Graphical abstract
Abbreviations: ML, Machine Learning; USML, Unsupervised Machine Learning; SML, Supervised Machine Learning; tSNE, t-distributed Stochastic Neighbor Embedding; PCoA, Principal Coordinate Analysis; ASV, Amplicon Sequence Variant; RF, Random Forests; SVM, Support Vector Machines; ANN, Artificial Neural Networks; AUC, Area Under the Curve; ROC, Receiver Operating Characteristic; GB, Gradient Boosting
Keywords: Machine learning, Marker genes, 16S rRNA, Metagenomics, Forensics
Abstract
Advances in nucleic acid sequencing technology have enabled expansion of our ability to profile microbial diversity. These large datasets of taxonomic and functional diversity are key to better understanding microbial ecology. Machine learning has proven to be a useful approach for analyzing microbial community data and making predictions about outcomes including human and environmental health. Machine learning applied to microbial community profiles has been used to predict disease states in human health, environmental quality and presence of contamination in the environment, and as trace evidence in forensics. Machine learning has appeal as a powerful tool that can provide deep insights into microbial communities and identify patterns in microbial community data. However, often machine learning models can be used as black boxes to predict a specific outcome, with little understanding of how the models arrived at predictions. Complex machine learning algorithms often may value higher accuracy and performance at the sacrifice of interpretability. In order to leverage machine learning into more translational research related to the microbiome and strengthen our ability to extract meaningful biological information, it is important for models to be interpretable. Here we review current trends in machine learning applications in microbial ecology as well as some of the important challenges and opportunities for more broad application of machine learning to understanding microbial communities.
1. Introduction
Environmental microbial communities are extremely diverse and play a role in driving many biogeochemical cycles and regulating human health. These environmental communities also have various applications in biotechnology. The ability to probe microbial diversity has been enabled through the increasing availability of high throughput sequencing (HTS) technologies. The microbial diversity of the human microbiome as well as soil and ocean microbial communities has been expanded through large-scale collaborative sequencing efforts such as the Human Microbiome Project [1] and the Earth Microbiome Project [2] as well as the TARA OCEANS project [3]. These large-scale efforts have provided baseline data for the microbial communities found in diverse settings. The low cost of sequencing now allows for large scale studies of systems and the generation of microbial community profiles for hundreds and thousands of samples. This scale of data necessitates methods capable of extracting meaningful information from these large datasets. Natural microbial communities have the potential to provide key insights into environmental phenomena and may be useful in predicting environmental phenomena. Machine learning (ML) has been employed to find patterns in data that can be predictive of various phenomena. In recent years machine learning has been applied to microbial community data to classify samples and predict various outcomes [4], [5], [6]. There is potential for expansion of the use of ML for microbial ecology studies. In this review, we seek to provide an overview of ML applications in microbial ecology and present some challenges and opportunities for the expansion of ML applications in the study of microbial communities.
2. Next generation sequencing methods in microbial ecology
Molecular methods have been used in microbial ecology for decades employing sequencing of ribosomal RNA genes to profile microbial diversity in settings ranging from soil and aquatic environments to hydrothermal vents and the built environment [7], [8], [9]. With the expansion of high throughput sequencing, the ability to generate thousands of sequences from hundreds of samples in a single sequencing run is possible [10], [11]. The application of next generation sequencing in microbial systems follows a pipeline that includes both wet lab and computational methods (Fig. 1). A goal of molecular profiling of microbial communities is to obtain a comprehensive assessment of the taxonomic and functional diversity of a community. In order to obtain this assessment, there are a number of important considerations that must be addressed during the analysis pipeline. The pipeline starts with wet lab methods for molecular profiling of microbial communities, which involves sample collection, extraction of nucleic acids from the environment or host and library preparation for sequencing Fig. 1. There are a number of biases that can be introduced with the wet lab portion of the methods [12]. In particular, extraction of DNA with different methods can result in differential extraction efficiencies for different taxa and the thus has the potential to skew diversity assessments. Often sequencing of DNA for microbial community profiling can take the form of marker gene surveys which profile the diversity of either taxa (small subunit rRNA such as the 16S rRNA for bacteria and archaea and 18S rRNA for eukaryotes) or of a particular functional gene. The choice of sequencing primers for marker gene surveys can also introduce bias as degenerate primers are not truly universal and may miss key microbial groups. These biases are important to consider in planning study design. Alternatively, shotgun metagenomic methods can be employed to profile the complement of genes that are present in a sample.
Fig. 1.
Illustrative pipeline for the investigation of microbial communities using metagenomics.
Sequencing depth is another key consideration in the process of profiling microbial communities. Sequencing is a sampling-based approach. Therefore, with increased sequencing depth, the diversity of reads is more completely sampled and thus diversity estimates are more reflective of the natural system. After sequencing, the primary analyses are computational. While the computational portion of the pipeline greatly depends on the goals of the study, often this portion is divided into sequence processing to generate a table of samples and taxonomic features in each sample followed by analysis methods to assess and link microbial diversity with various outcomes. A lot of work has been done related to processing of sequencing reads into meaningful data tables. This has included methods for binning marker genes into operational taxonomic units (OTUs). These OTUs are features that are representative of some biologically meaningful categories. Methods such as UCLUST [13] (often implemented in QIIME1 [14]) and mother [15] bin 16S rRNA reads based on percent identity to other reads in the dataset into OTUs. More recent methods have sought to cluster sequences into identical groups rather than cluster by some fixed percent identity. These approaches such as DADA2 [16] have employed denoising algorithms to correct sequencing errors and then dereplicate sequences into bins of identical sequences known as Amplicon Sequencing Variants (ASVs) or Exact Sequence Variants (ESVs). Each of these methods has advantages and limitations. They are all similar in that they are approaches for grouping marker gene sequences into biologically meaningful bins that result in feature tables for downstream analysis.
Much of the downstream analyses of feature tables generated from microbial community data has focused on the application of commonly used ecological measures for processing microbial community data. Methods such as alpha diversity assessments as well as beta diversity and multivariate statistics have been commonly used. A number of issues have been identified related to the application of methods designed for datasets with tens of features to highly dimensional datasets with thousands of features [17]. For example, specific diversity metrics have been shown to be highly impacted by the dimensionality and scale of the data, while others are less prone to errors resulting from highly dimensional data. Additional metrics, such as UniFrac distances, were developed that allowed researchers to more fully extract meaningful information from these marker gene surveys [18]. However, many of these methods seek to understand the data through decreasing dimensionality of the data and often can lose some of the important information that is contained within the rich datasets of microbial community profiles. For example, principal coordinate analysis (PCoA) is commonly used to assess overall differences in diversity. PCoA analysis is performed using distance or dissimilarity matrices of the microbial community profiles using metrics such as UniFrac distance or Bray-Curtis dissimilarity. While useful, these methods collapse the highly dimensional datasets and assess overall similarity or dissimilarity. This process can often lose important information and bias observations to highly abundant or highly prevalent features.
OTU-level analyses have also been important for analyzing the relationship between particular features in microbial data and specific outcomes. Indicator Species Analysis has been important for environmental monitoring. In Indicator Species Analysis, the prevalence of a species or OTU is linked to particular treatments or environmental states. Each OTU is given an indicator value (IndVal) which details how indicative that species is of the particular outcome. Additionally, differential abundance analysis has often been used to better understand differences between samples, categories, or particular outcomes on the level of particular features. Methods such as DESeq2 [19] and metagenomeSeq [20] have been used to identify which features are differentially abundant between different categories. DESeq in particular was originally developed to understand differentially expressed genes in RNASeq datasets. One advantage of the use of these methods for differential abundance analysis is that these approaches have been designed to work well with sequencing datasets and use normalization approaches tailored specifically to sequencing data. One of the limitations of differential abundance analysis is the ability to understand the importance of multiple features or the interaction of features in a particular outcome. Differential abundance analyses treat features as independent and it can be difficult to glean how increased or decreased abundance of groups of features may be a hallmark of a particular sample type or treatment category.
In addition to the methods described above, advanced computational methods are being employed to assist in analyzing the increasing amounts of data. In particular ML is being used with increasing frequency to use microbial communities to predict different outcomes. ML has advantages in that it is able to more fully appreciate the depth of data generated in microbiome studies as well as build predictive models for outcomes based on microbial community data. In the following sections we will provide an overview of commonly used ML methods, discuss key steps to be considered in the ML process and provide examples of how ML can be used in microbiome studies in the human microbiome, environmental monitoring, and forensics.
3. Machine learning and microbial community data analysis
In the context of microbial ecology, applied machine learning involves creating and evaluating models that use algorithms capable of recognizing, classifying and predicting specific outcomes from data. ML approaches take various forms including unsupervised, semi-supervised, reinforced, or supervised learning [21]. For example, often the goal of supervised machine learning (SML) applied to microbial community data is to construct a decision rule (i.e. a model) from a set of collected observations (i.e. samples) to predict the condition (i.e. response label (Y); such as an assigned category or value to each observation that have meaning to the model-operator) of an unlabeled sample using a set of measurements from next generation sequencing instruments. In microbiome studies this input data takes the form of a frequency count matrix of the observed microbial taxa from a sample (i.e., input variables (X) and their assigned values). Input variables are often referred to as features and samples as observations and will be used interchangeably throughout this review.
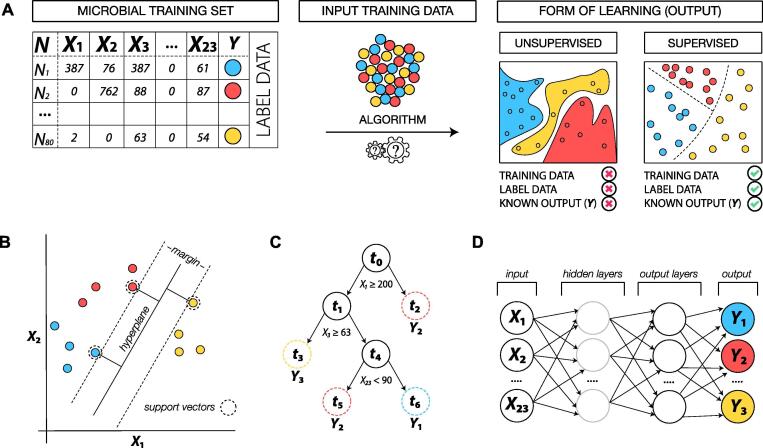
While there are other forms of learning that have been used on microbial community data, this manuscript discusses unsupervised techniques and supervised machine learning methods commonly applied to microbial datasets. The principal distinction between unsupervised (USML) and supervised machine learning (SML) is that in USML samples are segregated using features without any reference to response labels and the prediction is to which cluster a response may belong to, whereas the SML finds a best fit decision boundary between features and response labels [22] (Fig. 2A). A more precise overview of these methods is introduced below.
Fig. 2.
Schematic representation of unsupervised and supervised forms of learning and several ML methods predicting three conditional response labels (blue/red/yellow). (A) Depicts a common microbial frequency matrix containing observations or samples (N), features (X1, …, X23) and multiple class labels (Y). Input data are algorithmized and processed to either predict which cluster Y belongs to (unsupervised) or to find a best fit decision boundary between X and Y (supervised). (B) Linear SVM classifier demonstrating separation between class labels where the hyperplane maximizes the distance (margin) between the nearest data training points. Support vectors refer to the three position vectors drawn from the origin of the sample positions (dashed circle) with the goal of maximizing the distance between the optimal hyperplane and the support vectors (max-margin) so that a decision boundary can be drawn. (C) A decision tree constructed for the classification of samples into Y based on input feature values. Trees start from a root node (t0) and are grown to various leaf nodes (closed circle) to end at a terminal node (dashed circle) so that bootstrap aggregated predictions across terminal nodes are averaged across k-trees for best predictions of Ŷ. (D) A neural network displaying the structure of successive layers. Input values of X are transmitted to the proceeding hidden layer which passes weighted connections to the output layer for predictions of Ŷ. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.1. Unsupervised multivariate analysis common to marker-gene analysis
Unsupervised techniques are often employed for initial exploratory analysis of high-dimensional metagenomic data and for generating hypotheses for subsequent analysis as they aid in visualization and can search for structure in data that do not have predefined response labels assigned to observations. These methods operate with the goal of identifying homogenous subgroups by clustering data (hierarchical or centroid) or to detect anomalies by finding patterns through dimensionality reduction (DR) techniques. An example of DR by some unsupervised techniques (Principal Coordinate Analysis: PCoA and t-Distributed Stochastic Neighbor Embedding: t-SNE) is to take data points from a high-dimensional feature set and project them in low-dimensions to encapsulate the largest amount of statistical variance in a set of observations while preserving structure and minimizing information loss [23]. In USML, input data is either continuous data of features for each observation or a distance matrix of similarities between community composition. Observations that cluster together in USML represent microbial communities from samples that are more similar in composition.
3.1.1. K-means clustering (centroid)
The objective of K-means [24] is to cluster samples into a specified number of (k) non-overlapping subgroups (clusters) using distances calculated between features so each data point belongs to only one group. This technique assigns data points to a cluster such that the sum of squared distance between data points and the centroid (average of all data points represented by the geometric center of the cluster) is minimized. By reducing intra-cluster variation, data points are arranged to construct a cluster that assumes a spherical shape surrounding the centroid and allows different subgroups of data to remain as far apart as possible. A drawback of K-means is that it cannot construct clusters well on data points that are distanced to a more complex geometric shape. An additional constraint is that a pre-defined number of clusters is required, which necessitates assumptions to be placed on the structure of data prior to analysis.
3.1.2. Principal coordinate analysis (PCoA)
In PCoA analysis, data are decomposed into components to maximize the linear correlation between data points in a dissimilarity matrix, such as microbial taxa as input features [25]. Through a “coordinate transform”, x number of data points are replaced to newly derived y coordinates, thus reducing the dimensionality of a dataset by discarding the coordinates that may not capture a threshold of variance in the microbial community data. This technique preserves the global structure of the data while projecting it to low dimension. By mapping nearby points to each other and faraway points to each other, linear variance in the global relationships of the data are maximized to retain a faithful representation of the actual distances between original data points [23]. This method works from distance matrices calculated from biologically meaningful metrics such as UniFrac and is commonly employed in microbial analysis [26].
3.1.3. t-Distributed stochastic neighbor embedding (t-SNE)
In t-SNE analysis, data points are transformed and assigned a probability based on similarity to define relationships in high-dimension, guided by a Students t-distribution to help reduce crowding of data points during visual projection [27]. As the name entails, this method tries to identify close neighbors (samples with similar measurements) and tries to arrange these points in a low-dimension projection such that the close neighbors remain close and distant points remain distant. In contrast to PCoA that tends to preserve long distances for global retention of the original data, t-SNE tries to represent local relationships in the data, thus capturing non-linear variance and is not as faithful to the original state of the data [23]. Certain fields that use high-dimensional data currently benefit from the local non-linear structure of this method, such as single cell RNA-seq [28]. T-SNE is not commonly employed for metagenomic data despite its utility as a promising exploratory technique for the analysis of microbial communities [29], [30].
3.2. Supervised machine learning methods common to microbiome study
Supervised machine learning (SML) is a more elaborate form of exploring marker-gene datasets since unlike unsupervised methods, response labels (Y) are assigned to each sample in the dataset, grouping them into meaningful categories. A more targeted investigation of data can be achieved since the model is being trained to learn the structure of features (X) (training set) to create rules where they can serve as predictors of phenomena or outcome. In other words, which feature (X) maps to the response label (Y). Once trained, this model can intake new unlabeled samples with similar features (testing set) and predict their output (Y) based on what it has learned from the training set. SML can be used with continuous numerical outputs (regression: Y = .; continuous traits such age, blood pressure, concentration of contaminant) or categorical outputs (classification: X → Y; binary or symbolize grouped conditions such as ‘diseased’ or ‘healthy’). The following section seeks to provide an overview of some of the most common SML algorithms for microbiome-based prediction tasks (outlined as implemented methods in Table 1). We have focused our overview to primarily classification approaches, although this collection of methods could apply to regression as well.
Table 1.
Summary of ML techniques used for microbiome-based prediction tasks. This table briefly summarizes each technique, provides the source of the software, noteworthy ML implementations and interpretation of its result with reference to either the source study or specific studies that have applied these techniques for microbiome profiling. This table is not exhaustive but mentions current and commonly employed ML and ML related pipelines tailored to the characteristics of microbiome data or that are domain agnostic but relevant to research questions relating to the microbiome.
| Software name | Summary | Source | Example implementation | Remarks | URL |
|---|---|---|---|---|---|
| SIAMCAT (*) | Statistical Inference of Associations between Microbial Communities And host phenoTypes | R package ‘SIAMCAT’ https://siamcat.embl.de/ |
FS, ML, INTERP, VIS | Confounder analysis Enables cross-study comparison Advances visualization |
https://www.biorxiv.org/content/10.1101/2020.02.06.931808v2 |
| DeepMicro (*) | Deep representation learning for disease prediction based on microbiome data | Python: https://github.com/minoh0201/DeepMicro | DR, ML | Deep representation learning using autoencoders to handle high-dimensional data Accelerates model training and hyperparameter optimization |
https://www.nature.com/articles/s41598-020–63159-5 |
| MetAML (*) | Metagenomic prediction Analysis based on Machine Learning | Python: https://github.com/segatalab/metaml | FS, ML, INTERP, VIS | Enables cross study comparison of models on single cohorts, across stages of same the same study and across different studies | https://journals.plos.org/ploscompbiol/article?id=https://doi.org//10.1371/journal.pcbi.1004977 |
| mAML (*) | An automated machine learning pipeline with a microbiome repository for human disease classification | Python: https://github.com/yangfenglong/mAML1.0Web : http://lab.malab.cn/soft/mAML/ |
FS, ML, INTERP, VIS | Automates optimized, interpretable and reproducible models Deployed on a user-friendly web-based platform Advanced visuals |
https://pubmed.ncbi.nlm.nih.gov/32588040/ |
| BiomMiner (*) | An advanced exploratory microbiome analysis and visualization pipeline | Docker: https://mbac.gmu.edu/mbac_wp/biomminer-readme/ | FS, DR, ML, INTERP, VIS | Automatically tunes optimal hyper-parameters Tailored to clinical datasets Generates web-enabled visuals |
https://journals.plos.org/plosone/article?id=https://doi.org//10.1371/journal.pone.0234860 |
| MIPMLP (*) | Microbiome Preprocessing Machine Learning Pipeline | Python: https://github.com/louzounlab/microbiome/tree/master/PreprocessWeb : http://mip-mlp.math.biu.ac.il/Home |
FS, DR, ML, INTERP, VIS | Approaches for standardized ML preprocessing Consensus methods for optimal performance |
https://www.biorxiv.org/content/10.1101/2020.11.24.397174v1.full#ref-12 |
| MicrobiomeAnalystR (*) | Comprehensive statistical, functional, and meta-analysis of microbiome data | R package ‘MicrobiomeAnalystR’ Web: https://www.microbiomeanalyst.ca/ |
FS, DR, ML, INTERP, VIS | Comprehensive analysis reporting Real time feedback and recommendations Visual comparison with a public dataset |
https://www.nature.com/articles/s41596-019–0264-1 |
| Meta-Signer (*) | Metagenomic Signature Identifier based on Rank Aggregation of Features | Python: https://github.com/YDaiLab/Meta-Signer/tree/master/src | FS, ML, INTERP | Ensemble learning for feature ranking Identifies a robust set of highly informative taxa |
https://www.biorxiv.org/content/10.1101/2020.05.09.085993v1 |
| QIIME2 (*) | Quantitative Insights Into Microbial Ecology | https://qiime2.org/ | FS, DR, ML, INTERP, VIS | Automatic tracking of data provenance Multiple user interfaces Plugin support |
https://www.nature.com/articles/s41587-019–0209-9 |
| mothur (*) | Microbial community analysis pipeline | http://mothur.org/ | FS, DR, ML, INTERP, VIS | Can handle data from multiple sequencing platforms Encapsulates large elements of the pipeline in single command |
https://aem.asm.org/content/75/23/7537 |
| scikit-learn | Simple and efficient tools for predictive data analysis | Python: https://scikit-learn.org/stable/ | FS, DR, ML, INTERP, VIS | Robust machine learning library and support system Supports end-to-end projects with extensive documentation |
https://arxiv.org/abs/1201.0490 |
| Keras | Simple deep learning API | R package ‘keras’ Python: https://pypi.org/project/Keras/ |
FS, DR, ML, INTERP, VIS | High-level learning API that limits the number of user actions Multiple deployment capabilities Provides clear and actionable error messages |
https://link.springer.com/chapter/10.1007/978–1-4842–2766-4_7 |
| caret | Classification And REgression Training | R package ‘caret’ | FS, DR, ML, INTERP, VIS | Streamlines complex predictive tasks Large library of available models |
https://www.jstatsoft.org/article/view/v028i05 |
| mlr | Machine learning in R | R package ‘mlr3′ https://mlr3.mlr-org.com/ |
FS, DR, ML, INTERP, VIS | Modern and extensible ML framework for developers and practitioners Provides a unified interface to many learners |
https://joss.theoj.org/papers/10.21105/joss.01903 |
| H2O.ai | Fast scalable ML API | R package ‘h2o’ Python: http://h2o-release.s3.amazonaws.com/h2o/rel-zermelo/3/index.html |
FS, ML, DR, INTERP, VIS | End-to-end engine specialized for big data Parallel distributed ML algorithms Automatic ML interface |
https://journals.plos.org/plosone/article?id=https://doi.org//10.1371/journal.pone.0238648 |
| iml | Interpretable machine learning | R package ‘iml’ | FS, ML, INTERP, VIS | Feature effects on the influence of predictions | https://joss.theoj.org/papers/10.21105/joss.00786 |
| LIME | Local interpretable model-agnostic explanations | R package ‘lime’ Python: https://github.com/marcotcr/lime |
FS, ML, INTERP, VIS | Explains individual predictions of a black box ML model Model-agnostic |
https://arxiv.org/abs/1602.04938 |
| inTrees | Interpretable tree ensembles | R package ‘inTrees’ | FS, ML, INTERP | Extracts, measures, prunes, selects and summarizes rules from a tree ensemble Specific to decision trees |
https://link.springer.com/article/10.1007/s41060-018–0144-8 |
| dtreeviz | Decision Tree Visualization | Python: https://github.com/parrt/dtreeviz | FS, ML, INTERP | Advanced visualizations Provides user-friendly interpretations of prediction paths Specific to decision trees |
https://explained.ai/decision-tree-viz/index.html |
| ranger | RANdom forest GEneRator | R package ‘ranger’ | FS, ML, INTERP | Fast implementations of random forests optimized for high-dimensional data Has advanced and convenient functions for decision trees |
https://arxiv.org/abs/1508.04409 |
| partykit | A toolkit for recursive partitioning | R package ‘partykit’ | FS, ML, INTERP | Can coerce tree models from different sources into a unified infrastructure Contains a variety of novel decision tree implementations Parameterization requires expertise |
https://dl.acm.org/doi/10.5555/2789272.2912120 |
FS, Feature Selection; DR, Dimensionality Reduction; ML, Machine Learning; INTERP, Interpretation Measures; VIS, Visualization Outputs. (*) Denotes whether the software Is microbiome-specific (as opposed to domain agnostic).
3.2.1. Random forests (RF)
Random forests [31] have been extensively deployed to solve a variety of problems in microbiota analysis. This method constructs multiple forests composed of decision trees by using the information contained in input features (abundance of microbial taxa, for example) to successively split samples based on their assigned (Y) values. The forests are guided by bootstrapping (drawing a random subset of samples with replacement, to be drawn multiple times) and a node splitting criterion that uses the information contained in a random subset of features to decide how to split each node in each tree (Fig. 2C), where the best split is selected based on a node impurity estimate (the likelihood of misclassifying new samples as a classifier) or the prediction squared error (as a regressor) [32]. The fact that hundreds or thousands of decision trees are being constructed in each forest using a subset of both samples and features allows an aggregate average of the predictions made at each terminal node (Fig. 2C). The combination of bootstrapping and then aggregating is jointly known as bagging (bootstrap aggregating) and frames RF as an ensemble learning method, where multiple forests are leveraged to obtain better performance than any single decision tree alone [33]. By this effect, RF are an ideal framework for consistently identifying “true effects” in complex and heterogenous data (multiple feature types; numerical or categorical). Additional factors that make RF appealing in practice is that they are an off the shelf, computationally tractable and top performing classifier that are robust to outliers, inherently noisy and non-linear data (such as metagenomic), and errors in manually curated response labels [34], [4]. Using SML with highly dimensional data with limited numbers of observations, such as microbial community data, can lead to overfitting. The RF method is less prone to overfitting than other SML methods, which contributes to its appeal in microbial community analysis [35]. Lastly, decision tree models in general are considered interpretable in their evaluation as they aid in extracting meaningful information from RF models [36] (Fig. 3).
Fig. 3.
Depiction of performance-interpretability trade-off and random forests interpretation. Note that these figures are fictional and are not based on experimental quantification (the axes in this figure lack meaning). (A) Performance-interpretability tradeoff of commonly deployed algorithms in microbiome research. However, in practice, the models characterized here tend to varying degrees of accuracy and interpretability based on experimental procedure. Had a plot been generated from experiment, model choice and complexity could vary such that inconsistent illustrations could arise. By way of example: tuning models to become more accurate could result in the belief that more accurate models are less interpretable and may not respect whether the model infrastructure supports inherently easier interpretation. (B) Hypothetical extraction of ‘association’ rules that measure frequent microbial community member interactions from fictional decision tree ensembles (tree1, …, tree k) for low error predictions of Ŷ. Additionally diagramed is a feature ‘importance’ schematic that scores each feature on its relative importance in making predictions of Ŷ
3.2.2. Gradient boosting (GB)
Gradient boosting [37], when used for decision trees, is an ensemble method that uses a process called boosting to combine individual learning algorithms (decision trees) successively to arrive at a strong learner. Gradient boosted trees contrast RF as an ensemble learner in that each decision tree is constructed in series in attempt to reduce the errors of the preceding tree, rather than in parallel. In addition, each tree built in GB is a fixed size and is fit on the original data, instead of bootstrapping samples as done in RF. Similar to RF, both numerical and categorical features can be used, but may be harder in practice to find optimal tuning parameters for a good model fit, such as the number of tree estimators. This method in particular is sensitive to outliers but efficient for both classification and regression, with reports of achieving similar or better accuracy to RF [38].
3.2.3. Support vector machines (SVM)
The goal of an SVM [39] is to find the best generalized line separation of response labels (Y) through a hyper-plane that maximizes the margin between different values of Y (or each class) in the label data. A decision boundary is drawn such that each class is separated while keeping maximum distance from the closest samples as possible (called support vectors that dictate this decision boundary) (Fig. 2B). SVMs are in the category of linear discriminant SML techniques. Although, if a hyperplane cannot justify the separation between classes with a clear margin of separation (in the case of non-linear metagenomic data), a so called “kernel trick” for a nonparametric form of SVM can be introduced to transform the data and satisfy a non-linear separation [40]. Several factors contribute to the success of SVM in microbial community analysis in that it is effective in the high-dimensional nature of the data, where X > N (or the number of members of a microbial community as features is larger than the sample set) and that it is also computationally tractable since the decision function only uses a subset of the data. These models can handle various feature types but can be inherently hard to interpret as they do not directly provide probability estimates in their evaluation.
3.2.4. L2 regularized logistic regression
Regularization is a technique used to reduce overfitting. For example, if a model is parameterized to learn every small bit of information in the structure of the microbial community composition under a given set of labels in training, it may not generalize well to make predictions on samples collected and processed outside of the training set and is considered overfit. Ridge (L2) regression [41] satisfies a model that reduces variance without increasing bias and is achieved by placing restrictions on the complexity of parameters (i.e. where to ultimately draw the decision boundary to separate response labels). This technique adds information to features used in training the model and by adding a penalty term to a loss function (estimation of how wrong the relationship is between X and Y), enables a constraint on parameter complexity so as to not capture every specific detail of the training data. Ridge regression can be used for both classification and regression but can be computationally expensive in the case of large input feature space.
3.2.5. Neural networks
Neural networks [42] use a hierarchical model building architecture where multiple structured networks of interconnected nodes (neurons) are constructed with weights attached at each edge of the network to facilitate mapping inputs of X to responses Y (weights being parameters to define strength of connection, for example) (Fig. 2D). Networks are interconnected through a feed-forward propagation mechanism, where each neuron receives input from preceding neurons. The network starts from input layers (microbial taxa feature set; X1, X2, …, Xi), that are linked to each neuron in the one or many hidden layers that use a backpropagation algorithm to maximize the weights placed at each neuron to improve predictive power. This process is iterative, where the last hidden layer is met by an output layer to produce a predicted response output (Y) (Fig. 2 D). Neural networks are very dynamic in their ability to identify intricate structure in very high-dimensional and complex datasets, making them a tractable technique to investigate the role of microbes in complex settings [43]. Neural nets are often referred to as “black box” methods as it can be difficult to interpret how decisions are made.
3.2.6. Deep vs. shallow learning
Deep learning is a family of both unsupervised and supervised techniques that belong to the class of neural networks (Fig. 2D). Despite shallow networks (dependent on number of layers), all non-deep learning methods such as those summarized above can be qualified as shallow learners. Whereas deep learning methods automatically alter raw input features by successively extracting abstractions of the data to be used as more discriminative features to the learning process, shallow learners are more of a manual process that depend on domain knowledge for a reduced selection of features that would serve as good inputs for a model to make accurate predictions with (i.e., which microbial taxa are differentially abundant between response labels).
Shallow learners can also benefit from feature engineering, where new features are handcrafted as composites, or abstract representations of multiple raw features using heuristics of the domain problem (i.e., agglomerating multiple high-resolution taxonomic features (ASVs) into a single lower resolution feature (Phyla)).
Although deep learning has shown to create models that are more accurate compared with shallow learning methods for microbiome-based prediction tasks [44], the models often sacrifice interpretability or understanding of the inner logic behind the predictions, which, for microbial-based applications can be rewarding in addition to predictive accuracy. An example of learning performance-interpretability trade-off is displayed in Fig. 3A.
4. Advantages of machine learning vs. classical statistics for microbial community data
Microbial ecology has for long relied on traditional statistical analyses to summarize data, test hypotheses, and to interpret interactions between features and responses on microbial datasets [45]. However, researchers and developers are starting to realize the enormous potential for machine learning in the microbial realm. ML methods have some advantages over standard statistical methods. A principal distinction between statistical models and ML is that the goal of the former is to describe and infer the relationships between variables, whereas the latter is designed to optimize the ability to predict an outcome on an external dataset. For example, typically SML will use a training set (supplied labels) to learn patterns associated with an outcome and a test set (hidden labels) to determine the performance of the model. On the other hand, statistical models are primarily interested in determining the relationship of the values to the outcome and unlike many studies that use ML, most do not require partitioning the data to measure performance.
Classical statistical analysis presents microbial ecologists with a two major issues: (1) the assumption that features of metagenomic data are independent and identically distributed is often harmed through molecular methods of sample processing and sequencing; and (2) that data with NGS imposed characteristics [46] such as being high-dimensional (number of data points is large), sparse (contain a lot of zeros), and compositional (feature set of microbial taxa may be co-abundant and are a part of a unit sum) often cannot be met by specific assumptions in classical statistics [47]. Many machine learning methods, such as the ones summarized above, can accommodate these dynamics of marker-gene data for a robust interrogation of the complex association patterns in microbial communities.
Some of the benefits ML has over classical statistics is that it is particularly effective in identifying subtle variation in microbial community structure and can identify specific bacterial taxa that underlie prediction of a conditional outcome. Another strength of ML is its ability to model a non-linear combination of bacterial count data and environmental parameters (a feature space resembling the real-world system) that do not need to assume complex transformations or preprocessing, which are challenging to molecular data.
However, since ML can operate without explicit user-instruction, is highly configurable, and requires a considerable amount of data, the tendency of these methods to overfit data are often overlooked. ML interpretation is also model-specific, meaning that some ML algorithms have easily understandable metrics that can be used to evaluate how the model arrived at the prediction (random forests), while some only provide vague accuracy statistics (neural networks). A consequence of these less interpretable ‘black box’ machine learning methods is that they may leave the user without the utility to uncover associations that underlie predictions, or to access probability thresholds of why certain observations were grouped to a particular response output.
We urge that there is no best use scenario when it comes to ML, and that individual researchers should select methodologies that are consistent with the specific domain problem, the questions being asked, and based on available data. If the goal of the research is to build a predictive understanding of an outcome based on microbial community data, SML has appeal since these algorithms are tailored to optimize predictive accuracy. However, if the goal is to relate specific microbial groups to particular outcome, classical statistical models have utility as well. Statistical models have strengths in microbial community analysis, but SML can provide a research strategy that can be based on less a priori assumptions of the data (such as formulating decisions based on predefined significance level) and more emphasis can be placed on ML to identify intricate associations and confounding variables that may be hard to detect but are often responsible for cause-effect.
In practice, ML can perform surprisingly well on datasets that are sampled from and represent messy real-world systems, such as the human body, soil, and water [48], [49], [50] and demonstrates superiority over traditional multivariate statistics in analyzing metagenomic data. In addition to these benchmarks, there is an increase in the development of microbiome-specific 'pipelines' that have user-friendly ML implementation and can be accessed through web-interfaces, the statistical compute language R [51], or Python [52]. A collection of methodologies is described in Table 1 and although not exhaustive, mentions microbiome-specific or domain-agnostic procedural extensions of predictive data analysis, such as interpreting and visualizing model outputs, as will be described moving forward.
5. Optimizing model construction and evaluation
In most domains, input features can be challenging and economically expensive to obtain. In the case of marker-gene analysis there is often an overabundance of features as a result of how high-throughput sequencing platforms capture genetic diversity within samples. It is therefore the goal of those using machine learning on microbiome data to consider feature selection methods to identify and remove non-informative, noisy or redundant features. As opposed to using every available feature in training a model, carefully selecting features may lower the cost of computation, reduce the complexity of the model for easier interpretation, and in some cases improve generalized predictive performance of the model.
In most cases, it then becomes tractable to understand microbial community data at a deeper and more targeted level, since feature selection allows for easier evaluation of the relationship between each input feature (i.e., as a microbial taxa) to a response label, or whether any features are used together to drive predictions. In addition to its predictive capabilities, ML can be used as a powerful data mining tool and to access a translational component of data, such as assessing whether feature-response label linkages in a model correspond to similar conditions in the real-world system after which the model is constructed. A noteworthy caveat of using SML in translational research is that it would require subsequent testing and hypothesis validation independent of the modeling procedure to conclude such relationship, since this initial interpretation is at the level of the model only.
The use of ML in microbiome research is motivated by a range of research questions and expected outcomes of modeling. This makes ML a very dynamic approach to predictive and exploratory modeling with many user defined parameters to be considered for each objective. Many of the ‘pipelines’ described in Table 1 enforce optimal parameter tuning of ML and associated post-hoc analysis that enables more of an understanding of microbiome-specific research questions; however, it should be noted that the more informed a researcher is of how parameterization benefits their domain problem and research questions, the better. Likewise, as ‘pipelines’ offer more customization to allow more user-defined decision making, there calls for an increase in knowledge of the broadly applicable methodologies for predictive data analysis.
Accessibility of evaluation metrics that aid this interpretation may depend on which learning method is used. It is integral to consider this at the point of model selection in order to optimize ML for microbial community analysis. The remainder of this section will describe various techniques for feature selection, preferred model evaluation metrics and post-hoc model interpretations, with consideration of why particular methods may be better for certain problems.
5.1. Exploring feature selection methods
It is often the case that features in microbiome data greatly exceed the number of samples, which can lead to a model overfitting, provides overoptimistic model evaluations, and may limit cross-study comparison [53]. Feature selection methods generally dictate how well a model generalizes to novel input data by allowing for fewer and more discriminative features that maximize performance. This section discusses three main categories of feature selection: filter methods, wrapper methods, and embedded methods.
Filter methods are typically a pre-processing step performed outside of the modeling procedure that statistically measure and score correlations (i.e., univariate or multivariate: Spearman’s rank correlation [54], MANOVA [55]) between input features so that only those passing some relevant criteria can be considered for downstream modeling. Although filter methods are advantageous in that they are easy to parameterize, computationally inexpensive and scalable, they can be challenging for the following reasons: (1) choosing a specific method assumes prior assumptions about the relationships in the input feature space (2) filter methods become challenging when trying to satisfy a specific research question and account for potential feature heterogeneity or the multicollinearity and complex covariance structure of microbial community data and (3) since filter methods are done prior to modeling, they place no consideration on whether a specific ML model would maximize performance using the reduced set of features.
Wrapper methods repeatedly construct models (e.g., classifiers) by iteratively adding (forward selection), removing (backwards elimination) and ranking (recursive elimination) features to search for an optimized combination that improves or marginalizes performance of preceding models. Since wrapper methods are a repeated learning process that can exhaust through features, it is not as ideal as filter methods because it becomes computationally expensive with the high-dimensional structure of metagenomic data.
Embedded methods are a more computationally tractable approach to feature selection by relying on the algorithm itself to inform a ‘useful’ feature. As discussed earlier, decision tree algorithms GB and RF satisfy the objective of modeling a problem and inherently have a built-in feature selection method that operates during model training. Importantly, this provides embedded methods the ability to search the full feature space, that is, if the algorithm infrastructure is in place to handle such high-dimensional data. To this extent, many feature-response associations have the potential to be discovered that would otherwise have been disregarded had data been pre-processed with restrictive assumptions prior to modeling with a filter method, or if certain potentially important features were left out of a resulting wrapper method if not considered a part of the ‘optimal feature subset’. For these reasons, and on the basis of computational tractability, embedded methods are an ideal practical feature selection method for optimizing microbial-based ML models.
Despite not being as extensively reported in studies that profile the microbiome, new feature selection regimes that are more biologically motivated, such as taxonomy-aware hierarchical feature engineering (HFE) [53] are starting to gain traction and may be ideal for when embedded methods struggle with using the full search space when using very high-dimensional datasets.
5.2. Evaluating and interpreting estimator performance
For binary classification tasks (assigning samples to one of two response labels), receiver operating characteristic (ROC) [56] curves can be used to assess performance of the model at various decision thresholds by plotting TPR (true positive rate – sensitivity) as a function of the FPR (false positive rate – 1-specificity). By extension, computing the area under the ROC curve (AUC) [57] can provide a measure of how well the model could discriminate Ŷ. AUC can range from 0.5 (separation of Ŷ was no better than random chance) to 1.0 (perfect separation of Ŷ), assumes that the cost of misclassifying each response label is equal and is sensitive to when response labels are skewed.
For multiclass classification (assigning samples to more than two responses), we advocate that logistic loss (logloss), also known as cross-entropy loss be used, as it measures the quality of predictions using the probabilistic confidence of sample separation into respective Y labels and penalizes incorrect or uncertain predictions [58]. A low logloss is preferred and reflects the distribution of the certainty of predictions and like AUC, is also sensitive to when response labels are skewed.
When predicting continuous labels in regression, mean squared error (MSE) is a preferred metric that averages the squared difference of the known continuous Y value and the predicted value of Ŷ. This metric is desired because it is differentiable, which can be optimized better. A lower MSE is favorable as it measures how close a fitted line is to the data points.
Often in practice, these metrics are computed for predictions on a single cross-validated model rather than on separate models from splitting the same dataset into a training and testing set. Cross-validation is a method that holds out samples which are later used to validate prediction accuracy during the learning process and generally leads to models that are less biased and not as overoptimistic as compared to train/test splitting [36].
While accuracy measures as described above are useful, they cannot be used to explain why a model made a certain prediction. Typically, many algorithms have ad-hoc implementations for model interpretation, such as measuring the ‘importance’ of each feature or multiple features to response labels. In RF, for instance, this is usually done by permuting, or re-arranging the values of input features during the learning process, such that, if a feature is ‘important’, changing its values will lead to increased error rates in aggregated predictions. This process, also called variable importance, is often guided by model-specific information, such as the correlation structure between predictors, and usually scales features to have a maximum value of 100 to indicate the relative importance (Fig. 3B).
5.3. A use case summary of current software implementations
Table 1 describes recently developed and commonly employed toolkits designed to assist researchers through the steep learning curves of predictive data analysis. For instance, SIAMCAT [59] and BiomMiner [60] are comprehensive ML ‘pipelines’ tailored to clinical microbiome datasets. These pipelines include the ability to perform cross-study comparison, automatic tuning of optimal parameters for dimensionality reduction, feature selection and predictive modeling, provide post-hoc interpretable measures of feature ‘importance’, and can demonstrate the influence of different parameter choices on resulting classification accuracy.
Another variety includes web-based tools such as MicrobiomeAnalystR [61], which is an ML-toolkit deployed through a web-interface to assist users who may lack computational expertise or resources. MicrobiomeAnalystR provides real-time comprehensive analysis reporting, recommendations, and visual comparisons of an implemented model to public datasets. Moreover, commonly used analysis pipelines such as QIIME2 [62] and mother [15] include implementations of SML algorithms such as RF and SVMs.
Another implementation of ML is DeepMicro[ref], which has been shown to perform well when using the microbiome to predict various diseases through deep representation learning. This method uses autoencoders to transform high-dimensional microbiome data into low-dimensional representations, then applies classification algorithms on the various learned representations. This method accelerates model training and parameter tuning by significantly reducing dimensionality of the microbiome profiles.
Many re-implementations of the original RF, namely cforest [63] and ranger [64], include novel resampling schemes for more unbiased estimates of prediction accuracy, measures of feature importance, and for computational efficiency on high-dimensional data. By extension, tools like inTrees [65] and dtreeviz can be used for ad-hoc knowledge discovery, such as to interpret predictions of black box models. These systems are designed for extracting, measuring and summarizing rules that govern splitting criteria in decision tree ensembles. A brief schematic illustration of this process is displayed in Fig. 3B.
Other software such as LIME [66] and iml [67] seek to offer robust, model-agnostic explanations. These include measuring feature effects on the influence of predictions, and in the case of decision tree algorithms, approximating black box predictions by constructing less complex 'surrogate’ trees that provide accessible interpolations.
As comprehensive as some of the ML-toolkits described above may seem, they are still limited in their customization and cross-platform implementation. Given these constraints, more advanced users may consider domain agnostic end-to-end ML platforms with parallelized implementations for predictive data analysis, such as scikit-learn [68], keras [69], caret [70], and H2O.ai [71]. These ‘pipelines’ enable more customization for parameter tuning and parameter choices, allow multiple models to be built from scratch and ensembled using the same re-sampling parameters and provide more access to raw model contents (i.e., indexed predicted probabilities during cross-validation, as opposed to just an accuracy metric). Although less intuitive, these methods allow more in-depth analysis than the more automated, user-friendly microbiome-specific platforms that are built for execution efficiency on smaller ML workloads, rather than for scale.
Nevertheless, the domain specific tools described in Table 1 are useful for putting into context the biological relevance of the domain problem, allow fast and easy exploration, and serve as a good starting point for microbiome-based predictive data analysis. These ‘pipelines’ are also beneficial for those with a more advanced understanding of ML. While often the choice of pipeline comes down to optimization and comfortability as well as if visual outputs are necessary for data reporting, it is best practice to choose methodologies handle the characteristics of microbiome data and are interpretable, especially if the goal is to translate the research into diagnostics.
Aside from software implementations, it is worth mentioning that there are a few public repositories for curated microbiome datasets and related metadata from some of the most cited studies in the field of microbial ecology: GMRepo [72], MLrepo [73], curatedMetagenomicsData [74] and MicrobiomeHD. These public repositories can be used to practice ML, benchmark new approaches, and for cross-study comparison.
5.4. Machine learning for classification of human disease from microbiome data
Microbiome data has been used to link microbial community composition and disease state [75]. Diseases such as Inflammatory Bowel Disease, metabolic syndrome, obesity, hypertension, cancer, neurological diseases, among others have been linked to the human microbiome [76]. Many studies have sought to statistically link diversity metrics such as alpha diversity or abundance of particular taxonomic groupings with disease states [77]. However, as sample numbers have increased, these broad level relationships often do not hold up. For example, in studying obesity, it had been proposed that some taxonomic markers (Firmicutes and Bacteriodetes) [78] as well as decreased alpha diversity [79] were indicators of obesity. Reanalysis of this data, aggregating data across studies, demonstrated that some of these coarse measures for the microbiome did not adequately predict obesity across larger datasets [80]. The complexity and interpersonal variation within the microbiome of humans has complicated the use of the broad level metrics.
SML has been proposed as an alternative to other methods for associating microbiome with an outcome as SML may be a more robust analysis tool for predicting disease state based on microbial community profiles. Table 2 summarizes key studies employing SML to link microbial community data to a specific outcome to illustrate how SML has been previously used and highlight some considerations in employing SML to study microbial communities. One recent study used fecal microbial community profiles to predict the presence of colonic neoplasia [76]. The use of SML allows for models optimized for prediction of disease to be trained and validated on out of training set data that will enable more robust determination of the link between microbial communities and health states. This study explored multiple SML methods for this classification problem including L2 Regularized Linear Regression, RF, and SVM. This study found that many methods resulted in highly performing models, with RF performing the best (AUROC curve 0.695). Other models such as L2-regularized logistic regression, XGBoost, L2-regularized SVM with linear and radial basis function kernel all performed similarly with AUROC between 0.668 and 0.680. Interestingly, they found that while RF performed the best out of the tested models, some more interpretable approaches, such as L2-regularized logistic regression, had similarly high accuracies. These authors proposed that while more complicated models such as RF may result in higher accuracies, interpretability is an important factor in considering study design and the application of SML.
Table 2.
Studies using Machine learning in microbial ecology and microbiome studies.
| System | Classification | Input data | Number of samples | Method | Training and Validation | Reference |
|---|---|---|---|---|---|---|
| Human | Colonic screen relevant neoplasias | 16S rRNA | 172 patients with normal colonoscopies, 198 with adenomas, and 120 with carcinomas | L2-regularized logistic regression, L1- and L2-regularized SVM with linear and radial basis function kernels, a decision tree, RF, and gradient boosted trees | 80% Training, 20% Validation, 20% Test, Five-fold cross validation | Topçuoğlu et al 2020 [76] |
| Human | Personalized postprandial glycemic response | 16S rRNA | 900 samples, 800 in training100 in validation | Gradient boosted trees | 800 samples used and validated with a leave one out cross validation scheme, 100 sample validation cohort | Zeevi et al 2015 [82] |
| Environmental | Crop Productivity | Shotgun metagenomic | 12 samples | RF | 10 samples as training set, 2 samples as validation set (all combinations of the 12 samples) | Chang et al 2017 [89] |
| Environmental | DOC level | 16S rRNA | 302 samples | feed-forward neural network regression and RF | 257 samples as training set and 51 as test set | Thompson et al 2019 [90] |
| Environmental | Environmental quality status associated with salmon farms | SSU RNA (bacteria and ciliates) | 152 across seven salmon farms | RF and SVM | Models trained on six of the salmon farms and tested with the seventh | Cordier et al 2018 [91] |
| Environmental | Environmental impacts of marine aquaculture | SSU RNA (five marker genes – one bacterial, one foraminiferal, and three universal eukaryote) | 144 Sediment samples | RF | Models trained on four of the salmon farms and tested with the other farm | Frühe et al 2020 [92] |
| Environmental | Environmental quality status associated with salmon farms | Bacterial 16S rRNA | 12 sediment samples collected from six sites | RF | 12 samples validated with a leave one out cross validation scheme | Dully et al 2020 [93] |
| Environmental | Contamination state (uranium, nitrate, oil) | 16S rRNA | 93 samples for ground water contamination, 42 samples for oil contamination | RF | Performance metrics were determiened from a confusion matrix based on out-of-bag predictions | Smith et al 2015 [87] |
| Environmental | Glyphosate presence | 16S rRNA | 32 16S rRNA gene samples and 32 16S rRNA samples | ANN and RF | 32 samples used and validated with a leave one out cross validation scheme | Janßen et al 2019 [86] |
| Forensic | Postmortem Interval | 16S rRNA | 144 sample swabs were taken from a total of 21 cadavers | SVR, K-neighbor Regression, Ridge Regression, Lasso Regression, Elastic Net Regression, RF regression, Baysiean Ridge Regression | 80% of samples for training set and 20% of samples for validation set | Johnson et al 2016 [100] |
| Forensic | Postmortem Interval | 16S rRNA | 176 samples | RF, SVM, ANN | 70% for training and 30% for testing. Accuracy determined by mean absolute error and goodness of fit of 15 models | Liu et al 2020 [101] |
| Forensic | geospatial location (port of origin) | 16S rRNA | 1,218 samples | RF | repeated k-fold cross validation (k 10 with 3 repeats) | Ghannam et al 2020[50] |
The use of the microbiome to personalize treatment was further investigated in another study examining the interpersonal variation in the changes in blood glucose observed following meals (postprandial glycemic response (PPGR)). Previous studies have shown high interpersonal variability in PPGR in response to the same food [81]. This suggests that some foods might result in a high PPGR in some patients and low PPGR response in others. This finding coupled with the high interpersonal difference in microbiomes, led Zeevi et al (2015) to develop a classifier that could relate foods with the microbiome and other physiological data to accurately predict the PPGR of patients [82]. The authors used GB for regression to relate patient data, information about the meal, and the patient’s microbiome to predict the PPGR. This study revealed that the SML models that incorporated microbiome data were able to more accurately predict PPGR than meal carbohydrates or meal calories alone. The combined microbiome and patient data model’s prediction of PPGR was correlated with the measured PPGR with a Pearson correlation of 0.68. The model was trained using a cohort of 800 individuals and validated on a different 100 individuals. This type of analysis using ML with patient and microbiome information allows for a more tailored treatment that accounts for the high interpersonal variation that is often observed with human disease [83].
5.5. Machine learning for classification in environmental monitoring
In addition to prediction or disease state in the human system, coupling SML and microbial community profiling of microbial communities in the environment shows promise for the purpose of environmental monitoring [84]. Just like in the human environment, microbes in soil, water, or air can rapidly respond to changes in their environment. These changes in microbial community composition can often occur in a predictable manner. SML has been used in both natural and industrial settings to use microbial information to aid in predicting environmental quality [85], contamination state [86], [87] as well as rates of various processes including copper bioleaching [88], Previous studies have used microbial biomarkers as indicators of particular environmental processes or outcomes. Indicator species analysis has been used to identify taxa that are related to particular phenomena or treatments that could be used as biomarkers for that phenomena. However, like differential abundance analysis, indicator species is performed by analyzing the prevalence and abundance of individual features in different categories and is not able to identify complex interactions between microbes in the dataset and the possibility that large groups of microbes may respond to the treatment.
ML is gaining popularity in predicting environmental phenomena from environmental microbial community data to develop and predict environmental health indices. One such study used SML to relate the microbial community present in agricultural soil with crop productivity [89]. In this study the authors coupled SML with metagenome wide association studies to identify potential differences in the microbial communities that were related to crop productivity. RF models built from metagenomic data were able to predict the crop productivity with an accuracy of 0.79. Another study sought to relate dissolved organic carbon (DOC) with microbial community composition [90]. RF and artificial neural networks (ANN) were used to construct models to predict the DOC concentrations of leaf litter based on microbial community composition. The models from this study were reasonably accurate with the ability to predict DOC correlating with observed DOC with a Pearson correlation coefficient of 0.636 and 0.676 for the feed-forward ANN and the RF models respectively. Interestingly, these researchers compared the important features identified through SML with indicator species identified in indicator species analysis. While they found some overlap in the features identified in both methods, only about 30% of the features were shared between indicator species analysis and RF. This suggests that ML often uses distinct features for classification than what would be identified through differential abundance or indicator species analysis and may be able to more sensitively identify groups of features that are related to an outcome.
Biotic indices have been used to assess environmental health as biotic organisms are impacted by the overall ecological quality status of an environment and may be more sensitive than measurement of abiotic factors. Therefore, various organisms have been proposed as indicators of environmental health. SML can be used to associate environmental genomic profiles with environmental quality status, which is commonly used by regulators to guide decision making in restoration and environmental monitoring [85]. A number of studies have provided a framework for the use of SML to identify patterns in microbial eukaryote and bacterial communities to predict biotic indices and environmental quality status using salmon farming as a test case. Cordier et al. (2018) [91] used various marker genes targeting the small subunit rRNA for bacteria, ciliates, and universal eukaryotes to compare the performance of SML to predict the environmental quality status and biotic indices compared to using environmental DNA to measure known indicator taxa. They found that SML outperformed the use of metazoan-assigned OTUs. The predictions obtained from metazoan-assigned OTUs had kappa values between 0.211 and 0.569, whereas the SML models had kappa values raging from 0.755 to 0.881. Following on from this study, Frühe et al (2020) [92] compared the performance of SML with standard IndVal approach for prediction of environmental status. The indicator species approach directly from OTUs/ASVs has appeal due to there not being a need to assign taxonomy to the OTUs/ASVs like in the metazoan-assigned OTUs approach. This study found that SML outperformed the IndVal approach for prediction of environmental status. Furthermore, bacterial communities were better able to predict environmental quality status salmon farming compared to ciliates. These studies illustrated the utility of SML for environmental biomonitoring. However, these studies used training and validation data generated from the same lab. In order for this type of ML-coupled to molecular analysis approach to be used for environmental monitoring, there needs to be high replicability and generalization of the models. Therefore, Dully et al 2020 [93] performed an inter-lab validation study for the prediction of biological indices. In this study two series of samples were collected and split into technical replicates. From each site, biological replicates were also sampled. The authors of this study found that there was greater variability in diversity between biological replicates compared to technical replicates processed in each lab, which suggests that molecular methods can be standardized and have good replicability. Furthermore, SML models constructed from the two labs produced highly correlated data. These studies combine to demonstrate the promise, generalizability, and robustness of linking SML with environmental genomic data to assess environmental health status, which can guide decisions related to environmental health.
The prediction of environmental contamination is another growing area of interest in the application of SML and microbial ecology. Often contamination is identified in the environment through direct measurement of the contaminant of interest. While measuring of the contaminant is the gold standard for contaminant detection, often the contaminant may be present transiently. In these cases, the contaminant may not be detectable at the time of sampling. Smith et al (2015) demonstrated that RF could be used to predict the presence of uranium and nitrate contamination in groundwater [87]. This study demonstrates that a single set of microbial community profiles can be used to predict any number of response variables. Further, RF models were able to predict the presence of oil in the ocean with near perfect accuracy (F1 score of 0.98). Notably, RF could classify samples into no-oil, oil, and past oil contamination based on the microbial community alone. The past oil category contained samples that at one point in time had detectable levels of oil, but at the time of sampling, there was no detectable oil. This finding indicates that ML methods can identify patterns in the microbial community that are indicative of current and past contamination. The ability of the RF models to identify past contamination could be indicative of ecological resiliency and stability that allows microbial communities to maintain the signature of oil after the oil was no longer present.
The ability to predict contamination in the environment has been expanded to other systems including prediction of the herbicide glyphosate in the Baltic Sea [86]. In the Janßen et al (2019) study, the authors employed artificial neural networks and RF to predict the presence of glyphosate. Expanding on the previous work showing the ability of SML to predict contamination, Janßen et al (2019) identified important features through constructing a series of models leaving out individual features and monitoring the changes in accuracy of the models. This type of approach can be used to interrogate some of the more complex and less transparent approaches such as Artificial Neural Networks (ANNs). Another novel aspect of the work of Janßen et al (2019) is the use of a random forests proximity matrix as the dissimilarity measure in PCoA. This approach resulted in clearer separation of samples on the PCoA analysis compared to using a Bray Curtis dissimilarity matrix.
While these studies demonstrate the ability of ML to predict the presence of a specific contaminant in an environmental sample, other work has been used to predict more general properties such as environmental impacts of hydraulic fracturing as well as location and residence time of ballast water [94], [95], [96]. In both of these cases the specific relationship between the features and the output variable may not be clear. In other words, when predicting the presence of a specific contaminant, a single feature may increase or decrease in abundance in direct response to the contaminant due to the toxicity of the contaminant or ability of the contaminant to stimulate growth of the microorganism. These more generic phenomena may result in indirect impacts on the microbial community that are detectable using ML. These studies demonstrate that it is possible to detect and classify contamination both specifically and more generically using ML. Interrogation of the important features used in these classifiers may provide insights into specific biomarkers of contamination that could be used as tools for environmental monitoring.
In addition to the applied outcomes described above, SML has potential to be used to better understand the ecology of microorganism in the environment. Smith et al 2015 [87] demonstrated that microbial community composition as determined by 16S rRNA can be used to predict a diverse set of geochemical factors including pH, manganese and aluminum. Alneberg et al (2020) [97] also highlight the application of SML to predict the ecological niche of microbial groups with a focus on microbial communities from the Baltic Sea. The authors of this study use metagenomic binning to obtain 1962 metagenome assembled genomes (MAGs) representing the majority of prokaryotic diversity in in the Baltic Sea. These prokaryotic clusters demonstrated distinct ecological preferences along the various environmental gradients observed. Ridge Regression, RF, and GB were used to predict the niche gradient of the prokaryotic cluster based on the functional profile of genes found in each cluster. The authors of this study found that the predicted niche gradient agreed with the observed niche gradient with a Spearman’s rank correlation of 0.70 – 0.81. These studies highlight the fact that SML can be useful in identifying patterns in natural microbial communities and predicting the niche of an organism.
5.6. Microbial communities and machine learning for forensics
Microbes have been used for forensic applications for a long time. Normally microbial forensics is used to identify the source of particular organisms related to bioterrorism, disease, or contamination. However, it is possible to use microbial community composition as a tool for trace evidence [98], [99]. Previous work has shown the utility of microbial community composition in determining postmortem intervals (PMI). Various studies have examined the ability of the soil and skin microbiome to serve as a molecular clock for postmortem intervals. Other studies have used ML models constructed from skin microbiota to assess the PMI [100], [101]. Soil evidence has also been used as forensic information. In the same way that pollen can be used to identify the source of a particular soil sample, the microbial community in a soil sample may provide information about where that soil was derived. Metagenomic information from soils has been used to differentiate soil from different locations [102], [103]. These studies demonstrated that information contained within the microbial community from the soil sample could be used to identify the source of the soil. These studies used hierarchical clustering and non-metric multidimensional scaling (NMDS) to differentiate groups. More recently, SML has been applied to determining the geographic source of an ocean water sample based on the microbial community [50]. Ghannam et al (2020) [50] demonstrated that RF could be used to accurately differentiate the location of sampling of water from 20 different locations. This study is important in that it shows that SML can be used to identify important trace signals in the microbial community of water that can accurately distinguish between 20 diverse locations from around the world as well as specifically identify the location of collection within locations close in proximity to each other.
6. Summary and outlook
This review has sought to provide an overview of how ML has progressed the field of microbial ecology. Despite the unprecedented sophistication and promise of ML algorithms, there exist several outstanding issues that should be considered when applying ML to marker-gene datasets. Although ML models can be consistently constructed to produce high accuracy metrics on complex data, the underpinning decision support systems can often be largely black box methods of investigation where the rational and logic behind predictions are hidden behind layers that are challenging to interpret [104].
The large majority of studies using ML to investigate microbiome datasets gauge and validate hypotheses and report findings through performance and may apply post-hoc procedures to identify important biological taxa using variable importance metrics. However, due to the complexity of some modeling methods, inferring biological importance from feature importance could be problematic. Therefore, there is a need for increased interpretability in ML models used in microbial ecology studies. Often the learning algorithms applied to marker-gene datasets are developed and implemented for improved performance, rather than for model interpretation [104], [105]. In order to glean biologically meaningful data from these ML methods, it may be important to consider the choice of model with preference toward more interpretable algorithms as well as novel methods for interpreting models such as permutational approaches. Microbial ecology studies that demonstrate model transparency are limited to reporting single feature to response interaction or are overburdened by investigating feature contributions to each observation for accumulated local explanations of modeling procedures [48], [76], [106], [107].
There have been major improvements for model specific and model agnostic approaches for model interpretation [66], [108], [109], [110], [111], some were described in this review. However, these methods often cannot account for hidden heterogenous effects of the full feature space, which can reduce model fidelity and mislead researchers depending on algorithm selection.
Here, we argue that while methods for inferencing how single microbial community members influence single predictions are beneficial (local interpretations), appreciating the inner workings of multiple microbial community members and how they generally discern a group of the same response label is more robust and generalizable (global interpretation). In the context of microbial ecology, the lack of global interpretation techniques makes it challenging to inference on the basis of the full feature space and to identify all potential features that are interacting to most frequently to predict response labels with the least error. Often a condition is not attributable to a single feature, but multiple features. One of the strengths of ML is the ability to appreciate these groups of features in making a prediction. However, in interpreting a model, a focus on the importance of a single feature may limit the applicability to the real-world system that is being modeled (i.e., appreciating the full microbial community rather than subsets).
In high-risk domains like human health and biology, the ability to interpret and generalize a model has many downstream benefits, such as identifying biological relevance that support hypotheses of the system being investigated and the ability to extract actionable insights about the community of study. Many of the implementations described in this review seek to extract actional information from microbiome datasets that can be used in the clinic, environmental monitoring applications, and forensics. It is important that in implementing the use of ML-identified biomarkers in diagnostic application that there is a need for common acceptance and trust of the algorithms employed which lead to critical decisions relating to the microbiome [112], [113], [114], [115], [116].
While other disciplines of biology such as single-cell RNA seq, drug discovery and development, and neuroscience have attempted to bring interpretation to black box ML models [117], [118], [119], [120], [121], investigation into microbial ecology applying ML on marker-gene datasets is lagging behind. This is surprising since there has been a rapid expansion of microbiome related research that will continue to expand. With a lack of interpretation of ML in this field, fundamental dynamics of a microbial system will be left unreported.
It is notable to mention that as a result of the structure of marker-gene datasets from HTS platforms, Classification and Regression Trees (CART) algorithms continue to dominate the field of microbial ecology. However, deep learning is a promising approach to revolutionize how we investigate microbial communities. Considerations should be placed on whether deep learning is necessary to investigate metagenomic datasets, since, although the inner workings of neural networks are the focus of ongoing research [122], [123], they are some of the most notorious black box methods that lack interpretability. In some cases, it may be better to choose a model that can be more easily interpreted over a more complex model that has a higher performance metric.
There are still a number of open questions and considerations that need to be taken into account when considering the use of employing SML for monitoring and diagnostics. One of the first considerations is the need for sufficient replication in experimental design. Human microbiome studies have paved the way with high replication with hundreds of samples used in training algorithms. However, for environmental monitoring, sample collection is often costly, which can limit replication. In cases where sample replication is limited, some test and validation approaches may be more useful. For example, a leave-on-out validation strategy could be useful when replication is low and the splitting of samples into a training and test set would result in even less replication. Another consideration is sampling depth. As was discussed earlier, diversity estimates from sequencing data highly depends on sequencing depth. Therefore, it is important to ensure the diversity of the samples have been sufficient covered in constructing models to be used in SML. This is an example of how an understanding of ecological diversity measures and coverage estimates (e.g. Good's coverage) may be an important first step in determining if the obtained data is sufficient for development of SML models. Another important question that must be addressed is the level of accuracy that a model must obtain to be useful for its purpose. This question is a little more difficult to answer and depends highly on the domain problem. In certain domains higher accuracy may be required for a model to be of use. While 100% accuracy may not be achievable in noisy real-life environments, it is important to consider the level of accuracy that is needed. This may vary between medical diagnostics, forensics, and environmental monitoring applications.
If we are to move toward a translational framework for microbiome analysis where features extracted from ML models are used to inform development of particular treatments or monitoring approaches, it is important to have a thorough understanding of the interpretability of the models. It is also important to ensure that ML is used to complement other approaches for profiling microbial communities that confirm the choice of selected biomarkers. Overall, it is important to consider how ML models are interpreted and reported in situations where actionable insight can be extracted from modeling procedures and used to construct downstream molecular applications such as in health and environmental diagnostics.
CRediT authorship contribution statement
Ryan B. Ghannam: Conceptualization, Writing - original draft, Writing - review & editing, Visualization. Stephen M. Techtmann: Conceptualization, Writing - original draft, Writing - review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was sponsored by a DARPA Young Faculty Award D16AP00146.
References
- 1.Turnbaugh P.J., Ley R.E., Hamady M., Fraser-Liggett C.M., Knight R., Gordon J.I. The human microbiome project. Nature. 2007;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson L.R., Sanders J.G., McDonald D., Amir A., Ladau J., Locey K.J. A communal catalogue reveals Earth's multiscale microbial diversity. Nature. 2017;551(7681):457–463. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sunagawa S., Coelho L.P., Chaffron S., Kultima J.R., Labadie K., Salazar G. Ocean plankton. Structure and function of the global ocean microbiome. Science. 2015;348(6237) doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- 4.Knights D., Costello E.K., Knight R. Supervised classification of human microbiota. FEMS Microbiol Rev. 2011;35(2):343–359. doi: 10.1111/j.1574-6976.2010.00251.x. [DOI] [PubMed] [Google Scholar]
- 5.Larsen P.E., Field D., Gilbert J.A. Predicting bacterial community assemblages using an artificial neural network approach. Nat Methods. 2012;9(6):621. doi: 10.1038/nmeth.1975. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Y.H., Gallins P. A review and tutorial of machine learning methods for microbiome host trait prediction. Front Genet. 2019;10 doi: 10.3389/fgene.2019.00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lane D.J., Pace B., Olsen G.J., Stahl D.A., Sogin M.L., Pace N.R. Rapid-determination of 16s ribosomal-Rna sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stahl D.A., Lane D.J., Olsen G.J., Pace N.R. Analysis of hydrothermal vent-associated symbionts by ribosomal-rna sequences. Science. 1984;224(4647):409–411. doi: 10.1126/science.224.4647.409. [DOI] [PubMed] [Google Scholar]
- 9.Norman R. Pace, David A. Stahl, David J. Lane, Gary J. Olsen. The Analysis of Natural Microbial Populations by Ribosomal RNA Sequences. In: K.C. M, editor. Advances in Microbial Ecology Advances in Microbial Ecology. vol 9. Boston, MA: Springer; 1986.
- 10.Sogin M.L., Morrison H.G., Huber J.A., Mark Welch D., Huse S.M., Neal P.R. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA. 2006;103(32):12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Huntley J., Fierer N. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. Isme J. 2012;6(8):1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hazen T.C., Rocha A.M., Techtmann S.M. Advances in monitoring environmental microbes. Curr Opin Biotech. 2013;24(3):526–533. doi: 10.1016/j.copbio.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 13.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 14.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microb. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Preheim S.P., Perrotta A.R., Friedman J., Smilie C., Brito I., Smith M.B. Computational methods for high-throughput comparative analyses of natural microbial communities. Method Enzymol. 2013;531:353–370. doi: 10.1016/B978-0-12-407863-5.00018-6. [DOI] [PubMed] [Google Scholar]
- 18.Lozupone C., Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microb. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12) doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paulson J.N. metagenomeSeq: statistical analysis for sparse high-throughput sequencing. Bioconductor package. 2014;1 [Google Scholar]
- 21.Sathya R., Abraham A. Comparison of supervised and unsupervised learning algorithms for pattern classification. Int J Adv Res Artif Intell. 2013;2(2):34–38. [Google Scholar]
- 22.Hastie T., Tibshirani R., Friedman J. Springer Science & Business Media; 2009. The elements of statistical learning: data mining, inference, and prediction. [Google Scholar]
- 23.Silva V., Tenenbaum J. Global versus local methods in nonlinear dimensionality reduction. Adv Neural Inf Process Syst. 2002;15:721–728. [Google Scholar]
- 24.Forgy E.W. Cluster analysis of multivariate data: efficiency versus interpretability of classifications. Biometrics. 1965;21:768–769. [Google Scholar]
- 25.Ramette A. Multivariate analyses in microbial ecology. FEMS Microbiol Ecol. 2007;62(2):142–160. doi: 10.1111/j.1574-6941.2007.00375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lozupone C., Lladser M.E., Knights D., Stombaugh J., Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5(2):169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lvd M., Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9(Nov):2579–2605. [Google Scholar]
- 28.Kobak D., Berens P. The art of using t-SNE for single-cell transcriptomics. Nat Commun. 2019;10(1):1–14. doi: 10.1038/s41467-019-13056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belkina A.C., Ciccolella C.O., Anno R., Halpert R., Spidlen J., Snyder-Cappione J.E. Automated optimized parameters for T-distributed stochastic neighbor embedding improve visualization and analysis of large datasets. Nat Commun. 2019;10(1):1–12. doi: 10.1038/s41467-019-13055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu X., Xie Z., Yang Z., Li D., Xu X. A t-SNE based classification approach to compositional microbiome data. Front Genet. 2020;11:1633. doi: 10.3389/fgene.2020.620143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breiman L. Random forests. Machine Learn. 2001;45(1):5–32. [Google Scholar]
- 32.Biau G., Scornet E. A random forest guided tour. Test. 2016;25(2):197–227. [Google Scholar]
- 33.Louppe G. Understanding random forests: From theory to practice. arXiv preprint arXiv:14077502. 2014.
- 34.Mentch L, Zhou S. Randomization as regularization: A degrees of freedom explanation for random forest success. arXiv preprint arXiv:191100190. 2019.
- 35.Breiman L. Manual on setting up, using, and understanding random forests v3. 1. Statistics Department University of California Berkeley, CA, USA. 2002;1:58.
- 36.Probst P., Boulesteix A.-L., Bischl B. Tunability: importance of hyperparameters of machine learning algorithms. J Mach Learn Res. 2019;20(53):1–32. [Google Scholar]
- 37.Chen T, Guestrin C, editors. Xgboost: A scalable tree boosting system. Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining; 2016.
- 38.Wang X.-W., Liu Y.-Y. Comparative study of classifiers for human microbiome data. Med Microecol. 2020 doi: 10.1016/j.medmic.2020.100013. 100013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suykens J.A., Vandewalle J. Least squares support vector machine classifiers. Neural Process Lett. 1999;9(3):293–300. [Google Scholar]
- 40.Soman K., Loganathan R., Ajay V. Machine learning with SVM and other kernel methods. PHI Learning Pvt Ltd. 2009 [Google Scholar]
- 41.Hoerl A.E., Kennard R.W. Ridge regression: biased estimation for nonorthogonal problems. Technometrics. 1970;12(1):55–67. [Google Scholar]
- 42.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 43.Fiannaca A., La Paglia L., La Rosa M., Renda G., Rizzo R., Gaglio S. Deep learning models for bacteria taxonomic classification of metagenomic data. BMC Bioinf. 2018;19(7):198. doi: 10.1186/s12859-018-2182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qu K., Guo F., Liu X., Lin Y., Zou Q. Application of machine learning in microbiology. Front Microbiol. 2019;10:827. doi: 10.3389/fmicb.2019.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buttigieg P.L., Ramette A. A guide to statistical analysis in microbial ecology: a community-focused, living review of multivariate data analyses. FEMS Microbiol Ecol. 2014;90(3):543–550. doi: 10.1111/1574-6941.12437. [DOI] [PubMed] [Google Scholar]
- 46.Gloor G.B., Macklaim J.M., Pawlowsky-Glahn V., Egozcue J.J. Microbiome datasets are compositional: and this is not optional. Front Microbiol. 2017;8:2224. doi: 10.3389/fmicb.2017.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Økland R.H. Wise use of statistical tools in ecological field studies. Folia Geobotanica. 2007;42(2):123. [Google Scholar]
- 48.Aasmets O., Lüll K., Lang J.M., Pan C., Kuusisto J., Fischer K. Machine learning reveals time-varying microbial predictors with complex effects on glucose regulation. bioRxiv. 2020 doi: 10.1128/mSystems.01191-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Belk A., Xu Z.Z., Carter D.O., Lynne A., Bucheli S., Knight R. Microbiome data accurately predicts the postmortem interval using random forest regression models. Genes. 2018;9(2):104. doi: 10.3390/genes9020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghannam R.B., Schaerer L.G., Butler T.M., Techtmann S.M. Biogeographic patterns in members of globally distributed and dominant taxa found in port microbial communities. Msphere. 2020;5(1) doi: 10.1128/mSphere.00481-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Team RC. R: A language and environment for statistical computing. Vienna, Austria; 2013.
- 52.Van Rossum G., Drake F.L., Jr. Python tutorial. Centrum voor Wiskunde en Informatica Amsterdam. 1995 [Google Scholar]
- 53.Oudah M., Henschel A. Taxonomy-aware feature engineering for microbiome classification. BMC Bioinf. 2018;19(1):1–13. doi: 10.1186/s12859-018-2205-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mukaka M.M. A guide to appropriate use of correlation coefficient in medical research. Malawi Med J. 2012;24(3):69–71. [PMC free article] [PubMed] [Google Scholar]
- 55.O'Brien R.G., Kaiser M.K. MANOVA method for analyzing repeated measures designs: an extensive primer. Psychol Bull. 1985;97(2):316. [PubMed] [Google Scholar]
- 56.Bradley A.P. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recogn. 1997;30(7):1145–1159. [Google Scholar]
- 57.Ling CX, Huang J, Zhang H, editors. AUC: a statistically consistent and more discriminating measure than accuracy. Ijcai; 2003.
- 58.Bishop C.M. springer; 2006. Pattern recognition and machine learning. [Google Scholar]
- 59.Wirbel J., Zych K., Essex M., Karcher N., Kartal E., Salazar G. Microbiome meta-analysis and cross-disease comparison enabled by the SIAMCAT machine-learning toolbox. bioRxiv. 2020 doi: 10.1186/s13059-021-02306-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shamsaddini A., Dadkhah K., Gillevet P.M. BiomMiner: an advanced exploratory microbiome analysis and visualization pipeline. PLoS ONE. 2020;15(6) doi: 10.1371/journal.pone.0234860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chong J., Liu P., Zhou G., Xia J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat Protoc. 2020;15(3):799–821. doi: 10.1038/s41596-019-0264-1. [DOI] [PubMed] [Google Scholar]
- 62.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37(8):852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hothorn T., Zeileis A. partykit: A modular toolkit for recursive partytioning in R. J Machine Learn Res. 2015;16(1):3905–3909. [Google Scholar]
- 64.Wright MN, Ziegler A. ranger: A fast implementation of random forests for high dimensional data in C++ and R. arXiv preprint arXiv:150804409; 2015.
- 65.Deng H. Interpreting tree ensembles with intrees. Int J Data Sci Anal. 2019;7(4):277–287. [Google Scholar]
- 66.Ribeiro MT, Singh S, Guestrin C, editors. “Why should I trust you?” Explaining the predictions of any classifier. Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining; 2016.
- 67.Molnar C., Casalicchio G., Bischl B. iml: An R package for interpretable machine learning. J Open Sour Software. 2018;3(26):786. [Google Scholar]
- 68.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 69.Gulli A., Pal S. Packt Publishing Ltd; 2017. Deep learning with Keras. [Google Scholar]
- 70.Kuhn M. Building predictive models in R using the caret package. J Stat Softw. 2008;28(5):1–26. [Google Scholar]
- 71.Candel A, Parmar V, LeDell E, Arora A. Deep learning with H2O. H2O ai Inc; 2016.
- 72.Wu S., Sun C., Li Y., Wang T., Jia L., Lai S. GMrepo: a database of curated and consistently annotated human gut metagenomes. Nucleic Acids Res. 2020;48(D1):D545–D553. doi: 10.1093/nar/gkz764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vangay P., Hillmann B.M., Knights D. Microbiome Learning Repo (ML Repo): a public repository of microbiome regression and classification tasks. GigaScience. 2019;8(5) doi: 10.1093/gigascience/giz042. giz042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pasolli E., Schiffer L., Manghi P., Renson A., Obenchain V., Truong D.T. Accessible, curated metagenomic data through ExperimentHub. Nat Methods. 2017;14(11):1023. doi: 10.1038/nmeth.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Durack J., Lynch S.V. The gut microbiome: relationships with disease and opportunities for therapy. J Exp Med. 2019;216(1):20–40. doi: 10.1084/jem.20180448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Topçuoğlu B.D., Lesniak N.A., Ruffin M.T., Wiens J., Schloss P.D. A framework for effective application of machine learning to microbiome-based classification problems. Mbio. 2020;11(3) doi: 10.1128/mBio.00434-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reese A.T., Dunn R.R. Drivers of microbiome biodiversity: a review of general rules, feces, and ignorance. Mbio. 2018;9(4) doi: 10.1128/mBio.01294-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ley R.E., Backhed F., Turnbaugh P., Lozupone C.A., Knight R.D., Gordon J.I. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102(31):11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Turnbaugh P.J., Hamady M., Yatsunenko T., Cantarel B.L., Duncan A., Ley R.E. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–U7. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sze M.A., Schloss P.D. Looking for a signal in the noise: revisiting obesity and the microbiome. Mbio. 2016;7(4) doi: 10.1128/mBio.01018-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vrolix R., Mensink R.P. Variability of the glycemic response to single food products in healthy subjects. Contemp Clin Trials. 2010;31(1):5–11. doi: 10.1016/j.cct.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 82.Zeevi D., Korem T., Zmora N., Israeli D., Rothschild D., Weinberger A. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163(5):1079–1194. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 83.Pasolli E., Truong D.T., Malik F., Waldron L., Segata N. Machine learning meta-analysis of large metagenomic datasets: tools and biological insights. Plos Comput Biol. 2016;12(7) doi: 10.1371/journal.pcbi.1004977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Techtmann S.M., Hazen T.C. Metagenomic applications in environmental monitoring and bioremediation. J Ind Microbiol Biotechnol. 2016;43(10):1345–1354. doi: 10.1007/s10295-016-1809-8. [DOI] [PubMed] [Google Scholar]
- 85.Cordier T., Lanzen A., Apotheloz-Perret-Gentil L., Stoeck T., Pawlowski J. Embracing environmental genomics and machine learning for routine biomonitoring. Trends Microbiol. 2019;27(5):387–397. doi: 10.1016/j.tim.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 86.Janßen R., Zabel J., von Lukas U., Labrenz M. An artificial neural network and Random Forest identify glyphosate-impacted brackish communities based on 16S rRNA amplicon MiSeq read counts. Mar Pollut Bull. 2019;149 doi: 10.1016/j.marpolbul.2019.110530. [DOI] [PubMed] [Google Scholar]
- 87.Smith M.B., Rocha A.M., Smillie C.S., Olesen S.W., Paradis C., Wu L.Y. Natural bacterial communities serve as quantitative geochemical biosensors. Mbio. 2015;6(3) doi: 10.1128/mBio.00326-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Demergasso C., Véliz R., Galleguillos P., Marín S., Acosta M., Zepeda V. Decision support system for bioleaching processes. Hydrometallurgy. 2018;181:113–122. [Google Scholar]
- 89.Chang H.X., Haudenshield J.S., Bowen C.R., Hartman G.L. Metagenome-wide association study and machine learning prediction of bulk soil microbiome and crop productivity. Front Microbiol. 2017;8 doi: 10.3389/fmicb.2017.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thompson J., Johansen R., Dunbar J., Munsky B. Machine learning to predict microbial community functions: an analysis of dissolved organic carbon from litter decomposition. PLoS ONE. 2019;14(7) doi: 10.1371/journal.pone.0215502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cordier T., Forster D., Dufresne Y., Martins C.I., Stoeck T., Pawlowski J. Supervised machine learning outperforms taxonomy-based environmental DNA metabarcoding applied to biomonitoring. Mol Ecol Resour. 2018;18(6):1381–1391. doi: 10.1111/1755-0998.12926. [DOI] [PubMed] [Google Scholar]
- 92.Frühe L., Cordier T., Dully V., Breiner H.W., Lentendu G., Pawlowski J. Supervised machine learning is superior to indicator value inference in monitoring the environmental impacts of salmon aquaculture using eDNA metabarcodes. Mol Ecol. 2020 doi: 10.1111/mec.15434. [DOI] [PubMed] [Google Scholar]
- 93.Dully V, Balliet H, Frühe L, Däumer M, Thielen A, Gallie S, et al. Robustness, sensitivity and reproducibility of eDNA metabarcoding as an environmental biomonitoring tool in coastal salmon aquaculture–An inter-laboratory study. Ecol Indic 121:107049.
- 94.Ulrich N., Kirchner V., Drucker R., Wright J.R., McLimans C.J., Hazen T.C. Response of aquatic bacterial communities to hydraulic fracturing in northwestern pennsylvania: a five-year study. Sci Rep-Uk. 2018;8 doi: 10.1038/s41598-018-23679-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.See J.R.C., Ulrich N., Nwanosike H., McLimans C.J., Tokarev V., Wright J.R. Bacterial biomarkers of marcellus shale activity in Pennsylvania. Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.01697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gerhard W.A., Gunsch C.K. Microbiome composition and implications for ballast water classification using machine learning. Sci Total Environ. 2019;691:810–821. doi: 10.1016/j.scitotenv.2019.07.053. [DOI] [PubMed] [Google Scholar]
- 97.Alneberg J., Bennke C., Beier S., Bunse C., Quince C., Ininbergs K. Ecosystem-wide metagenomic binning enables prediction of ecological niches from genomes. Commun Biol. 2020;3(1):119. doi: 10.1038/s42003-020-0856-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Metcalf J.L., Xu Z.J.Z., Bouslimani A., Dorrestein P., Carter D.O., Knight R. Microbiome tools for forensic science. Trends Biotechnol. 2017;35(9):814–823. doi: 10.1016/j.tibtech.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 99.Hampton-Marcell J.T., Lopez J.V., Gilbert J.A. The human microbiome: an emerging tool in forensics. Microb Biotechnol. 2017;10(2):228–230. doi: 10.1111/1751-7915.12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Johnson H.R., Trinidad D.D., Guzman S., Khan Z., Parziale J.V., DeBruyn J.M. A machine learning approach for using the postmortem skin microbiome to estimate the postmortem interval. PLoS ONE. 2016;11(12) doi: 10.1371/journal.pone.0167370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu R.N., Gu Y.X., Shen M.W., Li H., Zhang K., Wang Q. Predicting postmortem interval based on microbial community sequences and machine learning algorithms. Environ Microbiol. 2020;22(6):2273–2291. doi: 10.1111/1462-2920.15000. [DOI] [PubMed] [Google Scholar]
- 102.Khodakova A.S., Smith R.J., Burgoyne L., Abarno D., Linacre A. Random whole metagenomic sequencing for forensic discrimination of soils. PLoS ONE. 2014;9(8) doi: 10.1371/journal.pone.0104996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Delgado-Baquerizo M., Oliverio A.M., Brewer T.E., Benavent-Gonzalez A., Eldridge D.J., Bardgett R.D. A global atlas of the dominant bacteria found in soil. Science. 2018;359(6373):320. doi: 10.1126/science.aap9516. [DOI] [PubMed] [Google Scholar]
- 104.Carvalho D.V., Pereira E.M., Cardoso J.S. Machine learning interpretability: a survey on methods and metrics. Electronics. 2019;8(8):832. [Google Scholar]
- 105.Fisher A., Rudin C., Dominici F. All models are wrong, but many are useful: learning a variable's importance by studying an entire class of prediction models simultaneously. J Mach Learn Res. 2019;20(177):1–81. [PMC free article] [PubMed] [Google Scholar]
- 106.Bogart E., Creswell R., Gerber G.K. MITRE: inferring features from microbiota time-series data linked to host status. Genome Biol. 2019;20(1):1–15. doi: 10.1186/s13059-019-1788-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Richardson M., Gottel N., Gilbert J.A., Lax S. Microbial similarity between students in a common dormitory environment reveals the forensic potential of individual microbial signatures. MBio. 2019;10(4):e01054–19. doi: 10.1128/mBio.01054-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lundberg SM, Lee S-I, editors. A unified approach to interpreting model predictions. Advances in neural information processing systems; 2017.
- 109.Goldstein A., Kapelner A., Bleich J., Pitkin E. Peeking inside the black box: visualizing statistical learning with plots of individual conditional expectation. J Comput Graphical Stat. 2015;24(1):44–65. [Google Scholar]
- 110.Zhao Q., Hastie T. Causal interpretations of black-box models. J Busin Econ Stat. 2019;1–10 doi: 10.1080/07350015.2019.1624293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Apley DW, Zhu J. Visualizing the effects of predictor variables in black box supervised learning models. arXiv preprint arXiv:161208468; 2016.
- 112.Mittelstadt B.D., Allo P., Taddeo M., Wachter S., Floridi L. The ethics of algorithms: mapping the debate. Big Data Soc. 2016;3(2) [Google Scholar]
- 113.Bathaee Y. The artificial intelligence black box and the failure of intent and causation. Harv JL & Tech. 2017;31:889. [Google Scholar]
- 114.Rudin C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat Mach Intell. 2019;1(5):206–215. doi: 10.1038/s42256-019-0048-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Doshi-Velez F, Kim B. Towards a rigorous science of interpretable machine learning. arXiv preprint arXiv:170208608; 2017.
- 116.Zerilli J., Knott A., Maclaurin J., Gavaghan C. Transparency in algorithmic and human decision-making: is there a double standard? Philos Technol. 2019;32(4):661–683. [Google Scholar]
- 117.Wu Y., Zhang K. Tools for the analysis of high-dimensional single-cell RNA sequencing data. Nat Rev Nephrol. 2020;1–14 doi: 10.1038/s41581-020-0262-0. [DOI] [PubMed] [Google Scholar]
- 118.Vamathevan J., Clark D., Czodrowski P., Dunham I., Ferran E., Lee G. Applications of machine learning in drug discovery and development. Nat Rev Drug Disc. 2019;18(6):463–477. doi: 10.1038/s41573-019-0024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zitnik M., Nguyen F., Wang B., Leskovec J., Goldenberg A., Hoffman M.M. Machine learning for integrating data in biology and medicine: principles, practice, and opportunities. Inform Fusion. 2019;50:71–91. doi: 10.1016/j.inffus.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Netzer M., Hackl W.O., Schaller M., Alber L., Marksteiner J., Ammenwerth E. Evaluating performance and interpretability of machine learning methods for predicting delirium in gerontopsychiatric patients. Stud Health Technol Inform. 2020;271:121–128. doi: 10.3233/SHTI200087. [DOI] [PubMed] [Google Scholar]
- 121.Fellous J.-M., Sapiro G., Rossi A., Mayberg H.S., Ferrante M. Explainable artificial intelligence for neuroscience: behavioral neurostimulation. Front Neurosci. 2019;13:1346. doi: 10.3389/fnins.2019.01346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Singla S, Wallace E, Feng S, Feizi S. Understanding impacts of high-order loss approximations and features in deep learning interpretation. arXiv preprint arXiv:190200407; 2019.
- 123.Montavon G., Samek W., Müller K.-R. Methods for interpreting and understanding deep neural networks. Digital Signal Process. 2018;73:1–15. [Google Scholar]