) were amplified at 65 °C for four time-increments (15, 20, 25, and 30 min) to examine the ability of a standard thermocycler reaction (without an emulsion component and without a saliva matrix) to amplify SARS-CoV-2 samples. The results of this can be found in Supplementary Figure S1, which shows that the two highest concentration samples (103 and 105 copies/μL) amplified at all time points, whereas the lower concentrations amplified only at the highest time point, and the bands (and prominent streaks throughout the lanes) for these samples show that the presence of amplification is likely due to non-specific amplification, giving evidence to how prominent this issue is when using LAMP.
3.2. Interfacial tension of conventional LAMP samples
Aqueous LAMP samples of E. coli O157:H7 (at 106 CFU/μL) were conventionally amplified for 0, 5, 10, and 20 min in a conventional thermocycler as a model for showing the impact increasing LAMP amplicon presence has on interfacial tension (IFT). Emulsions were not formed for IFT measurements. The IFT of these samples was measured via pendant droplet method, and results show a decreasing trend in IFT with increasing amplification time (Fig. 2 A). Essentially, decreasing IFT measurements with respect to amount of DNA is quantifying the stability of the droplet. As more DNA is present, the overall droplet's interfacial tension is destabilized due to the amplicon adsorption to the water-air interface of the hanging pendant droplet. This overarching phenomenon will be used as the means in which we justify the detection method in our emulsion platform.
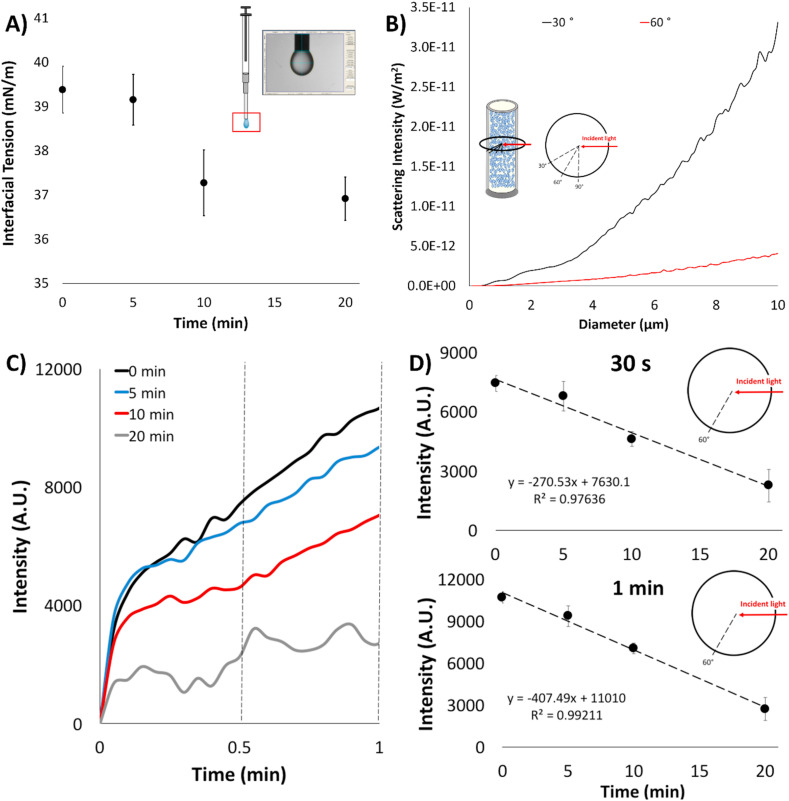
Fig. 2.
Proof-of-concept results with the pre-amplified E. coli O157:H7 samples (at 106 CFU/μL). A) Interfacial tension (IFT) measurements of the pre-amplified E. coli O157:H7 samples ran at varying conventional amplification times via pendant droplet analysis. Error bars show standard error with a sample size of 3. B) Mie scatter simulation of light scatter intensity of a normally distributed colloid size distribution in relation to the diameter at 30° and 60° with respect to a 650 nm incident light. Inset illustration depicts an artist's rendition of the emulsion. Blue spheres indicate aqueous droplets suspended in the bulk oil phase (not to scale). A red line shows where red incident light enters the system, and black lines indicate where scatter is measured. C) Emulsion light scatter intensities measured at 60° with respect to 650 nm incident light for the pre-amplified and emulsified suspensions of E. coli O157:H7 over time. D) 60° Light scatter intensities measured at 3 min for these pre-amplified E. coli O157:H7 and emulsified suspensions, showing the linear relationship between time amplified and light scatter intensity. Error bars show standard error with a sample size of 3. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Mie light scatter simulation with varying emulsion diameter
Within an emulsion are micron size aqueous droplets (confirmed from light microscope images suspended in a mineral oil and surfactant mixture). As seen in the previous section, the presence of DNA causes the destabilization of a droplet. Emulsions are a dynamic system with continuous agitation from a micro stir bar. Therefore, we expect that with increasing amount of DNA amplification, the size (diameter) distribution of emulsions will decrease. The method in which real-time change in emulsion size is monitored, is by collecting the bulk light scatter at various angles or angularly resolved light scattering with respect to an incident light. Based off the Mie theory, angularly resolved light scatter off a particle (in this case emulsion droplet) gives information to the overall size characteristics of the particle sample (Fu and Sun, 2001). The intensity of light scattered off a particle distribution in relation to various diameters (10 μm–0.05 μm) at 30° and 60° were simulated to model the decreasing diameter due to increasing presence of DNA amplicons that would be produced in an emulsion LAMP reaction. These angles were chosen because they represent two equally distant angles that are between 0 and 90°, thus allowing for adequate representation of forward scatter (which occurs between 0 and 90°). As seen in Fig. 2B, light intensity decreases as the diameter of emulsions decrease for both angles. The simulation also showed that 30° light scatter had higher intensity values, followed by 60°.
3.4. Light scatter measurements of the emulsions from pre-amplified samples
As a model sample matrix to simulate LAMP amplicon production, we again utilized the pre-amplified LAMP solutions of E. coli O157:H7 (at 106 CFU/μL). However, they were emulsified after the pre-amplification to ensure that amplification had occurred. Light scatter intensities of these emulsions were determined in relation to the time of pre-amplification (subsequently the amount of amplicon).
As demonstrated previously, light scatter, supported by the Mie theory, will change in intensity due to size dependencies. In Fig. 2C, the light scatter intensity profiles (experimentally measured using the pre-amplified E. coli O157:H7) at 60° for the varying samples is shown in relation to time (30° detection will also be demonstrated for later, in situ amplification of SARS-CoV-2). However, light scatter intensity at 30 s and 1 min in relation to pre-amplification time shows a linear relationship (R2 = 0.976, 0.992 respectively) (Fig. 2D). It can then be alluded, that the decrease in intensity is due to an increase amount of amplicon product due to a decrease distribution of emulsion diameter.
3.5. Emulsion diameter measurement of emulsion LAMP
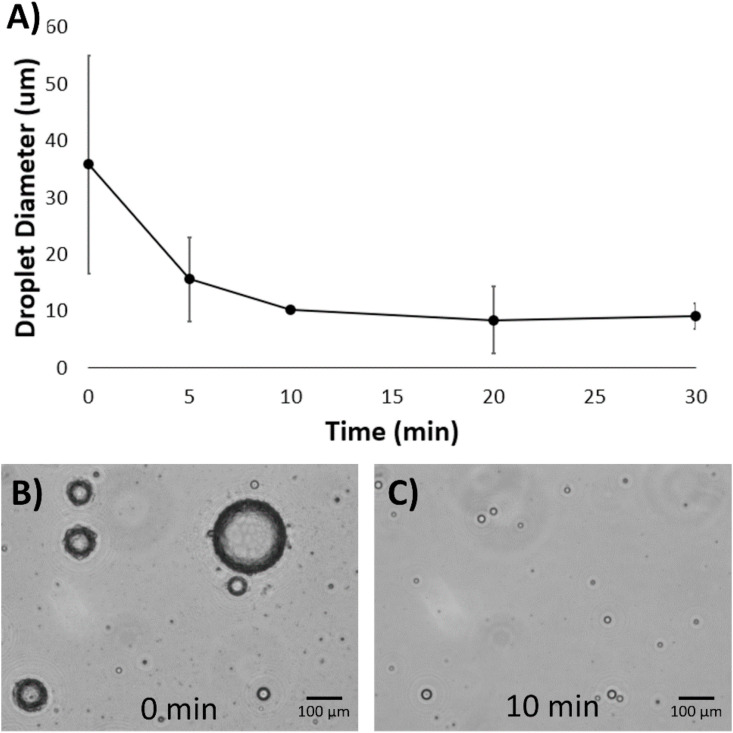
Emulsion diameters were measured from the microscopic images of the full emulsion LAMP reactions from 105 copies/μL SARS-CoV-2 samples. 10 μL samples were extracted from the emulsion at 0, 5, 10, 20, and 30 min. The average diameter for the emulsions ranged between 35.8 and 8.4 μm. Therefore, due to the increasing presence of LAMP amplicons, emulsion diameter size is decreasing due to decreasing interfacial tension at the water-oil interface, causing unstable emulsions and resulting in smaller diameters. Fig. 3 B and C shows the representative microscopic images of these emulsion suspensions at t = 0 min and t = 10 min. Together with the light scattering results shown in Fig. 2C and D, these results indicate that there were sufficient number of emulsion droplets that contained the amplicons with the reduced diameters.
Fig. 3.
A) Measured diameters from the light microscope images of 10 μL emulsion from the emulsion LAMP reactions of 105 copies/μL SARS-CoV-2, taken at 0, 5, 10, 20, and 30 min time points (n = 5). The average droplet counts are 29, 22, 44, 35, and 53 at 0, 5, 10, 20, and 30 min, respectively. B) Representative microscopic image of these emulsion suspensions at t = 0 min. C) The same at t = 10 min.
3.6. In situ light scatter measurements from emulsion LAMP of SARS-CoV-2 via a miniature spectrophotometer
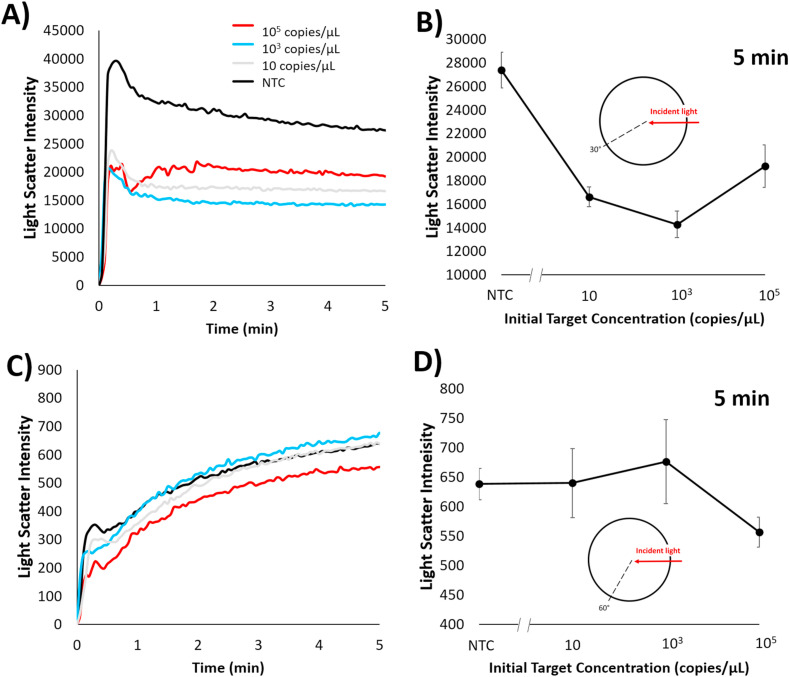
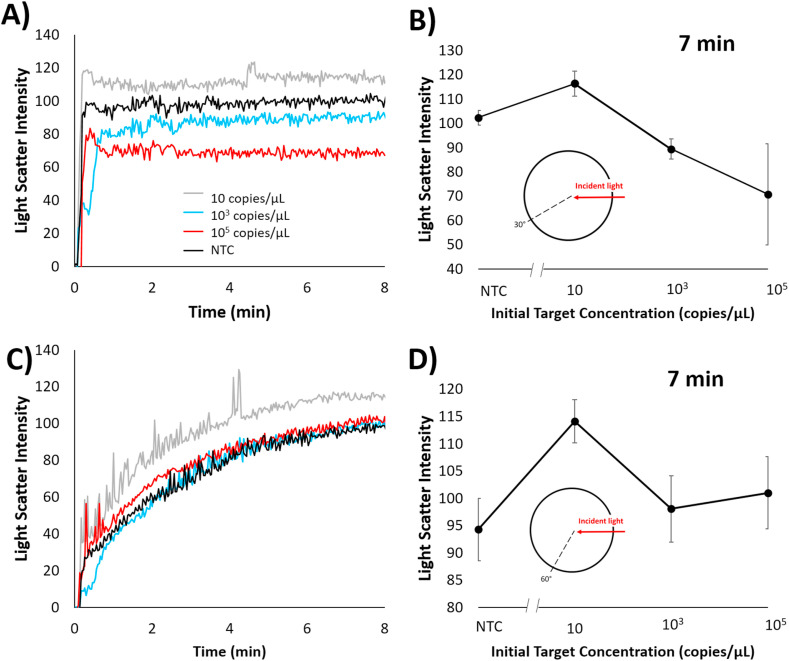
Light scatter intensities from fiber optic cables placed at 30° and 60° angles with respect to a 650 nm incident light was collected in situ from a miniature spectrophotometer of emulsion samples with LAMP reactions containing initial SARS-CoV-2 positive control concentrations of 105, 103, 10, and 0 copies/μL (Fig. 4 ). For both angles the underlying trend was that light scatter intensity decreased with increasing concentration. This trend is synonymous to trends found earlier with light scatter of emulsions containing E. coli O157:H7 samples that were conventionally amplified for a set time prior to emulsification. Light scatter intensities collected at the 30° angle showed the greatest difference in change for 103 copies/μL in comparison to no target control (NTC; 0 copies/μL with all other reagents), however all positive samples had significantly decreased intensities at the 5 min time point. The greatest percent change in light scatter intensity appeared to be 39.3% within 5 min on the emulsion platform between 0 and 103 copies/μL. 60° light scatter intensities at different initial concentration did not show significant differences other than 105 copies/μL. Light scatter intensity at 5 min shows a 12.84% intensity fluctuation amongst 0 to 105 copies/μL concentrations. These results indicate that the 30° angle may be the more sensitive angle for light scatter analysis, but the 60° angle can be utilized to detect whether the sample contains a high concentration of target. Compared to some existing LAMP assays used for SARS-CoV-2 detection, the time-to-detection is extremely fast (Ali et al., 2020; Thi et al., 2020; Zhu et al., 2020). This is due to the fact that the emulsion platform allows for compartmentalization of the LAMP reaction into micrometer-size droplets that change characteristics (droplet size namely) rapidly as the LAMP reaction occurs. These changes in droplet size cause a rapid change in light scatter intensity, allowing for rapid detection of the ongoing LAMP reaction.
Fig. 4.
In situ light scatter intensity changes for emulsion LAMP reaction of SARS-CoV-2 via spectrophotometer. Changes over time are shown at A) 30° and C) 60° angle with respect to 650 nm incident wavelength with varying initial SARS-CoV-2 positive control concentration of 105, 103, 10, and 0 copies per μL. A) and C) are representative plots; All other plots are available in Supplementary Figure S2. Light scatter intensities at 5 min for B) 30° and D) 60° angles are plotted against the SARS-CoV-2 positive control concentrations of 105, 103, 10, and 0 copies per μL. Error bars show standard error with a sample size of 3.
3.7. Fluorescent probe detection of SARS-CoV-2 amplification
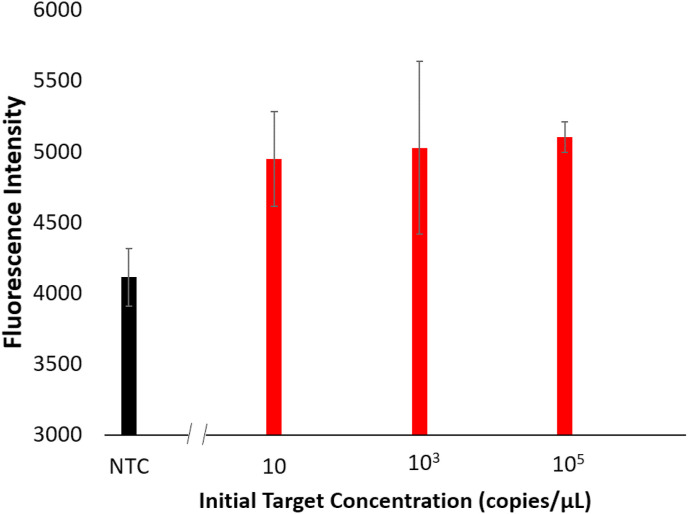
A fluorescence probe was added to the FIP primer and an end-point quantification of this probe was attempted for the aqueous phase broken from the emulsion after the emulsion LAMP reaction. A miniature spectrophotometer acquired the fluorescence spectrum and the emission at 520 nm was evaluated with 350 nm UV excitation (Fig. 5 ). The data shows a clear increase in fluorescence intensity between the NTC sample and the SARS-CoV-2 positive samples, with a maximum increase of 24.02% with the 105 copies/μL sample, and significant increases in fluorescence intensity for the 10 and 105 copies/μL samples, indicating that SARS-CoV-2 N protein gene amplification occurred within the aqueous droplets.
Fig. 5.
Fluorescent intensities for the broken aqueous solutions from emulsion LAMP reaction for SARS-CoV-2 positive control samples at varying concentrations. Error bars show standard error with a sample size of 3.
Additionally, gel electrophoresis experiments were also performed for the extracted aqueous phase from the broken emulsion (following the procedure described in sections 2.9-2.11). As the emulsion breaking and aqueous phase extraction are not efficient processes, only a tiny amount of aqueous phase could be extracted, which were insufficient to run an end-point gel electrophoresis analysis. Therefore, these extracted samples (from 30-min emulsion LAMP) were additionally amplified for 15 min in a conventional thermal cycler. We also conducted standard conventional LAMP reactions at the same initial target concentration for 15 min in parallel to these. If a sample had initially undergone an emulsion LAMP reaction before a conventional reaction, a higher band intensity could be observed. The results of these experiments are shown in Supplementary Figure S3. The samples that had undergone prior emulsion LAMP reactions contained clear bands at correct lengths, while those who underwent conventional amplification did not. This result indicates that the emulsion LAMP reaction did induce initial amplification of the target sequence.
3.8. In situ light scatter measurements from emulsion LAMP of SARS-CoV-2 via smartphone camera and 3D-printed enclosure
In order to test the efficacy of such a platform using a smartphone detection system, a 3D-printed enclosure (placed on a hot plate) was designed and fabricated to hold the emulsion reaction chamber and house 2 blinking red LEDs placed at 30° and 60° angles with respect to a smartphone camera. 10 μL LAMP reactions with varying initial SARS-CoV-2 positive sample concentrations of 105, 103, 10, and 0 copies/μL were placed into the emulsion platform in a similar fashion as the miniature spectrophotometer procedure. Images were taken every 1 s while the differently angled LEDs switched back and forth every 3 s (allowing 3 images per angle before switching) over the course of 15 min to characterize smartphone optical detection as replacement for a spectrophotometer and fiber optical cable experimental set up for a more user-friendly platform. From the captured images, red channel intensity was extracted and sorted from time-lapsed image sequence for both angles (Fig. 6 A and C). Intensity from these curves were taken in relation to the concentration of initial target concentration at 7 min (Fig. 6B and D). For the 60° angle light scatter trend over initial target concentration (Fig. 6D), no significant trend could be seen over concentration, which is similar to how the spectrophotometer platform performed (Fig. 4D). On the other hand, the 30° angle light scatter trend over initial target concentration (Fig. 6B) showed a decreasing trend over initial concentration, again similar to how the spectrophotometer platform performed (Fig. 4B). At this angle, the 103 copies/μL datapoint was statistically significant from the NTC at the 7-min time point, and with increasing time, both the 103 and 105 copies/μL become more differentiated from the NTC, indicating that these concentrations may be more significantly different at later times. The difference in detection limits between the smartphone and spectrophotometer platforms is likely due to the fact that the spectrophotometer platform focuses the scattered light in such a way that overall noise is reduced and slight changes in emulsion droplet size are more apparent. This results in the 30° angle being not as sensitive on this platform compared to the spectrophotometer platform. With the smartphone-based platform, we again see that this assay can potentially detect SARS-CoV-2 much faster than existing LAMP assays for the virus.
Fig. 6.
In situ red channel intensity changes for emulsion LAMP reaction of SARS-CoV-2 via smartphone camera. Changes over time are shown at A) 30° and C) 60° angle with respect to 650 nm incident wavelength with varying initial SARS-CoV-2 positive control concentrations of 105, 103, 10, and 0 copies/μL. A) and C) are representative plots; All other plots are available in Supplementary Figure S4. Representative raw smartphone images are also shown in Supplementary Figure S5. Red channel light scatter intensity changes at 7 min are shown for B) 30° and D) 60°. Error bars show standard error with a sample size of 3. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Conclusions
An emulsion platform was investigated to determine angle-dependent light scatter's potential to monitor in real-time the amplification of nucleic acids in an isothermal state via LAMP reaction. The phenomenon to attribute to light scatter intensity changes due to amplicon presence was verified via pendant droplet analysis for the measurement of interfacial tension (IFT). IFT measurements showed that with added LAMP amplicons there is a decrease in IFT. Amplicon presence destabilizes aqueous droplets due to the increased molecules at the interface. Therefore, we can allude that in an emulsion platform, solutions with less amplicon presence will have decreased diameter distribution. This was confirmed via light microscope images where the emulsion diameters with little-to-no presence of amplification (0 and 5 min of pre-amplification times) were smaller than the emulsion diameters with more extensive amplification (10 and 20 min of pre-amplification times).
Emulsions with various amounts of model amplicon solutions showed that the angle-dependent light intensities at 30° and 60° decreased in relation to increased number of amplicons. Mie scatter simulations further confirmed that light scatter intensity is diameter dependent. Smaller diameter emulsions will have higher intensity values than larger diameter emulsions.
In situ real-time monitoring of light scatter intensity from LAMP emulsions were performed with varying initial SARS-CoV-2 positive sample concentrations. Light scatter intensities at 5 min showed similar trends to the light scatter experiments with pre-amplified LAMP solutions. At 5 min, 30° light scatter intensity can statistically differentiate 10, 103 and 106 copies/μL initial concentrations in comparison to NTC (0 copies/μL). 5 min light scatter intensities collected at 60° can statistically differentiate 105 copies/μL initial concentrations in comparison to NTC (0 copies/μL). 30° light scatter shows high sensitivity but contains no concentration dependency, thus necessitating the need for the 60° data, which allows the user to differentiate between a high concentration and low concentration of initial target within the sample. The time-to-detection of this platform is therefore shown to be far faster than existing LAMP-based assays for SARS-CoV-2 detection.
Furthermore, a 3D-printed enclosure with an emulsion reaction chamber, 2 blinking LEDs placed at 30° and 60°, and smartphone holder was designed and utilized to simplify the emulsion platform from spectrophotometers and fiber optical cables to a more user-friendly platform. Similar light scatter intensities were achieved for both angles and in comparison to spectrophotometer collected data, thus demonstrating translatability of emulsion LAMP detection technologies toward field-deployability in resource-limited or clinical settings.
These conclusions indicate that this technology could be used to reduce the time-to-detection of SARS-CoV-2 in saliva samples from patients. These findings show that the hours-long process normally needed for RT-PCR tests could potentially be reduced to a minutes-long process, thus improving the overall detection times for patient samples and allowing for faster public health responses.
CRediT authorship contribution statement
Alexander S. Day: Conceptualization, Methodology, Investigation, Formal analysis, Software, Visualization, Writing – original draft. Tiffany-Heather Ulep: Conceptualization, Methodology, Investigation, Formal analysis, Software, Visualization, Writing – original draft. Babak Safavinia: Investigation, Data curation, Writing – review & editing. Tyler Hertenstein: Investigation, Data curation, Writing – review & editing. Elizabeth Budiman: Investigation, Data curation. Laurel Dieckhaus: Methodology, Investigation. Jeong-Yeol Yoon: Conceptualization, Methodology, Supervision, Project administration, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to thank Jokubas Ausra and Aarin Shah for their experimental assistance. This work was supported by The University of Arizona's Test All Test Smart Program and the Cardiovascular Biomedical Engineering Training Grant from the U.S. National Institute of Health (grant number T32HL007955).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113099.
Author contributions
Alexander S Day: Conceptualization, Methodology, Investigation, Formal Analysis, Software, Visualization, Writing – Original Draft; Tiffany-Heather Ulep: Conceptualization, Methodology, Investigation, Formal Analysis, Software, Visualization, Writing – Original Draft; Elizabeth Budiman: Investigation, Data Curation; Laurel Dieckhaus: Methodology, Investigation; Babak Safavinia: Investigation, Data Curation, Writing – Review & Editing; Tyler Hertenstein: Investigation, Data Curation, Writing – Review & Editing; Jeong-Yeol Yoon: Conceptualization, Methodology, Supervision, Project Administration, Writing – Review & Editing.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ali Z., Aman R., Mahas A., Rao G.S., Tehseen M., Marsic T., Salunke R., Subudhi A.K., Hala S.M., Hamdan S.M., Pain A. Virus Res. 2020;288 doi: 10.1016/j.virusres.2020.198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [Google Scholar]
- Cui F., Zhou H.S. Biosens. Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deguo W., Guicheng H., Fugui W., Yonggang L., Daxi R. Afr. J. Food Sci. 2008;1(7):83–86. doi: 10.5897/AJFS.9000079. [DOI] [Google Scholar]
- Fu Q., Sun W. Appl. Optic. 2001;40(9):1354–1361. doi: 10.1364/AO.40.001354. [DOI] [PubMed] [Google Scholar]
- Gadkar V.J., Goldfarb D.M., Gantt S., Tilley P.A. Sci. Rep. 2018;8:5548. doi: 10.1038/s41598-018-23930-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshman D.K., Rao B.M., McLain J.E., Watts G.S., Yoon J.Y. Sci. Adv. 2015;1(8) doi: 10.1126/sciadv.1400061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M., Komatsu J., Matsuura S.I., Takashima K., Katsura S., Mizuno A. J. Biotechnol. 2003;102(2):117–124. doi: 10.1016/S0168-1656(03)00023-3. [DOI] [PubMed] [Google Scholar]
- Nicolini A.M., Toth T.D., Kim S.Y., Mandel M.A., Galbraith D.W., Yoon J.Y. Adv. Biosyst. 2017;1(10) doi: 10.1002/adbi.201700098. [DOI] [PubMed] [Google Scholar]
- Ravi N., Cortade D.L., Ng E., Wang S.X. Biosens. Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thi V.L., Herbst K., Boerner K., Meurer M., Kremer L.P., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M.L., Boulant S. Sci. Transl. Med. 2020;12(556) doi: 10.1101/2020.06.15.152587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulep T.H., Day A.S., Sosnowski K., Shumaker A., Yoon J.Y. Sci. Rep. 2019;9:9629. doi: 10.1038/s41598-019-46028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R., Peisajovich S.G., Miller O.J., Magdassi S., Tawfik D.S., Griffiths A.D. Nat. Methods. 2006;3(7):545–550. doi: 10.1038/nmeth896. [DOI] [PubMed] [Google Scholar]
- Xu X.W., Wu X.X., Jiang X.G., Xu K.J., Ying L.J., Ma C.L., Li S.B., Wang H.Y., Zhang S., Gao H.N., Sheng J.F. Br. Med. J. 2020;369:606. [Google Scholar]
- Zhang Y., Odiwuor N., Xiong J., Sun L., Nyaruaba R.O., Wei H., Tanner N.A. 2020. MedRxiv 20028373. [DOI]
- Zhao X., Li Y., Wang L., You L., Xu Z., Li L., He X., Liu Y., Wang J., Yang L. Mol. Biol. Rep. 2010;37(5):2183–2188. doi: 10.1007/s11033-009-9700-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.