Abstract
Cullin-RING ligases (CRLs) recognize and interact with substrates for ubiquitination and degradation, and can be targeted for disease treatment when the abnormal expression of substrates involves pathologic processes. Phosphorylation, either of substrates or receptors of CRLs, can alter their interaction. Phosphorylation-dependent ubiquitination and proteasome degradation influence various cellular processes and can contribute to the occurrence of various diseases, most often tumorigenesis. These processes have the potential to be used for tumor intervention through the regulation of the activities of related kinases, along with the regulation of the stability of specific oncoproteins and tumor suppressors. This review describes the mechanisms and biological functions of crosstalk between phosphorylation and ubiquitination, and most importantly its influence on tumorigenesis, to provide new directions and strategies for tumor therapy.
KEY WORDS: Phosphorylation, Cullin-RING ligases, Ubiquitination, Tumorigenesis, Targeted therapy, Degradation, Crosstalk, Kinases
Abbreviations: AIRE, autoimmune regulator; AKT, AKT serine/threonine kinase; ATR, ataxia telangiectasia-mutated and Rad3-related; BCL2, BCL2 apoptosis regulator; BMAL1, aryl hydrocarbon receptor nuclear translocator like; CDK2/4, cyclin dependent kinase 2/4; CDT2, denticleless E3 ubiquitin protein ligase homolog; c-Fos, Fos proto-oncogene, AP-1 transcription factor subunit; CHK1, checkpoint kinase 1; Ci, cubitus interruptus; CK1/2, casein kinase I/II; TCN, triciribine hydrate; CLOCK, clock circadian regulator; COMMD1, copper metabolism domain containing 1; CRL, cullin-RING ligase; CRY1, cryptochrome circadian regulator 1; CSN, COP9 signalosome; DDB1, damage specific DNA binding protein 1; DYRK1A/B, dual-specificity tyrosine-phosphorylation-regulated kinases 1A/B; EMT, epithelial–mesenchymal transition; ERG, ETS transcription factor ERG; ERK, mitogen-activated protein kinase 1; EXO1, exonuclease 1; FBW7, F-box and WD repeat domain containing 7; FBXL3, F-box and leucine rich repeat protein; FBXO3/31, F-box protein 3/31; FZR1, fizzy and cell division cycle 20 related 1; HIB, Hedghog-induced MATH and BTB domain-containing protein; HIF1α, NF-κB and hypoxia inducible factor 1 subunit alpha; ID2, inhibitor of DNA binding 2; JAB1, c-Jun activation domain binding protein-1; KBTBD8, kelch repeat and BTB domain containing 8; KDM2B, lysine demethylase 2B; KEAP1, kelch like ECH associated protein 1; KLHL3, kelch like family member 3; KRAS, KRAS proto-oncogene, GTPase; MYC, MYC proto-oncogene, bHLH transcription factor; NEDD8, NEDD8 ubiquitin like modifier; NOLC1, nucleolar and coiled-body phosphoprotein 1; NRF2, nuclear factor, erythroid 2 like 2; p130Cas, BCAR1 scaffold protein, Cas family member; PDL1, programmed death ligand 1; PKC, protein kinase C; PKM2, pyruvate kinase M2 isoform; HCC, hepatocellular carcinomas; P-TEFb, positive transcription elongation factor b; PYGO2, pygopus 2; RA, retinoic acid; RARα, RA receptor α; RRM2, ribonucleotide reductase regulatory subunit M2; SNAIL1, snail family transcriptional repressor 1; SOCS6, suppressor of cytokine signaling 6; SPOP, speckle-type POZ protein; SRC-3, nuclear receptor coactivator 3; TCOF1, treacle ribosome biogenesis factor 1; TRF1, telomeric repeat binding factor 1; USP37, ubiquitin specific peptidase 37; VHL, von Hippel-Lindau tumor suppressor; Vps34, phosphatidylinositol 3-kinase catalytic subunit type 3; XBP1, X-box binding protein 1; ZBTB16, zinc finger and BTB domain containing 16
Graphical abstract
This review summarizes the crosstalk between phosphorylation and ubiquitination in different biological processes and diseases, mainly cancers, and highlights the therapeutic potential of the crosstalk in tumor treatments.
1. Introduction
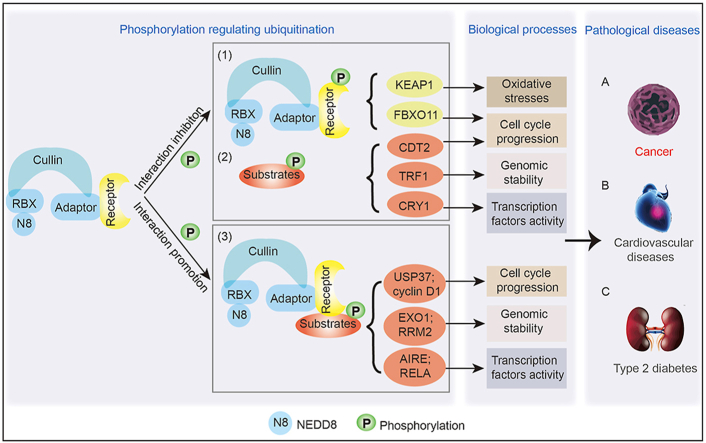
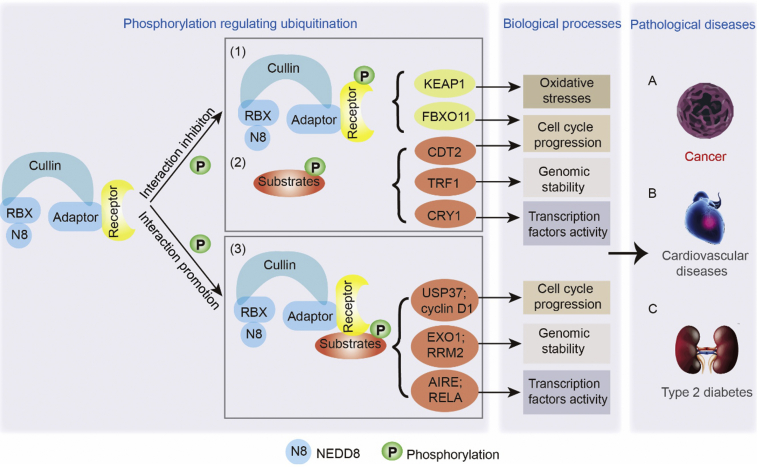
Cullin-RING ligases (CRLs) are essential components of the E3 ubiquitin ligase family in the ubiquitin-proteasome system, transferring ubiquitin to substrates and regulating the ubiquitination and degradation of approximately 20% of the proteins in mammalian cells1,2. CRLs consist of four components: one of eight cullin proteins that serve as a linker between an adaptor protein and a RING-finger protein, with the RING-finger protein responsible for binding to the E2 ubiquitin enzyme and the adaptor protein responsible for binding to a substrate receptor protein, essential for recognizing substrates3 (Fig. 1). Interestingly, cullin 3-RING ligases (CRL3s) do not have substrate receptor proteins; thus, adaptor proteins directly interact with substrates and mediate ubiquitination4,5. Considering that different types of adaptor and receptor proteins can interact with different substrates that are vital in physiology and pathological progression6, 7, 8, a strategy for disease treatment requires understanding how adaptor and receptor proteins recognize substrates and how to control the level of target substrates by altering the interaction between CRLs and substrates.
Figure 1.
The crosstalk between phosphorylation and ubiquitination regulates related biological and pathological processes. In most cases, CRLs are prone to recognize and interact with those proteins with Ser/Thr rich degrons which have been phosphorylated, but sometimes, it will abrogate the interaction between substrates and CRLs when anyone of them is phosphorylated. The crosstalk between phosphorylation and ubiquitination regulates different biological processes including oxidative stresses, cell cycle, genomic stability and transcription factors activity, subsequently inducing various diseases, mainly cancers.
Degrons existing in substrate proteins promote or inhibit the recognition of CRLs after being modified by different posttranslational modifications including phosphorylation, methylation and acetylation9,10. Phosphorylation has close crosstalk with ubiquitination, widely affecting the interaction between receptors and substrates11. The crosstalk also influences tumorigenesis and various other diseases, such as cardiovascular disease12,13. Phosphorylation itself is essential for cell growth and differentiation. It has been demonstrated that many signaling pathways participate in the phosphorylation-dephosphorylation cascade, including cyclin-dependent kinase tyrosine kinase, cadherin–catenin complex, and MAP kinase participating in the occurrence and development of various cancers14, 15, 16. In other words, regulating the activity of these kinases can restrain the overdegradation of substrates with important biological functions or promote the degradation of those substrates that can induce diseases. Ser/Thr-rich degrons in substrates can be modified by phosphorylation, and it has been demonstrated that partially phosphorylated substrates are easier or more difficult to be recognized and interact with CRLs17. Moreover, phosphorylation of receptors or adaptors also affects the recognition and interaction, indicating that phosphorylation deeply impacts the ubiquitin-proteasome system.
It is important to understand how the crosstalk between phosphorylation and ubiquitination mediates physiological and pathological events, and the biological pathways and diseases that are affected by phosphorylation-ubiquitination crosstalk. Kinase inhibitors and activators can be used to regulate phosphorylation of such substrates of cullin E3 ubiquitin ligases and control the levels of those proteins, which could be a new strategy for tumor treatment.
2. The mechanism of phosphorylation regulating the interaction between substrates and CRLs
Cullin-based ubiquitination is an acute enzyme cascade pathway, and phosphorylation mainly impacts the final segment: the interaction between substrates and CRLs. However, the effects of phosphorylation are diverse, and may depend on the sites of phosphorylation and the properties of substrates and CRLs. Here, we discuss how phosphorylation is involved in the ubiquitination process and the possible mechanisms.
2.1. Phosphorylation of substrates: Necessary for interactions under certain circumstances
Zou et al.18 found that the epithelial–mesenchymal transition (EMT) transcription factor snail family transcriptional repressor 1 (SNAIL1) could interact with F-box protein 31 (FBXO31) for ubiquitination and dependent degradation. SNAIL1 is a zinc-finger transcriptional factor and has two GSK-3β phosphorylation motifs. When the two phosphorylation sites of SNAIL1 are mutated, the interaction between FBXO31 and SNAIL1 is dramatically impaired. Phosphorylation at different sites of SNAIL1 determines whether it will be recognized by different E3 ubiquitin ligases, thus leading to different biological functions19,20. For example, GSK-3β mediating the phosphorylation of SNAIL1 promotes the ubiquitination and degradation of SNAIL1 induced by FBXO31, thus abrogating the migration of gastric cancer cells18.
Similarly, the p160 family of coactivator nuclear receptor coactivator 3 (SRC-3), which can bind to cullin 3-based speckle-type POZ protein (SPOP) E3 ubiquitin ligase complex for ubiquitination and degradation, contains serine-rich motifs and can be phosphorylated by p38MAPK12. Among four p38MAPK consensus phosphorylation sites, Ser860 is critical for the interaction between SRC-3 and cullin 3, and phosphorylation at Ser860 is the primary requirement for retinoic acid (RA)-induced ubiquitination of SRC-3. More importantly, SRC-3 regulates the transcription of the RA receptor α (RARα)-target genes, and overexpression of SRC-3 WT enhances transcription while SRC-3 (S860A) loses the capacity to facilitate transcription12. These results reveal that prior phosphorylation of substrates is necessary for subsequent ubiquitination and degradation.
2.2. Phosphodegron of substrates: Crucial sites in the interaction
Many receptor proteins or adaptor proteins of cullin E3 ligase recognize substrates through conserved phosphorylation sequence motifs, or Ser/Thr-rich degrons17. For example, cullin 3-RING ligases (CRL3s) are prone to bind substrates containing serine-rich domains for degradation, such as SRC-3, which can be phosphorylated in that domain. Substrates containing phosphotyrosine degrons are usually the targets of the cullin 5-RING ligases (CRL5) adaptor protein SOCS with SH2 domains for proteasome dependent degradation21. Moreover, phosphorylating residues often cause the substrate to become a negatively charged protein; adaptor proteins, such as kelch repeat and BTB domain containing 8 (KBTBD8) of CRL3s, have a complementary positively charged motif to bind the phosphorylated substrates22.
It has been confirmed that transcription factor cubitus interruptus (Ci) can be degraded by Cul3–Hedghog-induced MATH and BTB domain-containing protein (HIB) through Ser/Thr-rich motifs. Zhang et al.17 also reported that other proteins containing HIB binding sites such as Ci had similar Ser/Thr-rich motifs and similar Ser/Thr-rich motifs could serve as degrons of HIB binding proteins to be degraded by cullin 3–HIB-based SPOP E3 ligase, indicating that phosphorylation is essential for cullin 3-based E3 ligases to recognize substrates. Cullin 3-based KBTBD8 E3 ligase complex recognizes treacle ribosome biogenesis factor 1 (TCOF1) and nucleolar and coiled-body phosphoprotein 1 (NOLC1) for ubiquitination and degradation, which determines the function of cullin 3-based KBTBD8 E3 ligase for ribosome biogenesis and neural crest specification. Achim Werner et al.22 interrogated how cullin 3 E3 ligase recognizes its substrates and found that TCOF1 and NOLC1 have various phosphorylated casein kinase II (CK2) motifs and can thus be bound by KBTBD8, while CK2 inhibitor CX4945 can effectively prevent this process.
However, such phosphorylated motifs are also sometimes the main cause for the attenuation of CRLs recognition of substrates, such as multiple Ser/Thr-rich degrons of cubitus interruptus23. Moreover, adaptor proteins can lose the ability to interact with substrate proteins when the substrate-binding domain is phosphorylated, such as with the kelch domain of kelch like family member 3 (KLHL3)13.
2.3. Phosphorylation of CRLs can also impact its interaction with substrates
When receptors or adaptors of CRLs are phosphorylated, the interaction between substrates and CRLs can sometimes be interrupted. Jinfang Zhang et al.24 reported that SPOP could be phosphorylated by cyclin D1–CDK4 kinase, which inhibited its interaction with its E3 ligase fizzy and cell division cycle 20 related 1 (FZR1) for further degradation. Importantly, phosphorylation stabilized SPOP, thus promoting the degradation of programmed death 1 ligand 1 (PDL1), a newly found substrate of the cullin 3-based SPOP E3 ligase, which could be negatively regulated by CDK4 inhibitors such as palbociclib.
Apart from SPOP, two other adaptors of cullin 3-based E3 ligase, kelch-like ECH associated protein 1 (KEAP1) and zinc finger and BTB domain 16 (ZBTB16) can also be phosphorylated. Phosphorylation of KEAP1 at site 53 interrupts its interaction with nuclear factor, erythroid 2 like 2 (NRF2), protecting cells from oxidative stress. Additionally, GSK3β-mediated phosphorylation of ZBTB16 suppresses its interaction with Atg14L and then promotes autophagy25,26.
3. Regulation of the crosstalk between phosphorylation and ubiquitination in biological processes
The diversity of substrates and their specific function in organisms determine how crosstalk can influence and regulate different cellular biological functions, including regulation of genomic stability, cell cycle, the activity of transcription factors, and oxidative stresses. Crosstalk between phosphorylation and ubiquitination can stabilize the levels of some proteins so that they are not overexpressed or overactivated to avoid biological damage, such as exonuclease 1 (EXO1) and ribonucleotide reductase regulatory subunit M2 (RRM2), which are important for genomic stability. In other cases, the crosstalk is necessary for some proteins to exert functions, sometimes by inhibiting their degradation, such as transcriptional repressor cryptochrome circadian regulator 1 (CRY1), and sometimes by promoting their interaction or dissociation with other proteins, such as autoimmune regulator (AIRE) and KEAP1 (Table 112,18,21, 22, 23,27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45 and Table 224,25,31,46, 47, 48, 49).
Table 1.
Summary of the crosstalk between phosphorylation and ubiquitination and its biological function when phosphorylation exits in substrates.
| CRL | Substrate | Ubiquitinated site | Phosphorylated site | Kinase | How phosphorylation affects interaction between substrates and receptors | Biological function | Ref. |
|---|---|---|---|---|---|---|---|
| Cullin 1-based SCF E3 ligase | EXO1 | Lys796 | Ser714 | ATR | Promotion | Preservation of genomic stability | 27 |
| Cullin 1-based SCF E3 ligase | RRM2 | Lys796 | Thr33 | CDK | Promotion | Preservation of genomic stability | 28 |
| Cullin 1-based SCF E3 ligase | USP37 | Lys11 | Ser858 | CDK2 | Promotion | Regulation of cell cycle | 29 |
| Cullin 1-based FBX4 E3 ligase | TRF1 | Lys194/240 | Ser114 | NEK7 | Inhibition | Preservation of telomere chromatin integrity | 30 |
| Cullin 1-based FBXO11 E3 ligase | CDT2 | Lys48 | Thr464 | CDK | Inhibition | Regulation of cell cycle and cell differentiation | 31 |
| Cullin 1-based FBXO31 E3 ligase | SNAIL1 | Lys98/137/146 | Ser6 | GSK3β | Promotion | Tumor suppression | 18 |
| Cullin 1-based FBXO31 E3 ligase | Cyclin D1 | Lys48 | Thr286 | CDK4 | Promotion | Regulation of cell cycle | 32 |
| Cullin 1-based FBXO31 E3 ligase | MDM2 | Lys36 | Six sites | ATM | Promotion | Tumor suppression | 33 |
| Cullin 1-based FBW7 E3 ligase | XBP1 | Lys236 | Ser212/217 | – | Promotion | Tumor suppression | 34 |
| Cullin 1-based KDM2B E3 ligase | c-Fos | Lys113 | Ser374 | EGF | Inhibition | Tumor promotion | 35 |
| Cullin 1-based FBXL3 E3 ligase | CRY1 | Lys11/107/159/329/442/485 | Ser588 | DNA-PK | Inhibition | Regulation of transcription | 35 |
| Cullin 1-based FBXO3 E3 ligase | AIRE | – | Thr68/Ser156 | DNA-PK | Promotion | Regulation of transcription | 36 |
| Cullin 2-based COMMD1 E3 ligase | RELA | – | Ser468 | IκB | Promotion | Regulation of transcription | 37 |
| Cullin 2-based VHL E3 ligase | ID2 | Lys12 | Thr27 | DYRK | Promotion | Tumor suppression | 38 |
| Cullin 3-based SPOP E3 ligase | ERG | – | – | CKI | Promotion | Tumor suppression | 39 |
| Cullin 3-based SPOP E3 ligase | SRC-3 | Lys316 | Ser101/102/860 | CKIε/p38MAPK | Promotion | Tumor suppression | 12 |
| Cullin 3-based HIB E3 ligase | Ci/Gli | – | – | CK1 | Inhibition | Regulation of Hedgehog signaling | 23 |
| Cullin 3-based SPOP E3 ligase | PDX1 | – | Thr230/Ser231 | CK2 | Inhibition | Regulation of type 2 diabetes | 40 |
| Cullin 3-based KBTBD8 E3 ligase | TCOF1 | – | – | CK2 | Promotion | – | 22 |
| Cullin 3-based KBTBD8 E3 ligase | NOLC1 | Lys33/59 | – | CK2 | Promotion | – | 22 |
| Cullin 3 E3 ligase | BCL2 | – | Thr69 | PKM2 | Inhibition | Tumor promotion | 41 |
| Cullin 3 E3 ligase | NPR1 | – | Ser11/15 | – | Promotion | Regulation of plant immunity | 42 |
| Cullin 3-based KLHL20 E3 Ligase | ULK1 | – | Ser1042/Thr1046 | – | Promotion | Regulation of autophagy | 43 |
| Cullin 4A-based DDB1E3 ligase | CHK1 | Lys180/244/313/436 | Ser317/345 | ATR | Promotion | Tumor suppression | 44 |
| Cullin 4A E3 ligase | PYGO2 | – | Ser48 | AKT | Inhibition | Tumor promotion | 45 |
| Cullin 5-based SOCS6 E3 ligase | p130Cas | – | – | SRC | Promotion | Tumor suppression | 21 |
–Not applicable.
Table 2.
Summary of the crosstalk between phosphorylation and ubiquitination and its biological function when phosphorylation exits in CRLs.
| CRL | Substrate | Ubiquitinated site | Phosphorylated site | Kinase | How phosphorylation affects interaction between substrates and receptors | Biological function | Ref. |
|---|---|---|---|---|---|---|---|
| Cullin 1-based SCF E3 ligase | – | – | Thr31/Ser557 | AKT | Promotion | Regulation of cell cycle | 46 |
| Cullin 1-based FBW7 E3 ligase | – | – | Thr205 | ERK | – | Tumor promotion | 47 |
| Cullin 3-based SPOP E3 ligase | PDL1 | – | – | CDK4 | Promotion | Tumor promotion | 24 |
| Cullin 3-based KEAP1 E3 ligase | NRF2 | – | Ser53 | – | Inhibition | Response to oxidative stresses | 25 |
| Cullin 3-based KLHL3 E3 ligase | WNK4 | Lys157 | Ser433 | PKCα/PKCβ | Inhibition | Regulation of hypertension and cardiovascular disease | 48 |
| Cullin 4-based CDT2 E3 ligase | FBXO11 | Lys197 | Thr464 | CDK | Inhibition | Regulation of cell cycle | 31 |
| Cullin 5-based VACM-1 E3 ligase | – | – | Ser730 | PKA/PKC | The phosphorylated receptor is induced to be ubiquitinated | Tumor promotion | 49 |
–Not applicable.
3.1. Regulation of genome stability
The protein level of EXO1 is restrained by ataxia telangiectasia-mutated and Rad3-related (ATR)-mediated phosphorylation and SCF (Skp 1/Cul 1/F-box) E3 ligase complex-mediated ubiquitination, which restrains hyper-resection of DNA and preserves genomic stability27,50. Similarly, RRM2 binds to cyclin F, a subunit of SCF ubiquitin ligase complexes, for ubiquitination and degradation, which depends on CDK-mediated phosphorylation of RRM2 at Thr3328 to maintain DNA synthesis and repair and genome stability51,52. Tan et al.30 found that phosphorylation modification preserves telomeric repeat binding factor 1 (TRF1) stability through inhibiting the ubiquitin-mediated proteasome degradation pathway, thus protecting cells from telomeric DNA damage30,53.
3.2. Regulation of cell cycle
It has been reported that cyclin F46 and denticleless E3 ubiquitin protein ligase homolog (CDT2)54, both of which are substrate receptors for CRLs and important for the cell cycle, can be regulated by the crosstalk of phosphorylation and ubiquitination. Cyclin dependent kinase 4 (CDK4)-induced phosphorylation dependent ubiquitination regulates the protein level of cyclin D1, and its E3 ligase F-box protein 31 (FBXO31) functions as a regulator of the G1/S transition32. When the de-ubiquitinating enzyme ubiquitin specific peptidase 37 (USP37) is phosphorylated by cyclin dependent kinase 2 (CDK2) at Ser858 it can bind to SCF-βTrCP for ubiquitination and degradation in G2. The instability of USP37 in G2 is necessary for mitotic entry, while USP37 S858A-expressing cells lose the expression of mitotic markers.
3.3. Regulation of the activity of transcription factors
CRY1, a substrate of the F-box and leucine rich repeat protein 3 (FBXL3) and essential for the mammalian circadian clock, can be phosphorylated by DNA-dependent protein kinase at Ser588, which results in inhibition of FBXL3-mediated degradation, thus increasing the stability of the CRY1 and maintaining its function of repressing the activity of the transcriptional activators clock circadian regulator (CLOCK) and aryl hydrocarbon receptor nuclear translocator like (BMAL1)35,55. Phosphorylation at Thr68 and Ser156 of AIRE promotes F-box protein 3 (FBXO3) E3 ubiquitin ligase to recognize it and ubiquitylate it, which is necessary for AIRE binding to positive transcription elongation factor b (P-TEFb) and preserving its transcriptional activity36.
Interestingly, NF-κB-mediated transcription can be directly regulated by the crosstalk between phosphorylation and ubiquitination and indirectly regulated by the crosstalk between phosphorylation and deneddylation, which is another model of phosphorylation regulating ubiquitinated substrates. On the one hand, when RELA, a subunit of NF-κB, is phosphorylated by IκB kinase on Ser468, COMMD1-associated factor GCN5 interacts more avidly with RELA and promotes its degradation, ultimately repressing the NF-κB-mediated transcription37,56. Similarly, casein kinase 1 phosphorylates and promotes the degradation of NF-κB subunit p65 mediated by cullin 2-based copper metabolism domain containing 1 (COMMD1) E3 Ligase, thus regulating innate immune signaling37. On the other hand, Orel et al.57 found that IκB kinase could phosphorylate c-Jun activation domain binding protein-1 (JAB1), the subunit of COP9 signalosome (CSN), and induce its ubiquitination and degradation. Considering that the major function of CSN is deneddylation, IκB kinase-mediated degradation of JAB1 truly promotes the activation of CRLs and advances the ubiquitination and degradation of the NF-κB inhibitor, IκBα, which ultimately activates NF-κB signaling.
3.4. Regulation of the response to oxidative stresses
The KEAP1–NRF2–ARE signaling pathway is vital for the cytoprotective response to oxidative stresses58, 59, 60. Wei et al.25 reported that phosphorylation at S53 of KEAP1 dissociated the binding of NRF2 to KEAP1, subsequently leading to the improvement of NRF2 stability and translocation of NRF2 to the nucleus. Phosphorylation modification of KEAP1 often occurs under oxidative stress; thus, NRF2 could be released from the KEAP1–NRF2 complex to activate antioxidant enzymes and prevent free radical damage to cells. P62 is also a substrate of KEAP1 and phosphorylated P62 competes with NRF2 to interact with KEAP1, which often occurs under oxidative stress and not only results in the accumulation of NRF2 but also connects the KEAP1–NRF2 system with autophagy61.
4. Functional analysis of the crosstalk between phosphorylation and ubiquitination in tumorigenesis
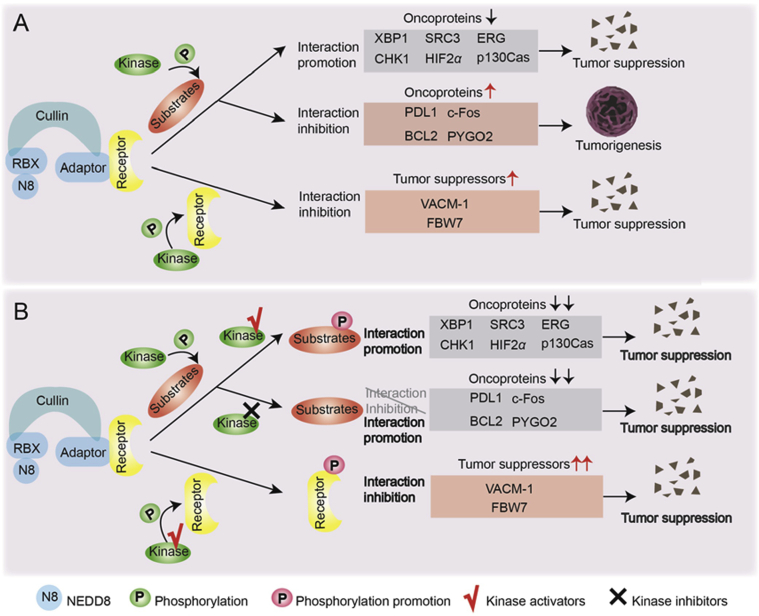
Some substrates of CRLs and the adaptors or receptors of CRLs themselves function as tumor suppressors or oncoproteins. For example, the CRL3 substrates ETS transcription factor ERG (ERG) and SRC-3 are encoded by oncogenes and overexpressed in prostate cancer and breast cancer respectively62,63. VACM-1, a receptor of CRL5, has been reported to be candidate tumor suppressor for its function of inhibiting cell growth49. In addition, the CRL1 receptor F-box and WD repeat domain containing 7 (FBW7) can also antagonize carcinogenesis by regulating the cell cycle64. The crosstalk between phosphorylation and ubiquitination regulates the stability of those key proteins involved in tumorigenesis, thus exerting an important role in tumor regulation (Fig. 2).
Figure 2.
The crosstalk between phosphorylation and ubiquitination influences tumorigenesis and related therapies (A) Crosstalk suppresses tumorigenesis by promoting the degradation of some oncoproteins or inhibiting the degradation of tumor suppressors, and promotes tumorigenesis by inhibiting the degradation of various oncoproteins. (B) Activators and inhibitors of kinases which can regulate the crosstalk between phosphorylation and ubiquitination can be used for tumor therapy. Kinase activators can be used to promote the phosphorylation and the ubiquitinated degradation of those oncoproteins when phosphorylation facilitates their interaction with CRLs, and promote the phosphorylation but inhibit the ubiquitinated degradation of those tumor suppressors when phosphorylation restrains their interaction with CRLs. Kinase inhibitors can be used to suppress the phosphorylation but promote the ubiquitinated degradation of those oncoproteins when phosphorylation restrains their interaction with CRLs.
4.1. Suppressing cancers: Augmenting the degradation of oncoproteins
In some cases, phosphorylation promotes the interaction between oncoproteins and CRLs, thus holding back the progression of cancer (Fig. 2A). Inducing the proteins overexpressed in tumors, such as ERG, SRC-3 and HIF2α, to degradation through the crosstalk can effectively inhibit tumor growth. Other proteins such as X-box binding protein 1 (XBP1) and checkpoint kinase 1(CHK1), which have important pathological functions in tumorigenesis, can also be phosphorylated and then induced to ubiquitin-proteasome degradation; thus, their functions of activating oncogenic pathways or regulating DNA damage repair will be blocked. Therefore, mediating the phosphorylation modification of these substrates, such as activating the related kinases, can be a strategy for cancer treatment (Fig. 2B).
4.1.1. XBP1
Chae et al.65 reported that XBP1 is a substrate of FBW7, a substrate recognition protein of cullin 1-RING E3 ligase (CRL1s). One of the mechanisms by which FBW7 suppresses tumorigenesis is by mediating the ubiquitination and degradation of XBP1 in a phosphorylation-dependent manner due to the close relationship of XBP1 with tumor progression. Phosphorylation at Ser212/217 sites of XBP1 promotes its interaction with Fbw7, thereby facilitating the degradation of XBP1, and inactivating XBP1 related oncogenic pathways such as MYC proto-oncogene, bHLH transcription factor (MYC), NF-κB and hypoxia inducible factor 1 subunit alpha (HIF1α), ultimately impairing cancer development66,67.
4.1.2. CHK1
The Ser/Thr protein kinase CHK1 is well-known for regulating DNA replication and DNA damage repair68,69. In addition, it is also regarded as an anticancer target, hinting that the protein level of CHK1 is crucial for drug discovery. Leung-Pineda et al.44 found that the stability of CHK1 was negatively regulated by cullin 4A-based DDB1 E3 ligase through ubiquitin-proteasome degradation, and phosphorylation of CHK1 at Ser317 and Ser345 promotes its interaction with cullin 4A-based damage specific DNA binding protein 1 (DDB1) E3 ligase. Therefore, regulating the ubiquitylation and phosphorylation of CHK1 may contribute to the treatment of cancer.
4.1.3. ERG
One of the mechanisms by which the adaptor protein SPOP suppresses prostate cancer involves promoting ERG ubiquitination and degradation70. After fusion with TMPRSS2, ERG protein is overexpressed and promotes the development of prostate cancer62. However, casein kinase I can phosphorylate ERG and facilitate its interaction with cullin 3-based SPOP E3 ligase, thus restraining the oncoprotein level of EGR in prostate cancer. Moreover, DNA damage drugs, such as topoisomerase inhibitors etoposide can promote SPOP/ERG interaction by activating casein kinase I (CKI), indicating that they could represent a potential effective therapy for prostate cancer39.
4.1.4. SRC-3
As mentioned above, SPOP facilitates phosphorylation of SRC-3 for proteasome degradation and activates transcription of RARα-target genes, indicating that phosphorylation is vital for the function of SRC-3. SRC3 is also well-known as an oncogene63,71. SPOP has been reported to interact with SRC-3 at Ser101/Ser102 in a degron-dependent manner, and can block SRC-3-induced oncogenic signaling pathways, such as estrogen and androgen receptor-dependent signals. Therefore, phosphorylation of SRCs promotes the tumor suppressing function of the cullin 3-based SPOP E3 ligase complex72.
4.1.5. HIF2α
Inducing HIF2α to ubiquitination and degradation is considered to be a potential therapy for cancer73. Lee et al.38 found that inhibitor of DNA binding 2 (ID2) protein, which is important for glioma stemness, can be phosphorylated by dual-specificity tyrosine-phosphorylation-regulated kinases 1A and 1B (DYRK1A and DYRK1B) kinases on Thr27 in normoxia. Phosphorylated ID2 cannot displace cullin 2-based von Hippel-Lindau tumor suppressor (VHL) E3 ligase from HIF2α as non-phosphorylated ID2 will do under the condition of hypoxia, thus ensuring the ubiquitylation of HIF2α and the inhibition of glioblastoma growth. It has been reported that high DYRK1 kinase activity is positively correlated with the clinical outcomes of glioblastoma patients74, during which phosphorylated ID2 may exert an important function through ensuring the degradation of HIF2α.
4.1.6. p130Cas
Cullin 5 can inhibit the transformation of human mammary epithelial cells and deficiency of cullin 5 often induces certain kinds of cancers, promoting the growth and migration of cancer cells75,76. Teckchandani et al.21 found that the cullin 5 adaptor suppressor of cytokine signaling 6 (SOCS6) could mediate BCAR1 scaffold protein, Cas family member (p130Cas) for proteasome-dependent degradation when Cas is phosphorylated by tyrosine kinase Src, hence promoting cullin 5 inhibition of the transformation of mammary epithelial cells. Therefore, phosphorylation-dependent protein ubiquitylation is implicated in the regulation of cullin 5-induced degradation of oncoproteins.
4.2. Promoting cancers: Inhibiting the degradation of oncoproteins
In certain cases, the progression of cancers is promoted when phosphorylation modification inhibits substrates, functioning as oncoproteins, binding to CRLs for degradation. Accumulation of c-Fos and NRF2 through the crosstalk between phosphorylation and ubiquitination is positively related with the proliferation of cancer cells. The crosstalk also facilitates the accumulation and tumor promotion function of some oncoproteins such as pygopus 2 (PYGO2) and BCL2 apoptosis regulator (BCL2), which mainly regulate WNT signaling and apoptosis, respectively. The famous drug target PDL1 is indirectly regulated by the crosstalk mediating the level of SPOP. Under this circumstance, blocking the phosphorylation could be a potential therapy (Fig. 2).
4.2.1. PDL1
As mentioned above, Zhang et al.24 reported that PDL1 was a new substrate of cullin 3-based SPOP E3 ligase. After being phosphorylated by cyclin D1–CDK4 kinase, SPOP cannot be ubiquitinated and degraded, which will inhibit the accumulation of PDL1, ultimately inducing tumors. As expected, they found that the combination of CDK4/6 inhibitor and anti-PD-1 therapy dramatically improved the overall survival of immunoproficient mice bearing CT26 tumors.
4.2.2. c-Fos
Phosphorylation of Fos proto-oncogene, AP-1 transcription factor subunit (c-Fos) protein, which is encoded by the proto-oncogene c-Fos, mediates its own stability and induces the proliferation of cancer cells77,78. Han et al.77 found that EGF-mediated phosphorylation at Ser374 of c-Fos compromises the binding of c-Fos to lysine demethylase 2B (KDM2B), a component of CRL1s, which inhibits c-Fos ubiquitination and degradation. Imitating the low-phosphorylation level mutant S374A of c-Fos suppresses cell proliferation, while imitating the high-phosphorylation level mutant S374D promotes progression, indicating that phosphorylation may result in the occurrence of cancer by impacting the normal ubiquitin degradation of oncoprotein.
4.2.3. PYGO2
Overexpression of PYGO2, a coactivator and chromatin effector in the WNT signaling pathway, is involved in the function of WNT signaling in tumorigenesis and has been found in various cancer cells79, 80, 81. However, the stability of PYGO2 can be mediated by the crosstalk of phosphorylation and ubiquitylation. AKT serine/threonine kinase (AKT) contributes to PYGO2 phosphorylation at Ser48 and impairs the ubiquitylation and degradation of PYGO2, during which process the cullin 4 E3 ligase functions as an ubiquitin E3 ligase, preserving the protein level of PYGO2 and leading to the ultimate occurrence of cancers45.
4.2.4. BCL2
BCL2 is characterized as an anti-apoptotic protein and is overexpressed in various cancer cells82,83. Phosphorylation can regulate the stability of BCL2. It has been found that the pyruvate kinase M2 isoform (PKM2) could phosphorylate BCL2 at Thr69 and subsequently prevent its interaction with cullin 3-based E3 ligase, which leads to the upregulation of BCL2 protein expression. When depleted of endogenous PKM2 or BCL2, cancer cells reconstituted with rPKM 2 WT or rBCL2 WT can rapidly cause tumorigenesis, and after injection in mice, the survival time of the mice can be significantly shortened. In contrast, expression of rPKM2 ILLL or rBCL2 T69A does not have such characteristics41. These results indicate that phosphorylation mediated inhibition of the ubiquitin proteasome system can result in the occurrence of cancers.
4.2.5. NRF2
As mentioned above, phosphorylated P62 can stabilize NRF2 by inhibiting its binding to KEAP161. Notably, the increase of phosphorylated P62-dependent NRF2 can cause the proliferation of hepatocellular carcinomas (HCC)84; thus treatments based on interrupting the phosphorylation of P62 and the interaction of P62 and KEAP1 may be useful for curing HCC. Moreover, it has been reported that phosphatidylinositol 3-kinase catalytic subunit type 3 (Vps34) can enhance protein kinase C (PKC)-δ-mediated phosphorylation of P62, resulting in the accumulation of NRF2 and the transcription of related oncogenes, ultimately promoting the growth of human breast cancer cells85.
4.3. Promoting cancers: Abrogating the functions of tumor suppressors
4.3.1. VACM-1
Notably, receptor proteins of CRLs, which are also characterized as tumor suppressors, can be phosphorylated. It has been reported that phosphorylated receptors will lose the anti-cancer activity or be degraded by the ubiquitin-proteasome system. For example, protein kinases A- and C-mediated phosphorylation inhibits NEDD8 ubiquitin like modifier (NEDD8) modification, mediating activation of cullin 5-based VACM-1 E3 ligase, and impairs its function of preventing cancer in cell lines49,86(Fig. 2).
4.3.2. FBW7
Another case that components of the CRLs complex are phosphorylated and then promote cancer progression is FBW764,87, a substrate recognition protein of the SCF ubiquitin ligase complex. KRAS proto-oncogene, GTPase (KRAS) mutations activate mitogen-activated protein kinase 1 (ERK) and then promote the degradation of FBW7 through the ubiquitin–proteasomes pathway in a phosphorylation-dependent manner, sequentially abrogating the function of FBW7 as a tumor suppressor. It was found that phospho-deficient mutation of FBW7 can inhibit tumorigenesis of pancreatic cancer, indicating it could be a potential therapy for cancer treatment47,88.
5. Tumor therapies based on kinases that regulate the crosstalk between phosphorylation and ubiquitination
The multiple means of regulation of phosphorylation towards cullin-based ubiquitination shed light on the therapies based on kinase activities for those diseases caused by dysregulation of ubiquitination substrates (Fig. 2B and Table 324,39,45,93, 94, 95, 96, 97, 98, 99, 100). Some kinase activators and inhibitors might exert more therapeutic effects in tumors, as they can regulate the stability of some oncoproteins and tumor suppressors through altering their phosphorylation and ubiquitination. Although the kinases may have other substrates that can also be influenced by inhibitors or activators, the successful clinical applications or clinical trials might prove their safety and efficacy, indicating that the regulation of kinases based on the crosstalk between phosphorylation and ubiquitination is a potential strategy. Three CDK 4/6 inhibitors have been approved by the FDA for treatment of hormone receptor (HR)-positive ERB-b2 receptor tyrosine kinase 2 (HER2)-negative advanced breast cancer97. Apart from the main mechanism of inhibiting the phosphorylation of the retinoblastoma tumor-suppressor protein Rb in tumor treatment, palbociclib was also found to be involved in the regulation of the crosstalk between phosphorylation and ubiquitination, and might exert a positive combined effect with immunotherapy24. Other kinase inhibitors which are involved in this crosstalk, such as the AKT inhibitor triciribine hydrate98 and the CKI activator etoposide99, are also approved or have already completed clinical trials. Apart from the main point provided in this review that the crosstalk between phosphorylation and ubiquitination is important in tumorigenesis, it also has to make allowance for the safety and efficacy of the kinases inhibitors and activators when they are put into use.
Table 3.
Summary of tumor therapies based on the kinases which regulate the crosstalk between phosphorylation and ubiquitination.
| Substrate | Kinase | Inhibitor/activator | Cancer | Ref. |
|---|---|---|---|---|
| SPOP | CDK4/6 | Palbociclib | Colon cancer | 24 |
| c-Fos | EGF | Dihydrocapsaicin | Neoplastic cell transformation | 89 |
| c-Fos | EGF | R1881 | Prostate cancer | 90 |
| PYGO2 | AKT | TCN | Various cancers | 45 |
| BCL2 | PKM2 | PKM2 389–405 peptide | Glioma | 91 |
| BCL2 | PKM2 | Benserazide | Melanoma | 92 |
| BCL2 | PKM2 | Shikonin | Various cancers | 93 |
| ERG | CKI | Etoposide | Prostate cancer | 39 |
| FBW7 | ERK | SCH772984 | Liver cancer | 94 |
| P62 | PKCδ | Rottlerin | Hepatocellular carcinomas | 97 |
| P62 | PKCδ | Sotrastaurin | Non-small cell lung cancer | 98 |
5.1. CDK4/6 inhibitors
Generally, CDK4/6 inhibitors are used for treating HR-positive HER2-negative advanced breast cancer by decreasing the phosphorylation of Rb and disrupting cell cycle progression100. A recent study reported that cyclin D1–CDK4 could also phosphorylate SPOP at Ser6. When the phosphorylation of SPOP is inhibited by palbociclib, a CDK4/6 inhibitor, SPOP poly-ubiquitination is increased and SPOP degradation is promoted. This study also found that PDL1 was a new substrate of cullin 3-based SPOP E3 ligase; thus palbociclib could increase the protein level of PDL1 by promoting the degradation of SPOP. The author combined palbociclib and anti-PD-1 therapy to treat immunoproficient mice bearing CT26 tumors and found that this combination could improve the overall survival relative to either treatment alone24. This work might broaden the use of CDK4/6 inhibitors through their function of indirectly regulating ubiquitinated substrates, and indicate their potential for being combined with immunotherapy in tumor treatment.
5.2. EGF inhibitors
Given that EGF phosphorylates the oncoprotein c-Fos and impairs its ubiquitination and degradation, Lee et al.89 reported that dihydrocapsaicin, a natural agent from chili pepper, could inhibit EGF-induced c-Fos, and ultimately suppress neoplastic cell transformation. Similarly, the androgen analogue R1881 represses EGF-induced c-Fos in prostate cancer cells, which promotes the ability of the combination of R1881 and TPA-induced prostate cancer cell death90.
5.3. AKT inhibitors
It has been confirmed that the AKT inhibitor triciribine hydrate (TCN) can disturb the stability of oncoprotein PYGO2 by suppressing its phosphorylation and ubiquitination. Considering that PYGO2 is important for the initiation of many kinds of cancers, TCN has potential for cancer treatment45. In fact, phase I clinical trials of TCN in the setting of solid tumors and hematological malignancies have been completed and have shown good anticancer effects with TCN98.
5.4. PKM2 inhibitors
PKM2 is important in tumorigenesis, and it has been recently reported that PKM2-mediated phosphorylation of BCL2 promotes glioma malignancy and prognosis. PKM2 389–405 peptide was used to antagonize the capacity of PKM2 to inhibit the phosphorylation and proteasome degradation of BCL2, which effectively suppressed gliomagenesis, indicating that it had a tumor therapeutic potential91. Moreover, benserazide and shikonin have been found to inhibit PKM2 and exert important functions in melanoma treatment and drug-sensitive and resistant cancer treatment respectively92,93.
5.5. CKI activators
It has been reported that the topoisomerase inhibitor etoposide can impair the migration of prostate cancer cells, the main mechanism of which is that etoposide activates CKI and then promotes the phosphorylation of ERG, contributing to its ubiquitination and degradation39. Etoposide plus estramustine showed efficacy and safety in a phase II trial in the setting of hormone-refractory prostate cancer101. SRC-3 is another oncoprotein substrate of SPOP, which can be phosphorylated by CKI and then promoted to ubiquitinated degradation; thus, it is reasonable that etoposide could enhance the function of SPOP, inhibiting oncogenic signaling mediated by SRC-3.
5.6. ERK inhibitors
ERK signaling plays an important role in various tumors. One of the mechanisms is that it phosphorylates FBW7, a tumor suppressor, and induces its ubiquitination and degradation. Morris et al.102 reported a novel ERK kinase inhibitor SCH772984, which inhibited cell proliferation and impaired tumor progression. Subsequently, Broutier et al.94 illustrated that SCH772984 could have a therapeutic effect on primary liver cancer. These results support the idea that regulating the kinases involved in the crosstalk between phosphorylation and ubiquitination could be a potential therapy for tumor treatment.
5.7. PKC δ inhibitors
It has been found that PKC δ-mediated phosphorylation of P62 causes the proliferation of HCC. Subsequently, Xia et al.95 reported that the PKC δ inhibitor rottlerin could suppress the progression of HCC, indicating that PKC δ is a potential target for tumor treatments through regulation of the crosstalk between phosphorylation and ubiquitination. In addition, PKC δ is involved significantly in tyrosine kinase inhibitor-resistant cancers, and the combination of EGFR inhibitors and the PKC δ inhibitor sotrastaurin can effectively regress resistant EGFR-mutant non-small cell lung cancer96.
5.8. Other potential therapies based on kinases
DYRK phosphorylates ID2 and suppresses its interaction with VHL, thus promoting the ubiquitinated degradation of HIF2α, which leads to tumor suppression and positive clinical outcomes in glioblastoma patients38. These findings indicate that DYRK activators are potential therapies for glioblastoma treatment. Similarly, ATM activator may have an important function in suppressing cancers, as it can promote the phosphorylation and ubiquitinated degradation of MDM2, thus increasing the level of the tumor suppressor P5333. Based on the understanding of the crosstalk between phosphorylation and ubiquitylation, it is important to determine which types of kinases play critical roles in the above progression and can be addressed for tumor treatments.
6. Perspectives
Different protein translational modifications can be affected by each other under various circumstances. Ubiquitylation, which utilizes E3 ligases that are mainly part of the CRLs family, has been reported to be regulated mainly by phosphorylation, acetylation and methylation. The crosstalk between phosphorylation and ubiquitylation exists in many biological processes and impacts the physiology and pathology by controlling the levels of targeted proteins.
The characteristic that CRLs are prone to recognize those proteins with Ser/Thr rich degrons may explain why phosphorylation can regulate ubiquitylation substrates. Phosphorylation-mediated proteasome degradation of some substrates preserves their function, hinting that the dysregulation of phosphorylation or ubiquitylation may cause the latter pathologies such as genomic instability, cell cycle arrest or disorders of transcription. Therefore, regulating phosphorylation may facilitate the biological function of ubiquitylation substrates, mainly by regulating the process by which receptors and adaptors recognize substrates and mediate the stability and activity of these substrates, as well as CRLs themselves.
Considering that the substrates of CRLs are diverse and consist of oncoproteins and tumor suppressors, it would be valuable to consider that kinase activators could be put into use to phosphorylate oncoproteins and cause them to undergo ubiquitylation and degradation. Kinase inhibitors can be used to inhibit the phosphorylation and ubiquitinated degradation of tumor suppressors. Therefore, learning more about the crosstalk may provide greater probability and specificity for kinase activators and inhibitors to be used in tumor treatment, promote the combination of kinase therapies and other therapies such as immunotherapies, and push forward the development of new therapies based on a new function, phosphorylation. The study of crosstalk between phosphorylation and ubiquitylation will extend the knowledge of the ubiquitin-proteasome system, including how it functions and how to control this system, providing new potential therapies for tumor treatment.
Acknowledgments
This work was supported by the State Key Program of National Natural Science Foundation of China (No. 81830107 to Qiaojun He) and grant from the National Natural Science Foundation of China (No. 81973354 to Meidan Ying).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Contributor Information
Qiaojun He, Email: qiaojunhe@zju.edu.cn.
Meidan Ying, Email: mying@zju.edu.cn.
Author contributions
Yifan Chen, Meidan Ying and Qiaojun He conceived and designed the conception of the review. Xuejing Shao and Ji Cao collected literatures. Hong Zhu, Bo Yang, Meidan Ying, Qiaojun He and Yifan Chen analyzed literatures and summarized results. Yifan Chen drafted the manuscript and drew the figures. Meidan Ying, Qiaojun He and Yifan Chen revised the manuscript.
Conflicts of interest
The authors have no conflicts of interest to declare.
References
- 1.Nalepa G., Rolfe M., Harper J.W. Drug discovery in the ubiquitin–proteasome system. Nat Rev Drug Discov. 2006;5:596–613. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- 2.Yu Q., Jiang Y., Sun Y. Anticancer drug discovery by targeting cullin neddylation. Acta Pharm Sin B. 2020;10:746–765. doi: 10.1016/j.apsb.2019.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petroski M.D., Deshaies R.J. Function and regulation of cullin–RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 4.Cheng J., Guo J., Wang Z., North B.J., Tao K., Dai X. Functional analysis of Cullin 3 E3 ligases in tumorigenesis. Biochim Biophys Acta Rev Canc. 2018;1869:11–28. doi: 10.1016/j.bbcan.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pintard L., Willems A., Peter M. Cullin-based ubiquitin ligases: Cul3–BTB complexes join the family. EMBO J. 2004;23:1681–1687. doi: 10.1038/sj.emboj.7600186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jang S.M., Redon C.E., Aladjem M.I. Chromatin-bound cullin-Ring ligases: regulatory roles in DNA replication and potential targeting for cancer therapy. Front Mol Biosci. 2018;5 doi: 10.3389/fmolb.2018.00019. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skaar J.R., Pagan J.K., Pagano M. SCF ubiquitin ligase-targeted therapies. Nat Rev Drug Discov. 2014;13:889–903. doi: 10.1038/nrd4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y., Jiang X., Feng F., Liu W., Sun H. Degradation of proteins by PROTACs and other strategies. Acta Pharm Sin B. 2020;10:207–238. doi: 10.1016/j.apsb.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koren I., Timms R.T., Kula T., Xu Q., Li M.Z., Elledge S.J. The eukaryotic proteome is shaped by E3 ubiquitin ligases targeting C-terminal degrons. Cell. 2018;173:1622–1635. doi: 10.1016/j.cell.2018.04.028. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng J., Guo J., North B.J., Tao K., Zhou P., Wei W. The emerging role for Cullin 4 family of E3 ligases in tumorigenesis. Biochim Biophys Acta Rev Canc. 2019;1871:138–159. doi: 10.1016/j.bbcan.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ni Z., He J., Wu Y., Hu C., Dai X., Yan X. AKT-mediated phosphorylation of ATG4B impairs mitochondrial activity and enhances the Warburg effect in hepatocellular carcinoma cells. Autophagy. 2018;14:685–701. doi: 10.1080/15548627.2017.1407887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferry C., Gaouar S., Fischer B., Boeglin M., Paul N., Samarut E. Cullin 3 mediates SRC-3 ubiquitination and degradation to control the retinoic acid response. Proc Natl Acad Sci U S A. 2011;108:20603–20608. doi: 10.1073/pnas.1102572108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishizawa K., Wang Q., Li J., Yamazaki O., Tamura Y., Fujigaki Y. Calcineurin dephosphorylates Kelch-like 3, reversing phosphorylation by angiotensin II and regulating renal electrolyte handling. Proc Natl Acad Sci U S A. 2019;116:3155–3160. doi: 10.1073/pnas.1817281116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humphrey S.J., James D.E., Mann M. Protein phosphorylation: a major switch mechanism for metabolic regulation. Trends Endocrinol Metab. 2015;26:676–687. doi: 10.1016/j.tem.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 15.Asghar U., Witkiewicz A.K., Turner N.C., Knudsen E.S. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov. 2015;14:130–146. doi: 10.1038/nrd4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu M., Ren Z., Wang X., Comer A., Frank J.A., Ke Z.J. ErbB2 and p38γ MAPK mediate alcohol-induced increase in breast cancer stem cells and metastasis. Mol Canc. 2016;15 doi: 10.1186/s12943-016-0532-4. 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q., Shi Q., Chen Y., Yue T., Li S., Wang B. Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proc Natl Acad Sci U S A. 2009;106:21191–21196. doi: 10.1073/pnas.0912008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou S., Ma C., Yang F., Xu X., Jia J., Liu Z. FBXO31 suppresses gastric cancer emt by targeting snail 1 for proteasomal degradation. Mol Canc Res. 2018;16:286–295. doi: 10.1158/1541-7786.MCR-17-0432. [DOI] [PubMed] [Google Scholar]
- 19.Zheng H., Shen M., Zha Y.L., Li W., Wei Y., Blanco M.A. PKD1 phosphorylation-dependent degradation of SNAIL by SCF-FBXO11 regulates epithelial-mesenchymal transition and metastasis. Canc Cell. 2014;26:358–373. doi: 10.1016/j.ccr.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lander R., Nasr T., Ochoa S.D., Nordin K., Prasad M.S., Labonne C. Interactions between Twist and other core epithelial-mesenchymal transition factors are controlled by GSK3-mediated phosphorylation. Nat Commun. 2013;4:1542. doi: 10.1038/ncomms2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teckchandani A., Laszlo G.S., Simo S., Shah K., Pilling C., Strait A.A. Cullin 5 destabilizes Cas to inhibit Src-dependent cell transformation. J Cell Sci. 2013;127:509–520. doi: 10.1242/jcs.127829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Werner A., Baur R., Teerikorpi N., Kaya D.U., Rape M. Multisite dependency of an E3 ligase controls monoubiquitylation-dependent cell fate decisions. Elife. 2018;7 doi: 10.7554/eLife.35407. e35407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi Q., Li S., Li S., Jiang A., Chen Y., Jiang J. Hedgehog-induced phosphorylation by CK1 sustains the activity of Ci/Gli activator. Proc Natl Acad Sci U S A. 2014;111:E5651–E5660. doi: 10.1073/pnas.1416652111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J., Bu X., Wang H., Zhu Y., Geng Y., Nihira N.T. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553:91–95. doi: 10.1038/nature25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei S., Pei Y., Wang Y., Guan H., Huang Y., Xing T. Role of human Keap 1 S53 and S293 residues in modulating the binding of Keap1 to Nrf 2. Biochimie. 2019;158:73–81. doi: 10.1016/j.biochi.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Zhang T., Dong K., Liang W., Xu D., Xia H., Geng J. G-protein-coupled receptors regulate autophagy by ZBTB16-mediated ubiquitination and proteasomal degradation of Atg14L. Elife. 2015;4 doi: 10.7554/eLife.06734. e06734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomimatsu N., Mukherjee B., Harris J.L., Boffo F.L., Hardebeck M.C., Potts P.R. DNA-damage-induced degradation of EXO1 exonuclease limits DNA end resection to ensure accurate DNA repair. J Biol Chem. 2017;292:10779–10790. doi: 10.1074/jbc.M116.772475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D'Angiolella V., Donato V., Forrester Frances M., Jeong Y.T., Pellacani C., Kudo Y. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 2012;149:1023–1034. doi: 10.1016/j.cell.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burrows A.C., Prokop J., Summers M.K. Skp 1-Cul 1-F-box ubiquitin ligase (SCFβTrCP)-mediated destruction of the ubiquitin-specific protease USP37 during G2-phase promotes mitotic entry. J Biol Chem. 2012;287:39021–39029. doi: 10.1074/jbc.M112.390328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan R., Nakajima S., Wang Q., Sun H., Xue J., Wu J. Nek7 protects telomeres from oxidative DNA damage by phosphorylation and stabilization of TRF1. Mol Cell. 2017;65:818–831. doi: 10.1016/j.molcel.2017.01.015. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossi M., Duan S., Jeong Y.T., Horn M., Saraf A., Florens L. Regulation of the CRL4Cdt2 ubiquitin ligase and cell-cycle exit by the SCFFbxo11 ubiquitin ligase. Mol Cell. 2013;49:1159–1166. doi: 10.1016/j.molcel.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santra M.K., Wajapeyee N., Green M.R. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature. 2009;459:722–725. doi: 10.1038/nature08011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malonia S.K., Dutta P., Santra M.K., Green M.R. F-box protein FBXO31 directs degradation of MDM2 to facilitate p53-mediated growth arrest following genotoxic stress. Proc Natl Acad Sci U S A. 2015;112:8632–8637. doi: 10.1073/pnas.1510929112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chae U., Lee H., Kim B., Jung H., Kim B.M., Lee A.H. A negative feedback loop between XBP1 and Fbw7 regulates cancer development. Oncogenesis. 2019;8 doi: 10.1038/s41389-019-0124-4. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao P., Yoo S.H., Lee K.J., Rosensweig C., Takahashi J.S., Chen B.P. Phosphorylation of the cryptochrome 1 C-terminal tail regulates circadian period length. J Biol Chem. 2013;288:35277–35286. doi: 10.1074/jbc.M113.509604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shao W., Zumer K., Fujinaga K., Peterlin B.M. FBXO3 protein promotes ubiquitylation and transcriptional activity of AIRE (autoimmune regulator) J Biol Chem. 2016;291:17953–17963. doi: 10.1074/jbc.M116.724401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mao X., Gluck N., Li D., Maine G.N., Li H., Zaidi I.W. GCN5 is a required cofactor for a ubiquitin ligase that targets NF-κB/RelA. Genes Dev. 2009;23:849–861. doi: 10.1101/gad.1748409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee S.B., Frattini V., Bansal M., Castano A.M., Sherman D., Hutchinson K. An ID2-dependent mechanism for VHL inactivation in cancer. Nature. 2016;529:172–177. doi: 10.1038/nature16475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gan W., Dai X., Lunardi A., Li Z., Inuzuka H., Liu P. SPOP promotes ubiquitination and degradation of the ERG oncoprotein to suppress prostate cancer progression. Mol Cell. 2015;59:917–930. doi: 10.1016/j.molcel.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ostertag M.S., Messias A.C., Sattler M., Popowicz G.M. The structure of the SPOP-Pdx 1 interface reveals insights into the phosphorylation-dependent binding regulation. Structure. 2019;27:327–334.e3. doi: 10.1016/j.str.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 41.Liang J., Cao R., Wang X., Zhang Y., Wang P., Gao H. Mitochondrial PKM2 regulates oxidative stress-induced apoptosis by stabilizing Bcl 2. Cell Res. 2016;27:329–351. doi: 10.1038/cr.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spoel S.H., Mou Z., Tada Y., Spivey N.W., Genschik P., Dong X. Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell. 2009;137:860–872. doi: 10.1016/j.cell.2009.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu C.C., Lin Y.C., Chen Y.H., Chen C.M., Pang L.Y., Chen H.A. Cul3-KLHL20 ubiquitin ligase governs the turnover of ULK1 and VPS34 complexes to control autophagy termination. Mol Cell. 2016;61:84–97. doi: 10.1016/j.molcel.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 44.Leung-Pineda V., Huh J., Piwnica-Worms H. DDB1 targets Chk1 to the Cul 4 E3 ligase complex in normal cycling cells and in cells experiencing replication stress. Cancer Res. 2009;69:2630–2637. doi: 10.1158/0008-5472.CAN-08-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Q., Li Y., Gu B., Fang L., Zhou P., Bao S. Akt phosphorylates Wnt coactivator and chromatin effector Pygo2 at Serine 48 to Antagonize its ubiquitin/proteasome-mediated degradation. J Biol Chem. 2015;290:21553–21567. doi: 10.1074/jbc.M115.639419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choudhury R., Bonacci T., Wang X., Truong A., Arceci A., Zhang Y. The E3 ubiquitin ligase SCF(Cyclin F) transmits AKT signaling to the cell-cycle machinery. Cell Rep. 2017;20:3212–3222. doi: 10.1016/j.celrep.2017.08.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ji S., Qin Y., Shi S., Liu X., Hu H., Zhou H. ERK kinase phosphorylates and destabilizes the tumor suppressor FBW7 in pancreatic cancer. Cell Res. 2015;25:561–573. doi: 10.1038/cr.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shibata S., Arroyo J.P., Castañeda-Bueno M., Puthumana J., Zhang J., Uchida S. Angiotensin II signaling via protein kinase C phosphorylates Kelch-like 3, preventing WNK4 degradation. Proc Natl Acad Sci U S A. 2014;111:15556–15561. doi: 10.1073/pnas.1418342111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bradley S.E., Johnson A.E., Le I.P., Oosterhouse E., Hledin M.P., Marquez G.A. Phosphorylation of VACM-1/Cul 5 by protein kinase A regulates its neddylation and antiproliferative effect. J Biol Chem. 2010;285:4883–4895. doi: 10.1074/jbc.M109.085225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nimonkar A.V., Genschel J., Kinoshita E., Polaczek P., Campbell J.L., Wyman C. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen C.W., Li Y., Hu S., Zhou W., Meng Y., Li Z. DHS (trans-4,4′-dihydroxystilbene) suppresses DNA replication and tumor growth by inhibiting RRM2 (ribonucleotide reductase regulatory subunit M2) Oncogene. 2019;38:2364–2379. doi: 10.1038/s41388-018-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Foskolou I.P., Jorgensen C., Leszczynska K.B., Olcina M.M., Tarhonskaya H., Haisma B. Ribonucleotide reductase requires subunit switching in hypoxia to maintain DNA replication. Mol Cell. 2017;66:206–220. doi: 10.1016/j.molcel.2017.03.005. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L., Tu Z., Liu C., Liu H., Kaldis P., Chen Z. Dual roles of TRF1 in tethering telomeres to the nuclear envelope and protecting them from fusion during meiosis. Cell Death Differ. 2018;25:1174–1188. doi: 10.1038/s41418-017-0037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terai K., Abbas T., Jazaeri A.A., Dutta A. CRL4(Cdt2) E3 ubiquitin ligase monoubiquitinates PCNA to promote translesion DNA synthesis. Mol Cell. 2010;37:143–149. doi: 10.1016/j.molcel.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patke A., Murphy P.J., Onat O.E., Krieger A.C., Özçelik T., Campbell S.S. Mutation of the human circadian clock gene CRY1 in familial delayed sleep phase disorder. Cell. 2017;169 doi: 10.1016/j.cell.2017.03.027. 203-15.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mu P., Akashi T., Lu F., Kishida S., Kadomatsu K. A novel nuclear complex of DRR1, F-actin and COMMD1 involved in NF-κB degradation and cell growth suppression in neuroblastoma. Oncogene. 2017;36:5745–5756. doi: 10.1038/onc.2017.181. [DOI] [PubMed] [Google Scholar]
- 57.Orel L., Neumeier H., Hochrainer K., Binder B.R., Schmid J.A. Crosstalk between the NF-κB activating IKK-complex and the CSN signalosome. J Cell Mol Med. 2010;14:1555–1568. doi: 10.1111/j.1582-4934.2009.00866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoshizaki Y., Mori T., Ishigami-Yuasa M., Kikuchi E., Takahashi D., Zeniya M. Drug-repositioning screening for Keap1-Nrf2 binding inhibitors using fluorescence correlation spectroscopy. Sci Rep. 2017;7 doi: 10.1038/s41598-017-04233-3. 3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baird L., Llères D., Swift S., Dinkova-Kostova A.T. Regulatory flexibility in the Nrf 2-mediated stress response is conferred by conformational cycling of the Keap 1–Nrf2 protein complex. Proc Natl Acad Sci U S A. 2013;110:15259–15264. doi: 10.1073/pnas.1305687110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abed D.A., Goldstein M., Albanyan H., Jin H., Hu L. Discovery of direct inhibitors of Keap 1–Nrf 2 protein–protein interaction as potential therapeutic and preventive agents. Acta Pharm Sin B. 2015;5:285–299. doi: 10.1016/j.apsb.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ichimura Y., Waguri S., Sou Y.S., Kageyama S., Hasegawa J., Ishimura R. Phosphorylation of p62 activates the Keap 1-Nrf 2 pathway during selective autophagy. Mol Cell. 2013;51:618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 62.Tomlins S.A., Rhodes D.R., Perner S., Dhanasekaran S.M., Mehra R., Sun X.W. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 63.Zhou X.E., Suino-Powell K.M., Li J., He Y., Mackeigan J.P., Melcher K. Identification of SRC3/AIB1 as a preferred coactivator for hormone-activated androgen receptor. J Biol Chem. 2010;285:9161–9171. doi: 10.1074/jbc.M109.085779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yokobori T., Mimori K., Iwatsuki M., Ishii H., Onoyama I., Fukagawa T. p53-altered FBXW7 expression determines poor prognosis in gastric cancer cases. Cancer Res. 2009;69:3788–3794. doi: 10.1158/0008-5472.CAN-08-2846. [DOI] [PubMed] [Google Scholar]
- 65.Chae U., Lee H., Kim B., Jung H., Kim B.M., Lee A.H. A negative feedback loop between XBP1 and Fbw7 regulates cancer development. Oncogenesis. 2019;8 doi: 10.1038/s41389-019-0124-4. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang L.Y., Zhao J., Chen H., Wan L., Inuzuka H., Guo J. SCF(FBW7)-mediated degradation of Brg1 suppresses gastric cancer metastasis. Nat Commun. 2018;9 doi: 10.1038/s41467-018-06038-y. 3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davis R.J., Gönen M., Margineantu D.H., Handeli S., Swanger J., Hoellerbauer P. Pan-cancer transcriptional signatures predictive of oncogenic mutations reveal that Fbw7 regulates cancer cell oxidative metabolism. Proc Natl Acad Sci U S A. 2018;115:5462–5467. doi: 10.1073/pnas.1718338115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou X., Liu W., Hu X., Dorrance A., Garzon R., Houghton P.J. Regulation of CHK1 by mTOR contributes to the evasion of DNA damage barrier of cancer cells. Sci Rep. 2017;7 doi: 10.1038/s41598-017-01729-w. 1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Menolfi D., Jiang W., Lee B.J., Moiseeva T., Shao Z., Estes V. Kinase-dead ATR differs from ATR loss by limiting the dynamic exchange of ATR and RPA. Nat Commun. 2018;9 doi: 10.1038/s41467-018-07798-3. 5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carver B.S., Tran J., Gopalan A., Chen Z., Shaikh S., Carracedo A. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu J., Wu R.C., O'Malley B.W. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Canc. 2009;9:615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li C., Ao J., Fu J., Lee D.F., Xu J., Lonard D. Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene. 2011;30:4350–4364. doi: 10.1038/onc.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pietras A., Hansford L.M., Johnsson A.S., Bridges E., Sjölund J., Gisselsson D. HIF-2 alpha maintains an undifferentiated state in neural crest-like human neuroblastoma tumor-initiating cells. Proc Natl Acad Sci U S A. 2009;106:16805–16810. doi: 10.1073/pnas.0904606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pozo N., Zahonero C., Fernández P., Liñares J.M., Ayuso A., Hagiwara M. Inhibition of DYRK1A destabilizes EGFR and reduces EGFR-dependent glioblastoma growth. J Clin Invest. 2013;123:2475–2487. doi: 10.1172/JCI63623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ju Kazi, Sun J., Phung B., Zadjali F., Flores-Morales A., Rönnstrand L. Suppressor of cytokine signaling 6 (SOCS6) negatively regulates Flt 3 signal transduction through direct binding to phosphorylated tyrosines 591 and 919 of Flt3. J Biol Chem. 2012;287:36509–36517. doi: 10.1074/jbc.M112.376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ehrlich E.S., Wang T., Luo K., Xiao Z., Niewiadomska A.M., Martinez T. Regulation of Hsp 90 client proteins by a Cullin 5-RING E3 ubiquitin ligase. Proc Natl Acad Sci U S A. 2009;106:20330–20335. doi: 10.1073/pnas.0810571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Han X.R., Zha Z., Yuan H.X., Feng X., Xia Y.K., Lei Q.Y. KDM2B/FBXL10 targets c-Fos for ubiquitylation and degradation in response to mitogenic stimulation. Oncogene. 2016;35:4179–4190. doi: 10.1038/onc.2015.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Muhammad N., Bhattacharya S., Steele R., Phillips N., Ray R.B. Involvement of c-Fos in the promotion of cancer stem-like cell properties in head and neck squamous cell carcinoma. Clin Canc Res. 2017;23:3120–3128. doi: 10.1158/1078-0432.CCR-16-2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Watanabe K., Fallahi M., Dai X. Chromatin effector Pygo2 regulates mammary tumor initiation and heterogeneity in MMTV-Wnt 1 mice. Oncogene. 2014;33:632–642. doi: 10.1038/onc.2012.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lu X., Pan X., Wu C.J., Zhao D., Feng S., Zang Y. An in vivo screen identifies PYGO2 as a driver for metastatic prostate cancer. Cancer Res. 2018;78:3823–3833. doi: 10.1158/0008-5472.CAN-17-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Z.M., Wu J.F., Luo Q.C., Liu Q.F., Wu Q.W., Ye G.D. Pygo2 activates MDR1 expression and mediates chemoresistance in breast cancer via the Wnt/β-catenin pathway. Oncogene. 2016;35:4787–4797. doi: 10.1038/onc.2016.10. [DOI] [PubMed] [Google Scholar]
- 82.Correia C., Schneider P.A., Dai H., Dogan A., Maurer M.J., Church A.K. BCL2 mutations are associated with increased risk of transformation and shortened survival in follicular lymphoma. Blood. 2015;125:658–667. doi: 10.1182/blood-2014-04-571786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y., Liu C., Barbier O., Smalling R., Tsuchiya H., Lee S. Bcl 2 is a critical regulator of bile acid homeostasis by dictating Shp and lncRNA H19 function. Sci Rep. 2016;6 doi: 10.1038/srep20559. 20559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bartolini D., Dallaglio K., Torquato P., Piroddi M., Galli F. Nrf 2-p62 autophagy pathway and its response to oxidative stress in hepatocellular carcinoma. Transl Res. 2018;193:54–71. doi: 10.1016/j.trsl.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 85.Jiang X., Bao Y., Liu H., Kou X., Zhang Z., Sun F. VPS34 stimulation of p62 phosphorylation for cancer progression. Oncogene. 2017;36:6850–6862. doi: 10.1038/onc.2017.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Johnson A.E., Le I.P., Buchwalter A., Burnatowska-Hledin M.A. Estrogen-dependent growth and estrogen receptor (ER)-alpha concentration in T47D breast cancer cells are inhibited by VACM-1, a cul 5 gene. Mol Cell Biochem. 2007;301:13–20. doi: 10.1007/s11010-006-9392-3. [DOI] [PubMed] [Google Scholar]
- 87.Kurashige J., Watanabe M., Iwatsuki M., Kinoshita K., Saito S., Hiyoshi Y. Overexpression of microRNA-223 regulates the ubiquitin ligase FBXW7 in oesophageal squamous cell carcinoma. Br J Canc. 2012;106:182–188. doi: 10.1038/bjc.2011.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu P., Wang Y., Li X. Targeting the untargetable KRAS in cancer therapy. Acta Pharm Sin B. 2019;9:871–879. doi: 10.1016/j.apsb.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee J.S., Kim Y.A., Jang Y.J., Oh Y., Byun S. Dihydrocapsaicin inhibits epithelial cell transformation through targeting amino acid signaling and c-Fos expression. Nutrients. 2019;11:1269. doi: 10.3390/nu11061269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shankar E., Song K., Corum S.L., Bane K.L., Wang H., Kao H.Y. A signaling network controlling androgenic repression of c-Fos protein in prostate adenocarcinoma cells. J Biol Chem. 2016;291:5512–5526. doi: 10.1074/jbc.M115.694877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liang J., Cao R., Wang X., Zhang Y., Wang P., Gao H. Mitochondrial PKM2 regulates oxidative stress-induced apoptosis by stabilizing Bcl 2. Cell Res. 2017;27:329–351. doi: 10.1038/cr.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhou Y., Huang Z., Su J., Li J., Zhao S., Wu L. Benserazide is a novel inhibitor targeting PKM2 for melanoma treatment. Int J Canc. 2019;147:139–151. doi: 10.1002/ijc.32756. [DOI] [PubMed] [Google Scholar]
- 93.Chen J., Xie J., Jiang Z., Wang B., Wang Y., Hu X. Shikonin and its analogs inhibit cancer cell glycolysis by targeting tumor pyruvate kinase-M2. Oncogene. 2011;30:4297–4306. doi: 10.1038/onc.2011.137. [DOI] [PubMed] [Google Scholar]
- 94.Broutier L., Mastrogiovanni G., Verstegen M.M., Francies H.E., Gavarró L.M., Bradshaw C.R. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med. 2017;23:1424–1435. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xia J., Ozaki I., Matsuhashi S., Kuwashiro T., Takahashi H., Anzai K. Mechanisms of PKC-mediated enhancement of HIF-1α activity and its inhibition by vitamin K2 in hepatocellular carcinoma cells. Int J Mol Sci. 2019;20:1022. doi: 10.3390/ijms20051022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee P.C., Fang Y.F., Yamaguchi H., Wang W.J., Chen T.C., Hong X. Targeting PKCδ as a therapeutic strategy against heterogeneous mechanisms of EGFR inhibitor resistance in EGFR-mutant lung cancer. Canc Cell. 2018;34:954–969.e4. doi: 10.1016/j.ccell.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Klein M.E., Kovatcheva M., Davis L.E., Tap W.D., Koff A. CDK4/6 inhibitors: the mechanism of action may not be as simple as once thought. Canc Cell. 2018;34:9–20. doi: 10.1016/j.ccell.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brown J.S., Banerji U. Maximising the potential of AKT inhibitors as anti-cancer treatments. Pharmacol Ther. 2017;172:101–115. doi: 10.1016/j.pharmthera.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.O'Dwyer P.J., Leyland-Jones B., Alonso M.T., Marsoni S., Wittes R.E. Etoposide (VP-16-213). Current status of an active anticancer drug. N Engl J Med. 1985;312:692–700. doi: 10.1056/NEJM198503143121106. [DOI] [PubMed] [Google Scholar]
- 100.Teh J.L., Aplin A.E. Arrested developments: CDK4/6 inhibitor resistance and alterations in the tumor immune microenvironment. Clin Canc Res. 2019;25:921–927. doi: 10.1158/1078-0432.CCR-18-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Spitaleri G., Matei D.V., Curigliano G., Detti S., Verweij F., Zambito S. Phase II trial of estramustine phosphate and oral etoposide in patients with hormone-refractory prostate cancer. Ann Oncol. 2009;20:498–502. doi: 10.1093/annonc/mdn650. [DOI] [PubMed] [Google Scholar]
- 102.Morris E.J., Jha S., Restaino C.R., Dayananth P., Zhu H., Cooper A. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Canc Discov. 2013;3:742–750. doi: 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]