Abstract
Evolutionary biologists typically envision a trait’s genetic basis and fitness effects occurring within a single species. However, traits can be determined by and have fitness consequences for interacting species, thus evolving in multiple genomes. This is especially likely in mutualisms, where species exchange fitness benefits and can associate over long periods of time. Partners may experience evolutionary conflict over the value of a multi-genomic trait, but such conflicts may be ameliorated by mutualism’s positive fitness feedbacks. Here, we develop a simulation model of a host–microbe mutualism to explore the evolution of a multi-genomic trait. Coevolutionary outcomes depend on whether hosts and microbes have similar or different optimal trait values, strengths of selection and fitness feedbacks. We show that genome-wide association studies can map joint traits to loci in multiple genomes and describe how fitness conflict and fitness feedback generate different multi-genomic architectures with distinct signals around segregating loci. Partner fitnesses can be positively correlated even when partners are in conflict over the value of a multi-genomic trait, and conflict can generate strong mutualistic dependency. While fitness alignment facilitates rapid adaptation to a new optimum, conflict maintains genetic variation and evolvability, with implications for applied microbiome science.
Keywords: host-microbiome, mutualism, quantitative genetics, coevolution
1. Introduction
Evolutionary biologists have known for decades that traits of one individual can affect both its own fitness as well as the fitness of interacting con- or hetero-specific individuals. A host’s immune response affects the fitness of its pathogenic and beneficial microbes [1]; an individual’s behaviour impacts the fitness of other group members [2] and so on. The idea that a trait has fitness effects beyond the trait-bearer is enshrined in the concept of an extended phenotype [3] and is necessary for coevolution, defined as reciprocal adaptation between species [4]. Extended phenotypes have renewed relevance with the revelation from microbiome science that many so-called plant and animal traits are actually partly determined by microbial genomes [5–7]. Thus, phenotypes may map to genes in the genome of a completely different species—which raises the question: whose trait is it anyways? And how do these joint phenotypes evolve?
Nowhere is the importance of extended phenotypes more obvious than mutualisms, reciprocally beneficial interactions between species. In host–microbe mutualisms and other symbioses, prolonged physical association may link trait-fitness relationships across species especially tightly [8]. Even in non-symbiotic mutualisms, reciprocal benefits can result in selection for traits that enhance the interaction and allow their partner to influence trait expression, in contrast to antagonisms (which can be interpreted as joint traits with selection for reduced or increased interactions, respectively, in victims and exploiters [9]). In mutualisms, an individual’s fitness will be determined by its own genome’s influence on traits, the influence of its partner’s genome on traits, and also by the fitness costs or benefits accrued through interacting with its partner, which may in turn depend on how expressed traits alter partner fitness via fitness feedbacks. Thus, the fitnesses of both partners are jointly determined by both partners’ genomes, which coevolve. However, the genetic basis and evolutionary trajectories of mutualism-related traits have received relatively little attention from evolutionary quantitative geneticists.
Understanding the evolutionary quantitative genetics of joint traits in mutualisms is pressing in light of the debate over the extent of conflict in these otherwise cooperative interactions. Although mutualisms are defined by mutual benefit, they can be viewed equivalently as mutual exploitation [10]. Mutualists are widely expected to be under selection to ‘cheat’ by over-exploiting or under-provisioning their partners, although there is currently only weak evidence for cheating [11,12] and conflicts may be more usefully viewed as over jointly determined traits [12]. When partner fitnesses are positively genetically correlated (i.e. aligned), natural selection favours more cooperation between species, but when this correlation is negative, increased fitness in one comes at the expense of fitness in the other [11].
Here, we describe mounting evidence for the existence of joint traits and their evolutionary relevance and develop a simulation framework to explore their evolutionary dynamics, focusing on two themes. First, we track coevolutionary trajectories of traits, alleles and patterns of genetic variation, considering impacts of trait-fitness relationships in both species and fitness conflict between species. Second, we sample simulated genomes for in silico genome-wide association study (GWAS) to validate methods for inferring the genetic basis and selective history of a multi-genomic trait.
2. Background
(a). Traits with a multi-genomic basis
There is a growing literature on intraspecific indirect genetic effects [13] and their contributions to genetic variance (e.g. [14]). However, though the multi-genomic basis of traits is also a research theme in both ‘niche construction’ [15] and community genetics [16], identifying loci underlying multi-genomic traits and the evolutionary forces acting on them remains relatively rare, especially in mutualisms.
When we consider how traits evolve in mutualisms, the traits we consider are often those that directly mediate the interaction. For example, legumes produce host-specific flavonoids, inducing compatible rhizobial bacteria to produce Nod factors that initiate the formation of root nodules where plant and bacterial metabolism interact to fuel the fixation of atmospheric nitrogen into plant-accessible forms [17]. Both Nod factor secretion and nitrogen fixation by rhizobia are bacterial traits encoded by bacterial genes whose expression is affected by the plant host. Likewise, nodule number and leaf nitrogen content are legume traits altered by rhizobia [6]. Thus, these are joint phenotypes encoded by loci in both legume and rhizobium genomes.
However, not all multi-genomic traits are directly involved in the host–microbe interaction [5,6]. Flowering time has a major genetic basis in plant genomes and is under strong selection [18], yet is also impacted by microbes [19,20]. Abiotic stress is also often modulated by microbes, such as the fungi associated with Pinus edulis that confer drought tolerance [21]. Similarly, microbial symbionts regulate many traits in animal hosts, including maturation, reproduction, neurodevelopment and obesity in model animals [5,7,22] and even social status in a non-model animal, the hyena [23].
Thus, microbial and host genomes can act together to produce a wide variety of joint phenotypes. Indeed, some have proposed that microbiomes may help resolve the ‘missing heritability’ paradox: since (host) loci identified via GWAS incompletely explain heritable trait variation, some causal variants may reside in genomes of inherited microbes [24] (but see other possible explanations [25]). If this view is correct, then we need to map jointly determined phenotypes to multiple genomes to study trait evolution. The combination of GWAS to detect loci underlying traits and population genetic tests to distinguish among selection regimes on those traits has proven highly effective in single species [26]. With sequence data from interacting genomes and joint phenotype measurements, we may have the power to infer coevolutionary scenarios.
(b). The joint fitness effects of joint phenotypes
For multi-genomic traits that impact one or both partner’s fitnesses, alleles underlying the trait may evolve in both genomes. For example, plant flowering time can evolve in bacteria, demonstrated by artificial selection on the rhizosphere microbiome of Arabidopsis thaliana shifting flowering time [19]. Such change can feed back to the host genome: microbial variation in effects on maturation in Drosophila melanogaster induced evolutionary change at loci in the fly genome [27].
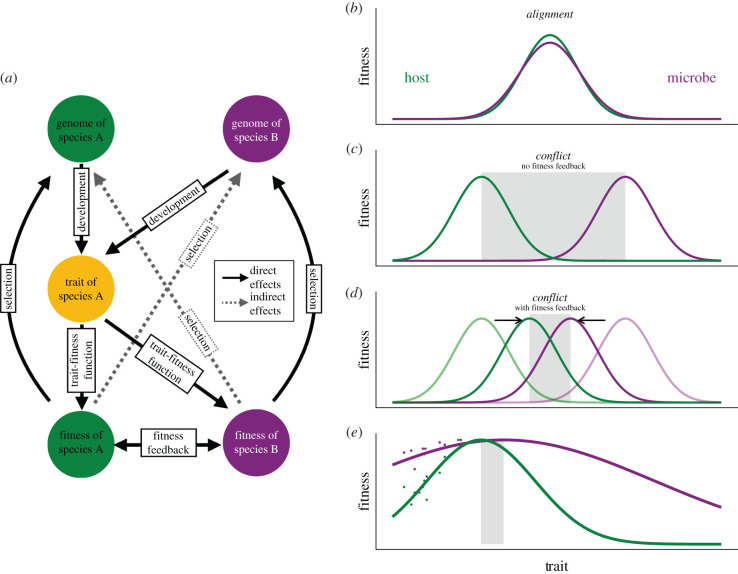
To predict the selective response of a multi-genomic trait, we must understand how the trait influences both partners’ fitness. First, a trait may have direct effects on each partner’s fitness; i.e. varying the trait influences each partner’s survival and reproduction (figure 1a), in similar (figure 1b) or different (figure 1c–e) ways. For instance, plant and microbial fitness could both be maximized by the same flowering time and under the same strength of selection, as in figure 1b, or plant fitness may be maximized by an earlier flowering time, as in figure 1c, or plants may experience stronger selection on traits (yielding a pattern like figure 1e). When the two species have the same trait optimum, their fitnesses are aligned and there is no conflict over the expressed trait value. If the trait is far from the optimum, selection will favour alleles in both species that move the trait closer. Otherwise, selection will purge alleles underlying extreme phenotypes. In contrast, when partners have different optima for the joint trait, adaptation in one partner generally comes at the expense the other, producing fitness conflict. Yet, how the trait evolves under conflict can also depend on fitness feedbacks between partners and how far the phenotype is from the optima (figure 1 and below).
Figure 1.
(a) The genomes of two species, A (host, green) and B (microbe, purple), affect a joint trait (yellow) that can affect the fitness of both. Fitness of A and B results in direct selection on the genomes of A and B, respectively. (b) The relationship between the trait and fitnesses of hosts and microbes when both experience stabilizing selection for the same trait value. This aligns fitness interests regardless of expressed trait values in the population. (c) A difference between species A and B in the optimal value of the trait. This results in conflict across the range of trait values between the two optima (grey shading). (d) Such conflicts can be ameliorated or resolved by indirect effects of traits on each species’ fitness via positive fitness feedbacks (see text), which effectively move the trait optima together (indicated by arrows). (e) Data (points) from Haney et al. [28] for host (Arabidopsis thaliana) and microbe (Pseudomonas fluorescens) fitness as a function of a joint trait, plant root branching. Lines show data fit to a Gaussian curve for each species, which predict different optima, potentially creating conflict (grey-shaded region) and stronger direct selection on plants (curve steepness).
An important determinant of the amount of evolutionary conflict between species is what we here call partner fitness feedbacks. Positive fitness feedbacks occur in mutualisms if fitness benefits provided by species A to species B eventually feed back to benefit species A [29]. This occurs when A increases the condition or overall vigour of B, which then directly benefits A. Equivalently, if A ‘skimps’ on providing fitness benefits to B, A gets fewer fitness benefits in return (commonly called ‘sanctions’, among other names [29–31]). There is empirical evidence of these feedbacks in many mutualisms: when mycorrhizae provide more phosphorus to plants, plants reward them with more carbon [31]; when yucca moths or fig wasps do not pollinate yucca or fig flowers, plants selectively abort their offspring ([30] and references therein); etc. While many mutualisms are horizontally transmitted, another source of fitness feedback can be vertical transmission, which links symbiont and host reproduction and therefore fitness interests [32].
Our simulation model imagines a general case where species B can benefit from shifting the joint trait value closer to the optimum for species A, because the higher fitness of species A then feeds back to benefit species B (figure 1d). For example, nitrogen fixation by rhizobia is energetically costly, so the direct trait-fitness function for rhizobia might have a low optimum—just enough to sustain the rhizobia themselves. However, nitrogen-starved plants are small and do not support much microbial growth. Because microbes on fit plants have much greater fitness than microbes on unfit plants, both partners can benefit from more exchange of carbon for nitrogen [33] and the optimal amount of nitrogen fixation by microbes may instead be near the plant’s optimal phenotype. Thus, fitness feedbacks can shift the microbe’s effective optimal trait value towards the host’s optimum (figure 1d).
Most past work has emphasized directional selection in mutualisms and simply asked whether partner fitnesses are positively or negatively correlated overall [11,12]. Yet, in reality, we expect most traits to be under stabilizing selection [34], especially traits involved in mutualisms. If the fitness of species A has heritable, genetic variation in species B, then mutualistic fitness feedbacks between A and B imply that higher fitness of species A will increase the fitness of species B. However, this cannot continue indefinitely. May [35] famously derided this possibility by calling unbounded, positive feedback between mutualistic partners ‘an orgy of reciprocal benefaction’ because at some point, more mutualism is not better. At some point, rhizobia have fixed sufficient nitrogen for their plant hosts and plant growth is limited by other resources [36]. Furthermore, the point at which continuing to invest in the mutualism becomes maladaptive may differ between partners. Thus, even with fitness feedbacks, differences in optimal values across partners can lead to evolutionary conflicts [37]. In short, we have an a priori expectation that traits in mutualisms should generally be under stabilizing selection with weak conflicts.
3. A simulated case study of joint trait evolution in host–microbe mutualisms
(a). Simulation details
We simulated coevolving loci in host and microbial genomes that impact a joint trait with fitness consequences for one or both species in R (further details in electronic supplementary material; code: see ‘Data accessibility’) [38]. Selection acts initially on a trait with no segregating variation (e.g. a joint trait in a mutualism that has recently formed or invaded a novel habitat, thereby resulting in an initially narrow base of genetic variation and a trait mean far from the optimum).
For each simulation step, we draw from the populations of hosts and microbes (both of size n = 2000) such that they interact at random, assuming for simplicity that each host interacts with one microbe (therefore, n also equals the number of host–microbe pairs). Our model incorporates the fundamentally different genetic architectures of macrobes and microbes: plants and animals are most often diploid, while their microbial partners are generally haploid. The joint trait is quantitative, and the phenotypic value expressed is the sum of the additive effects (a) at each allele (a1 and a2 with no dominance in diploid hosts, e.g. dominance coefficients all 0.5; but a single allele in haploid microbes) at each locus (l) across all loci (totalling LM and LH) within both host (H) and microbe (M) individuals. LM = 40 and LH = 20 to equalize mutational target size across partners. The phenotypic value (z) of the nth host–microbe pair is given by
| 3.1 |
in a similar, additive, fashion as models of joint phenotypes in evolutionary conflicts [9].
Next, direct fitness effects of trait values are determined by the difference between the expressed phenotype and each partner’s phenotypic optimum (θ) with a Gaussian function with variance ω2 [39]. For hosts,
| 3.2 |
where the direct fitness component (CH) derived from the trait is relative to trait values expressed by all host–microbe pairs,
| 3.3 |
Previous theoretical work on the evolution of quantitative traits in mutualisms has mainly considered independent traits that must match between species for the interaction to happen (e.g. phenology, the length of pollinator tongues and flower corolla tubes, etc.) and often incorporates stabilizing selection similarly to equation (3.2) [40]. Low fitness from failed interactions forces these non-joint traits to evolve away from their ‘direct’ optima (i.e. the optima when not interacting as mutualists) and towards values enabling interactions. In contrast, we assume all individuals interact, but that the jointly determined trait values affect interaction outcomes. Specifically, fitness feedbacks occur when host and microbe direct components (equation (3.3)) contribute as weighted indirect components to each other’s fitnesses, as in indirect genetic effects models (see [41]). Fitness is weighted between each component via α, which sets the relative importance of direct and indirect effects of a trait on fitness. Lower values give more weight to indirect effects via partner fitness feedbacks and interact with the variance (ω2, inverse ‘strength’ of selection) of the direct trait–fitness function in both hosts and microbes to determine the effective optimal trait value and combined selection strength. Thus, the relative fitness of the host in the nth combination of a host and microbe is
| 3.4 |
Microbial direct trait fitness component (CM) and relative fitness (wM) are calculated similarly, substituting microbe parameters for host parameters and vice versa.
Because each fitness component depends on the distance to the optima of all hosts and microbes in the populations (equation (3.3)), the fitness landscape and effects of fitness feedbacks can change through time (electronic supplementary material, figures S4 and S5). The next generation of individuals are then sampled from parents according to parent relative fitness, with free recombination between loci in hosts and clonal reproduction in microbes, mimicking real-world eukaryotes and microbes.
Next, mutations occur with binomial probability , effect size exponentially distributed with λ = 25, an equal chance of being negative or positive (both genomes, electronic supplementary material, figure S1), and are added to the previous value of alleles (initially, all a = 0; n × ploidy or evolutionary dynamics limit the number of alleles per locus). While parameters are equal here for simplicity, in practice, mutational inputs may differ between hosts and microbes; diploid, eukaryotic genomes more often generate point mutations [42], while microbes readily lose and acquire portions of genomes through horizontal gene transfer (HGT) [43]. We allow HGT of single microbial loci with binomial probability phrz = 0.2, wherein another individual’s genotype is drawn at random and copied to the focal. This occurs simultaneously across all individuals. Finally, the cycle begins again and repeats for 300 generations. Note that included features of microbes (clonality and haploidy) can reduce the efficacy of selection and evolutionary rates, because beneficial mutations compete with one another [44]. HGT may partially counteract this, but for simplicity, we do not model other potentially counteracting forces: different generation duration between hosts and microbes, and dominance.
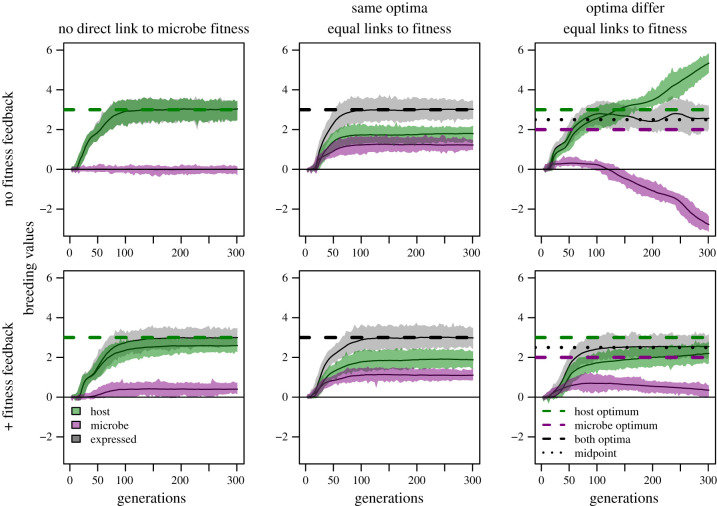
We explored six scenarios of joint trait evolution in interacting species. In the first three, there is no fitness feedback, so αH = αM = 1. These three scenarios are as follows. (1) A null model. The trait directly affects the fitness of only the host, though loci in both genomes affect the trait. To achieve this, we flattened the direct trait-fitness curve for microbes (ωM = 10 much greater than both ωH = 0.75 and the range of phenotypic values during simulations; figure 1). (2) A joint trait under fitness alignment. The trait directly affects both host and microbe fitness equally (ωM = ωH = 0.75) and partners have the same optimum. (3) A joint trait under conflict. We shifted the microbe trait optimum (θM = 2) away from the host trait optimum (θH = 3), creating conflict. In scenarios 4–6 we added fitness feedbacks by setting αH = αM = 0.6 but keeping other parameters the same as in scenarios 1–3. Scenarios 4–6 are more realistic models of mutualism than 1–3, which lack the exchange of fitness benefits. We further explored outcomes beyond these six scenarios, including flipping host and microbe parameters for scenarios 1, 3, 4 and 6 (electronic supplementary material, figures S2 and S3), and independent and interactive influence of parameters on outcomes in replicated simulations across a range of values (see electronic supplementary material, figures S6–S8).
(b). Joint trait responses to selection depend on conflict and fitness feedbacks
When a joint trait is only linked to host fitness and there are no fitness feedbacks (scenario 1), adaptation occurs in hosts alone. The host’s mean breeding value moves rapidly to the host optimum, θH, while the microbe’s breeding value takes a random walk (figure 2, top left). When the trait also directly affects microbe fitness, and hosts and microbes have identical trait optima (scenario 2; figure 2, top middle), both partners evolve increased breeding values until the joint trait reaches the shared optimum. Despite equal selection strength in direct trait-fitness components (ωM = ωH) in scenario 2, breeding values respond more in the host than the microbe, reflecting more efficient selection in hosts (see also flipped scenarios, electronic supplementary material, figures S2 and S3).
Figure 2.
Simulation results for different scenarios. Mean and range of breeding values for host (green) and microbes (purple), and expressed phenotype (grey). Trait optima or midpoints at dashed lines (see legend). Top left: a trait is directly linked to host fitness only. Top middle: a trait is directly linked to host and microbe fitness, with identical optima. Top right: trait is directly linked to both microbe and host fitness, at separate optima. Bottom row: as above, but host and microbe fitnesses are linked via positive fitness feedbacks.
When hosts and microbes have conflicting phenotypic optima (scenario 3; figure 2, top right; see also electronic supplementary material, figure S3) both partners’ breeding values initially increase, because the joint trait is below both optima (dashed lines). As soon as the joint trait value passes the microbe optimum, microbes experience selection for lower breeding values while the host still benefits from higher trait values. Host and microbe breeding values evolve in opposite directions indefinitely, while the expressed phenotype remains approximately equidistant from host and microbe optima and neither partner ‘wins’. The near-constancy in the expressed phenotype at the midpoint between optima is similar to coevolutionary outcomes when mutualisms depend on trait matching [40], while the dynamics of breeding value evolution are similar to coevolution in antagonistic interactions (e.g. [45]), yet this interaction is neither inherently mutualistic nor antagonistic per se. Instead, partners are in conflict over the joint phenotype.
In most scenarios (2–6), hosts become dependent on microbes for optimal phenotype expression. Counter-intuitively, under greater conflict, hosts become more dependent: in the top right panel of figure 2, after 300 generations, a host with no microbe expresses a trait value far beyond its optimum. Phenotypic expression often goes awry in novel environments, where development can be miscued; this is particularly true for hosts that develop gnotobiotically [46]. Our simulation shows how host evolution in response to microbial influences on joint traits could underlie this. In nature, this outcome would be classified as a mutualism, since both partners have higher fitness together; in fact, evolved dependence may be a common way mutualism originates [47].
Fitness feedbacks (scenarios 4–6) cause strikingly different simulation outcomes, even when the trait only indirectly affects microbe fitness (in scenario 4; figure 2, bottom left; see also electronic supplementary material, figure S2). In scenario 4, positive fitness feedbacks cause the effective microbe optimum to match the host optimum, albeit with a shallower fitness landscape (see electronic supplementary material, figure S4). Microbe breeding values thus initially increase and maintain contributions to the joint trait. This result could explain why microbiota affect such a wide variety of host traits, including traits unlikely to directly affect microbe fitness [6,7]; partner fitness feedbacks mean that any joint trait under selection in the host can evolve in microbes, even if the trait has little to do with the host–microbe interaction. Adding fitness feedbacks to direct selection with a shared optimum (compare scenarios 2 and 5; figure 2, middle) causes a lag in reaching the optimum, because segregating loci in multiple partners adds error and weakens selection relative to drift (electronic supplementary material, figure S9). The efficacy of selection may generally be lower whenever fitness benefits accrue indirectly: work on social evolution similarly finds that selection via indirect fitness effects is weaker than via direct fitness effects [48].
The most realistic scenario we modelled is scenario 6, wherein the joint trait has different fitness optima for hosts and microbes, but they experience positive fitness feedbacks. In this scenario (figure 2, bottom right), moderate fitness feedbacks weaken conflict. Each genome continues shifting trait values towards its own optimum indefinitely, but much more slowly than without fitness feedbacks (compare scenarios 3 and 6, figure 2, right; see also electronic supplementary material, figure S3). Further simulations of fitness conflicts with feedbacks reveal that larger differences in optima and more similar selection strengths drive rapid evolution, while increasing fitness feedbacks (smaller αs) slow evolutionary conflict (see electronic supplementary material, figures S6 and S8). These results help resolve an apparent paradox about the tempo of mutualism evolution, which is alternately expected to be slow due to widespread stabilizing selection [49] or fast because of underlying conflict [37].
How much conflict actually occurs in mutualisms and whether mutualists ‘cheat’ remain unresolved questions, despite much attention [12,30,31]. Previous studies have taken negative fitness correlations as evidence of selection for cheating [11,12]. However, our simulations show that evolutionary conflict sometimes occurs alongside positive fitness correlations (electronic supplementary material, figures S6, S8). Thus, mutualism evolution does not follow a dichotomy between selection for ‘cheating’ or ‘cooperation’. When hosts and microbes have equal trait optima, their fitnesses are perfectly correlated, although as selection reduces genetic variance, detecting correlations would become difficult (electronic supplementary material, figure S5). When partners have different trait optima, there is a trait value on either side of θH that results in the same host fitness but generates very different microbial fitnesses (figure 1). As a result, rather fit hosts can have low- or high-fitness microbes, explaining the horseshoe shape of the relationship between partner fitnesses under conflict (electronic supplementary material, figure S5, bottom right panel). Thus, while evolutionary conflicts in mutualisms are often invoked as evidence for cheating (e.g. [37]), our simulations show that conflict is compatible with positive, negative or non-linear fitness correlations, depending on evolutionary trajectories.
(c). Evolution shapes the multi-genomic architecture of joint traits and multi-genomic architecture affects evolution
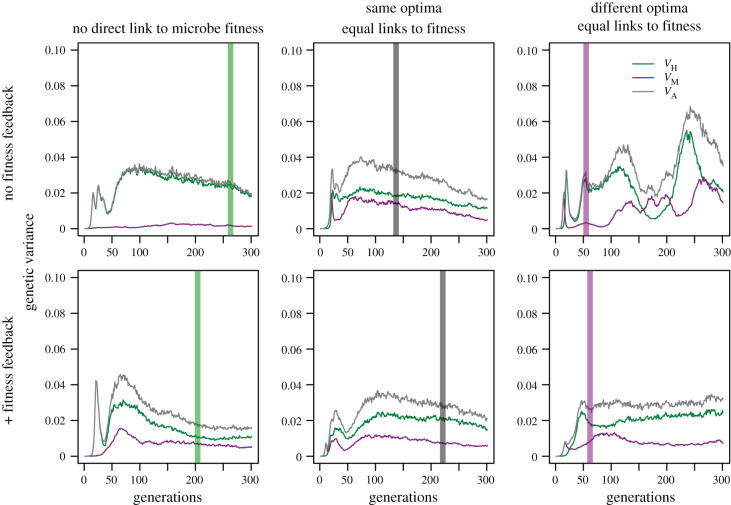
Genetic variance is necessary for natural selection to act, yet selection often erodes variance. After decades of research into what prevents mutualists from cheating, Heath et al. [50] recast the fundamental problem of mutualism evolution: hosts often preferentially associate with or reward more beneficial symbionts and thus select for cooperative symbionts [11,29–31], so it is puzzling that ample genetic variation for symbiont quality persists in natural populations [50]. Our simulation results show how genetic variance for a joint trait is eroded or maintained across partner genomes, depending on the evolutionary scenario.
In scenario 1, where the trait only affects host fitness and there are no fitness feedbacks, host genomes’ additive genetic variance initially spikes as mutations with large, positive effects rise in frequency (figure 3, top left). As the trait approaches the optimum, unfixed large-effect alleles more often produce phenotypes overshooting the optimum so they decrease in frequency [51]; the additive genetic variance declines as segregating alleles are removed. In contrast, the microbial genome slowly accumulates variation, approaching mutation-drift equilibrium (figures 3; electronic supplementary material, S9 and S10, top left).
Figure 3.
Genetic variance through time for each scenario in figure 2. Vertical green (host), grey (both), purple (microbe) lines on each panel mark when the average trait value reaches (or passes, right-hand panels) the respective trait optimum.
In scenarios 2 and 3, which add alignment or conflict over traits, respectively, genetic variance initially spikes then falls in both partners. The spike is larger for hosts (figure 3), as more alleles initially escape drift to rise in frequency (compare electronic supplementary material, figures S10 and S9). Scenarios 2 and 3 diverge after the joint trait value passes the microbe optimum in scenario 3 and the partners begin experiencing conflict. When the joint trait value is between the two partners’ optima, large-effect alleles again rise in frequency, but only positive-effect alleles in hosts and only negative-effect alleles in microbes (electronic supplementary material, figures S9 and S10). The joint trait value remains in this conflict zone and alleles accumulate in both partners. Points in time when many or especially large effect alleles are segregating correspond to the largest peaks in genetic variance (compare top right panels of figure 3 and electronic supplementary material, S9 and S10). Hosts generally retain segregating variants for longer (compare electronic supplementary material, figures S9 and S10, right panels, including alleles ultimately lost), increasing genetic variance (figure 3). This resembles common findings of victim–exploiter coevolutionary models, in which coevolution often increases genetic variation [52]. Adding fitness feedbacks in scenarios 4–6 generally diminishes the host’s relative contribution to genetic variance (figure 3, bottom panels, not scenario 5). Even when the trait only indirectly affects microbe fitness through feedbacks (comparing scenarios 1 and 4), the initial uptick in total trait variance is quicker, and the microbial genome contributes substantially (figure 3, bottom left), because microbial loci also respond to selection (electronic supplementary material, figures S9 and S10). When partner fitness feedbacks increase the indirect component of fitness for microbes (scenario 5), the decrease in variance is slower than in scenario 2, reflecting that indirect selection is weaker than direct [48].
While partner fitness feedbacks strengthen correlated selection in the positive direction (scenarios 4 and 5), they weaken correlated selection in opposing directions (scenario 6). As feedbacks shift effective optima closer together (figure 1c; electronic supplementary material, figure S4), the expressed phenotype at the midpoint is closer to the effective optima and selection weakens. Alleles that rise in frequency after the microbe optimum is passed have smaller effects (electronic supplementary material, figures S9 and S10), dampening peaks in genetic variance (compare scenarios 3 and 6, figure 3).
(d). GWAS can detect causal variants in multiple genomes, which may permit inferences about trait selective history
Rather than an all-or-nothing view of ‘cheaters’ in a mutualism, mutualist genomes are probably mosaics of loci, with some encoding traits under conflict, but most encoding traits with a shared optimum. Population genetic analyses of loci [53] can reveal the history of selection acting on quantitative traits [54]. We seek to verify whether the loci underlying multi-genomic traits can be identified with GWAS, and if loci that GWAS identifies have different selective histories among evolutionary scenarios.
We designed an in silico GWAS ‘experiment’, similar to real empirical GWAS. Briefly, we simulated neutral, unlinked loci in the six simulation scenarios, sampled 800 hosts and microbes each from the 300th generation, redefined our multi-allelic quantitative loci as sets of linked biallelic sites and interacted one host genotype with all microbes and one microbe genotype with all hosts in two GWAS ‘experiments’ (see electronic supplementary material for details).
GWAS detects coarsely similar proportions of causal loci across most scenarios and host and microbial genomes (generally 30–60%), and the number of false positives reflects the false positive rate (set to 5%) and number of neutral loci (electronic supplementary material, figures S15 and S16). It is hard to detect loci with alleles that are small-effect, rare or both (electronic supplementary material, figure S14) [25]. Yet, estimated and known effect sizes are positively correlated (mostly ρ > 0.3; electronic supplementary material, figure S13), and large-effect loci provide more information about evolutionary scenarios.
When the trait only affects host fitness (scenario 1), the microbe genome retains few segregating variants and those that are retained will resemble neutral sites in diversity (electronic supplementary material, figure S11, upper left). Across conflicts with weak or no fitness feedbacks, selection remains directional through time and more larger effect, younger alleles with rapid trajectories remain segregating (electronic supplementary material, figures S9–S12, right panels). Therefore, when evolving under conflict, large effect loci in both genomes are likely to exhibit characteristic sweep or partial sweep patterns, where the selected site carries adjacent, linked variants to high frequency, reducing nearby diversity [55]. With fitness alignment, selection is stabilizing, there are fewer segregating variants, and common variants of large effect tend to be older than under even weak fitness conflict (electronic supplementary material, figures S11 and S12, right panels). Under alignment (but also drift in microbe genomes), segregating loci at appreciable frequencies are likely to have had slower trajectories and retained or accumulated linked diversity [56].
While recent shifts in optima under alignment might also generate sweep signals, and weak conflict will have softer sweeps, younger derived alleles in one species will have different directions of effects from younger derived alleles in the other in conflict only (electronic supplementary material, figures S11 and S12). With alignment, younger alleles will have concordant or equally variable effects in hosts and microbes. Polarizing derived alleles relative to phylogenetic outgroups can identify effect signs of younger alleles and help distinguish purifying selection from sweeps [53,56]. Distinguishing among scenarios where fitness interests are aligned (scenarios 2, 4 and 5) would be harder because segregating variants have similar trajectories (electronic supplementary material, figures S9–S12). Yet these scenarios are also more biologically similar, and perhaps distinguishing them is less urgent.
In summary, GWAS appears to be effective for identifying loci underlying multi-genomic traits and selective forces acting on them. Indeed, the greatest challenge we expect is distinguishing strong selection when fitness interests are aligned from low-powered GWAS when no segregating large effect alleles are identified and from cases where traits simply do not have a genetic basis in both genomes, though others have overcome analogous hurdles [54].
4. Closing remarks
Joint traits with a multi-genomic basis can affect the fitness of multiple species, changing how we think about evolution. The extent of evolutionary conflict in mutualisms will be driven simultaneously by differences in fitness optima and by the strength of direct selection relative to partner fitness feedbacks (figure 2; electronic supplementary material, figures S6–S8). Furthermore, conflict leaves a signature on alleles in the genome, and we show that this can be evaluated with sequence data and GWAS (electronic supplementary material, figures S9–S16).
The scope of host traits affected by microbes is vast, going far beyond traits directly involved in host–microbe interactions [6,7,22], so almost any trait could have a multi-genomic basis. Transcriptomics might help identify phenotypes whose expression depends on multiple genomes [57] and can be paired with GWAS through an eQTL approach to identify loci [54].
Conflicts over multi-genomic traits underlaid by genetic constraints could be resolved by breaking them. For example, if late-flowering plants have frost-damaged fruits, but earlier flowering reduces microbe fitness, constraint between flowering time and fruiting success would drive conflict, and the evolution of frost-tolerant fruit would resolve it. Conflict resolution may occur through the microbiome via altered trait covariances, such as between root mass and phenology [20]. Much work has focused on whether the microbiome itself is heritable (e.g. [58]), yet whether the microbiome possesses genetic variation for host traits may matter more: in our simulations, microbe genotype is not heritable in hosts, yet multi-genomic traits still evolve.
Our model could be expanded to consider additional genomic or ecological complexity. Whereas we modelled purely additive traits, in nature, dominance and epistasis—both within and across genomes—affect evolutionary rates [59]. Genomic complexity could influence genetic variance and evolutionary trajectories, as in multi-nucleate arbuscular mycorrhizal fungi [60]. Furthermore, our model comprises a panmictic population in a constant environment; yet environments fluctuate in space and time, especially for horizontally transmitted microbes [61], and such variation in selection could alter evolution [62] or generate spatial correlations between host and symbiont breeding values. In our model, partnerships form at random, so genetic correlations do not arise pre-selection through spatial structure or other mechanisms (e.g. partner choice, vertical transmission). Models including pre-selection correlations could leverage quantitative genetic theory for phenotypes influenced by interacting conspecifics [13]. Lastly, our model assumes one microbe interacts with each host, but real host-associated microbiomes comprise diverse microbial taxa (e.g. [58]). Nonetheless, GWAS methods for communities exist [63] and diffuse coevolution across communities sometimes resembles pairwise coevolution [4]. Simulations of mutualistic networks based on trait-matching show coevolution causes trait convergence [64], according with patterns in nature [65].
Binary traits such as whether an interaction occurs may also map to multiple genomes and thus be considered joint traits. Many mutualisms begin with one partner producing a signal that is received by the other; these independently expressed traits jointly determine whether partners interact [66]. Cleaner fish eat parasites attached to client fish, and signals emitted by cleaner fish and received by client fish initiate interactions [67]. Likewise, floral scents and insect odorant receptors presumably evolve only in their respective genomes but co-determine whether insect pollinators visit plants [66]. Signal emitters and signal receivers do not always have the same fitness interests (e.g. visits to fully fertilized flowers may benefit pollinators, but not plants). Screening mechanisms that filter out ineffective partners can also be considered joint traits. For example, the squid Euprymna scolopes depends on the horizontally acquired bacterium Vibrio fischeri to produce light the squid uses for predator evasion. Light is an emergent property of host screening traits and bacterial traits: mutualistic V. fisheri catabolize toxic reactive oxygen species synthesized by the host with the light-producing enzyme luciferase [68]. Thus, interaction traits in both host–microbe and macrobe–macrobe mutualisms can also be considered through the multi-genomic trait framework we present.
Finally, multi-genomic trait evolution has implications for global change biology. Widespread, rapid environmental change can lead to mutualism breakdown [69], but mutualism can also facilitate rapid adaptation [19]. The degree of mutualism disruption and evolutionary rescue will hinge on whether trait optima match under historical and new environmental conditions. Fitness alignment facilitates, and fitness conflict constrains, adaptation to a new optimum (figure 2). However, traits historically under conflict retain the most genetic variation (figure 3) and thus evolvability. Understanding the evolution of multi-genomic traits thus enhances our ability to predict how mutualisms will adapt in a changing world.
Supplementary Material
Acknowledgements
We acknowledge support from an NSERC Discovery Grant, the University of Toronto, and a Radcliffe Fellowship to MEF; NSF DEB-1943628, NSF DEB-1821892, NSF 1753917 and USDA NIFA Hatch project 1014527 to MLF. MEF and MLF thank J. Bronstein and R. Ferrière for organizing Exploitation and Cheating in Mutualisms: Syntheses, Challenges, and New Directions, Paris, France, 2016, where we had the first inklings of these ideas. We thank the Frederickson and Wright laboratories at the University of Toronto, Susan Johnston, and anonymous reviewers for helpful comments.
Data accessibility
Code available at: https://github.com/amob/whosetrait-is-it-anyway---sims.
Authors' contributions
All authors conceived of the ideas, drafted and revised the manuscript, gave final approval for publication and agree to be held accountable for the work performed therein. AMO wrote the code.
Competing interests
We declare we have no competing interests.
References
- 1.Stecher B et al. 2007. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 5, e244–e240. ( 10.1371/journal.pbio.0050244) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamilton WD 1964. The genetical evolution of social behaviour. II. J. Theor. Biol. 7, 17–52. ( 10.1016/0022-5193(64)90039-6) [DOI] [PubMed] [Google Scholar]
- 3.Dawkins R et al. 1982. The extended phenotype, vol. 8 Oxford, UK: Oxford University Press. [Google Scholar]
- 4.Thompson JN 2005. The geographic mosaic of coevolution. Chicago, IL: University of Chicago Press. [Google Scholar]
- 5.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. 2008. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host & Microbe 3, 213–223. ( 10.1016/j.chom.2008.02.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friesen ML, Porter SS, Stark SC, von Wettberg EJ, Sachs JL, Martinez-Romero E. 2011. Microbially mediated plant functional traits. Ann. Rev. Ecol. Evol. Syst. 42, 23–46. ( 10.1146/annurev-ecolsys-102710-145039) [DOI] [Google Scholar]
- 7.Gould AL et al. 2018. Microbiome interactions shape host fitness. Proc. Natl Acad. Sci. USA 115, E11951–E11960. ( 10.1073/pnas.1809349115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bordenstein SR, Theis KR. 2015. Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 13, e1002226 ( 10.1371/journal.pbio.1002226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Queller DC 2014. Joint phenotypes, evolutionary conflict and the fundamental theorem of natural selection. Phil. Trans. R. Soc. B 369, 20130423 ( 10.1098/rstb.2013.0423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bronstein JL 1994. Conditional outcomes in mutualistic interactions. Trends Ecol. Evol. 9, 214–217. ( 10.1016/0169-5347(94)90246-1) [DOI] [PubMed] [Google Scholar]
- 11.Friesen ML 2012. Widespread fitness alignment in the legume–rhizobium symbiosis. New Phytol. 194, 1096–1111. ( 10.1111/j.1469-8137.2012.04099.x) [DOI] [PubMed] [Google Scholar]
- 12.Jones EI et al. 2015. Cheaters must prosper: reconciling theoretical and empirical perspectives on cheating in mutualism. Ecol. Lett. 18, 1270–1284. ( 10.1111/ele.12507) [DOI] [PubMed] [Google Scholar]
- 13.Moore AJ, Brodie ED III, Wolf JB. 1997. Interacting phenotypes and the evolutionary process: I. Direct and indirect genetic effects of social interactions. Evolution 51, 1352–1362. ( 10.1111/j.1558-5646.1997.tb01458.x) [DOI] [PubMed] [Google Scholar]
- 14.Bergsma R, Kanis E, Knol EF, Bijma P. 2008. The contribution of social effects to heritable variation in finishing traits of domestic pigs (Sus scrofa). Genetics 178, 1559–1570. ( 10.1534/genetics.107.084236) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Odling-Smee FJ, Laland KN, Feldman MW. 1996. Niche construction. Am. Nat. 147, 641–648. ( 10.1086/285870) [DOI] [Google Scholar]
- 16.Whitham TG et al. 2003. Community and ecosystem genetics: a consequence of the extended phenotype. Ecology 84, 559–573. ( 10.1890/0012-9658(2003)084[0559:CAEGAC]2.0.CO;2) [DOI] [Google Scholar]
- 17.Clúa J, Roda C, Zanetti ME, Blanco FA. 2018. Compatibility between legumes and rhizobia for the establishment of a successful nitrogen-fixing symbiosis. Genes 9, 125–120. ( 10.3390/genes9030125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor MA, Wilczek AM, Roe JL, Welch SM, Runcie DE, Cooper MD, Schmitt J. 2019. Large-effect flowering time mutations reveal conditionally adaptive paths through fitness landscapes in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 116, 17 890–17 899. ( 10.1073/pnas.1902731116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panke-Buisse K, Poole AC, Goodrich JK, Ley RE, Kao-Kniffin J. 2015. Selection on soil microbiomes reveals reproducible impacts on plant function. ISME J. 9, 980–989. ( 10.1038/ismej.2014.196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Brien AM, Sawers RJ, Strauss SY, Ross-Ibarra J. 2019. Adaptive phenotypic divergence in an annual grass differs across biotic contexts. Evolution 73, 2230–2246. ( 10.1111/evo.13818) [DOI] [PubMed] [Google Scholar]
- 21.Gehring CA, Sthultz CM, Flores-Rentería L, Whipple AV, Whitham TG. 2017. Tree genetics defines fungal partner communities that may confer drought tolerance. Proc. Natl Acad. Sci. USA 114, 11 169–11 174. ( 10.1073/pnas.1704022114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buffington SA, Di Prisco GV, Auchtung TA, Ajami NJ, Petrosino JF, Costa-Mattioli M. 2016. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell 165, 1762–1775. ( 10.1016/j.cell.2016.06.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theis KR, Venkataraman A, Dycus JA, Koonter KD, Schmitt-Matzen EN, Wagner AP, Holekamp KE, Schmidt TM. 2013. Symbiotic bacteria appear to mediate hyena social odors. Proc. Natl Acad. Sci. USA 110, 19 832–19 837. ( 10.1073/pnas.1306477110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandoval-Motta S, Aldana M, Martínez-Romero E, Frank A. 2017. The human microbiome and the missing heritability problem. Front. Genet. 8, 80 ( 10.3389/fgene.2017.00080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rockman MV 2012. The QTN program and the alleles that matter for evolution: all that’s gold does not glitter. Evolution 66, 1–17. ( 10.1111/j.1558-5646.2011.01486.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Josephs EB, Stinchcombe JR, Wright SI. 2017. What can genome-wide association studies tell us about the evolutionary forces maintaining genetic variation for quantitative traits? New Phytol. 214, 21–33. ( 10.1111/nph.14410) [DOI] [PubMed] [Google Scholar]
- 27.Rudman SM et al. 2019. Microbiome composition shapes rapid genomic adaptation of Drosophila melanogaster. Proc. Natl Acad. Sci. USA 116, 20 025–20 032. ( 10.1073/pnas.1907787116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haney CH, Samuel BS, Bush J, Ausubel FM. 2015. Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nat. Plants 1, 1–9. ( 10.1038/nplants.2015.51) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sachs JL, Mueller UG, Wilcox TP, Bull JJ. 2004. The evolution of cooperation. Q Rev. Biol. 79, 135–160. ( 10.1086/383541) [DOI] [PubMed] [Google Scholar]
- 30.Frederickson ME 2013. Rethinking mutualism stability: cheaters and the evolution of sanctions. Q Rev. Biol. 88, 269–295. ( 10.1086/673757) [DOI] [PubMed] [Google Scholar]
- 31.Kiers ET et al. 2011. Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 333, 880–882. ( 10.1126/science.1208473) [DOI] [PubMed] [Google Scholar]
- 32.Doebeli M, Knowlton N. 1998. The evolution of interspecific mutualisms. Proc. Natl Acad. Sci. USA 95, 8676–8680. ( 10.1073/pnas.95.15.8676) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoeksema JD, Schwartz MW. 2003. Expanding comparative–advantage biological market models: contingency of mutualism on partner’s resource requirements and acquisition trade-offs. Proc. R. Soc. Lond. B 270, 913–919. ( 10.1098/rspb.2002.2312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonamour S, Teplitsky C, Charmantier A, Crochet PA, Chevin LM. 2017. Selection on skewed characters and the paradox of stasis. Evolution 71, 2703–2713. ( 10.1111/evo.13368) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.May RM 1982. Mutualistic interactions among species. Nature 296, 803–804. ( 10.1038/296803a0) [DOI] [Google Scholar]
- 36.Menge DN, Wolf AA, Funk JL. 2015. Diversity of nitrogen fixation strategies in Mediterranean legumes. Nat. Plants 1, 1–5. ( 10.1038/nplants.2015.64) [DOI] [PubMed] [Google Scholar]
- 37.Sachs JL, Quides KW, Wendlandt CE. 2018. Legumes versus rhizobia: a model for ongoing conflict in symbiosis. New Phytol. 219, 1199–1206. ( 10.1111/nph.15222) [DOI] [PubMed] [Google Scholar]
- 38.R Core Team. 2019. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
- 39.Haldane J 1954. The measurement of natural selection. In Proceedings of the 9th International Congress of Genetics (Caryologia, Supplement to Volume 6) 1, pp. 480–487.
- 40.Nuismer S. 2017. Introduction to coevolutionary theory. New York, NY: WH Freeman & Co. [Google Scholar]
- 41.Bailey NW 2012. Evolutionary models of extended phenotypes. Trends Ecol. Evol. 27, 561–569. ( 10.1016/j.tree.2012.05.011) [DOI] [PubMed] [Google Scholar]
- 42.Lynch M 2010. Evolution of the mutation rate. Trends Genet. 26, 345–352. ( 10.1016/j.tig.2010.05.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EP. 2010. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74, 434–452. ( 10.1128/MMBR.00020-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maddamsetti R, Lenski RE, Barrick JE. 2015. Adaptation, clonal interference, and frequency-dependent interactions in a long-term evolution experiment with Escherichia coli. Genetics 200, 619–631. ( 10.1534/genetics.115.176677) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Valen L 1974. Molecular evolution as predicted by natural selection. J. Mol. Evol. 3, 89–101. ( 10.1007/BF01796554) [DOI] [PubMed] [Google Scholar]
- 46.Partida-Martinez LPP, Heil M. 2011. The microbe-free plant: fact or artifact? Front. Plant Sci. 2, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Mazancourt C, Loreau M, Dieckmann U. 2005. Understanding mutualism when there is adaptation to the partner. J. Ecol. 93, 305–314. ( 10.1111/j.0022-0477.2004.00952.x) [DOI] [Google Scholar]
- 48.Linksvayer TA, Wade MJ. 2009. Genes with social effects are expected to harbor more sequence variation within and between species. Evolution 63, 1685–1696. ( 10.1111/j.1558-5646.2009.00670.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chomicki G, Renner SS. 2017. Partner abundance controls mutualism stability and the pace of morphological change over geologic time. Proc. Natl Acad. Sci. USA 114, 3951–3956. ( 10.1073/pnas.1616837114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heath KD, Stinchcombe JR. 2014. Explaining mutualism variation: a new evolutionary paradox? Evolution 68, 309–317. ( 10.1111/evo.12292) [DOI] [PubMed] [Google Scholar]
- 51.Orr HA 1998. The population genetics of adaptation: the distribution of factors fixed during adaptive evolution. Evolution 52, 935–949. ( 10.1111/j.1558-5646.1998.tb01823.x) [DOI] [PubMed] [Google Scholar]
- 52.Nuismer SL, Ridenhour BJ, Oswald BP. 2007. Antagonistic coevolution mediated by phenotypic differences between quantitative traits. Evolution 61, 1823–1834. ( 10.1111/j.1558-5646.2007.00158.x) [DOI] [PubMed] [Google Scholar]
- 53.Oleksyk TK, Smith MW, O’Brien SJ. 2010. Genome-wide scans for footprints of natural selection. Phil. Trans. R. Soc. B 365, 185–205. ( 10.1098/rstb.2009.0219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Josephs EB, Lee YW, Stinchcombe JR, Wright SI. 2015. Association mapping reveals the role of purifying selection in the maintenance of genomic variation in gene expression. Proc. Natl Acad. Sci. USA 112, 15 390–15 395. ( 10.1073/pnas.1503027112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith JM, Haigh J. 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23, 23–35. ( 10.1017/S0016672300014634) [DOI] [PubMed] [Google Scholar]
- 56.Messer PW, Petrov DA. 2013. Population genomics of rapid adaptation by soft selective sweeps. Trends Ecol. Evol. 28, 659–669. ( 10.1016/j.tree.2013.08.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burghardt LT, Guhlin J, Chun CL, Liu J, Sadowsky MJ, Stupar RM, Young ND, Tiffin P. 2017. Transcriptomic basis of genome by genome variation in a legume-rhizobia mutualism. Mol. Ecol. 26, 6122–6135. ( 10.1111/mec.14285) [DOI] [PubMed] [Google Scholar]
- 58.Goodrich JK et al. 2014. Human genetics shape the gut microbiome. Cell 159, 789–799. ( 10.1016/j.cell.2014.09.053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carter AJ, Hermisson J, Hansen TF. 2005. The role of epistatic gene interactions in the response to selection and the evolution of evolvability. Theor. Popul. Biol. 68, 179–196. ( 10.1016/j.tpb.2005.05.002) [DOI] [PubMed] [Google Scholar]
- 60.Scott TW, Kiers ET, Cooper GA, Dos Santos M, West SA. 2019. Evolutionary maintenance of genomic diversity within arbuscular mycorrhizal fungi. Ecol. Evol. 9, 2425–2435. ( 10.1002/ece3.4834) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burghardt LT, Epstein B, Tiffin P. 2019. Legacy of prior host and soil selection on rhizobial fitness in planta. Evolution 73, 2013–2023. ( 10.1111/evo.13807) [DOI] [PubMed] [Google Scholar]
- 62.Takahata N, Ishii K, Matsuda H. 1975. Effect of temporal fluctuation of selection coefficient on gene frequency in a population. Proc. Natl Acad. Sci. USA 72, 4541–4545. ( 10.1073/pnas.72.11.4541) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang J, Jia H. 2016. Metagenome-wide association studies: fine-mining the microbiome. Nat. Rev. Microbiol. 14, 508–522. ( 10.1038/nrmicro.2016.83) [DOI] [PubMed] [Google Scholar]
- 64.Nuismer SL, Jordano P, Bascompte J. 2013. Coevolution and the architecture of mutualistic networks. Evolution 67, 338–354. ( 10.1111/j.1558-5646.2012.01801.x) [DOI] [PubMed] [Google Scholar]
- 65.Bascompte J, Jordano P. 2007. Plant-animal mutualistic networks: the architecture of biodiversity. Annu. Rev. Ecol. Evol. Syst. 38, 567–593. ( 10.1146/annurev.ecolsys.38.091206.095818) [DOI] [Google Scholar]
- 66.Schiestl FP 2015. Ecology and evolution of floral volatile-mediated information transfer in plants. New Phytol. 206, 571–577. ( 10.1111/nph.13243) [DOI] [PubMed] [Google Scholar]
- 67.Caves EM, Green PA, Johnsen S. 2018. Mutual visual signalling between the cleaner shrimp Ancylomenes pedersoni and its client fish. Proc. R. Soc. B 285, 20180800 ( 10.1098/rspb.2018.0800) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ruby EG, McFall-Ngai MJ. 1999. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends Microbiol. 7, 414–420. ( 10.1016/S0966-842X(99)01588-7) [DOI] [PubMed] [Google Scholar]
- 69.Kiers E, Ives A, Kawakita A. 2015. Global change and mutualisms. In Mutualism (ed. J Bronstein), pp. 241–267. Oxford University Press.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code available at: https://github.com/amob/whosetrait-is-it-anyway---sims.