Summary
Sepsis is a life-threatening condition with high mortality rates and expensive treatment costs. Early prediction of sepsis improves survival in septic patients. In this paper, we report our top-performing method in the 2019 DII National Data Science Challenge to predict onset of sepsis 4 h before its diagnosis on electronic health records of over 100,000 unique patients in emergency departments. A long short-term memory (LSTM)-based model with event embedding and time encoding is leveraged to model clinical time series and boost prediction performance. Attention mechanism and global max pooling techniques are utilized to enable interpretation for the deep-learning model. Our model achieved an average area under the curve of 0.892 and was selected as one of the winners of the challenge for both prediction accuracy and clinical interpretability. This study paves the way for future intelligent clinical decision support, helping to deliver early, life-saving care to the bedside of septic patients.
Keywords: DII challenge, sepsis prediction, emergency department, deep learning, interpretability
Graphical Abstract
Highlights
-
•
We present benchmark results of sepsis-onset prediction in emergency department
-
•
An LSTM-based model captures irregular time intervals with time encodings
-
•
Our deep-learning model shows superior performance compared with existing methods
-
•
Model interpretation enables real-world clinical applications
The bigger picture
Sepsis is the leading cause of death worldwide and has become a global epidemiological burden. Early prediction of sepsis enables early treatment and increases the likelihood of survival for septic patients. The broad adoption of electronic health records (EHRs) provides an opportunity for sepsis prediction. However, most existing prediction approaches do not consider irregular time intervals between neighboring clinical events in EHRs. Besides, many deep-learning models suffer from black-box problems and are not trusted in clinical settings. We propose a deep-learning model with time encodings, offering both high accuracy and high transparency as well as clinical interpretability. We have already made our code and its detailed documentations publicly available, enabling colleagues to apply it to their applications and eventually make clinical impacts.
Electronic health records contain valuable temporal information for sepsis prediction. However, irregular time intervals between neighboring events are typically neglected. Besides, transparency and interpretability of deep-learning models with increasing complexity and superior performance has become a barrier to the models' clinical adoption. To this end, we propose an interpretable deep-learning model that better captures time information and achieves promising performance on sepsis prediction in the emergency department.
Introduction
Sepsis, a life-threatening illness caused by the body's response to an infection, is the leading cause of death worldwide and has become a global epidemiological burden. Sepsis occurs at all ages and increases mortality rates. In the United States, for example, over 1.7 million adults develop sepsis and nearly 270,000 patients die as a result of sepsis each year.1 Besides, sepsis is the costliest among all disease states and accounted for $24 billion of United States hospital costs in 2013.2 Without timely and adequate treatment, sepsis can progress to severe sepsis and septic shock, which lead to higher mortality rates.3 Several studies suggest that early prediction of sepsis enables early treatment and is able to significantly improve patient outcomes.4,5 However, common signs and symptoms of sepsis, such as fever, chills, rapid respiration, and high heart rate, are the same as in other conditions, making sepsis difficult to diagnose in its early stages. Besides, it is clinically meaningless to predict sepsis minutes before onset even with high prediction accuracy. A good predictive model should be able to trigger alerts as early as possible and present increasingly stronger signals as it approaches the actual event.
Electronic health records (EHRs) are longitudinal electronic records of patients' health information. The rapid growth in volume and diversity of EHRs during the last decades makes it possible to apply machine-learning and data-mining methods to the early prediction of sepsis. Screening tools have been used clinically to recognize sepsis, including quick Sequential (Sepsis-Related) Organ Failure Assessment (qSOFA),4 Modified Early Warning Score (MEWS),6 National Early Warning Score (NEWS),7 and Systemic Inflammatory Response Syndrome (SIRS).8 However, those tools were designed to screen existing symptoms as opposed to explicitly predicting sepsis prior to its onset, and their efficacy in sepsis diagnosis is limited. For example, prior studies show that qSOFA had low sensitivities in identifying sepsis in both prehospital and emergency department (ED) settings.9,10
With recent advances and success, machine-learning methods have shown great potential in unlocking insights from EHRs. Various methods have been developed for accurate sepsis prediction.11,12 Faisal et al.13 developed a logistic regression model (CARS) to predict the risk of sepsis using a patient's firstly recorded vital signs and blood test results, which are usually available within a few hours of emergency admission. Horng et al.14 constructed a machine-learning model using a linear support vector machine and demonstrated the incremental benefit of using free text data in addition to vital signs and demographic data for sepsis clinical decision support at the ED. Mollura et al.15 trained a bagged tree classifier using the recorded electrocardiogram and arterial blood pressure waveforms, showing that the waveform monitoring information may help in detecting sepsis within the first hour of stay in the intensive care unit (ICU). Kamaleswaran et al.16 showed that artificial intelligence can be used to predict the onset of severe sepsis as early as 8 h ahead using physiomarkers in critically ill children. Lyra et al.17 used an optimized random forest to predict sepsis for imbalanced clinical data from ICUs in the PhysioNet Computing in Cardiology Challenge 2019.12 Mao et al.18 validated a machine-learning algorithm with gradient-boosting trees, InSight, which used only six vital signs for the prediction of sepsis, severe sepsis, and septic shock and showed that InSight outperformed existing sepsis-scoring systems. Using 65 features from a combination of EHRs and high-frequency physiological data, Nemati et al.19 developed and validated an interpretable machine-learning model based on a modified Weibull-Cox proportional hazards algorithm for making an accurate and interpretable prediction of sepsis. Recently, deep-learning methods have achieved improving performances over traditional models and have shown unprecedented potential in the healthcare domain.20 Deep-learning models automatically learn the data representation with improved performance and do not require conventional feature-extraction steps. Recurrent neural networks (RNNs) are commonly used network architectures in modeling multivariate series prediction.21, 22, 23 Kam and Kim21 proposed a sepsis-detection model with long short-term memory (LSTM), which showed better performance than InSight and superior capability for sequential patterns. However, deep-learning models usually suffer from black-box problems and are not trusted in clinical settings. RETAIN24 and Dipole25 proposed to introduce attention mechanisms and interpret the models' output risks based on the learned attention weights, which is helpful for models' application to real-world clinical settings.
Most existing approaches11,12,17,21 focus on the sepsis prediction for ICU settings and may suffer from performance decrease for predicting sepsis onset for patients in EDs with low resolution of medical observations, while many patients have been diagnosed with sepsis at ICU admission.26 Moreover, most of the aforementioned existing methods do not or only consider the relative order of events and ignore the irregular time intervals between neighboring events while modeling time-series EHR data. Besides, the increasing complexity of deep-learning models has brought superior model performances at the price of lack of transparency and interpretability, which has become a barrier to the models' clinical adoption. To this end, we address these problems with our proposed interpretable LSTM-based deep-learning model that can achieve state-of-the-art sepsis-onset prediction in the ED.
Our proposed deep-learning model handles irregular time intervals with time encodings, and leverages attention mechanism and global max pooling techniques to help interpret the model's behavior. Our team, BuckeyeAI, participated in the 2019 DII challenge with the proposed deep-learning method and ranked second out of 30 teams on the early prediction of sepsis onset in the ED, with an average area under the receiver-operating characteristic curve (AUC) score of 0.892. The goal of the 2019 DII challenge is the early prediction of sepsis using a patient's demographic and physiological data in the ED. Different from the PhysioNet Computing in Cardiology Challenge 2019 on sepsis prediction in the ICU,12 the 2019 DII challenge focused on sepsis prediction in the ED where the environment is more chaotic.27 In this paper, we present our methods, results, and analyses. To summarize, the contributions are as follows.
-
•
We present benchmark results of sepsis-onset prediction in the ED. We show that our model outperforms four early-warning scores and three baseline machine-learning models.
-
•
We propose an LSTM-based model for sepsis-onset prediction, which handles irregular time intervals with time encodings.
-
•
We leverage the attention mechanism and global max pooling techniques to help interpret our model.
Results
Study design
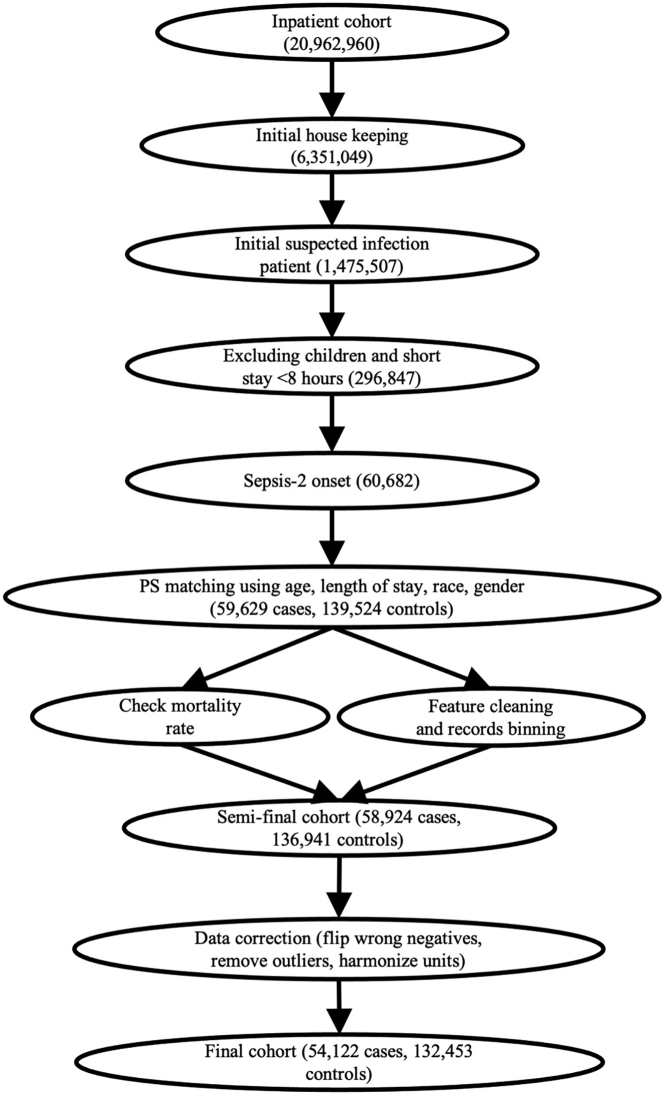
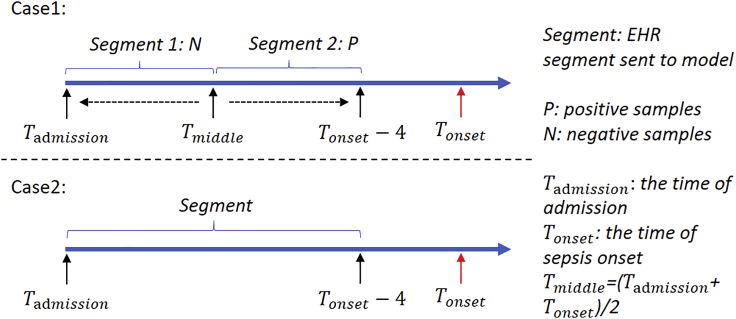
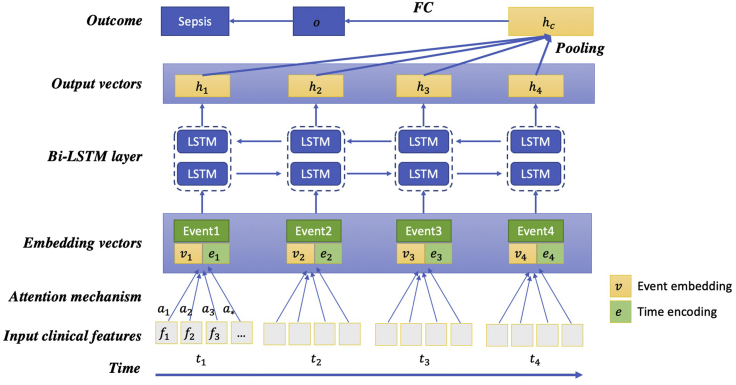
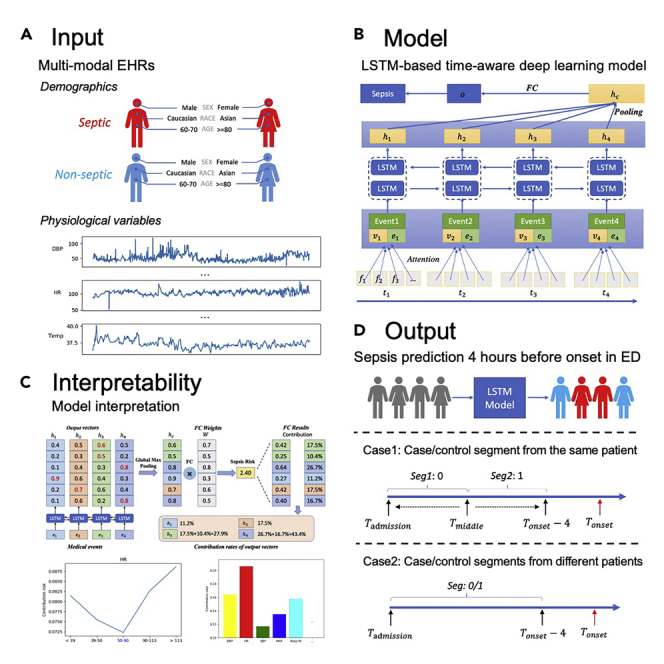
Definition of Sepsis-2, the presence of proven or suspected infection together with two or more SIRS criteria,28 is used to define ground truth in the ED. The inclusion and exclusion diagram of the 2019 DII challenge data preparation pipeline is shown in Figure 1. A summary of patient characteristics is provided in Table 1. Distribution of length of stay until sepsis onset is shown in Figure S1. Two use cases of sepsis-onset prediction 4 h before it occurs is demonstrated in Figure 2. The proposed deep-learning model's architecture is illustrated in Figure 3. Our proposed model handles irregular time intervals with time encodings, and the model is interpretable due to the attention mechanism and global max pooling techniques.
Figure 1.
Inclusion and exclusion diagram of DII challenge data preparation pipeline
After filtering and correction, the final cohort has a sepsis prevalence of 29.0%.
Table 1.
Label statistics and characteristics of the final cohort
| Sepsis-2 patients (n = 52,802) (29.5%) | Sepsis-2 controls (n = 126,041) (70.5%) | Risk ratio | |
|---|---|---|---|
| Gender | |||
| Female | 25,936 (49.1%) | 65,523 (52.0%) | 0.92 |
| Male | 26,866 (50.9%) | 60,518 (48.0%) | 1.08 |
| Race | |||
| African American | 11,084 (21.0%) | 20,556 (16.3%) | 1.24 |
| Asian | 1,085 (2.1%) | 1,627 (1.3%) | 1.36 |
| Caucasian | 35,059 (66.4%) | 95,657 (75.9%) | 0.73 |
| Others/unknown | 5,574 (10.5%) | 8,201 (6.5%) | 1.41 |
| Age | |||
| 18–20 | 1,602 (3.0%) | 1,776 (1.4%) | 1.63 |
| 20–40 | 8,100 (15.3%) | 15,288 (12.1%) | 1.20 |
| 40–60 | 15,654 (29.6%) | 34,295 (27.2%) | 1.09 |
| 60–80 | 20,241 (38.3%) | 51,914 (41.2%) | 0.92 |
| 80–100 | 7,205 (13.6%) | 22,768 (18.1%) | 0.78 |
Figure 2.
Two use cases of sepsis-onset prediction 4 h before it occurs
Figure 3.
Architecture of proposed LSTM-based model
The concatenation of the medical event embedding vectors () and the corresponding time encoding vectors () are inputs to the BiLSTM model, which generates output vectors (). All the output vectors are concatenated, then a global max pooling operation is performed to produce the patient representation vector. Finally, a fully connected layer and the sigmoid function are used to predict the probability of sepsis onset in the next 4 h.
We implemented and evaluated four early-warning scores, three traditional machine-learning methods, and four deep-learning models as baselines. The four early-warning scores comprised MEWS,6 NEWS,7 SIRS,8 and qSOFA.4 For traditional machine-learning methods, we considered logistic regression, random forest, and gradient-boosting trees. Because these standard machine-learning methods cannot work directly with multivariate time-series sequences, the element-wise aggregation (i.e., count, mean value, minimum value, maximum value, and standard deviation of events) of temporal features are used as model inputs. For the deep-learning baselines, two classical RNN models (i.e., GRU29 and LSTM30) and two state-of-the-art interpretable RNN models (i.e., RETAIN24 and Dipole25) are selected. The RNN models cannot handle the missing values of EHR data. We mapped the feature variables into vectors via an embedding layer. The concatenation of the embedding vectors and the observed feature values were then input to the RNN models.
Classification results
Table 2 summarizes the performance of various models for sepsis-onset prediction. From Table 2, our model outperforms baseline models. The main reasons why our model works better are 2-fold: (1) our model can automatically learn better patient representations as the network grows deeper and yield more accurate predictions with sufficient data; (2) our LSTM-based model can better capture temporal information, while logistic regression, random forest, and gradient-boosting trees simply aggregate time-series features and hence suffer from information loss.
Table 2.
AUC scores of sepsis-onset prediction task
| Method | Case 1 | Case 2 | Average |
|---|---|---|---|
| MEWS | 0.54 | 0.72 | 0.63 |
| NEWS | 0.52 | 0.72 | 0.62 |
| SIRS | 0.56 | 0.69 | 0.62 |
| qSOFA | 0.53 | 0.65 | 0.59 |
| Logistic regression | 0.89 | 0.79 | 0.84 |
| Random forest | 0.90 | 0.81 | 0.85 |
| Gradient-boosting trees | 0.91 | 0.81 | 0.86 |
| GRU | 0.88 | 0.80 | 0.84 |
| LSTM | 0.89 | 0.80 | 0.85 |
| RETAIN | 0.90 | 0.80 | 0.85 |
| Dipole | 0.90 | 0.81 | 0.86 |
| Proposed model | 0.94 | 0.84 | 0.89 |
We found that machine-learning-based algorithms outperformed early-warning scores on both cases. All three machine-learning methods achieved similar performance on both Case 1 and Case 2. MEWS and NEWS were shown to perform better than SIRS and qSOFA on Case 2. However, the result suggested little discrimination of four scores on Case 1 with low AUC scores. The deep-learning models outperformed the early-warning scores and performed comparably with the machine-learning algorithms. We speculate the reason for this is that the feature engineering (e.g., minimum and maximum feature values) is effective, and both machine-learning and deep-learning methods can capture the abnormal values from EHRs. With the help of attention mechanisms, RETAIN and Dipole can focus on the abnormal values better, and thus outperform GRU and LSTM.
On the private test dataset, our proposed model achieved AUC scores of 0.940 and 0.845 for two use cases, respectively. The official score is . Compared with attention-based models (i.e., RETAIN and Dipole), the proposed model still achieves better prediction accuracy. Our model considers the whole history of a patient's EHRs with a global pooling operation rather than attention, which is useful for relieving the long-term dependency problem of RNN. Moreover, the time embedding can capture the temporal information more efficiently, which further improves the proposed model's performance.
Ablation study
To measure the effectiveness of different components (i.e., event embeddings, time encodings, and global max pooling), we adopt an ablation study to gain a better understanding of the proposed model by removing one component each time. The results of ablation study on Case 1 sepsis-onset prediction are reported in Table 3. Based on the results from Table 3, the most influential component is event embeddings. By removing event embeddings, the AUC score decreases by 0.11. By handling irregular time intervals using time encoding, the model performance increases from 0.89 to 0.94. Moreover, incorporating global max pooling causes an AUC score increase of 0.03.
Table 3.
Ablation study of different components (i.e., event embeddings, time encodings, and global max pooling) on Case 1 sepsis prediction
| Model | AUC |
|---|---|
| Proposed model without event embeddings | 0.83 |
| Proposed model without time encodings | 0.89 |
| Proposed model without global max pooling | 0.91 |
| Proposed model | 0.94 |
Discussion
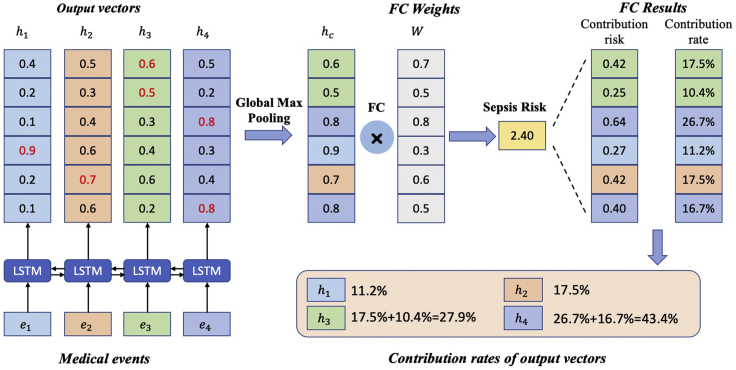
Generally, linear models and tree-based models can be easily interpreted because of their intuitive way of predicting output from inputs, but these models are quite simple. Although deep-learning models can usually yield more accurate predictions, they usually operate as black boxes and make it unclear why the models make specific predictions. However, due to the attention mechanism and global max pooling operation, our deep-learning model is interpretable as shown in Figure 6. At patient level, we are able to calculate the contribution rate of each medical event for sepsis risk according to Equation 5. Medical events with higher contribution rates contribute most to the clinical outcome (i.e., sepsis onset in the next 4 h).
Figure 6.
Interpretability of the proposed model with global max pooling: a toy example
Here we display four medical events () and their corresponding output vectors (). After a global max pooling layer and a fully connected layer, the model predicts the risk of sepsis onset in the next 4 h for an individual patient. Each output vector's contribution is then calculated by summing the corresponding dimensions' contribution risks. Finally, the contribution of each medical event is calculated according to Equation 5.
While patient-level interpretation reveals medical events that are most influential to sepsis onset for an individual patient, population-level analysis is needed to determine the most influential medical events as well as clinical features over the entire EHR dataset. Therefore, to better understand the model's behavior, we try to interpret our model at the population level in two ways, namely medical event importance and influential clinical features.
Medical event importance
As we can calculate the contribution rate of each medical event of each patient, we can compute each medical event's importance at the population level. For each medical event, event importance is calculated by averaging its contribution rates for all patients whose EHR data contain this event.
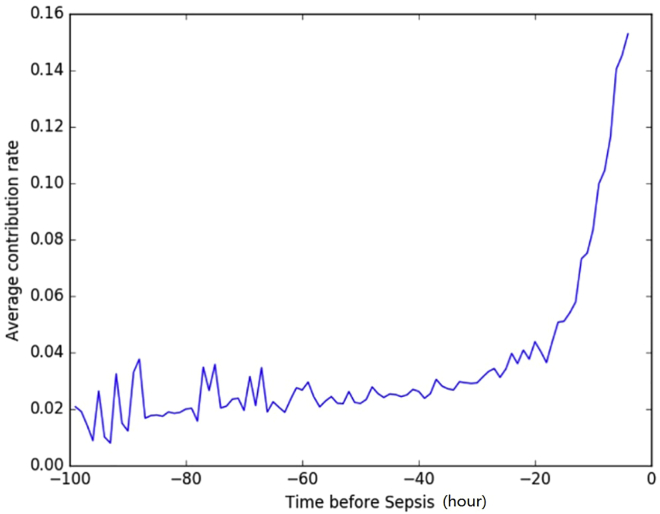
Figure 4 shows the medical event importance (average contribution rate) over time for all patients. This plot shows an overall upward trend, which meets our expectation that the medical events closer to sepsis onset are more important for our model to make predictions.
Figure 4.
Average contribution rate of medical events over time for patients in test set
Note that when computing the average contribution rate for a specific time point, we only consider the patients who have medical events at the time point.
Clinical feature importance
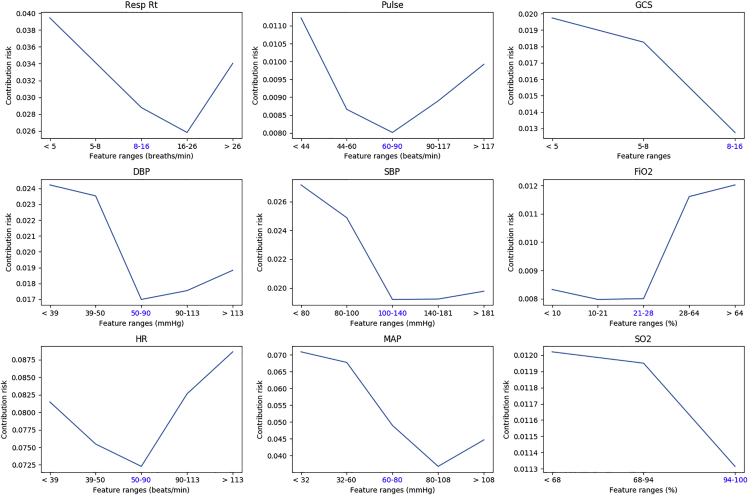
Apart from medical event importance, we also want to know which clinical features are most important for sepsis-onset prediction. Similar to medical event importance, for each clinical feature we compute its importance over all medical events across the entire population according to Equation 6. The top influential features found by the deep-learning model are shown in Figure 5. The full contribution rate list of clinical features can be found in Table S1. The clinical features with the highest contribution to sepsis prediction are easily attainable clinical values. Thus, our model suggests that the development of sepsis can be predicted easily based on items within the EHR. Interestingly, lab values traditionally associated with sepsis prediction (e.g., white blood cell count and renal function) were not predictive.
Figure 5.
Contribution risks of top influential features found by the deep-learning model
Blue-colored tick label on x axis is the corresponding normal range of each feature.
Model performance across subpopulations
From this perspective, we compare the model performance across various subpopulations and report the results on Case 1 sepsis prediction as an example in Table 4. The results show that our model achieves high prediction performance (AUC ≥0.929) across all subpopulations. Confidence intervals are calculated at the 95% level. We also test paired p values for model performance between subgroups, the results of which are reported in Table S2. Concerning gender, the model seems to perform better on female patients compared with male patients, with higher AUC scores (p = 0.025). For race subgroups, performance on the African American patients is the most discriminatory, with relatively lower p values compared with other combinations. The model's AUC on Asian patients is lower with large variance, perhaps because the proportion of Asian patients is small. With respect to age subgroups, the model achieves higher performance for patients whose age is lower than 20 years while the result shows large variance due to the low proportion of such patients. Model performances on patient pairs aged 20–30 and 30–40, 50–60, and 60–70 years are quite similar. The reason for this could be that the distributions of features of these pairs are closer.
Table 4.
Model performance (AUC) with 95% confidence interval across various subpopulations on Case 1 sepsis prediction
| AUC | |
|---|---|
| Total | 0.942 (0.938, 0.946) |
| Gender | |
| Female | 0.946 (0.939, 0.953) |
| Male | 0.938 (0.935, 0.941) |
| Race | |
| African American | 0.950 (0.941, 0.959) |
| Asian | 0.929 (0.894, 0.963) |
| Caucasian | 0.937 (0.934, 0.941) |
| Others/unknown | 0.933 (0.925, 0.941) |
| Age | |
| 18–20 | 0.966 (0.949, 0.983) |
| 20–30 | 0.933 (0.919, 0.946) |
| 30–40 | 0.931 (0.923, 0.939) |
| 40–50 | 0.947 (0.941, 0.953) |
| 50–60 | 0.939 (0.933, 0.946) |
| 60–70 | 0.940 (0.928, 0.952) |
| 70–80 | 0.931 (0.921, 0.941) |
| 80–100 | 0.947 (0.938, 0.955) |
Conclusion
Our team, BuckeyeAI, participated in the 2019 DII Challenge and ranked #2 out of 30 teams on the early prediction of sepsis onset task. In this paper, we present our solution to sepsis-onset prediction 4 h before it occurs. For sepsis-onset prediction, our proposed deep-learning model achieved an AUC score of 0.892 and outperformed four early-warning scores and three baseline machine-learning models. By incorporating event embeddings, time encodings, and global max pooling, our model yields more accurate predictions. Time encodings help to handle irregular time intervals. The global pooling operation enables the model to associate the contribution of each medical event with the final clinical outcome, paving the way for interpretable clinical risk predictions.
Although we mainly focus on sepsis-onset prediction in this challenge, our model is general and can be applied to other multivariate time-series prediction problems. In addition to the superior performance, our proposed model is interpretable from an individual patient to the whole population.
Experimental procedures
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Ping Zhang, PhD (zhang.10631@osu.edu).
Materials availability
This study did not generate any new materials.
Data and code availability
Protected Health Information restrictions apply to the availability of the 2019 DII Challenge dataset. As a result, the dataset is not publicly available. The source code is provided and is available at https://github.com/yinchangchang/DII-Challenge.
Ethical statement
The challenge data are extracted from the Cerner Health Facts database. All challenge entrants signed an enforceable data use agreement as part of the competition registration process. Regarding the use of Cerner Health Facts, all challenge publications authors are covered under IRB protocol HSC-SBMI-13-0549, approved by the UT Health Committee for the Protection of Human Subjects.
Data
The challenge data are extracted from the Cerner Health Facts database. Cerner Health Facts is a database that comprises de-identified EHR data from over 600 participating Cerner client hospitals and clinics in the United States and represents over 106 million unique patients. With this longitudinal, relational database reflecting data from 2000 to 2016, researchers can analyze detailed sets of de-identified clinical data at the patient level. Types of data available include demographics, encounters, diagnoses, procedures, lab results, medication orders, medication administration, vital signs, microbiology, surgical cases, other clinical observations, and health systems attributes.
The goal of 2019 DII challenge is the early prediction of sepsis with demographic and physiological data provided. Sepsis-2 is diagnosed as the presence of proven or suspected infection together with two or more SIRS criteria. The SIRS criteria are defined as:
-
•
Heart rate >90 beats/min
-
•
Body temperature >38°C or <36°C
-
•
Respiratory rate >20 breaths/min or PaCO2 <32 mm Hg
-
•
White blood cell count cells/L or cells/L
Sepsis-2 definition is used to define the ground truth. Patients who are <18 years old or do not have enough observation data are excluded. The whole data preparation pipeline diagram is shown in Figure 1. The label statistics and characteristics of the final cohort are provided in Table 1. Descriptions and statistics of clinical features are available in Table S1.
Predictive tasks
In this challenge, we aim to predict sepsis 4 h before onset for hospitalized adult patients. There are two use cases, as demonstrated in Figure 2.
Case 1
In this case, patients are sampled from septic patients, and the goal is to find out whether a model can tell if a patient is likely to have high sepsis risk a few hours before the onset. For each patient, the patient records is split into two segments at the middle point, segment close to sepsis onset (= 4 h) is labeled as 1, another segment (>4 h before sepsis onset) is labeled 0. We randomly pick either the former or latter segment to build the Case 1 cohort. The introduction of case 1 is to measure the model in terms of time-sensitive prediction to ensure models are indeed clinically useful and relieve warning fatigue as alarm burden. Given patient records either from to or from to , our model is required to distinguish these two kinds of records.
Case 2
In this case, case and control segments are from different patients who have sepsis onset in the next 4 h, as well as those who do not have sepsis. Given patient records from to , we are going to predict whether sepsis occurs in the following 4 h.
Neural network architecture
The proposed neural network architecture is shown in Figure 3. This model is inspired by DG-RNN.31 Although we focus on the early prediction of sepsis onset in this challenge, our proposed model is general and can be applied to other multivariate time-series prediction tasks, such as mortality prediction for septic patients.
Event embeddings
For each temporal feature, we sort the values from low to high and use the order to replace the original values. We then divide the orders into ten groups (i.e., 0.0–0.1, 0.1–0.2, …, 0.9–1.0) and each event is then embedded into a 512-day vector.
Time encodings
When modeling time-series EHR data, most existing LSTM-based models do not or only consider the relative order of events. However, these methods typically ignore the irregular time intervals between neighboring events. Similar to position encodings in Transformer,32 we infuse time information using time encodings. Time encodings are sent to LSTM together with event embeddings. We compute each event's relative time to the criterion operation date and the time interval relative to the last event. We then use sine and cosine functions of the different time intervals to represent the time encoding for the tth event:
| (Equation 1) |
where denotes the criterion operation date, denotes the tth event's date, denotes the time encoding vector, and j is the dimension of EHRs event embeddings. Both the event embeddings and time encodings are then input to LSTM.
To better align patient records at their last recorded medical event, the time of each event is mapped from to .
LSTM and attention mechanism
RNNs are popular and suitable for sequential EHR data modeling. Given medical event embedding and time encoding vectors, we build our model based on LSTM30 for its ability to recall long-term information. The LSTM model can be described as follows:
| (Equation 2) |
where σ is the sigmoid function, t denotes the tth step of LSTM, and is the corresponding cell state, and is the output vector. is the input event embedding and is the input time encoding. , , , and are learnable parameters. Attention mechanism is used to automatically identify influential clinical features.
Global max pooling
RNN-based models are sometimes inefficient due to their long-term dependency. When the input sequence is too long, it is easy for the models to forget the earlier data. Therefore, we adopt a global pooling operation to shorten the distance between the earlier inputs and the final outputs. As is shown in Figure 3, all the outputs of the LSTM are concatenated, then a global pooling operation is followed. The output is fed through the fully connected layer to produce the clinical risk of patient i, which is defined as
| (Equation 3) |
where and are the learnable parameters and denotes the predicted probability for sepsis onset. Because of the shortened distance between the inputs and the outputs, the pooling operation makes it more efficient to propagate the gradients. Besides, the global pooling operation is useful to compute the contribution rates of the outputs and their corresponding input medical events.
Objective function
For binary classification, the objective function is defined as the binary cross-entropy loss between ground truth and predicted probability y:
| (Equation 4) |
Interpretability
Interpretability is very important for machine-learning models of clinical applications. The global pooling operation leveraged in our architecture can associate the contribution of each input medical event to the final clinical outcome, paving the way for interpretable clinical risk predictions.
In Figure 3, given the output vectors, the global max pooling operation is followed and produced the final patient feature vector , which is used to predict risk of sepsis onset. We can track the output vectors which constitutes specific element of . After the fully connected layer, we can calculate every dimension's contribution rate. For a case patient, the contribution rate of output vector for the tth input event is calculated as
| (Equation 5) |
To illustrate the interpretability of our model clearly, we display four input events and four corresponding six-dimensional output vectors () in Figure 6. Given patient feature vector () and fully connected parameters (), the output risk is computed (). For example, the first dimension's contribution risk is 0.42 and the contribution rate is 17.5%, which comes from the third output vector . Similarly, the second dimension's contribution rate also comes from . Thus, the contribution rate of the third vector is computed by summing the two contribution rates. We thus compute the contribution rate of the input event as . For feature j in event i, we can compute its contribution rate with attention weight as
| (Equation 6) |
Implementation and evaluation
The four early-warning scores (MEWS, NEWS, SIRS, and qSOFA) are calculated based on the worst value for each physiological variable within the past 24 h before (i.e., the last observed time points). Logistic regression and random forest are implemented with the scikit-learn toolkit.33 We implement gradient-boosting trees using LightGBM.34 For the proposed LSTM-based model we use PyTorch,35 and the number of time steps for LSTM is set to 100. For evaluation, 80% of the data are used for training, 10% for validation, and 10% for testing. The competition was hosted on Amazon Web Services, and experiments were conducted on a limited secure server to protect data privacy. GPUs are available to accelerate computing.
To evaluate the performance and discrimination of binary classifier, for each use case we use the AUC as the evaluation metric. The arithmetic average of AUC scores of two use cases is used for final performance comparison.
Acknowledgments
This work was supported by the National Institute on Aging (R03AG064379 for K.M.H., R01AG066749 for X.J.), the National Science Foundation (CBET-2037398 for C.Y. and P.Z.), the National Center for Advancing Translational Research (UL1TR002733 for P.Z.), the Cancer Prevention and Research Institute of Texas (RR180012 for X.J.), and a sponsored research agreement from Lyntek Medical Technologies Inc (for D.Z., C.Y., and P.Z.). The authors would like to thank Dr. Lawrence Lynn for the weekly discussions of sepsis during the 2019 DII National Data Science Challenge.
Author contributions
Conceptualization, P.Z.; Resources, X.J. (organizing the 2019 DII Challenge); Methodology, C.Y., D.Z., and P.Z.; Investigation, D.Z. and C.Y.; Formal Analysis, D.Z., C.Y., K.M.H., J.M.C., and P.Z.; Writing – Original Draft, D.Z., C.Y., and P.Z.; Writing – Review & Editing, D.Z., C.Y., K.M.H., X.J., J.M.C., and P.Z.; Supervision, P.Z.
Declaration of interests
The authors declare no competing interests.
Published: January 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.patter.2020.100196.
Supplemental information
References
- 1.Centers for Disease Control and Prevention Sepsis: data and reports. 2016. https://www.cdc.gov/sepsis/datareports/index.html
- 2.Torio C.M., Moore B.J. National inpatient hospital costs: the most expensive conditions by payer, 2013. Healthcare Cost and Utilization Project (HCUP) Statistical Briefs. 2016. https://www.hcup-us.ahrq.gov/reports/statbriefs/sb204-Most-Expensive-Hospital-Conditions.jsp [PubMed]
- 3.Martin G.S. Sepsis, severe sepsis and septic shock: changes in incidence, pathogens and outcomes. Expert Rev. Anti Infect. Ther. 2012;10:701–706. doi: 10.1016/j.afjem.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., Bellomo R., Bernard G.R., Chiche J.-D., Coopersmith C.M. The third international consensus definitions for sepsis and septic shock (sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paoli C.J., Reynolds M.A., Sinha M., Gitlin M., Crouser E. Epidemiology and costs of sepsis in the United States—an analysis based on timing of diagnosis and severity level. Crit. Care Med. 2018;46:1889–1897. doi: 10.1097/CCM.0000000000003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Subbe C.P., Kruger M., Rutherford P., Gemmel L. Validation of a modified early warning score in medical admissions. Q. J. Med. 2001;94:521–526. doi: 10.1093/qjmed/94.10.521. [DOI] [PubMed] [Google Scholar]
- 7.Smith G.B., Prytherch D.R., Meredith P., Schmidt P.E., Featherstone P.I. The ability of the national early warning score (NEWS) to discriminate patients at risk of early cardiac arrest, unanticipated intensive care unit admission, and death. Resuscitation. 2013;84:465–470. doi: 10.1016/j.resuscitation.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 8.Bone R.C., Balk R.A., Cerra F.B., Dellinger R.P., Fein A.M., Knaus W.A., Schein R.M., Sibbald W.J. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Chest. 1992;101:1644–1655. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- 9.Dorsett M., Kroll M., Smith C.S., Asaro P., Liang S.Y., Moy H.P. qSOFA has poor sensitivity for prehospital identification of severe sepsis and septic shock. Prehosp. Emerg. Care. 2017;21:489–497. doi: 10.1080/10903127.2016.1274348. [DOI] [PubMed] [Google Scholar]
- 10.Usman O.A., Usman A.A., Ward M.A. Comparison of SIRS, qSOFA, and NEWS for the early identification of sepsis in the emergency department. Am. J. Emerg. Med. 2019;37:1490–1497. doi: 10.1016/j.ajem.2018.10.058. [DOI] [PubMed] [Google Scholar]
- 11.Islam M.M., Nasrin T., Walther B.A., Wu C.-C., Yang H.-C., Li Y.-C. Prediction of sepsis patients using machine learning approach: a meta-analysis. Comput. Methods Programs Biomed. 2019;170:1–9. doi: 10.1016/j.cmpb.2018.12.027. [DOI] [PubMed] [Google Scholar]
- 12.Reyna, M.A., Josef, C.S., Jeter, R., Shashikumar, S.P., Westover, M.B., Nemati, S., Clifford, G.D., and Sharma, A. (2019). Early prediction of sepsis from clinical data: the PhysioNet/Computing in Cardiology Challenge 2019. 2019 computing in Cardiology (CinC).DOI: 10.23919/CinC49843.2019.9005736. [DOI] [PMC free article] [PubMed]
- 13.Faisal M., Scally A., Richardson D., Beatson K., Howes R., Speed K., Mohammed M.A. Development and external validation of an automated computer-aided risk score for predicting sepsis in emergency medical admissions using the patient’s first electronically recorded vital signs and blood test results. Crit. Care Med. 2018;46:612–618. doi: 10.1097/ccm.0000000000002967. [DOI] [PubMed] [Google Scholar]
- 14.Horng S., Sontag D.A., Halpern Y., Jernite Y., Shapiro N.I., Nathanson L.A. Creating an automated trigger for sepsis clinical decision support at emergency department triage using machine learning. PLoS One. 2017;12 doi: 10.1371/journal.pone.0174708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mollura, M., Mantoan, G., Romano, S., Lehman, L.-W., Mark, R.G., and Barbieri, R. (2020). The role of waveform monitoring in sepsis identification within the first hour of intensive care unit stay. In 2020 11th Conference of the European Study Group on Cardiovascular Oscillations (ESGCO). DOI: 10.1109/ESGCO49734.2020.9158013.
- 16.Kamaleswaran R., Akbilgic O., Hallman M.A., West A.N., Davis R.L., Shah S.H. Applying artificial intelligence to identify physiomarkers predicting severe sepsis in the PICU. Pediatr. Crit. Care Med. 2018;19:495–503. doi: 10.1097/PCC.0000000000001666. [DOI] [PubMed] [Google Scholar]
- 17.Lyra, S., Leonhardt, S., and Antink, C.H. (2019). Early prediction of sepsis using random forest classification for imbalanced clinical data. In 2019 Computing in Cardiology (CinC). DOI: 10.23919/CinC49843.2019.9005769.
- 18.Mao Q., Jay M., Hoffman J.L., Calvert J., Barton C., Shimabukuro D., Shieh L., Chettipally U., Fletcher G., Kerem Y. Multicentre validation of a sepsis prediction algorithm using only vital sign data in the emergency department, general ward and ICU. BMJ Open. 2018;8:017833. doi: 10.1136/bmjopen-2017-017833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nemati S., Holder A., Razmi F., Stanley M.D., Clifford G., Buchman T. An interpretable machine learning model for accurate prediction of sepsis in the ICU. Crit. Care Med. 2018;46:547–553. doi: 10.1097/CCM.0000000000002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miotto R., Wang F., Wang S., Jiang X., Dudley J.T. Deep learning for healthcare: review, opportunities and challenges. Brief. Bioinformatics. 2018;19:1236–1246. doi: 10.1093/bib/bbx044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kam H.J., Kim H.Y. Learning representations for the early detection of sepsis with deep neural networks. Comput. Biol. Med. 2017;89:248–255. doi: 10.1016/j.compbiomed.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 22.Choi E., Schuetz A., Stewart W.F., Sun J. Using recurrent neural network models for early detection of heart failure onset. J. Am. Med. Inform. Assoc. 2016;24:361–370. doi: 10.1093/jamia/ocw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Che Z., Purushotham S., Cho K., Sontag D., Liu Y. Recurrent neural networks for multivariate time series with missing values. Sci. Rep. 2018;8:1–12. doi: 10.1038/s41598-018-24271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi E., Bahadori M.T., Sun J., Kulas J., Schuetz A., Stewart W. RETAIN: an interpretable predictive model for healthcare using reverse time attention mechanism. In: Lee D., Sugiyama M., Luxburg U., Guyon I., Garnett R., editors. vol. 29. 2016. pp. 3504–3512.https://proceedings.neurips.cc/paper/2016/file/231141b34c82aa95e48810a9d1b33a79-Paper.pdf (Advances in Neural Information Processing Systems). [Google Scholar]
- 25.Ma, F., Chitta, R., Zhou, J., You, Q., Sun, T., and Gao, J. (2017). Dipole: diagnosis prediction in healthcare via attention-based bidirectional recurrent neural networks. In Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. KDD ’17. pp. 1903–1911. DOI: 10.1145/3097983.3098088.
- 26.Sakr Y., Jaschinski U., Wittebole X., Szakmany T., Lipman J., Ñamendys Silva S.A., Martin-Loeches I., Leone M., Lupu M.-N., Vincent J.-L., ICON Investigators Sepsis in intensive care unit patients: worldwide data from the intensive care over nations audit. Open Forum Infect. Dis. 2018;5:ofy313. doi: 10.1093/ofid/ofy313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor R.A., Pare J.R., Venkatesh A.K., Mowafi H., Melnick E.R., Fleischman W., Hall M.K. Prediction of in-hospital mortality in emergency department patients with sepsis: a local big data–driven, machine learning approach. Acad. Emerg. Med. 2016;23:269–278. doi: 10.1111/acem.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levy M.M., Fink M.P., Marshall J.C., Abraham E., Angus D., Cook D., Cohen J., Opal S.M., Vincent J.-L., Ramsay G. 2001 SCCM/ESICM/ACCP/ATS/SIS international sepsis definitions conference. Intensive Care Med. 2003;29:530–538. doi: 10.1007/s00134-003-1662-x. [DOI] [PubMed] [Google Scholar]
- 29.Cho, K., van Merriënboer, B., Bahdanau, D., and Bengio, Y. (2014). On the properties of neural machine translation: encoder–decoder approaches. In Proceedings of SSST-8, Eighth Workshop on Syntax, Semantics and Structure in Statistical Translation. pp. 103–111. DOI: 10.3115/v1/W14-4012.
- 30.Hochreiter S., Schmidhuber J. Long short-term memory. Neural Comput. 1997;9:1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 31.Yin, C., Zhao, R., Qian, B., Lv, X., and Zhang, P. (2019). Domain knowledge guided deep learning with electronic health records. In 2019 IEEE International Conference on Data Mining (ICDM). pp. 738–747. DOI: 10.1109/ICDM.2019.00084.
- 32.Vaswani A., Shazeer N., Parmar N., Uszkoreit J., Jones L., Gomez A.N., Kaiser Ł., Polosukhin I. Advances in Neural Information Processing Systems 30 (NIPS 2017) 2017. Attention is all you need; pp. 5998–6008. [DOI] [Google Scholar]
- 33.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V. Scikit-learn: machine learning in Python. J. Machine Learn. Res. 2011;12:2825–2830. doi: 10.5555/1953048.2078195. [DOI] [Google Scholar]
- 34.Ke G., Meng Q., Finley T., Wang T., Chen W., Ma W., Ye Q., Liu T.-Y. Advances in Neural Information Processing Systems 30 (NIPS 2017) 2017. Lightgbm: a highly efficient gradient boosting decision tree; pp. 3146–3154. [DOI] [Google Scholar]
- 35.Paszke A., Gross S., Massa F., Lerer A., Bradbury J., Chanan G., Killeen T., Lin Z., Gimelshein N., Antiga L. Advances in Neural Information Processing Systems 32 (NeurIPS 2019) 2019. PyTorch: an imperative style, high-performance deep learning library; pp. 8024–8035.https://papers.nips.cc/paper/2019/file/bdbca288fee7f92f2bfa9f7012727740-Paper.pdf [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Protected Health Information restrictions apply to the availability of the 2019 DII Challenge dataset. As a result, the dataset is not publicly available. The source code is provided and is available at https://github.com/yinchangchang/DII-Challenge.