Abstract
Background
Despite recent advances in the understanding of the genetic architecture of osteoarthritis (OA), only two genetic loci have been identified for OA of the hand, in part explained by the complexity of the different hand joints and heterogeneity of OA pathology.
Methods
We used data from the Rotterdam Study (RSI, RSII and RSIII) to create three hand OA phenotypes based on clustering patterns of radiographic OA severity to increase power in our modest discovery genome-wide association studies in the RS (n=8700), and sought replication in an independent cohort, the Framingham Heart Study (n=1203). We used multiple approaches that leverage different levels of information and functional data to further investigate the underlying biological mechanisms and candidate genes for replicated loci. We also attempted to replicate known OA loci at other joint sites, including the hips and knees.
Results
We found two novel genome-wide significant loci for OA in the thumb joints. We identified WNT9A as a possible novel causal gene involved in OA pathogenesis. Furthermore, several previously identified genetic loci for OA seem to confer risk for OA across multiple joints: TGFa, RUNX2, COL27A1, ASTN2, IL11 and GDF5 loci.
Conclusions
We identified a robust novel genetic locus for hand OA on chromosome 1, of which WNT9A is the most likely causal gene. In addition, multiple genetic loci were identified to be associated with OA across multiple joints. Our study confirms the potential for novel insight into the genetic architecture of OA by using biologically meaningful stratified phenotypes.
Keywords: osteoarthritis, polymorphism, genetic, epidemiology
Key messages.
What is already known about this subject?
Hand osteoarthritis (OA) is one of the most prevalent forms of OA and has a large genetic component, yet only two common genetic loci have been found.
Lack of findings may be attributed to the modest samples sizes in previous genome-wide association studies (GWAS) and the high disease heterogeneity, which can negatively affect statistical power to robustly identify genetic loci.
What does this study add?
Using three distinct hand OA phenotypes (hand, finger, thumb OA), based on clustering patterns of radiographic OA severity, we have increased the power of our GWAS (n~10 000), and robustly identified a novel genetic locus for thumb OA.
Functional genomic data from OA disease relevant tissue identified a potential causal variant, predicted to be located in a gene regulatory element, which through chromatin looping interacts with the WNT9A promoter to influence WNT9A expression.
How might this impact on clinical practice or future developments?
Our results provide the first evidence for WNT9A, a non-canonical Wnt ligand, in human thumb OA.
Introduction
Osteoarthritis (OA) is a serious destructive joint disorder and the third most rapidly rising condition associated with disability.1 Despite this, no effective treatments that target OA are available. Current treatments only manage pain, not the underlying mechanisms of disease aetiology. An estimated 5% of the world population is affected by OA, with hand OA as one of its most prevalent forms. Given the high prevalence with age and high estimated lifetime risk rates for symptomatic hand OA (39.8%), the number of individuals affected will only continue to increase.2 3 Hand OA has a high clinical burden, involving considerable pain, deformity and impaired function.4–6 In addition, hand joints are non-weight bearing, and therefore may reflect the systemic aspects of the disease more than knee and/or hip OA, where mechanical loading is a dominant risk factor.7 A better understanding of hand OA, its causes and pathophysiological mechanisms is therefore urgently needed.
Hand OA is a complex multifactorial disorder. It shares risk factors, such as repetitive movements and obesity, with OA at other joint sites.8 9 Hand OA also has a strong genetic component, with heritability estimates ranging from 39% to 84% depending on the hand joint affected.10 11 Recently, major advances were made in elucidating the genetic background of hip and knee OA, using large (n>400 000 individuals) genome-wide association studies (GWAS).12–14 Yet, for hand OA only two common genetic loci have been found near the MGP and ALDH1A2 genes.15 16 The lack of findings for hand OA may be attributed to the modest samples sizes (n<9000 individuals) in previous GWAS and disease heterogeneity,17 which is known to reduce power to robustly detect genetic loci.18
OA in the hand may be present in any joint, but is most prevalent in the distal interphalangeal joints (DIP), first carpometacarpal (CMC1) and trapezioscaphoid (TS) joints, followed by the proximal interphalangeal joints (PIP). OA is least prevalent in metacarpophalangeal joints (MCP).19 20 Diverse definitions of hand OA have been used including nodal hand OA (interphalangeal (IP) joints), thumb base OA (CMC1/TS) and generalised hand OA (DIP/PIP/CMC1)19 20 with varying results, suggesting that disease aetiology may differ between the joints. Moreover, OA affects multiple tissues within the joint including cartilage and bone. This further contributes to disease heterogeneity and warrants the assessment of hand OA by endophenotypes such as joint space narrowing (JSN) and osteophytes (OST) that may capture separate biological processes underlying OA pathology. The use of endophenotypes, quantifiable biological phenotypes intermediate to the genes and the disease, has been successfully used in OA for the detection of novel genetic loci16 21 22 in knee and hip OA, and may provide new insights into hand OA. For heterogeneous diseases such as OA, stratification of OA phenotypes into different dimensions of disease is one way of increasing power in GWAS.18
There are few GWAS of hand OA and none has examined hierarchically defined clusters of OA joint presentation in the hands or hand OA endophenotypes that may provide new insights into hand OA pathogenesis. We therefore set out to examine the occurrence of OA across hand joints and conduct a GWAS stratified by hand OA patterns23 to identify novel genetic loci for hand OA.
Methods
GWAS, discovery, replication and meta-analysis
We conducted GWAS on radiographic structural phenotypes for OA of the hand using data from the Rotterdam Study (RS) (n~8700).24 For a detailed description of the RS, subcohorts and GWAS methods, see online supplemental text and table 1. Briefly, genotypes were imputed to the Haplotype Reference Consortium reference panel (V.1.0) using the Michigan Imputation Server.25 We assessed genetic associations in each RS sub-cohort using linear regression models adjusted for age, sex and the first four genetic principal components. RVtests26 was used for the GWAS analyses and results were quality controlled using EasyQC.27 Variants with an imputation quality <0.3, minor allele frequency <0.05 or effective allele count <5 were excluded and genomic control correction was applied to all SE and p values. Meta-analysis between the discovery cohorts was performed using fixed-effects inverse variance weighting with METAL.28 Manhattan plots and QQ plots were generated using R and R package qqman.29 Independent variants with a p<1×10−6 and a χ2 statistic test of heterogeneity p>1×10−6 were selected for replication in the Framingham Heart Study (FHS) (n=1203).30 For a detailed description of the FHS, see the online supplemental text. Summary level data from the discovery and replication stage were combined in a joint meta-analysis (METAL).28 Variants met criteria for replication if the association reached a p<0.05, had the same direction of effect as the discovery sample and reached a joint meta-analysis p<5×10−8. Replicated variants were also examined for association with clinical OA (ie, hospital diagnosed OA) based on GWAS summary statistics from a large-scale OA meta-analysis of data from the UK Biobank and Icelandic deCODE populations.14 15 For a detailed description of the UK Biobank and deCODE, see the online supplemental text. Associations that reached a p value <0.01 were considered statistically significant.
Table 1.
General characteristics of the study population
| Cohort | RSI | RSII | RSIII |
| Hand | |||
| n (OA cases) | 4829 (1830) | 1791 (688) | 2071 (526) |
| Female (%) | 2773 (0.57) | 964 (0.54) | 1180 (0.57) |
| Age (SD) | 67.6 (7.9) | 64.6 (7.9) | 57.1 (7.0) |
| KLsum (aver. (SD)) | 8.4 (9.9) | 6.9 (4.0) | 4.7 (6.5) |
| Osteophytes (aver. (SD)) | 7.1 (7.8) | 6.8 (8.2) | 4.6 (6.3) |
| JSN (aver. (SD)) | 0.84 (2.4) | 0.34 (1.3) | 0.2 (0.8) |
| Finger | |||
| n (OA cases) | 4839 (1244) | 1803 (474) | 2072 (298) |
| Female (%) | 2779 (0.57) | 972 (0.54) | 1181 (0.57) |
| Age (SD) | 67.6 (7.9) | 64.6 (7.9) | 57.1 (7.0) |
| KLsum (aver. (SD)) | 5.8 (7.4) | 4.5 (6.1) | 3.0 (4.7) |
| Osteophytes (aver. (SD)) | 4.7 (5.5) | 4.3 (5.8) | 2.9 (4.5) |
| JSN (aver. (SD)) | 0.6 (1.9) | 0.25 (1.1) | 0.1 (0.7) |
| Thumb | |||
| n (OA cases) | 4882 (916) | 1813 (255) | 2083 (166) |
| Female (%) | 2785 (0.57) | 972 (0.54) | 1184 (0.57) |
| Age (SD) | 67.6 (7.9) | 64.6 (7.9) | 57.1 (7.0) |
| KLsum (aver. (SD)) | 2.1 (3.3) | 1.2 (2.3) | 0.8 (1.7) |
| Osteophytes (aver. (SD)) | 1.3 (2.1) | 0.93 (1.7) | 0.6 (1.3) |
| JSN (aver. (SD)) | 0.4 (1.0) | 0.18 (0.63) | 0.1 (0.4) |
aver., average; JSN, joint space narrowing; KL, Kellgren-Lawerence; OA, osteoarthritis; RS, Rotterdam Study.
annrheumdis-2020-217834supp001.pdf (213.8KB, pdf)
Detailed phenotype descriptions
For each participant, all hand joints (16 joints per hand, 32 joints per individual) were scored for Kellgren and Lawrence (KL) grade31 based on hand radiographs. KL grade is a semi-quantitative score ranging from 0 to 4, where higher scores indicate more severe disease. Radiographic OA was defined as KL grade ≥2 (definite JSN and definite OST). Each joint was also scored for individual radiographic features including JSN and OST.31 The JSN and OST scores are semi-quantitative scores ranging from 0 to 3, where 0=none, 1=possible, 2=definite and 3=marked.
We conducted hierarchical clustering of KL grade across all hand joints to identify patterns of disease occurrence defined by location and disease severity (see online supplemental text). This yielded three semi-quantitative hand OA phenotypes for analysis: (1) hand KLsum=sum of KL grades across all DIP, PIP, MCP, IP and CMC1 joints in both hands (15 joints per hand, 30 joints per individual, hand KLsum score range: 0–120), (2) finger KLsum=sum of KL grades across all DIP and PIP joints in both hands (8 joints per hand, 16 joints per individual, KLsum score range: 0–64) and (3) thumb KLsum=sum of KL grades across the CMC1 and TS joint (2 joints per hand, 4 joints per individual, KLsum score range: 0–16). Individuals with a missing KL grade in one or more hand joints were excluded from the analysis of phenotypes that required scoring of the missing joint(s). Also, individuals with missing age, sex or genetic principal components were excluded from all analyses (table 1).
Post-GWAS analysis
Post-GWAS analysis consisted of multiple bioinformatic and functional analyses (online supplemental text). Briefly, all GWAS variants were annotated using HaploReg (V.4.1) and FUMA.32 33 Intersection of variants with epigenetic markers, proteins, transcription factor (TF) motifs and binding and chromatin interactions was done using data from ROADMAP, ENCODE, HaploReg (V.4.1) and the three-dimensional (3D) genome browser.34–37 Functional studies included expression quantitative trait loci (eQTL) analysis, methylation expression quantitative trait loci (meQTL) analysis, ATAC-seq analysis and differential gene expression analysis. All functional analyses were performed in human articular cartilage. Details are provided in the online supplemental text.
Results
Patterns of osteoarthritis severity in joints of the hand
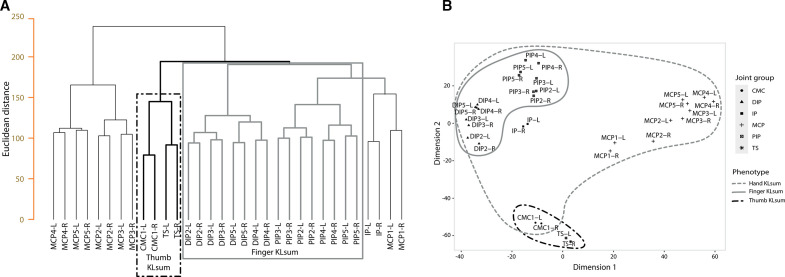
We used hierarchical cluster analysis on the KL grades of all 32 hand joints of the left hand and right hand to identify clusters (figure 1). Tight symmetric clustering between the left and right joints was seen, in addition to clustering based on joint group, which is in line with previous findings19 38 39 (figure 1A, B). Clustering based on individual radiographic features, JSN and number of OST produced similar symmetric and joint group clusters (online supplemental figures S1 and 2). We observed consistent clustering of the PIP with the DIP joints and the TS with the CMC joints (figure 1, online supplemental figures S1 and 2). Based on these analyses, we created three semi-quantitative hand OA phenotypes: hand KLsum, thumb KLsum and finger KLsum (table 1).
Figure 1.
Tree-dendrogram and multidimensional scaling (MDS) plot of KL scores in the joints of the hand. (A) Tree-dendogram of complete hierarchical clustering of Euclidean distance matrix of KL scores of all hand joints left (L) and right (R). (B) MDS plot of KL scores for all hand joints left (L) and right (R). Data consisted of all RSI, RSII and RSIII (n=8691) participants of whom radiographic X-rays of the hands were made. Selected phenotypes are depicted by the different (dashed) lines. TS, trapezioscaphoid joints; CMC, carpometacarpal joints; DIP, distal interphalangeal joints; IP, interphalangeal joints; KL, Kellgren-Lawrence OA severity score; L/R, left or right joint; MCP, metacarpophalangeal joints; OA, osteoarthritis; PIP, proximal interphalangeal joints; IP, interphalangeal joints; number denotes which joint, ie, PIP2-L, the second PIP joint at the left hand. See online supplemental figures 2 and 3 for tree dendrogram and MDS plots for joint space narrowing and osteophytosis scores.
annrheumdis-2020-217834supp002.pdf (108.5KB, pdf)
Identification of genetic hand osteoarthritis loci
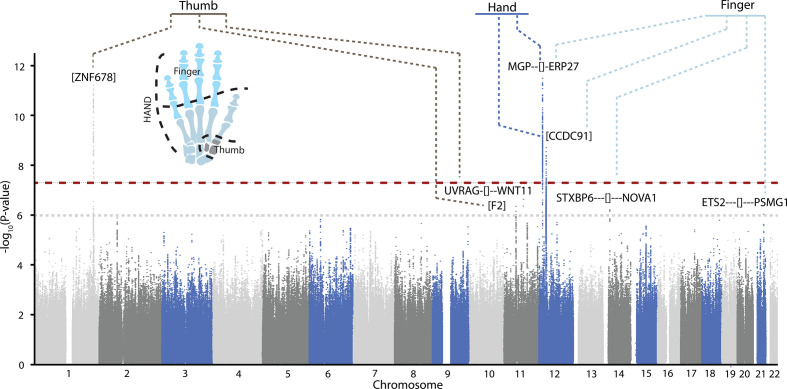
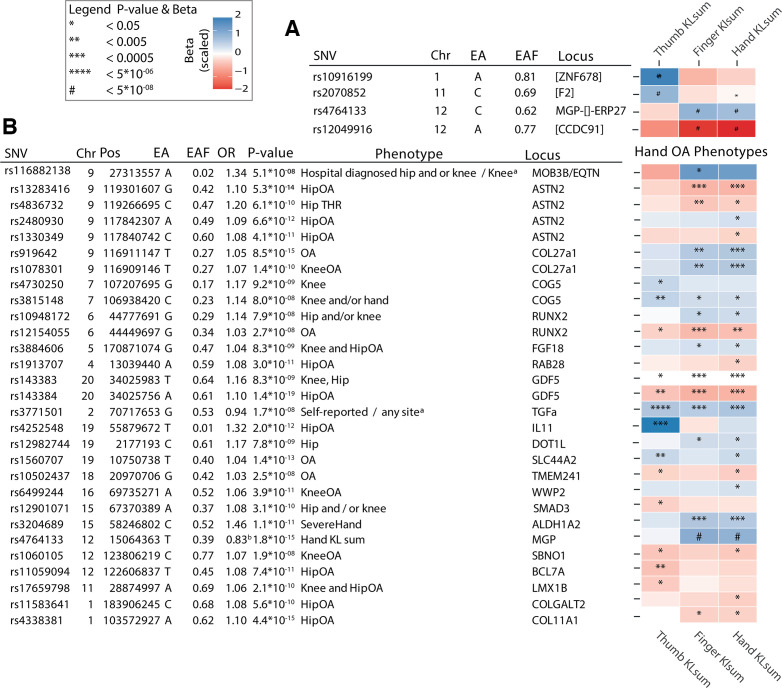
We conducted GWAS on each of the three identified hand OA phenotypes in a discovery sample that included RSI, RSII and RSIII cohorts (n~8700) (online supplemental table S1). In total, we identified seven independent signals with genome-wide suggestive association (p<1×10−6), which were taken forward for replication in the FHS (n=1203) (figure 2 and table 2). In total, four independent signals were genome-wide significant in the meta-analysis (p≤5×10−8), of which three were significantly replicated (p<0.05) (table 2). Two of these signals were novel OA associated loci. The first and most significant novel locus was located on chromosome 1 near the ZNF678, WNT3A and WNT9A genes, with rs10916199 as the lead single nucleotide variant (SNV). This signal is replicated (p<0.05) and genome-wide significantly associated with thumb KLsum (beta=−0.31, p=2.36×10−13). The second replicated novel locus is located on chromosome 11 containing the F2, LRP4 and CREB3L1 genes, and is associated with thumb KLsum (beta=−0.19, p=4.7×10−8). We also identified two known OA associated loci. The first locus was located near the MGP gene, with rs4767133 as the lead SNV. This locus was previously found to be associated with hand KLsum.16 The second known OA locus, also located on chromosome 12, is the CCDC91 locus, which was also previously found to be associated with hand KLsum, although it did not reach genome-wide significance.16 Here, the lead variant rs12049916, was genome-wide significantly associated with hand KLsum (beta=0.78, p=1.5×10−8) and finger KLsum (beta=0.58, p=2.0×10−8), but did not reach nominal significance in the replication cohort, though the direction of effect was the same between discovery and replication cohorts (table 2).
Figure 2.
Combined Manhattan plot of genome-wide association studies (GWAS) discovery results of all radiographic hand OA structural phenotypes. Thumb=thumb KLsum score, finger=finger KLsum score and hand=hand KLsum score. GWAS discovery consists of RSI, RSII and RSIII, and was adjusted for age, sex and the first four genetic principle components. The −log10 p values, for each of the ~11 million single nucleotide polymorphisms (SNPs) analysed (remaining after EASYQC quality control) from the association studies is plotted against their position per chromosome. The dotted red horizontal line corresponds to the genome-wide significant threshold (p=5×10−8). The dotted grey line corresponds to the selection for replication threshold (p=1×10−6). Lead SNP location is represented by [] (intronic), if the SNP is localised intergenic the dashes denotes the distance, - ≤10 kb, -- ≤100 kb, ---- ≤1 Mkb, ---- ≥1 Mkb. For plots of the individual GWAS, see online supplemental figures 3–5.
Table 2.
Summary of radiographic hand OA structural phenotypes GWAS results
| rsID | Chr | Pos (hg19) |
Discovery* | Replication† | Meta-analysis‡ | ||||||||||
| EA | NEA | EAF | Beta | SE | P value | Beta | SE | P value | Beta | SE | P value | Locus§ | |||
| Thumb KLsum | |||||||||||||||
| rs10916199 | 1 | 227 902 472 | A | G | 0.81 | −0.31 | 0.04 | 2.1×10−12 | −0.27 | 0.13 | 3.8×10−2 | −0.31 | 0.04 | 2.4×10−13 | (ZNF678) |
| rs2070852 | 11 | 46 744 925 | C | G | 0.69 | −0.19 | 0.04 | 4.5×10−7 | −0.22 | 0.11 | 3.4×10−2 | −0.19 | 0.04 | 4.7×10−8 | (F2) |
| rs621457 | 11 | 75 858 695 | A | G | 0.52 | −0.18 | 0.03 | 2.4×10−7 | −0.07 | 0.11 | 5.2×10−1 | −0.17 | 0.03 | 3.3×10−7 | UVRAG-[]--WNT11 |
| Finger KLsum | |||||||||||||||
| rs4764133 | 12 | 15 064 363 | T | C | 0.38 | 0.61 | 0.09 | 5.7×10−12 | 1.38 | 0.38 | 2.7×10−4 | 0.65 | 0.09 | 4.8×10−14 | MGP—[]-ERP27 |
| rs12049916 | 12 | 28 359 985 | G | A | 0.22 | 0.57 | 0.11 | 7.5×10−8 | 0.73 | 0.44 | 9.9×10−2 | 0.58 | 0.10 | 2.0×10−8 | (CCDC91) |
| rs1950427 | 14 | 25 955 502 | T | C | 0.12 | 0.66 | 0.13 | 6.2×10−7 | 0.63 | 0.53 | 2.4×10−1 | 0.66 | 0.13 | 3.0×0−7 | STXBP6 ---[]---NOVA1 |
| rs1029003 | 21 | 40 309 122 | A | G | 0.46 | 0.43 | 0.09 | 9.3×10−7 | 0.35 | 0.37 | 3.4×10−1 | 0.42 | 0.09 | 5.9×10−7 | ETS2---[]---PSMG1 |
| Hand KLsum | |||||||||||||||
| rs4764133 | 12 | 15 064 363 | T | C | 0.38 | 0.75 | 0.12 | 3.0×10−10 | 1.76 | 0.49 | 2.9×10−4 | 0.81 | 0.12 | 2.9×10−12 | MGP—[]-ERP27 |
| rs12049916 | 12 | 28 359 985 | A | G | 0.77 | 0.77 | 0.14 | 5.3×10−8 | 0.90 | 0.56 | 0.11 | 0.78 | 0.14 | 1.5×10−8 | (CCDC91) |
*Discovery consists of RSI, RSII, RSIII, samples sizes per phenotype are: thumb KLsum n=8778; finger KLsum n=8714; hand KLsum n=8629.
†Replication cohorts consist of Framingham Heart Study n=1203.
‡Meta-analysis is discovery and replication cohorts using inverse variance weighted meta-analysis using METAL.
§SNP location represented by [], if the SNP is localised intergenic the dashes denotes the distance, - ≤10 kb, -- ≤100 kb, --- ≤1 Mkb, ----- >1 Mkb.
EA, effect allele; EAF, effect allele frequency; GWAS, genome-wide association studies; NEA, non-effect allele; RS, Rotterdam Study; SNP, single nucleotide polymorphism.
annrheumdis-2020-217834supp003.pdf (356.4KB, pdf)
To examine if replicated loci were also associated with clinically defined OA, we looked up findings in GWAS summary statistics from a recent large-scale OA meta-analysis that included the UK Biobank and deCODE populations14 15 (table 3). Of the novel signals, only rs10916199 (thumb KLsum) was significantly associated with its matching clinical OA phenotype (thumb OA: OR=0.9, 95% CI=0.86 to 0.95, p=5.7×10−5). Of the known signals, rs4764133 was significantly associated with multiple clinical OA phenotypes: thumb OA (OR=1.07, 95% CI=1.03 to 1.12, p=6.3×10−4), finger OA (OR=1.12, 95% CI=1.07 to 1.17, p=5.7×10−7), hand OA (OR=1.09, 95% CI=1.04 to 1.13, p=6.7×10−5) and nominal significantly with knee OA (OR=0.99, 95% CI=0.96 to 1.00, p=1.8×10−2) (table 3). In addition, we also performed a sensitivity analysis in our discovery cohorts to see if the association of rs10916199 with thumb KLsum was driven by body mass index (BMI. After additional adjustment for BMI, rs10916199 remained strongly associated with thumb KLsum (beta=−0.37, SE=0.05, p=5.8×10−13).
Table 3.
Association with hip, knee, hand, finger and thumb OA
| rsID | EA | EAF | Thumb OA (7280 cases; 605 132 controls) |
Finger OA (7037 cases; 222 772 controls) |
Hand OA (8591 cases; 224 326 controls) |
Hip OA (17 151 cases, 613 790) |
Knee OA (24 919 cases, 613 702 controls |
||||||||||
| OR | 95% CI | P value | OR | 95% CI | P value | OR | 95% CI | P value | OR | 95% CI | P value | OR | 95% CI | P value | |||
| rs10916199 | A | 0.81 | 0.91 | 0.86 to 0.95 | 5.7×10−5 | 0.98 | 0.93 to 1.04 | 0.57 | 1.03 | 0.98 to 1.08 | 0.32 | 1.00 | 0.98 to 1.03 | 0.95 | 0.99 | 0.97 to 1.02 | 0.59 |
| rs2070852 | C | 0.69 | 0.97 | 0.89 to 1.05 | 0.47 | 0.99 | 0.94 to 103 | 0.58 | 0.99 | 0.96 to 103 | 0.80 | 0.98 | 0.96 to 1.01 | 0.22 | 0.99 | 0.97 to 1.01 | 0.40 |
| rs4764133 | T | 0.39 | 1.07 | 1.03 to 1.12 | 6.3×10−4 | 1.12 | 1.07 to 1.17 | 5.7×10−7 | 1.09 | 1.04 to 1.13 | 6.7×10−4 | 0.99 | 0.96 to 1.01 | 0.30 | 0.98 | 0.96 to 1.00 | 0.018 |
| rs12049916 | A | 0.77 | 1.00 | 0.96 to 1.05 | 0.87 | 0.99 | 0.94 to 1.04 | 0.67 | 0.99 | 0.94 to 1.04 | 0.68 | 0.96 | 0.93 to 0.99 | 2.9×10−3 | 0.99 | 0.97 to 1.01 | 0.27 |
Thumb OA, finger OA, hand OA GWAS summary statistics are from the deCODE cohort. Hip OA and knee OA summary statistics are from a meta-analysis of deCODE and UK Biobank cohorts.
EA, effect allele; EAF, effect allele frequency; NEA, non-effect allele; OA, osteoarthritis.
WNT9A as potential causal gene for thumb OA
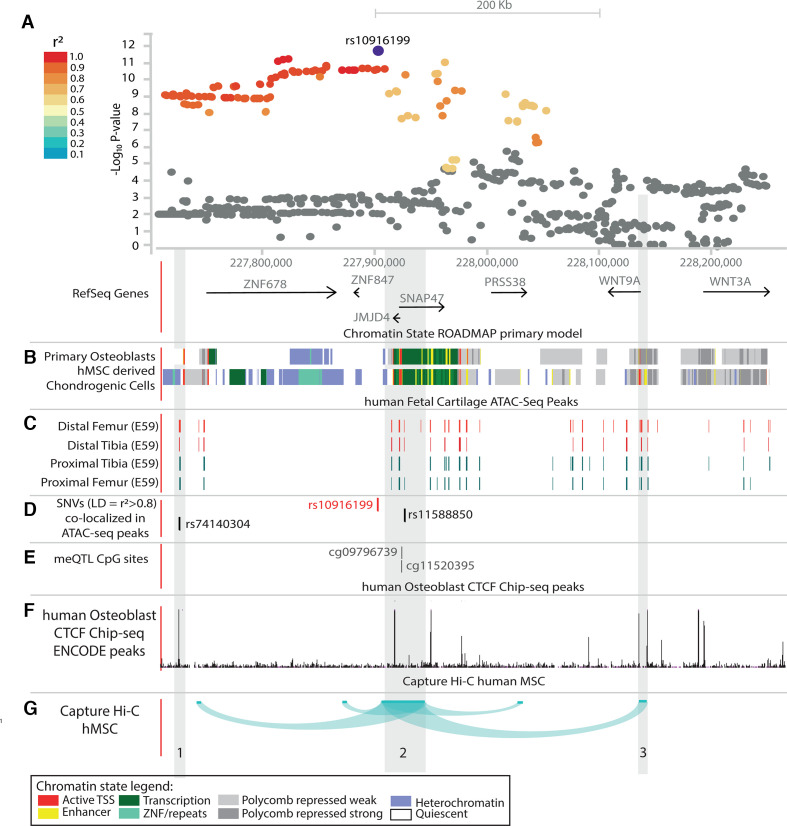
We examined the rs10916199 locus in more detail given the strong and consistent association with thumb OA and thumb KLsum. Different levels of information were leveraged for all genes within 1 Mb surrounding rs10916199 in order to prioritise a putative causal gene (figure 3 and online supplemental text). First, we meta-analysed two human osteoarthritic cartilage expression quantitative trait loci (eQTL) datasets (n=116, hip/knee joints, online supplemental table S1)40 41 and found no significant effect of rs10916199 on gene expression in these datasets. Next, there were significant methylation quantitative trait loci (meQTL) associated with rs10916109 and two CpG sites in human osteoarthritic cartilage (knee/hip joints): CpG09796739 (beta=0.39, FDR p=8.3×10−7) and CpG11520395 (beta=0.21, FDR p=5.5×10−3) (figure 3E, zone 2). These methylation sites are located in a region flanking an active transcriptional start site in primary osteoblasts and chondrogenic cells (figure 3B).
Figure 3.
Schematic overview of part of the rs10916199 locus. (A) LocusZoom plot of rs10916199 locus, the Y-axis depicts the −log10 p value of the single nucleotide variant (SNV) from the thumb Kellgren-Lawrence (KL)sum genome-wide association studies (GWAS). Colours depict the linkage disequilibrium (LD) (r2) between the variant and the LD SNV rs10916199. The X-axis depicts the relative genomic location, depict are the protein coding genes at those genomic locations. For this genomic region depicted in figure (B–G) are several epigenetic annotations are plotted. (B) Chromatin state, as predicted by the ROADMAP 15-state model on histone modifications, for primary osteoblasts and human mesenchymal stem cell-derived chondrocytes. Colours depict chromatin states, legend at bottom of full figure. (C). ATAC-seq peaks from human embryonic cartilage at different bone development sites at gestation day(E) 59. (D) Location of the lead SNV, rs10916199 and two putative causal SNVs which co-locative with ATAC-seq peaks. (E) Genomic location of rs10916199 meQTL CpG sites. (F) Chip-seq CTCF protein binding peaks from human primary osteoblasts from ENCODE. (G) Capture Hi-C chromatin interactions from three-dimensional (3D) genome browser for human mesenchymal stem cells (hMSC) and mesoderm. Depicted are the chromatin interactions from the promoters of the queried gene to the genomic location of interaction, this was done for the JMJD4/SNAP47 transcription start site (TSS), WNT9A TSS and the WNT3A TSS. For details on methods and underlying data, see online supplemental methods.
To further assess co-localisation of the identified genetic loci with regulatory function during cartilage differentiation, as many OA loci haven been linked to skeletal development,42 we intersected the GWAS signals in the locus with accessible chromatin regions (ATAC-seq peaks) in rare human fetal cartilage acquired from proximal and distal long bones from gestational day(E) 59 of development.43 Two of the SNVs in high LD (r2 ≥0.8, figure 3A) with rs10916199 (rs74140304 and rs11588850) intersected with accessible chromatin regions across multiple human long bone cartilage (figure 3C and 3D). In addition, rs74140304 also intersected with an active transcription start site in osteoblast and chondrogenic cells (figure 3B).28 Next, we examined 3D chromatin conformation in the locus. Since no chromatin conformation capture data were available for chondrogenic or bone cells, we examined data from human mesenchymal stem cells, which are stem cell progenitors for chondrocytes and osteoblasts (figure 3G).28 The genomic location of rs1158850 (figure 3G, zone 2) appears to come into close proximity with the promoter region of WNT9A (figure 3G, zone 3). In addition, CTCF binding peaks in osteoblasts also intersect with the WNT9A promoter region (figure 3F, zone 3), and are located near rs11588850 (figure 3F, zone 2).
Next, differential expression analysis between OA lesioned and preserved cartilage (hip/knee joints) identified WNT9A as the most significant result, with increased expression in OA lesioned cartilage (n=21, fold change=2.42, p=9.4×10−8, online supplemental table S2). Lastly, we examined whether rs11588850 may significantly affect (p<4.0×10−8) the regulatory TF binding motifs37 located within the WNT9A promoter.35 The minor allele (G) of rs11588850 is in high linkage disequilibrium (r2 >0.8) with the OA risk increasing allele (G) of rs10916199, which significantly increases the binding affinity of the TF binding motif for RAD21 (G-allele logarithm of odds (LOD)=11.2, A-allele LOD=9.8). The RAD21 protein has been previously shown to bind to the WNT9A promoter region (online supplemental table S3). Thus, our results indicate WNT9A as novel OA associated gene, where rs1158850 is a potential regulatory variant for WNT9A (figure 3, zones 2–3).
Additional hand osteoarthritis associated loci
OA is highly heritable and co-occurrence of OA in multiple joint sites is well recognised44. As the hand joints are non-weight bearing, causes of OA in these joints may reflect effects of systemic risk factors, unlike the hip and knee joint where mechanical loading is a dominant risk factor.7 Thus, we examined whether other known OA loci may also confer risk for hand OA (figure 4). For 29 of the previously reported OA associated SNVs,13 14 nominally significant associations (p<0.05) were observed for one or more hand OA phenotypes. Strong associations were seen for known hand OA loci: ALDH1A2-locus (rs3204689), MGP-locus (rs4767133) (figure 4A, B) and COG5 (rs3815148),45 an SNV identified from a candidate gene study for hand OA. Interestingly, the MGP-loci and ALDH1A2-loci, were only associated with finger and/or hand OA phenotypes, but not with thumb OA. In contrast, the BCL7A-locus (rs11059094), was only associated with thumb OA. Strikingly, several reported knee and hip OA loci were also associated with hand OA phenotypes in our study (Bonferroni, p<5.8×10−4): RUNX2-locus (rs12154055), COL27A1-locus (rs919642), ASTN2-locus (rs13253416), IL11-locus (rs4252548), TGFa-locus (rs3771501) and GDF5-locus (rs143384) (figure 4B). Since some of these known loci were previously found to be associated with knee and/or hip OA, they may reflect common mechanisms across all joints in OA.
Figure 4.
Heatmap depicting the effect of osteoarthritis (OA) associated single nucleotide variants (SNVs) in each hand OA phenotype. (A) The found associated lead SNVs of the hand OA phenotypes and their beta and p value in the other phenotypes. (B) Depicted are all known OA SNVs which had a nominal significant effect in at least one stratified hand OA phenotype. All betas were calculated for the reported effect allele and scaled. Colours represent the scaled beta of the effect allele, which is here the minor allele. P values are represented by * in the box. Chr, chromosome; EA, effect allele; EAF, effect allele frequency; Gene, reported gene from the GWAs study; KL, Kellgren-Lawrence score; Pos, base pair position on the chromosome Hg19.
Discussion
We identified four genome-wide significant loci associated with hand OA phenotypes, of which two were novel and specific for thumb OA. Integration of multiple lines of data provided cumulative evidence that WNT9A may be a causal gene for thumb OA. We first conducted a cluster analysis of hand joints to identify less heterogeneous clusters of joints that served as the basis of the hand OA phenotypes assessed in this GWAS. With this approach of using radiographically defined biologically relevant OA phenotypes to reduce phenotype heterogeneity and increase statistical power, we were able to robustly identify known and novel OA loci despite our modest sample size (n~9900).18 22 This indicates that assessment of stratified phenotypes in OA may be warranted to improve GWAS statistical power and provide novel insight into OA biology.
Using bioinformatic analysis and functional genomics datasets, we were able to identify rs1158850 as potential causal variant. This SNV is nearby meQTL CpGs and the G allele of this variant is predicted to increase RAD21 (RAD21 Cohesin Complex Component) binding affinity in a region that has chromatin interactions with the WNT9A promoter. RAD21 is a part of the cohesion complex, involved in the formation of chromatin loops with CTCF.46 Both RAD21 and CTCF bind to the WNT9A promoter region. In line with these findings, WNT9A expression was significantly increased in OA lesioned cartilage compared with preserved OA cartilage. Combining all results, we postulate that rs1158850 is located in a gene regulatory element, increases RAD21 binding, and mediated by CTCF, interacts with the WNT9A promoter to influence WNT9A expression.
WNT9A (wingless-type MMTV integration site family, member 9A) previously known as WNT14, is a member of the WNT gene family, and has been shown to play a central role in synovial joint formation.47 48 Knockout WNT9A mice have severe skeletal developmental defects, and are neonatal lethal.49 Expression of WNT members by chondrocytes leads to the destruction of the cartilage matrix by the upregulation of Wnt/β-catenin signalling. Inhibition of WNT members has been suggested as a plausible OA therapeutic strategy, with recent success in a murine model of OA.50 51 However, this is the first evidence for WNT9A, a non-canonical Wnt ligand, in human OA.
In addition, to identifying novel OA loci, we also provide evidence for a subset of generalised OA genetic risk loci: GDF5 (rs143384), TGFα (rs2862851/rs3771501), RUNX2 (rs12154055), ASTN2 (rs2480930, rs13823416), COL27A1 (rs919642) and IL11 (rs4252548). These loci should be given priority as potential therapeutic targets since genetically supported drug targets have been shown to double the success rate of therapeutics in clinical development and intervention at these target loci may be beneficial regardless of which joint site is affected by OA.52
Although collectively our findings implicate WNT9A in thumb OA, there are several limitations. First, age is the most predominant risk factor for OA, yet the genetic background may determine the age of onset, rather than the lifetime risk for OA. Therefore, future genetic studies may benefit from examining the age of onset rather than adjust for age.53 Second, our functional findings are based on chondrogenic data sourced from several different tissues that did not include tissues from the hand joints. However, consistent results were found across the available chondrogenic source material from several different origins (primary, cell culture, developmental), indicating a more general role for the WNT9A locus in chondrocyte functional pathways. Given the complex nature of OA susceptibility and the fact that pathophysiological causes are not uniform across skeletal sites, alterations in WNT9A expression may be seen in other joints, but may have a more marked detrimental effect on the thumb joints. The lack of genetic association of WNT9A variants for knee and hip OA might be due to the phenotype definition of these GWAS studies: self-reported OA and 10th revision of the International Statistical Classification of Diseases codes, which do not necessarily correspond or have enough statistical power to identify genetic variants associated to radiographic phenotypes.
In summary, by examining the distribution of radiographic OA features in the hand joints, we identified three distinct hand OA phenotypes that provided the basis for identification of a novel locus for thumb OA despite our modest sample size. We identified WNT9A as a plausible causal gene for thumb OA, providing new insights into the genetic architecture of hand OA and a new candidate for OA therapeutic development.
Acknowledgments
The authors would like to thank the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists. The generation and management of GWAS genotype data for the Rotterdam Study (RSI, RSII, RSIII) was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, Rotterdam, The Netherlands. The authors would like to thank Pascal Arp, Mila Jhamai, Marijn Verkerk, Lizbeth Herrera and Marjolein Peters, MSc, and Carolina Medina-Gomez, MSc, for their help in creating the GWAS database, and Linda Broer PhD, for the creation of the imputed data.
Footnotes
Handling editor: Josef S Smolen
Twitter: @CurlyGeneticist, @TDCapellini
Contributors: CGB designed the study, performed the analyses, made the figures and tables and wrote the manuscript. MSY performed the replication analysis. SJR, RCdA, KC and LS performed functional analyses and look-ups (eQTL, meQTL and differential expression), MY and TDC performed ATAC-seq analysis, US provided GWAS data of deCODE and UK Biobank, LB provided analysis help. AGU provided access to the Rotterdam study dataset, DF provided replication data, EZ, JL, TDC and IM provided functional data. JBJvM designed the study and supervised this work. All authors critically assessed the manuscript.
Funding: The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII) and the Municipality of Rotterdam. The Rotterdam Study GWAS datasets are supported by the Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012), the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) Netherlands Consortium for Healthy Aging (NCHA), project nr. 050-060-810. The Framingham Heart Study of the National Heart, Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine was supported by the National Institutes of Health (contract no. HHSN268201500001I, N01-HC-25195, AG18393, AR47785) and its contract with Affymetrix, Inc. for genotyping services (N02‐HL‐6‐4278). Analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. MSY was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) and the National Institute on Aging (NIA) (R01AR075356). Rice, Cheung and Loughlin were supported by vs Arthritis (grant 20771), by the Medical Research Council and Arthritis Research UK as part of the MRC-Arthritis Research UK Centre for Integrated Research into Musculoskeletal Ageing (CIMA, grant references JXR 10641, MR/P020941/1 and MR/R502182/1) and by the European Union’s Seventh Framework Programme for research, technological development and demonstration under grant agreement number no. 305 815 (D-BOARD). The research leading to the RAAK biobank and the current results has received funding from the Dutch Arthritis Association (DAA 2010_017) and the European Union’s Seventh Framework Programme (FP7/2007-2011) under grant agreement no. 259 679. ATAC-seq datasets generated by MY and TDC were funded by a Harvard University Dean’s Competitive Award.
Competing interests: None declared.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available on reasonable request. GWAS summary statistics are available through the GWAS catalogue. All data relevant to the study are included in the article or uploaded as supplementary information. All relevant data supporting the key findings of this study are available within the article and its supplementary information files. Other data are available from the corresponding author on reasonable request. Due to ethical and legal restrictions, individual-level data of the Rotterdam Study (RS) cannot be made publicly available. Data are available on request to the data manager of the Rotterdam Study Frank van Rooij (f.vanrooij@erasmusmc.nl) and subject to local rules and regulations. This includes submitting a proposal to the management team of RS, where upon approval, analysis needs to be done on a local server with protected access, complying with GDPR regulations.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
References
- 1. GBD 2017 Disease and Injury Incidence and Prevalence Collaborators Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the global burden of disease study 2017. Lancet 2018;392:1789–858. 10.1016/S0140-6736(18)32279-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barbour KE, Lui L-Y, Nevitt MC, et al. Hip osteoarthritis and the risk of all-cause and disease-specific mortality in older women: a population-based cohort study. Arthritis Rheumatol 2015;67:1798–805. 10.1002/art.39113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Qin J, Barbour KE, Murphy LB, et al. Lifetime risk of symptomatic hand osteoarthritis: the Johnston County osteoarthritis project. Arthritis Rheumatol 2017;69:1204–12. 10.1002/art.40097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bijsterbosch J, Watt I, Meulenbelt I, et al. Clinical burden of erosive hand osteoarthritis and its relationship to nodes. Ann Rheum Dis 2010;69:1784–8. 10.1136/ard.2009.125435 [DOI] [PubMed] [Google Scholar]
- 5. Kwok WY, Vliet Vlieland TPM, Rosendaal FR, et al. Limitations in daily activities are the major determinant of reduced health-related quality of life in patients with hand osteoarthritis. Ann Rheum Dis 2011;70:334–6. 10.1136/ard.2010.133603 [DOI] [PubMed] [Google Scholar]
- 6. Kjeken I, Dagfinrud H, Slatkowsky-Christensen B, et al. Activity limitations and participation restrictions in women with hand osteoarthritis: patients' descriptions and associations between dimensions of functioning. Ann Rheum Dis 2005;64:1633–8. 10.1136/ard.2004.034900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hunter DJ, Bierma-Zeinstra S. Osteoarthritis. Lancet 2019;393:1745–59. 10.1016/S0140-6736(19)30417-9 [DOI] [PubMed] [Google Scholar]
- 8. Jiang L, Xie X, Wang Y, et al. Body mass index and hand osteoarthritis susceptibility: an updated meta-analysis. Int J Rheum Dis 2016;19:1244–54. 10.1111/1756-185X.12895 [DOI] [PubMed] [Google Scholar]
- 9. Jensen V, Bøggild H, Johansen JP. Occupational use of precision grip and forceful gripping, and arthrosis of finger joints: a literature review. Occup Med 1999;49:383–8. 10.1093/occmed/49.6.383 [DOI] [PubMed] [Google Scholar]
- 10. Spector TD, Cicuttini F, Baker J, et al. Genetic influences on osteoarthritis in women: a twin study. BMJ 1996;312:940–3. 10.1136/bmj.312.7036.940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ishimori ML, Altman RD, Cohen MJ, et al. Heritability patterns in hand osteoarthritis: the role of osteophytes. Arthritis Res Ther 2010;12:R180. 10.1186/ar3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zengini E, Hatzikotoulas K, Tachmazidou I, et al. Genome-wide analyses using UK Biobank data provide insights into the genetic architecture of osteoarthritis. Nat Genet 2018;50:549–58. 10.1038/s41588-018-0079-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tachmazidou I, Hatzikotoulas K, Southam L, et al. Identification of new therapeutic targets for osteoarthritis through genome-wide analyses of UK Biobank data. Nat Genet 2019;51:230–6. 10.1038/s41588-018-0327-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Styrkarsdottir U, Lund SH, Thorleifsson G, et al. Meta-analysis of Icelandic and UK data sets identifies missense variants in SMO, IL11, COL11A1 and 13 more new loci associated with osteoarthritis. Nat Genet 2018;50:1681–7. 10.1038/s41588-018-0247-0 [DOI] [PubMed] [Google Scholar]
- 15. Styrkarsdottir U, Thorleifsson G, Helgadottir HT, et al. Severe osteoarthritis of the hand associates with common variants within the ALDH1A2 gene and with rare variants at 1p31. Nat Genet 2014;46:498–502. 10.1038/ng.2957 [DOI] [PubMed] [Google Scholar]
- 16. den Hollander W, Boer CG, Hart DJ, et al. Genome-wide association and functional studies identify a role for matrix Gla protein in osteoarthritis of the hand. Ann Rheum Dis 2017;76:2046–53. 10.1136/annrheumdis-2017-211214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kloppenburg M, Kwok W-Y. Hand osteoarthritis--a heterogeneous disorder. Nat Rev Rheumatol 2011;8:22–31. 10.1038/nrrheum.2011.170 [DOI] [PubMed] [Google Scholar]
- 18. Manchia M, Cullis J, Turecki G, et al. The impact of phenotypic and genetic heterogeneity on results of genome wide association studies of complex diseases. PLoS One;8:82013 10.1371/journal.pone.0076295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Egger P, Cooper C, Hart DJ, et al. Patterns of joint involvement in osteoarthritis of the hand: the Chingford study. J Rheumatol 1995;22:1509–13. [PubMed] [Google Scholar]
- 20. Dahaghin S, Bierma-Zeinstra SMA, Ginai AZ, et al. Prevalence and pattern of radiographic hand osteoarthritis and association with pain and disability (the Rotterdam study). Ann Rheum Dis 2005;64:682–7. 10.1136/ard.2004.023564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Castaño-Betancourt MC, Evans DS, Ramos YFM, et al. Novel genetic variants for cartilage thickness and hip osteoarthritis. PLoS Genet 2016;12:e1006260. 10.1371/journal.pgen.1006260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Panoutsopoulou K, Thiagarajah S, Zengini E, et al. Radiographic endophenotyping in hip osteoarthritis improves the precision of genetic association analysis. Ann Rheum Dis 2017;76:1199–206. 10.1136/annrheumdis-2016-210373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bierma-Zeinstra SM, van Middelkoop M. Osteoarthritis: in search of phenotypes. Nat Rev Rheumatol 2017;13:705–6. 10.1038/nrrheum.2017.181 [DOI] [PubMed] [Google Scholar]
- 24. Ikram MA, Brusselle GGO, Murad SD, et al. The Rotterdam study: 2018 update on objectives, design and main results. Eur J Epidemiol 2017;32:807–50. 10.1007/s10654-017-0321-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Das S, Forer L, Schönherr S, et al. Next-generation genotype imputation service and methods. Nat Genet 2016;48:1284–7. 10.1038/ng.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhan X, Hu Y, Li B, et al. RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics 2016;32:1423–6. 10.1093/bioinformatics/btw079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Winkler TW, Day FR, Croteau-Chonka DC, et al. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc 2014;9:1192–212. 10.1038/nprot.2014.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Willer CJ, Li Y, Abecasis GR. Metal: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–1. 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Team RC R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2014. http://www.R-project.org/ [Google Scholar]
- 30. Haugen IK, Englund M, Aliabadi P, et al. Prevalence, incidence and progression of hand osteoarthritis in the general population: the Framingham osteoarthritis study. Ann Rheum Dis 2011;70:1581–6. 10.1136/ard.2011.150078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kellgren JH, Lawrence JS. Radiological assessment of osteo-arthrosis. Ann Rheum Dis 1957;16:494–502. 10.1136/ard.16.4.494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res 2016;44:D877–81. 10.1093/nar/gkv1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Watanabe K, Taskesen E, van Bochoven A, et al. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017;8:1826. 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature 2015;518:317–30. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Y, Song F, Zhang B, et al. The 3D genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol 2018;19:151. 10.1186/s13059-018-1519-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012;40:D930–4. 10.1093/nar/gkr917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kalichman L, Cohen Z, Kobyliansky E, et al. Patterns of joint distribution in hand osteoarthritis: contribution of age, sex, and handedness. Am J Hum Biol 2004;16:125–34. 10.1002/ajhb.20007 [DOI] [PubMed] [Google Scholar]
- 39. Marshall M, van der Windt D, Nicholls E, et al. Radiographic hand osteoarthritis: patterns and associations with hand pain and function in a community-dwelling sample. Osteoarthritis Cartilage 2009;17:1440–7. 10.1016/j.joca.2009.05.009 [DOI] [PubMed] [Google Scholar]
- 40. den Hollander W, Pulyakhina I, Boer C, et al. Annotating transcriptional effects of genetic variants in disease-relevant tissue: transcriptome-wide allelic imbalance in osteoarthritic cartilage. Arthritis Rheumatol 2019;71:561–70. 10.1002/art.40748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Steinberg J, Ritchie GRS, Roumeliotis TI, et al. Integrative epigenomics, transcriptomics and proteomics of patient chondrocytes reveal genes and pathways involved in osteoarthritis. Sci Rep 2017;7:8935. 10.1038/s41598-017-09335-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reynard LN, Loughlin J. Insights from human genetic studies into the pathways involved in osteoarthritis. Nat Rev Rheumatol 2013;9:573–83. 10.1038/nrrheum.2013.121 [DOI] [PubMed] [Google Scholar]
- 43. Richard D, Liu Z, Cao J, et al. Evolutionary selection and constraint on human knee chondrocyte regulation impacts osteoarthritis risk. Cell 2020;181:362–81. 10.1016/j.cell.2020.02.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vaes RBA, Rivadeneira F, Kerkhof JM, et al. Genetic variation in the GDF5 region is associated with osteoarthritis, height, hip axis length and fracture risk: the Rotterdam study. Ann Rheum Dis 2009;68:1754–60. 10.1136/ard.2008.099655 [DOI] [PubMed] [Google Scholar]
- 45. Hämäläinen S, Solovieva S, Vehmas T, et al. Genetic influences on hand osteoarthritis in Finnish women--a replication study of candidate genes. PLoS One 2014;9:e97417. 10.1371/journal.pone.0097417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell 2009;137:1194–211. 10.1016/j.cell.2009.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Später D, Hill TP, O'sullivan RJ, et al. Wnt9a signaling is required for joint integrity and regulation of Ihh during chondrogenesis. Development 2006;133:3039–49. 10.1242/dev.02471 [DOI] [PubMed] [Google Scholar]
- 48. Hartmann C, Tabin CJ. Wnt-14 plays a pivotal role in inducing synovial joint formation in the developing appendicular skeleton. Cell 2001;104:341–51. 10.1016/S0092-8674(01)00222-7 [DOI] [PubMed] [Google Scholar]
- 49. Maupin KA, Droscha CJ, Williams BO. A comprehensive overview of skeletal phenotypes associated with alterations in Wnt/β-catenin signaling in humans and mice. Bone Res 2013;1:27–71. 10.4248/BR201301004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou Y, Wang T, Hamilton JL, et al. Wnt/β-catenin Signaling in Osteoarthritis and in Other Forms of Arthritis. Curr Rheumatol Rep 2017;19:53. 10.1007/s11926-017-0679-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lietman C, Wu B, Lechner S, et al. Inhibition of Wnt/β-catenin signaling ameliorates osteoarthritis in a murine model of experimental osteoarthritis. JCI Insight 2018;3. 10.1172/jci.insight.96308. [Epub ahead of print: 08 Feb 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nelson MR, Tipney H, Painter JL, et al. The support of human genetic evidence for approved drug indications. Nat Genet 2015;47:856–60. 10.1038/ng.3314 [DOI] [PubMed] [Google Scholar]
- 53. Mars N, Koskela JT, Ripatti P, et al. Polygenic and clinical risk scores and their impact on age at onset and prediction of cardiometabolic diseases and common cancers. Nat Med 2020;26:549–57. 10.1038/s41591-020-0800-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
annrheumdis-2020-217834supp001.pdf (213.8KB, pdf)
annrheumdis-2020-217834supp002.pdf (108.5KB, pdf)
annrheumdis-2020-217834supp003.pdf (356.4KB, pdf)