Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic represents a global threat, and the interaction between the virus and angiotensin-converting enzyme 2 (ACE2), the primary entry receptor for SARS-CoV-2, is a key determinant of the range of hosts that can be infected by the virus. However, the mechanisms underpinning ACE2-mediated viral entry across species remains unclear. Using infection assay, we evaluated SARS-CoV-2 entry mediated by ACE2 of 11 different animal species. We discovered that ACE2 of Rhinolophus sinicus (Chinese rufous horseshoe bat), Felis catus (domestic cat), Canis lupus familiaris (dog), Sus scrofa (wild pig), Capra hircus (goat), and Manis javanica (Malayan pangolin) facilitated SARS-CoV-2 entry into nonsusceptible cells. Moreover, ACE2 of the pangolin also mediated SARS-CoV-2 entry, adding credence to the hypothesis that SARS-CoV-2 may have originated from pangolins. However, the ACE2 proteins of Rhinolophus ferrumequinum (greater horseshoe bat), Gallus gallus (red junglefowl), Notechis scutatus (mainland tiger snake), or Mus musculus (house mouse) did not facilitate SARS-CoV-2 entry. In addition, a natural isoform of the ACE2 protein of Macaca mulatta (rhesus monkey) with the Y217N mutation was resistant to SARS-CoV-2 infection, highlighting the possible impact of this ACE2 mutation on SARS-CoV-2 studies in rhesus monkeys. We further demonstrated that the Y217 residue of ACE2 is a critical determinant for the ability of ACE2 to mediate SARS-CoV-2 entry. Overall, these results clarify that SARS-CoV-2 can use the ACE2 receptors of multiple animal species and show that tracking the natural reservoirs and intermediate hosts of SARS-CoV-2 is complex.
Keywords: SARS-CoV-2, ACE2, susceptibility, cross-species, pangolin
Abbreviations: ACE2, angiotensin-converting enzyme 2; BSA, bovine serum albumin; COVID-19, coronavirus disease 2019; DAPI, 4′,6-diamidino-2-phenylindole; DMEM, Dulbecco’s modified Eagle’s medium; FBS, fetal bovine serum; HA, hemagglutinin; ICTV, International Committee on Taxonomy of Viruses; MOI, multiplicity of infection; PBS, phosphate-buffered saline; RT, room temperature; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2
In December 2019, a novel pneumonia, termed coronavirus disease 2019 (COVID-19) by the World Health Organization (WHO), emerged from Wuhan, China, and the causative agent was soon identified as a novel coronavirus, which was named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the International Committee on Taxonomy of Viruses (ICTV) (1, 2). The SARS-CoV-2 outbreak has been speculatively associated with a seafood market where sales also include various wild animals (3). Bats are recognized as a potential natural reservoir for SARS-CoV-2 (1, 3). However, recent studies have indicated that pangolins are also considered possible natural reservoirs of this coronavirus (4, 5). Discovering the potential intermediate animal hosts of SARS-CoV-2 and evaluating their possible cross-species transmissibility are scientifically very important. Unfortunately, we know little about this. A recent study revealed that ferrets and cats are sensitive to SARS-CoV-2 infection; however, these animals showed no clinical symptoms (6). Whether other animals exist as a candidate SARS-CoV-2 infection model should be further explored.
The interaction between receptors and viruses is a key determinant of the host range. SARS-CoV-2 resembles severe acute respiratory syndrome coronavirus (SARS-CoV), and both use angiotensin-converting enzyme 2 (ACE2) as the primary cell entry receptor (1, 7, 8, 9). When we retrace the origin of a virus, the cell susceptibility to viruses conferred by receptors of speculated animals is preferentially investigated (10, 11). Before clarifying that Rhinolophus sinicus (Chinese rufous horseshow bat) is the natural reservoir of SARS-CoV, scientists first evaluated the susceptibility provided by ACE2 from different bat species to SARS-CoV. They found that the ACE2 of R. sinicus was responsible for the susceptibility to SARS-CoV and subsequently confirmed that R. sinicus was the natural reservoir of SARS-CoV (10, 12, 13). The Middle East respiratory syndrome coronavirus (MERS-CoV) was also recognized as having a bat origin because MERS-CoV and two MERS-CoV-related viruses from bats could utilize human or bat dipeptidyl peptidase 4 (DPP4) for cell entry (14, 15, 16).
Therefore, in this study, we systemically evaluated the ability of SARS-CoV-2 to infect nonsusceptible HEK293T cells utilizing ACE2 proteins from nine different animal species and humans to determine its possible origin and to further explore its cross-species transmission. Our findings provide evidence that SARS-CoV-2 is able to engage the ACE2 receptor of different species, which poses a very large challenge to determine the natural reservoir of SARS-CoV-2 for the control and prevention of coronaviruses in the future. Furthermore, we found a natural isoform of ACE2 in Macaca mulatta (rhesus monkey; RhACE2) with the Y217N mutation, which was resistant to SARS-CoV-2 infection, and this amino acid residue is a novel critical determinant of the ability of ACE2 to mediate SARS-CoV-2 entry.
Result
Cell entry of SARS-CoV-2 conferred by ACE2 of different species
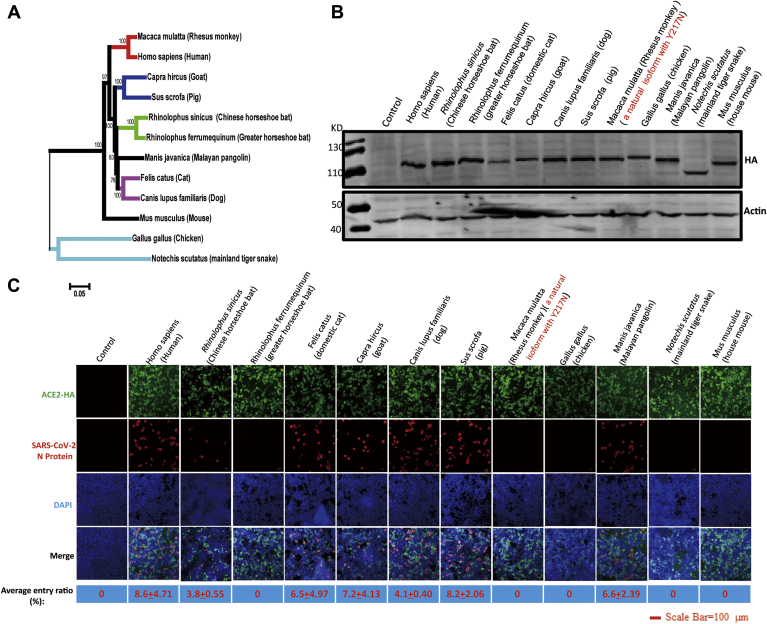
To investigate which animal ACE2 proteins could facilitate SARS-CoV-2 entry, we synthesized full-length cDNA fragments of ACE2 from 11 animal species as well as from humans. These species were R. sinicus (Chinese rufous horseshoe bat), Rhinolophus ferrumequinum (greater horseshoe bat), Felis catus (domestic cat), Capra hircus (goat), Canis lupus familiaris (dog), Sus scrofa (wild pig), Manis javanica (Malayan pangolin), Gallus gallus (red junglefowl), Notechis scutatus (mainland tiger snake), Mus musculus (house mouse), M. mulatta (rhesus monkey), and Homo sapiens (human). Synthesized cDNA fragments were then subcloned into the pCAGGS-HA vector for expression in eukaryotic cells. The origins and GenBank accession numbers of these ACE2 molecules are listed in the Table 1. We first compared the nucleotide sequence of the ACE2 coding region of these 11 animals with that of humans. The sequence similarities of these ACE2 cDNAs are exhibited in the Table 1. Among these sequences, the sequence of RhACE2 was the most similar to that of human ACE2 (hACE2), and in contrast, the sequence of mainland tiger snake ACE2 was the least similar (Fig. 1A). It has been reported that two virus-binding hotspots, K31 and K353 in hACE2, are critical for SARS-CoV infection (17, 18). In this study, we found that K31 was not conserved in ACE2 of all 11 animal species observed in this study. However, K353 was conserved in all ten animal species except mouse (Table 1).
Table 1.
Nucleotide sequence similarity of the ACE-2 proteins of various animals to human ACE-2
| ACE-2 origin | Length of coding sequence (bp) | Similarity to human ACE-2 (%) | Position 31 | Position 353 | GenBank accession number |
|---|---|---|---|---|---|
| Homo sapiens (Human) | 2418 | 100 | K | K | AB046569.1 |
| Rhinolophus ferrumequinum (Greater horseshoe bat) | 2418 | 86.2 | D | K | AB297479.1 |
| Rhinolophus sinicus (Chinese horseshoe bat) | 2418 | 85.5 | E | K | GQ262791.1 |
| Macaca mulatta (Rhesus monkey)a | 2418 | 96.6 | K | K | NM_001135696.1 |
| Sus scrofa (Pig) | 2418 | 84.5 | K | K | NM_001123070.1 |
| Canis lupus familiaris (Dog) | 2415 | 87 | K | K | NM_001165260.1 |
| Capra hircus (Goat) | 2415 | 85.5 | K | K | KF921008.1 |
| Felis catus (Cat) | 2418 | 86.8 | K | K | AY957464.1 |
| Gallus gallus (Chicken) | 2427 | 68.1 | E | K | MK560199.1 |
| Manis javanica (Malayan pangolin) | 2418 | 86.5 | K | K | XM_017,650,257.1 |
| Notechis scutatus (Mainland tiger snake) | 2487 | 66.5 | Q | K | XM_026674969 |
| Mus musculus (Mouse) | 2418 | 85.2 | N | H | NM_001130513.1 |
NOTE: The sequence for Macaca mulatta (Rhesus monkey) provided here contains the natural mutation Y217N.
Figure 1.
Susceptibility of ACE2 proteins from different species to SARS-CoV-2 in HEK 293T cells.A, phylogenetic analysis of the ACE2 cDNA sequences of 11 animal species and humans. B, HEK 293T cells were transfected with plasmids expressing the indicated ACE2 protein. Cells were lysed for western blotting 36 h after being transfected. C, HEK 293T cells were transfected with plasmids expressing the indicated ACE2 protein. Cells were infected at a 0.5 MOI of SARS-CoV-2, and 24 h after transfection, IFA was used to detect the entry of SARS-CoV-2 in cells. The average entry ratio was analyzed by ImageJ software.
Next, we tested whether ACE2 of the 11 animal species was able to facilitate SARS-CoV-2 entry to nonsusceptible HEK 293T cell lines. Different ACE2 proteins were expressed in HEK 293T cells (Fig. 1, B and C). Plasmids expressing hACE2 and mouse ACE2 were applied as the positive and negative controls of the entry assay, respectively. The infection ratio varied according to the ACE2 protein expressed (Fig. 1C). As expected, hACE2 supported SARS-CoV-2 entry, whereas mouse ACE2 did not (Fig. 1C). One previous study indicated that the SARS-CoV outbreak 17 years ago originated from Rhinolophus affinis (intermediate horseshoe bat) (12). A recent study further demonstrated that ACE2 of R. sinicus (Chinese rufous horseshoe bat) allowed both SARS-CoV-2 and SARS-CoV entry (1). In this study, we were not able to synthesize the ACE2 cDNA of R. affinis due to the absence of its sequence. Therefore, we synthesized the ACE2 cDNA of R. sinicus and R. ferrumequinum (greater horseshoe bat) to test whether ACE2 of other bat species was responsible for the susceptibility to SARS-CoV-2. Interestingly, we found that the ACE2 of R. ferrumequinum did not support SARS-CoV-2 entry but R. sinicus did (Fig. 1C), suggesting that not all species of bat are sensitive to SARS-CoV-2 infection.
A recent study indicated that SARS-CoV-2 did not replicate and shed in dogs, pigs, chickens, and ducks but replicated fairly well in ferrets and replicated effectively in cats (6). Our study demonstrated that cat ACE2 supported viral entry of SARS-CoV-2 (Fig. 1C). Although pigs, dogs, and chickens are not sensitive to SARS-CoV-2 infection, we know little about the molecular mechanisms and role of receptor avidity in resistance. Our data demonstrated that the ACE2 protein of dogs and pigs supported SARS-CoV-2 entry similar to that of cats.
There is a debate regarding if SARS-CoV-2 originated from bats or pangolins (1, 4, 5). It has been demonstrated that bat ACE2 mediates SARS-CoV-2 entry (1). Therefore, we expressed ACE2 in M. javanica (Malayan pangolins) and tested its role in conferring susceptibility to SARS-CoV-2. We demonstrated that pangolin ACE2 could mediate the entry of SARS-CoV-2 (Fig. 1C).
Finally, we demonstrated that ACE2 of N. scutatus (mainland tiger snake) could not support SARS-CoV-2 entry as previously predicted (19). Snakes may not be the natural reservoir of SARS-CoV-2.
Y217 of ACE2 is a novel critical determinant residue on ACE2 that mediates SARS-CoV-2 entry
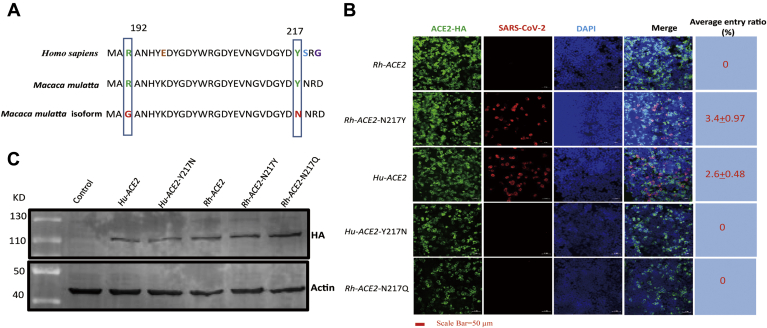
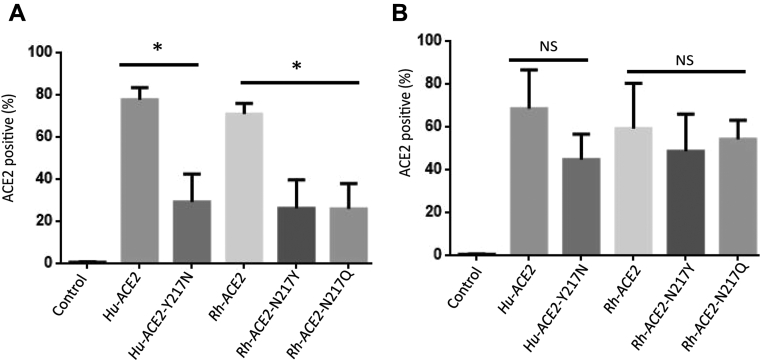
Old world monkeys (M. mulatta (rhesus monkey) and Macaca fascicularis (crab-eating macaque)) were used as experimental animal models for SARS-CoV-2 infection (20). Surprisingly, we found that RhACE2 in our study did not support SARS-CoV-2 entry, as expected (Fig. 1C). By investigating the monkey ACE2 sequence, we found that the ACE2 isoform of rhesus monkey cloned in our study contained two natural variations (R192G and Y217N) (Fig. 2A). When we reverted the Y217N mutation to N217Y (wild-type sequence of ACE2) in RhACE2, RhACE2 with N217Y had the ability to support SARS-CoV-2 infection (Fig. 2B). We noticed that Y217 was conserved in the other species investigated in this study (data not shown), which suggests that residue Y217 is a key residue for SARS-CoV-2 entry. To further confirm this hypothesis, we constructed hACE2 with the Y217N mutation and found that this mutation completely blocked SARS-CoV-2 entry, as expected (Fig. 2B).
Figure 2.
Sequence composition of RhACE2 cloned in this study with that of the wild-type RhACE2 and the susceptibility of RhACE2 proteins to SARS-CoV-2.A, two sites of natural variation (R192G and Y217N) were identified in the cDNA of RhACE2 cloned in this study, and wild-type RhACE2, mutant RhACE2, and hACE2 were compared. B, HEK 293T cells were transfected with plasmids expressing the indicated ACE2 protein. Cells were infected at a 0.5 MOI of SARS-CoV-2, and 24 h after transfection, IFA was used to detect the replication of SARS-CoV-2 in cells. The average entry ratio was analyzed by ImageJ software. C, HEK 293T cells were transfected with plasmids expressing the indicated ACE2 protein. Cells were lysed for western blotting 36 h after being transfected.
Potential asparagine (N)-linked glycosylation may influence virus and receptor binding; however, when we analyzed the sequence, we found that ACE2 N217 was not a potential N-linked glycosylation site (N-X-T/S motif). We further constructed an RhACE2 with the N217Q mutation, and this mutation did not facilitate SARS-CoV-2 entry (Fig. 2B). This further demonstrated that the ability of N217 blocking viral entry is not due to N-linked glycosylation of this residue. A previous study indicated that the natural mutation of Y217N in RhACE2 dramatically alters RhACE2 expression and reduces the efficiency of SARS-CoV entry (21). In the current study, we illustrated that both hACE2 and RhACE2 Y217N expressed as well as the corresponding wild-type ACE2 proteins (Fig. 2C).
ACE2 Y217N significantly reduced the binding ability of the receptor-binding domain (RBD) and spike protein
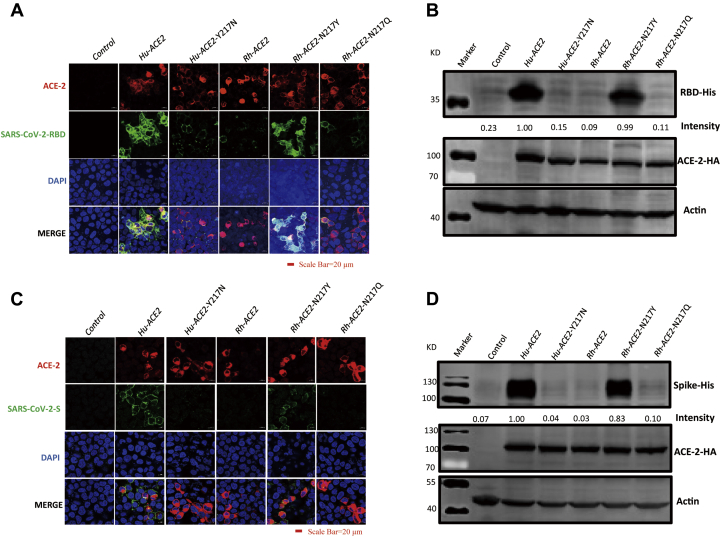
Next, we used the RBD as a probe to examine receptor-binding ability by using IFA. We performed an RBD binding assay and found that wild-type hACE2 potently bound the RBD, as expected (Fig. 3A); however, hACE2 Y217N, RhACE2 Y217N, and RhACE2 Y217Q almost lost the ability to bind the RBD (Fig. 3A). Receptor-binding ability was further confirmed and quantified by western blotting (Fig. 3B). Since the conformation of a recombinant RBD may not completely reflect its native conformation within an entire envelope glycoprotein, we also investigated ACE2 binding with more relevant systems of the full-length S (spike) protein, and we obtained a similar result as for the RBD (Fig. 3, C and D). The above data demonstrated that the Y217 residue of ACE2 significantly reduced the binding ability of the RBD and spike protein.
Figure 3.
The Y217N mutation in ACE2 significantly reduced the ability of ACE2 to bind the RBD and spike protein.A, HEK 293T cells were transfected with plasmids expressing the indicated ACE2 protein. At 24 hpt, the transfected cells were incubated with 5 μg RBD for 1 h at 37 °C, and then IFA and (B) western blotting were performed and RBD binding was quantified by densitometry scanning of western blot images by Image J software. Intensity of RBD binding for Human ACE2 was set as 1. C, HEK 293T cells were transfected with plasmids expressing the indicated ACE2 protein. At 24 hpt, the transfected cells were incubated with 5 μg S protein for 1 h at 37 °C, and then IFA and (D) western blotting were performed and S protein binding was quantified by densitometry scanning of western blot images by Image J software. Intensity of S protein binding for Human ACE2 was set as 1.
Structural modeling of ACE2 Y217N shows no changes in ACE2 and RBD binding
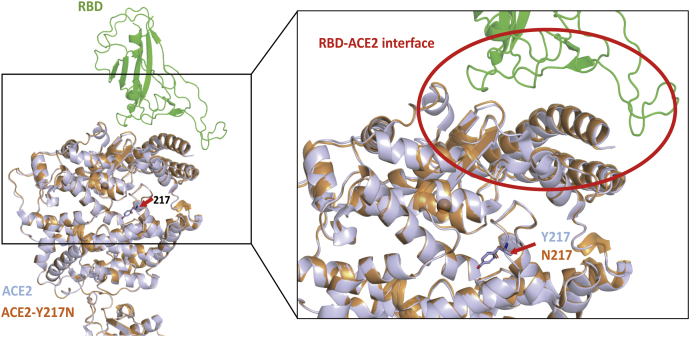
To test whether position 217 of ACE2 influenced its RBD binding activities is due to N217 disrupting the interaction between ACE2 and RBD, we used homology-based structural modeling to analyze the effects of residue substitutions at position 217. Structural models of Y217N were generated based on the crystal structure of the SARS-CoV-2 RBD/ACE2 complex (22, 23). We found that position 217 was not located at the interaction domain with RBD; furthermore, the predicted model of residue substitutions at position 217 seemed to have no influence on the interaction between ACE2 and RBD (Fig. 4). Overall, structural modeling analysis demonstrated that the substitutions at position 217 in ACE2 did not influence its corresponding receptor activities.
Figure 4.
Structural modeling of ACE2 Y217N shows no changes in binding between ACE2 and RBD. Structural models of Y217N were generated based on the crystal structure of the SARS-CoV-2 RBD/ACE2 complex. Green is SARS-CoV-2 RBD, gray is wild-type hACE2, and orange is hACE2 Y217N. Blue indicates hydrophobic residues.
The Y217N mutation in ACE2 alters its cell surface localization
To enter into cells, viruses need to first interact with the corresponding receptor at the cell surface; therefore, we tested the possibility that ACE2 Y217N fails to mediate SARS-CoV-2 entry due to a change in the cell surface localization of ACE2 induced by the Y217N mutation. To test this hypothesis, HEK 293T cells were first transfected with the indicated plasmids. Forty-eight hours later, the transfected cells were detached from the plate and then stained with a FITC-labeled anti-ACE2 antibody. We found that both wild-type hACE2 and RhACE2 Y217N were strongly positive for FITC, whereas the other mutants were weakly positive (Fig. 5A). To confirm that this difference was not due to transfection efficacy, the transfected cells were permeabilized with Triton X-100 and then stained with ACE2. The results indicated that all cells transfected with different ACE2 molecules were labeled with FITC at a high level (Fig. 5B).This demonstrated that RhACE2 Y217N failed to mediate SARS-CoV-2 entry, mainly due to its cell surface localization.
Figure 5.
The Y217N mutation in ACE2 alters the cell surface localization of ACE2.A, HEK 293T cells were plated in 6-well plates and transfected with the indicated plasmids when the cells were 90% confluent. Then, the transfected cells were detached and treated without or (B) with Triton X-100. Then, the cells were stained with an anti-ACE2 antibody (FITC-conjugated). Each experiment was repeated at least three times. Error bars represent the standard error (SD); ∗ = p < 0.05; NS = no significant differences.
Discussion
The spike (S) protein features of coronaviruses and lysosomal proteases of hosts determine the tropism of coronaviruses (24). To date, the bat coronavirus RaTG13 in R. affinis (intermediate horseshoe bat) from Yunnan Province exhibits the highest sequence similarity to SARS-CoV-2 (1). In this study, we found that ACE2 of R. ferrumequinum (greater horseshoe bat) failed to mediate SARS-CoV-2 entry, whereas ACE2 of R. sinicus (Chinese rufous horseshoe bat) facilitated SARS-CoV-2 entry to nonsusceptible cells. In fact, in contrast to genetically homologous hACE2, bat ACE2 proteins have great genetic diversity (13). A number of ACE2 molecules isolated from different bat species failed to mediate SARS-CoV entry (13). A study has reported that R. sinicus serves as a natural reservoir of SARS-CoV, and an isolated bat-origin SARS-CoV-like virus is able to use ACE2 proteins from humans and civets for cell entry (12). These results suggest that analysis of the receptor-conferred susceptibility to virus entry is important before investigating the origin of SARS-CoV-2.
Recently, pangolins have also been considered a possible natural reservoir of SARS-CoV-2 (4, 5). Coronaviruses with high sequence homology were identified in M. javanica (Malayan pangolins), suggesting that Malayan pangolin is a possible source for the emergence of SARS-CoV-2 (4). We demonstrated that ACE2 of Malayan pangolin supported SARS-CoV-2 entry in nonsusceptible cells. SARS-CoV and MERS-CoV engage receptors of both human and natural animal hosts (12, 16). Similarly, the ability of pangolin ACE2 to confer susceptibility to SARS-CoV-2 entry increases the possibility that SARS-CoV-2 originated from pangolins. This was further confirmed by two recent studies (25, 26).
In this study, we demonstrated that ACE2 of pigs and dogs facilitated SARS-CoV-2 entry in nonsusceptible cells. However, a recent study reported that SARS-CoV-2 replicated poorly in dogs and pigs (6). We speculate that there other factors that determine host tropism in addition to receptor interactions. A recent study demonstrated that pigs and dogs exhibit relatively low levels of ACE2 in the respiratory tract, which may be the reason that SARS-CoV-2 replicated poorly in dogs and pigs (27). Because in the current study, we did not use cells derived from different animals as host cells for the infection assay, we cannot completely exclude the possibility that cellular factors other than ACE2 play (additional) roles in supporting SARS-CoV-2 entry and cross-species transmission.
Based on structural analysis of hACE2 and the S protein of SARS-CoV-2, the RBD of the S protein of SARS-CoV-2 has a more compact conformation than that of SARS-CoV, implicating a relation to the higher transmission of SARS-CoV-2 than SARS-CoV (23, 28). However, in mouse ACE2, position 353 is a His not a Lys, and H353 in mouse ACE2 does not fit into the virus–receptor binding interface as tightly as K353 of hACE2. Consequently, this may result in the failure of mouse ACE2 to confer susceptibility to SARS-CoV-2 entry. A recent publication also demonstrated that the substitution of K353A in hACE2 was sufficient to abolish the interaction between hACE2 and the S protein of SARS-CoV-2 (29). In addition, although the residue at position 217 of ACE2 is not directly in contact with the RBD, this site in RhACE2 is still critical for SARS-CoV-2 entry. It was observed that the natural isoform of Y217N at this site of RhACE2 significantly reduced the susceptibility to SARS-CoV entry, which demonstrated that this substitution is responsible for the downregulation of ACE2 expression (21). However, our results showed that Y217N RhACE2 was expressed at a similar level as hACE2 in transfected cells. Therefore, the failure of this RhACE2 isoform to allow SARS-CoV-2 entry is not due to the poor expression of the receptor, as previously speculated (21). We also demonstrated that ACE2 Y217N had a significantly decreased SARS-CoV-2 entry ability due to its cell surface localization, in our previous study, we found that virus receptor abundance of cell surface is a pivotal switch for virus efficient infection, this may be the reason that ACE2 Y217N failed to mediate SARS-CoV-2 entry (30). We suspect that the Y217N of ACE2 may affect ACE2 transport in the cells, and this needs further study. We also speculated that the Y217N mutation may alter the folding of ACE2; this needs to be investigated further. It is also important to detect the expression of Y217N RhACE2 in rhesus monkeys recruited for studies on SARS-CoV-2 infection.
Experimental procedures
Cells and SARS-CoV-2
HEK 293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA) with 10% fetal bovine serum (FBS, HyClone, USA). The SARS-CoV-2 used in this study (GenBank: MT123290) was isolated from a patient's throat swab and stored at the BSL-3 Laboratory of Guangzhou Customs Technical Center.
Plasmids
The full-length cDNA fragments of ACE2 of different species were synthesized at either Sangon Biotech (Shanghai, China) or TsingKe Biotech (Nanjing, China). The species and GenBank accession numbers of ACE2 sequences are listed in the table. Synthesized cDNA fragments were then subcloned into the eukaryotic expression vector pCAGGS-HA for expression in human cell lines.
Sequence analysis
ACE2 sequences of 12 different species were acquired from NCBI, and the alignment of these sequences was assessed using the ClustalW method in Lasergene software (Version 7.1) (DNASTAR Inc, USA).
Entry assay
HEK 293T cells were plated in 48-well plates and transfected with the indicated plasmids by the X-tremeGENE HP DNA Transfection Reagent (Roche, USA) when the cells were 90% confluent. Cells were infected with 0.5 multiplicity of infection (MOI) of SARS-CoV-2 24 h after transfection. The detection of infected cells was performed 12 h later by using an IFA as described previously (31). An HA-Alexa Fluor 488 monoclonal antibody (Thermo Fisher Scientific, USA) was used to stain ACE2 with a hemagglutinin (HA) tag. The infection and replication of the virus were determined by detecting the nucleoprotein (N) of SARS-CoV-2 using an N-specific polyclonal antibody (Sino Biological, China), and a donkey anti-rabbit IgG (H + L) labeled with Cy3 (Jackson Laboratory, USA) was used as the secondary antibody. All the cells were stained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma, USA) for nuclear visualization. The average entry ratio was analyzed by ImageJ software, in brief, DAPI stained cells were calculated as total cells, and N-protein stained cells were calculated as infected cells. The average entry ratio was quantified by N-protein stained cells/DAPI stained cells. In total, 5–10 different fields were selected for analysis, and this experiment was performed three times, and a representative result is shown.
Flow cytometry (FCM)
HEK 293T cells were plated in 6-well plates and transfected with the indicated plasmids with X-tremeGENE HP DNA Transfection Reagent (Roche, USA) or PolyJet (SignaGen, China)when the cells were 90% confluent. The transfected cells were detached and then treated with or without Triton X-100. An anti-ACE2 antibody (FITC-conjugated) (Sino Biological, China, cat. 10,108-MM37-F) was used to label ACE2. The samples were inserted into and analyzed by a Beckman Coulter Cytomics FC 500 flow cytometer.
RBD and S protein binding assay
HEK 293T cells were seeded in 20-mm glass-bottom cell culture dishes and then transfected with the indicated plasmids. At 24 h posttransfection (hpt), the transfected cells were incubated with 5 μg RBD (Qianxun Bio, China) or 5 μg full-length spike protein (Genscript, China) for 1 h at 37 °C. After being washed with phosphate-buffered saline (PBS) three times, the cells were then subjected to IFA or western blotting. For IFA, the cells were fixed in 4% paraformaldehyde for 1 h at 4 °C, permeabilized with 0.2% Triton X-100 for 1 h, and then blocked in 3% bovine serum albumin (BSA) at 37 °C for 1 h. Subsequently, the cells were incubated with a primary antibody: rabbit anti-HA tag (cat #51064-2-AP, 1:1000) or mouse anti-His tag (cat. #66005, 1:1000) at 37 °C for 2 h. The cells were washed three times with PBS and incubated with Alexa Fluor 488-labeled goat anti-mouse IgG (H + L) (Invitrogen cat. #A11029), goat anti-rabbit IgG (whole molecule), or F(ab')2 fragment-Cy3 (Sigma cat. #A2306) diluted in PBS (1:1000) for 1 h at room temperature (RT). All the cells were stained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma, USA) for nuclear visualization. Confocal microscopy and sequential confocal analyses were performed using Zeiss confocal laser scanning microscopy.
Western blotting
HEK 293T cells were plated in 12-well plates, and when confluency reached 80%, cells were transfected with the indicated plasmids (2 μg for each). Cells were harvested at 48 hpt and lysed in radioimmunoprecipitation assay lysis buffer (50 mM Tris, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS) with 1 mM phenylmethylsulfonyl fluoride for 30 min on ice. Cell lysates were centrifuged at 12,000×g for 10 min at 4 °C, and the obtained supernatants were mixed with 35 μl of loading buffer (62.5 mM Tris pH 6.8, 1% SDS, 10% glycerol, 5 mM dithiothreitol, and 0.001% bromophenol blue) and heated at 100 °C for 10 min. Equal amounts of cell lysate samples were analyzed by western blotting. Briefly, protein lysate samples from cells were separated on a 12% SDS-PAGE gel and transferred to a polyvinylidene difluoride membrane. The membrane was blocked in 5% skim milk containing PBS and Tween 20 (PBST) for 2 h at RT, followed by washing with PBST. Then, the membrane was incubated with a primary antibody at 4 °C overnight. Subsequently, the membrane was washed three times in PBST (20 min/wash) and incubated with the corresponding secondary antibody for 1 h in the dark box with continuous shaking at RT. The membrane was washed three times with PBST (20 min/wash). This was followed by detection with an infrared imaging system (Odyssey CLX). The aforementioned antibodies were diluted in western blotting buffer (PBS, 5% BSA, and 0.05% Tween), mouse anti-beta-actin (Sigma cat. #A1978, 1:10,000), mouse anti-HA tag (Sigma cat. #H9658, 1:4000), and anti-mouse secondary DyLight 800-labeled antibodies (cat. #5230-0415, diluted 1:10,000).
Structural models
The hACE2/RBD complex was used as a template for homology modeling (22, 23). The mutations in the models were aligned, and the interactions between the SARS-CoV-2 RBD and ACE2 proteins were compared in PyMOL.
Statistical analysis
All data are presented as the mean ± SD from three or more independent experiments, unless otherwise stated. Different treatments were compared with Student's t-test.
Data availability
All data pertinent to this work are contained within this article or available upon request. For requests, please contact Yan-Dong Tang (tangyandong2008@163.com).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors are grateful for Prof. Jian-Hua Zhou, who kindly revised the manuscript. The authors thank Xin Yin for their constructive suggestions in this study. We are also grateful to Zeli Zhang, who kindly helped us with the structural analysis.
Author contributions
Y.-D. T., J. Z., and X. C. conceived the project; Y.-D. T. designed the experiment and wrote the article; H.-L. Z., Y.-M. L., J. S., Y.-Y. Z., T.-Y. W., M.-X. S., M.-H. W., Y.-L. Y., X.-L. H. performed the experiment.
Funding and additional information
This study was funded by grants from The National Key Research and Development Program of China (2018YFC1200100, 2018ZX10301403), the emergency grants for prevention and control of SARS-CoV-2 of Ministry of Science and Technology (2020YFC0841400) and Guangdong Province (2020B111108001, 2018B020207013).
Edited by Phyllis Hanson
Footnotes
Ethical approval: This article does not contain any studies with human participants.
Contributor Information
Yan-Dong Tang, Email: tangyandong2008@163.com, tangyandong@caas.cn.
Jincun Zhao, Email: zhaojincun@gird.cn.
Xuehui Cai, Email: caixuehui139@163.com.
References
- 1.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam T.T., Shum M.H., Zhu H.C., Tong Y.G., Ni X.B., Liao Y.S., Wei W., Cheung W.Y., Li W.J., Li L.F., Leung G.M., Holmes E.C., Hu Y.L., Guan Y. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 5.Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020;30:1346–1351.e2. doi: 10.1016/j.cub.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., Zhao Y., Liu P., Liang L., Cui P., Wang J. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry Depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: An analysis based on Decade-Long structural studies of SARS coronavirus. J. Virol. 2020;94:e00127-20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W., Wong S.K., Li F., Kuhn J.H., Huang I.C., Choe H., Farzan M. Animal origins of the severe acute respiratory syndrome coronavirus: Insight from ACE2-S-protein interactions. J. Virol. 2006;80:4211–4219. doi: 10.1128/JVI.80.9.4211-4219.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren W., Qu X., Li W., Han Z., Yu M., Zhou P., Zhang S.Y., Wang L.F., Deng H., Shi Z. Difference in receptor usage between severe acute respiratory syndrome (SARS) coronavirus and SARS-like coronavirus of bat origin. J. Virol. 2008;82:1899–1907. doi: 10.1128/JVI.01085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., Zhang Y.J., Luo C.M., Tan B., Wang N., Zhu Y. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hou Y., Peng C., Yu M., Li Y., Han Z., Li F., Wang L.F., Shi Z. Angiotensin-converting enzyme 2 (ACE2) proteins of different bat species confer variable susceptibility to SARS-CoV entry. Arch. Virol. 2010;155:1563–1569. doi: 10.1007/s00705-010-0729-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raj V.S., Mou H., Smits S.L., Dekkers D.H., Muller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A., Thiel V., Drosten C., Rottier P.J., Osterhaus A.D., Bosch B.J. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau S.K.P., Fan R.Y.Y., Luk H.K.H., Zhu L., Fung J., Li K.S.M., Wong E.Y.M., Ahmed S.S., Chan J.F.W., Kok R.K.H., Chan K.H., Wernery U., Yuen K.Y., Woo P.C.Y. Replication of MERS and SARS coronaviruses in bat cells offers insights to their ancestral origins. Emerging microbes & infections. 2018;7:209. doi: 10.1038/s41426-018-0208-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y., Du L., Liu C., Wang L., Ma C., Tang J., Baric R.S., Jiang S., Li F. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc. Natl. Acad. Sci. U. S. A. 2014;111:12516–12521. doi: 10.1073/pnas.1405889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li F. Structural analysis of major species barriers between humans and palm civets for severe acute respiratory syndrome coronavirus infections. J. Virol. 2008;82:6984–6991. doi: 10.1128/JVI.00442-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu K., Peng G., Wilken M., Geraghty R.J., Li F. Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2012;287:8904–8911. doi: 10.1074/jbc.M111.325803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji W., Wang W., Zhao X., Zai J., Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J. Med. Virol. 2020;92:433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu S., Zhao Y., Yu W., Yang Y., Gao J., Wang J., Kuang D., Yang M., Yang J., Ma C., Xu J., Qian X., Li H., Zhao S., Li J. Comparison of SARS-CoV-2 infections among 3 species of non-human primates. bioRxiv. 2020 [Google Scholar]
- 21.Chen Y., Liu L., Wei Q., Zhu H., Jiang H., Tu X., Qin C., Chen Z. Rhesus angiotensin converting enzyme 2 supports entry of severe acute respiratory syndrome coronavirus in Chinese macaques. Virology. 2008;381:89–97. doi: 10.1016/j.virol.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 23.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng Y., Shang J., Yang Y., Liu C., Wan Y., Geng Q., Wang M., Baric R., Li F. Lysosomal proteases are a determinant of coronavirus tropism. J. Virol. 2018;92:e01504-18. doi: 10.1128/JVI.01504-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yinghui Liu G.H., Wang Y., Zhao X., Ji F., Ren W., Gong M., Ju X., Li C., Hong J., Zhu Y., Cai X., Wu J., Lan X., Xie Y., Wang X. Functional and genetic analysis of viral receptor ACE2 Orthologs Reveals a Broad potential host range of SARS-CoV-2. bioRxiv. 2020 doi: 10.1073/pnas.2025373118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao X., Chen D., Szabla R., Zheng M., Li G., Du P., Zheng S., Li X., Song C., Li R., Guo J.T., Junop M., Zeng H., Lin H. Broad and Differential animal angiotensin-converting enzyme 2 receptor usage by SARS-CoV-2. J. Virol. 2020;94 doi: 10.1128/JVI.00940-20. e00940-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhai X., Sun J., Yan Z., Zhang J., Zhao J., Zhao Z., Gao Q., He W.T., Veit M., Su S. Comparison of SARS-CoV-2 spike protein binding to ACE2 receptors from human, pets, farm animals, and putative intermediate hosts. J. Virol. 2020;94 doi: 10.1128/JVI.00831-20. e00831-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang T.Y., Liu Y.G., Li L., Wang G., Wang H.M., Zhang H.L., Zhao S.F., Gao J.C., An T.Q., Tian Z.J., Tang Y.D., Cai X.H. Porcine alveolar macrophage CD163 abundance is a pivotal switch for porcine reproductive and respiratory syndrome virus infection. Oncotarget. 2018;9:12174–12185. doi: 10.18632/oncotarget.24040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y.L., Meng F., Qin P., Herrler G., Huang Y.W., Tang Y.D. Trypsin promotes porcine deltacoronavirus mediating cell-to-cell fusion in a cell type-dependent manner. Emerging microbes & infections. 2020;9:457–468. doi: 10.1080/22221751.2020.1730245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data pertinent to this work are contained within this article or available upon request. For requests, please contact Yan-Dong Tang (tangyandong2008@163.com).