Abstract
Organoids are in vitro miniaturized and simplified model systems of organs that have gained enormous interest for modelling tissue development and disease, and for personalized medicine, drug screening and cell therapy. Despite considerable success in culturing physiologically relevant organoids, challenges remain to achieve real-life applications. In particular, the high variability of self-organizing growth and restricted experimental and analytical access hamper the translatability of organoid systems. In this Review, we argue that many limitations of traditional organoid culture can be addressed by engineering approaches at all levels of organoid systems. We investigate cell surface and genetic engineering approaches, and discuss stem cell niche engineering based on the design of matrices that allow spatiotemporal control of organoid growth and shape-guided morphogenesis. We examine how microfluidic approaches and lessons learnt from organs-on-a-chip enable the integration of mechano-physiological parameters and increase accessibility of organoids to improve functional readouts. Applying engineering principles to organoids increases reproducibility and provides experimental control, which will, ultimately, be required to enable clinical translation.
Subject terms: Stem cells, Morphogenesis, Organogenesis, Tissue engineering
Organoids are cellular 3D models of organs, which provide powerful in vitro platforms for the investigation of tissue and disease biology. In this Review, the authors investigate engineering approaches, including cellular engineering, designer matrices and microfluidics, to improve the reproducibility and physiological relevance of organoids.
Introduction
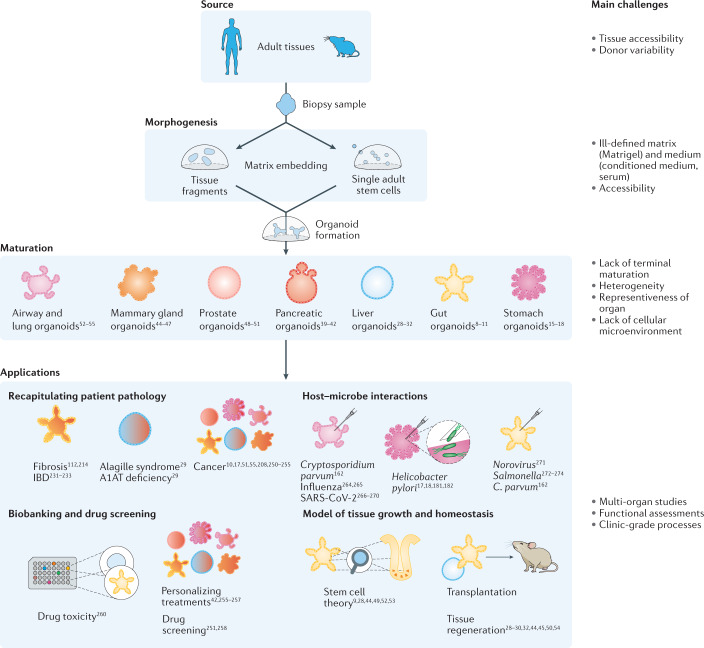
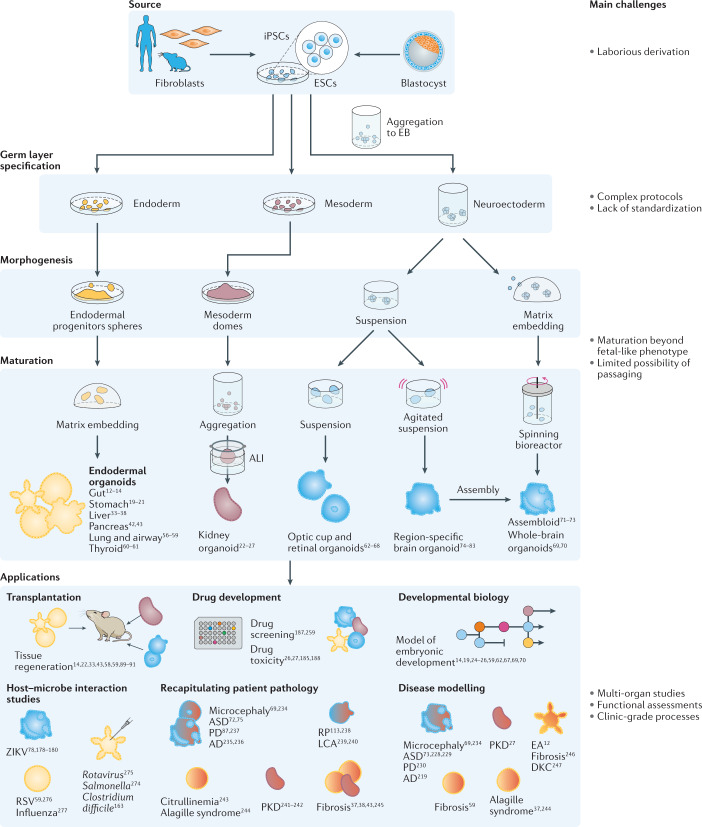
Advances in our understanding of stem cell behaviour have led to the establishment of complex 3D culture systems, termed organoids1–7 (Box 1). Indeed, providing stem cells with a set of biochemical and biophysical cues mimicking the in vivo stem cell niche allows them to maintain their capacity to differentiate into the different cell types of their tissue of origin, and to constantly renew themselves to replenish the stem cell pool, even when cultured in vitro. In these artificial conditions, stem cells cannot only proliferate but also self-organize into complex structures. Compared with traditional 2D culture systems, 3D organoids better resemble the native organ in terms of gene and protein expression, metabolic function and microscale tissue architecture. Hence, similar to well-established organ-on-a-chip systems (Box 2), organoids provide a promising tool to advance personalized medicine and next-generation drug screening, and to limit the need for animal experimentation. Organoids can be established for an increasing variety of organs, including but not limited to gut8–14, stomach15–21, kidney22–27, liver28–38, pancreas39–43, mammary glands44–47, prostate48–51, upper and lower airways52–59, thyroid60,61, retina62–68 and brain69–83 — either from tissue-resident adult stem cells (ASCs), directly sourced from biopsy samples (Fig. 1), or from pluripotent stem cells (PSCs), such as embryonic stem cells (ESCs) or induced PSCs (iPSCs) (Fig. 2), obtained by reprogramming differentiated cell types.
Fig. 1. Tissue-derived organoids.
Organoids can be generated from tissue samples for a variety of organs. A1AT, alpha-1 antitrypsin; IBD, inflammatory bowel disease; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Fig. 2. Pluripotent stem-cell-derived organoids.
Organoids can be generated from pluripotent stem cells for a variety of organs. AD, Alzheimer disease; ALI, air–liquid interface; ASD, autism spectrum disorders; DKC, dyskeratosis congenita; EA, enteric anendocrinosis; EB, embryoid bodies; ESCs, embryonic stem cells; iPSCs, induced pluripotent stem cells; LCA, Leber congenital amaurosis; PD, Parkinson disease; PKD, polycystic kidney disease; RP, retinitis pigmentosa; RSV, respiratory syncytial virus; ZIKV, Zika virus.
In a standard ASC-derived organoid culture system, single stem cells (or small clusters of cells) are seeded into a matrix that mimics essential aspects of the niche in which the cells normally reside1–3. Under appropriate signalling conditions that target key regulatory pathways, the cells undergo proliferation, differentiation, migration and selection, and, despite the lack of spatially organized cues, they grow into structures that can undergo symmetry breaking and acquire a remarkable degree of architectural complexity, to some extent mirroring the organization and cell type diversity of their in vivo organ counterpart. ASC-derived intestinal organoids typically first grow into monolayered spheres, which develop crypt-like protrusions reminiscent of the glandular epithelium of the native tissue. In vivo, morphogen gradients are required to pattern crypts and villi along their axis; remarkably, intestinal organoids acquire a similar organization of cell types in homogeneous culture conditions9 (that is, without any defined external gradients). Organoids further surpass 2D cell cultures in terms of structural resemblance to the corresponding organ and recapitulation of function. For example, intestinal organoids can produce mucus, absorb and secrete biomolecules, and exhibit epithelial barrier integrity84. Owing to stem cell maintenance, ASC-derived organoids can be expanded seemingly indefinitely, making them an incredibly valuable source of untransformed primary cells.
PSC-derived organoids are promising platforms for modelling organs, such as the brain, from which tissue samples cannot be easily obtained, and for modelling the developmental processes of organogenesis. Akin to the development of organs in vivo, PSCs grow, differentiate and eventually form organoids with structural and functional resemblance to the adult organ. For example, protocols for the generation of brain organoids generally start with aggregates of PSCs, which form so-called embryoid bodies. After a first suspension culture step, these embryoid bodies are embedded in an extracellular matrix (ECM)-protein-rich matrix and placed in a spinning bioreactor, in which they grow to self-organized neuroepithelial structures with compartments corresponding to a variety of brain regions69. However, this development is dependent on the stochastic nature of differentiation processes, which, ultimately, leads to high heterogeneity among organoids. Several protocols were developed to better control the differentiation processes, with the aim to generate organoids of a specific brain region85,86; for example, brain organoids resembling the cerebral cortex74,78,83, the cerebellum80,82 or the midbrain78,79. In addition, the generation of midbrain organoids has been achieved starting also from more differentiated neuroepithelial precursor cells87,88, which, ultimately, led to more functional organoids.
Endoderm-derived epithelial organoids, such as gastrointestinal and respiratory organoids, can also be generated from PSCs6,7. PSCs can be differentiated into endodermal progenitors, which are then embedded in ECM-protein-rich matrices, in which they mature under appropriate stimuli to form structures similar to native organs. In contrast to their ASC-derived analogues, they also contain a mesenchymal layer and are usually larger, but, once established, they cannot be easily propagated by simple passaging6,7.
Organoids have proven useful for many applications in basic research and contributed to biomedical advances (Figs 1,2 and Box 3). However, organoids have not yet been widely used for translational studies, such as drug screening or regenerative medicine. Organoids were widely suggested to empower medical transplantation, by the use of healthy cells to replace diseased tissues, and, indeed, several studies in mice justify this hope14,22,28–30,32,33,43–45,50,54,58,59,89–91; however, the road to real-life applications is long, and several limitations have to be overcome to exploit the full promise of organoids.
Box 1 History of organoid systems.
The first in vitro models for organs were based on cancer cells, because cancer cells are immortalized and are easily maintainable. However, the molecular characteristics of cancer cell lines are highly altered, owing to a positive selection for procancerous genetic mutations. Thus, central pathways, including cell cycle regulator genes, growth factors, receptors, tumour suppressors and oncogenes, are dysregulated. In addition, cancer cell lines cannot be established from every patient sample and, therefore, they are not representative for the genetic variation within a population. Moreover, these models were cultured in 2D and, thus, lack spatial organization and fail to recapitulate the whole set of healthy differentiated cell types. Alternatively, primary explanted cells can be used to model mammalian tissues in vitro. These cultures recapitulate cellular heterogeneity and, therefore, are physiologically more relevant. However, they lack the ability to self-renew, and their culture relies on fresh surgical tissues, which limits their applicability.
In 1975, James Rheinwald and Howard Green initiated the field of organoids by showing that co-culture of primary human keratocytes and irradiated mouse fibroblasts leads to the formation of stratified self-organized squamous epithelium, with cell division confined to the basal layer and terminally differentiated, keratinized cells present in the upper layers, reminiscent of stratified skin221. This work was the first long-term culture of untransformed human cells. The importance of culturing cells in 3D, that is, in a hydrogel, was first highlighted by the group of Mina Bissel in 1987, who demonstrated functional differentiation to alveolus-like structures and recapitulation of a pathological state in primary mammary epithelial cells embedded in an extracellular-matrix-like environment, pioneering the field of in vitro morphogenesis222.
Adult stem cells can give rise to all cell types of a tissue in vivo, and they constantly renew themselves to replenish the stem cell pool223. In a landmark study published in 2009, the laboratory of Hans Clevers reported seeding of single intestinal stem cells in an extracellular matrix substitute with tissue-like stiffness and essential soluble niche factors9. The stem cells proliferated and formed complex 3D organized structures containing stem cells and various differentiated cells types, which are now considered the first organoids1–3. These intestinal and other epithelial organoids, such as gastric or hepatic organoids, self-organize into an architecture partially resembling the original tissue9,17,28. The stem cell population in these organoids is maintained, making them a valuable source of untransformed cells. Epithelial organoids are cystic by nature, that is, they are composed of a cellular monolayer with a closed architecture surrounding a central lumen, which makes experimental access challenging and the necessity of reseeding hinders long-term studies.
Organoids developed from human pluripotent stem cells (PSCs) are particularly useful for engineering tissues for which cell retrieval is impractical, such as the brain or retina4,7. PSC-derived organoids were pioneered by the group of Yoshiki Sasai, who formed cortical tissues and optic-cup-like structures from embryonic stem cells in vitro62,83 by mimicking developmental pathways7. Adult-stem-cell-derived and PSC-derived organoids rely on the intrinsic ability of stem cells and their progeny to self-organize and form 3D structures, resembling tissues in vivo, which makes them a promising model system for drug screening and disease modelling.
Box 2 Organoids and organs-on-a-chip.
Organs-on-a-chip are alternative (yet complementary) approaches to organoids for modelling human organs95,224. Organoids rely on the self-organization of growing cell aggregates, whereas organs-on-a-chip are based on a reductionist engineering approach. Organs-on-a-chip aim to capture key functions that are indispensable for the physiological functioning of a specific organ by mimicking the tissue elements that perform these functions in vivo. Thus, organs-on-a-chip usually comprise specialized cell types and the key biochemical and biophysical properties of the in vivo microenvironment. The field of organs-on-a-chip has been landmarked by the lung-on-a-chip device established by Dongeun Huh et al.164, which was then further adapted to model other organs, such as the intestine and kidneys165,166. These devices are amenable to easy readouts and allow precise control of environmental parameters. In addition, organs-on-a-chip are generally reproducible and have longer lifespans compared with organoid systems. However, they often lack the complex cellular architecture of tissues. Moreover, organ-on-a-chip platforms require a complex experimental setup, including tubing, pumps and the requirement of microfabrication equipment and expertise. Finally, the use of cell lines reduces the physiological relevance of organs-on-a-chip compared with organoids.
Box 3 Applications of organoids.
Organoids are widely used to model and investigate tissue development, homeostasis and regeneration, for example, to identify factors required for stem cell differentiation into specific lineages or the molecular mechanisms underlying cell fate programs1–4. Organoids also enable knockout studies of essential genes, which is not feasible in vivo, owing to embryonic lethality. Pluripotent stem cell (PSC)-derived organoids are an invaluable model system for developmental biology; for example, human-specific features of neurodevelopmental processes were discovered by analysing brain organoids from human and non-human primates at a single-cell level219,220.
Modelling diseases in mice is associated with technical and ethical challenges, and murine cells can often not recapitulate all aspects of human pathologies. Alternatively, human organoids can be applied; for example, cancer-promoting genes can be mutated in gastrointestinal organoids114–116, liver organoids225, pancreatic organoids226 or mammary gland organoids227 to investigate the relationship between acquired mutations and growth factor independencies in cancer. For many neurological diseases, including Alzheimer disease (AD), Parkinson disease (PD) and autism spectrum disorders, accurate animal models are lacking. Brain organoids are a valuable model for studying the cellular and molecular processes involved in these neurological diseases75,217,228–230; for example, overexpression of disease-specific mutated forms of the β-amyloid precursor protein and presenilin-1 in an organoid-like 3D culture system allowed the phenotypic recapitulation of AD and revealed the implication of glycogen synthase kinase 3 (GSK-3)217. Similarly, introducing mutations in the gene coding for leucine-rich repeat kinase 2 (LRRK2) enables the generation of a PD model from midbrain organoids230, which recapitulates key pathological features of PD. This model led to new mechanistic insights, such as the involvement of the thioredoxin interacting protein (TXNIP) in LRRK2 mutation-dependent dopaminergic neuron death230.
Organoids can also be generated from cells from patients2,5,6. For example, intestinal organoids derived from patients with cystic fibrosis show aberrant chloride channel function, which can be rescued by correcting the disease-causing mutation112,212. Similarly, organoids from patients with inflammatory bowel disease (IBD) capture pathophysiological aspects of the disease and provide information on transcriptional and methylation alterations231–233. The disease phenotype of individual patients can also be studied using induced PSC (iPSC)-derived organoids2,5,6; for example, brain organoids have been generated from iPSCs established from patients with microcephaly69,234, AD235,236, PD87,237 or autism spectrum disorders72,75. Of note, in the iPSC-derived brain organoids of patients with AD, amyloid pathology was observed before tau hyperphosphorylation, which sheds light on the controversy of AD phenotypes235. Patient-specific iPSC-derived retinal organoids allow the modelling of retinitis pigmentosa113,238 and Leber congenital amaurosis239,240, contributing to the knowledge needed to establish effective gene-editing therapies. Kidney organoids developed from patients with polycystic kidney diseases241,242 exhibit a significant increase in cyst formation compared with healthy control organoids and enabled the discovery of the crucial role of the microenvironment in this disease. Indeed, many other pathologies were also recapitulated in iPSC organoids37,38,43,243–247.
Cancer organoids can be obtained from tumour biopsy samples248,249, including from the gastrointestinal tract10,17,250, liver251,252, breast206, prostate51,253 and lung55,254. Cancer organoids capture the disease heterogeneity and, thus, present an excellent tool for personalized medicine to predict the outcome of clinical treatments42,255–257. They also allow mid-throughput to high-throughput screening of therapeutics185,251,258,259 and assessment of drug toxicity26,27,185,188,260,261.
Organoids are also useful models in infection biology6,262,263; for example, epithelial organoids can be applied to study host–microorganism interactions17–19,59,78,161,176–180,264–277. Gastric organoids were used to study Helicobacter pylori infection17–19,179,180, and brain organoids were used to model and investigate the mechanisms of Zika virus infection78,176–178. Organoids are further valuable research tools in response to the SARS-CoV-2 pandemic. Lung, capillary, kidney and intestinal organoids could be infected with the virus, providing insight into tissue tropism and replication sites266–270.
Limitations of current organoid systems
Limited level of maturity and function
An impressive degree of physiologic functionality has been achieved in intestinal (mucus production and absorptive activities), gastric (histamine-inducible acidification), hepatic (albumin expression, glycogen accumulation and low-density lipoprotein uptake) and mammary gland organoids (milk production)21,30,47,84; however, none of the established organoid systems reproduces the full functional repertoire of their respective organ. Organoids often lack key specialized cell types and fail to recapitulate the complexity of native organs, owing to the (partial) absence of a mesenchymal compartment, vascularization and/or microbiome. Even though multi-compartment organoids have been established, they lack consistent cellular organization, which hinders faithful and robust experimental readouts. The application of flow, an air interface or mechanical stimuli can improve terminal maturation of cells in vitro92–94; however, integration of such features remains technically challenging95. An important drawback of organoid systems is the limited time span for which they can be maintained in culture. Epithelial organoids have lifespans on the order of one week, which is often insufficient to robustly differentiate ASCs into the full set of differentiated cell types expected in vivo. This culture time restriction is even more problematic in PSC-derived organoids, whose lifespans are in vast disagreement with the timing of in vivo organogenesis, especially in human systems. In consequence, these organoids generally fail to mature beyond a fetal phenotype1,2. Accordingly, brain organoids, for instance, are mimicking a fetal brain phenotype and further efforts reinforcing maturation are required to obtain a faithful model of the adult brain70,96.
Accessibility
The limited lifespan of organoids is often a direct consequence of restricted accessibility. As organoids grow in size, diffusion-dependent nutrient supply and waste removal become less efficient. For example, in cystic epithelial organoids, dead cells accumulate in the hollow lumen and, thus, the organoids have to be fragmented and reseeded. In brain organoids, which can grow to several millimetres in size, nutrient inaccessibility leads to necrosis of the inner core, which can only partly be resolved by shaking cultures69,78. In addition to problems with nutrient supply, inaccessibility also poses an issue for controlling the different compartments of the organoid. In epithelial organoids, such as intestinal organoids, experiments often require access to the inner and outer sides of the epithelium. For example, in studies focusing on host–pathogen interactions, microbes need to be delivered to the apical side of the organoid (usually lumen-facing), whereas metabolites and cytokines would preferentially be delivered from the basal side.
Heterogeneity
Organoid systems also suffer from considerable variability in organoid formation efficiency, end-point morphology and function, which is often inherent to the stochastic nature of in vitro self-organization and cell fate choices. Reducing this variability will be essential to fully capitalize on the potential of organoids in disease modelling, drug screening and regenerative medicine. Engineering strategies, such as increasing the degree of automation, the use of defined media and matrices, and exact live assessments, need to be further explored to reduce variability in organoid development. Moreover, the initial conditions of organoid growth contribute to organoid variability, including the starting cell population, their positioning and aggregation. A key limitation to automatization is also the complexity of protocols for organoid generation, which often require multiple experimental steps, in particular, for PSC-derived organoids. For example, simplifying the protocol for the generation of dorsal forebrain organoids substantially increased reproducibility96. Engineering techniques — from engineering cells to whole-organism engineering — are expected to increase the robustness of organoid protocols.
Readouts
Routinely used setups mainly use optical monitoring as readout, which provides only little information about the functionality of the organoids. In situ monitoring of metabolites, secreted peptides or electric potentials is challenged by variabilities related to organoid formation. For example, measuring the trans-epithelial electric resistance provides a standard technique in 2D transwell epithelial culture systems to assess epithelial barrier integrity; however, such measurements are technically challenging in organoids97. Similarly, the functionality of hepatic in vitro systems can be assessed by analysing metabolites of exogenous compounds and endogenous substrates, or the synthesis of bile and of proteins, such as albumin or transferrin98; however, only small amounts of these analytes are usually present in single organoids, which makes their analysis difficult. The integration of miniature biosensors into organoid systems could address this problem, which would require an adaption of the culturing setup. Indeed, typical hydrogel domes containing many organoids are rather impractical for the incorporation of biosensor electrodes. The consistent measurement of analytes would require setups in which the location of organoids can be controlled and the organoid shape can be constrained. Furthermore, high-throughput measurements, which are relevant for drug screening applications, require the implementation of automated functional measurements.
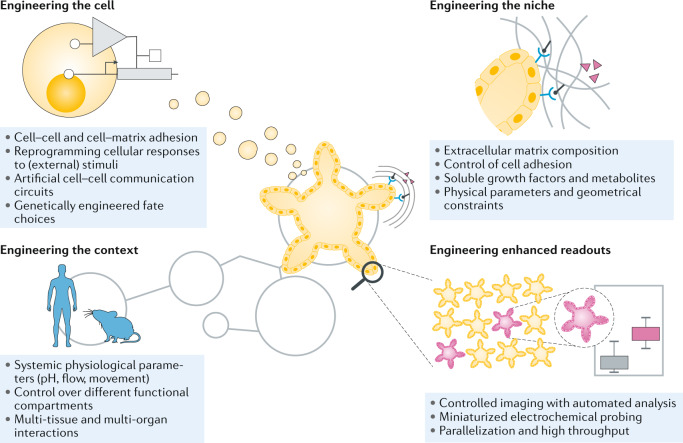
These issues, which limit the translatability of organoids, are inherent to their very design principle. Thus, only the fine-tuning of controllable parameters (for example, choice of cytokines and matrix composition) may not suffice to overcome these limitations. The design of organoid systems needs to be re-evaluated to increase the number of controllable parameters. Engineering approaches, often explored by proof-of-concept studies on organ-on-a-chip technologies, offer the possibility to address many limitations of organoid platforms. In this Review, we discuss engineering strategies at different scales, including engineering of (sub-)cellular behaviour, local (niche) tissue engineering and engineering attempts to create holistic models of entire organisms. Finally, we examine new approaches for the readout of functional development in organoids to allow more thorough characterization and incorporation in high-throughput assay pipelines (Fig. 3).
Fig. 3. Engineering approaches for organoids.
Engineering approaches can be applied at several levels to organoids, including at the cellular level, niche level, multi-tissue level and to improve functional readouts.
Engineering the cell
As the building blocks of organoids, cells are a promising point of manipulation. Indeed, changing the intrinsic properties of cells offers a powerful approach to increase organoid robustness and to tailor them for specific applications.
Surface engineering
Poor robustness of growth and patterning are major limitations of organoids, resulting in highly heterogeneous end-point collections. Indeed, the initial culture conditions determine whether a cell (or cell clusters) will form an organoid by self-organization99,100. In particular, the size and shape of the initial cell aggregates are important starting conditions, and many protocols fail, owing to fine deviations in the initial number of cells. Controlled cell aggregation is usually achieved by microwell structures or microfluidic devices; alternatively, the surface of cells, that is, the cell membrane, can be modified to improve or initiate cell clustering101. For example, cell membranes can be decorated with specific attachment peptides, proteins, nanoparticles, polymers or bio-orthogonal chemical species, usually by bio-orthogonal chemical ligation (for example, using N-hydroxysuccinimide (NHS) esters, sialic acid, oxyamines and derivates thereof), by electrostatic interactions or by liposomal delivery102–104. Bio-orthogonal oxime formation reaction between functionalized cell surfaces (that is, ketone or oxyamine groups on cells bind to each other) then leads to the formation of multilayered microtissues (for example, human mesenchymal stem cells and fibroblasts) with light-mediated control of size and density104. Binding of synthetic DNA fragments to cell surfaces has also been extensively explored; for example, decoration of cell surfaces with 3D DNA origami nanostructures enables the programming of cell–cell adhesion105. Another elegant way of controlling the spatial arrangement of cells in vitro is the application of external magnetic fields to cells magnetized by membrane-binding nanoparticles106. We suggest that such surface engineering approaches could also be applied in the organoid field to preorganize different starting cell types, for example, to align endothelial cells within a larger cluster of hepatocytes, which would allow the generation of more physiologically relevant and long-lived, vascularized liver organoids.
Genetic engineering
In addition to modifying cellular surfaces, a wealth of genetic engineering strategies are available to control the intrinsic properties of a cell107,108. At the genetic level, cells can be engineered in two conceptually different ways: by targeting components of pathways that control stem cell differentiation and/or key niche signals, or by integrating artificial logic circuits inspired by synthetic biology. Organoid systems often lack terminally differentiated cell types, which is related to our limited understanding of the drivers of stem cell fate, and poor recapitulation of in vivo cues by exogenous, broadly applied, signals in vitro. Targeted genome editing through, for example, CRISPR–Cas9 technology has proven its enormous potential for basic research and clinical applications109, despite remaining concerns regarding off-target effects.
Genome editing could be employed to modify the intrinsic response of cells in an organoid to external stimuli with the aim of generating cell types that would otherwise be absent. Also, inducible expression of specific differentiation drivers could be used to favour terminal differentiation of cells that may otherwise only exist in more immature states. For example, targeted transcriptional activation of a protein of the cytochrome P450 superfamily (here, CYP3A4) enhances cellular maturation in PSC-derived liver organoids110. Conversely, genetic engineering could be deployed to knock down relevant signalling pathways in specific subsets of cells to make them unresponsive to the corresponding stimulus, which could address the lack of spatial signalling control in organoids. However, such a targeted approach requires knowledge of the regulatory networks underlying cell fate decisions, which often remains limited. Moreover, gene-editing technology is complex, and genetically engineered organoids are inherently less physiologically relevant, which is especially undesirable for investigations of cell–cell interactions and self-organizational processes.
Gene editing can also be performed in a corrective paradigm111, that is, to correct single disease-causing mutations. For example, gene-corrected patient-derived organoids could provide a source for autologous transplantation to replenish diseased tissue. Gene correction in organoids was first performed on intestinal organoids derived from patients with cystic fibrosis112. Here, the causal mutation (in the CFTR gene) was corrected by CRISPR–Cas9, and the resulting organoids showed functional restoration as assessed in forskolin-induced swelling assays. Retinal organoids, derived from iPSCs from patients with retinitis pigmentosa, could also be gene-corrected to rescue defects in photoreceptor morphology and function113. Although these gene-corrected organoids were not transplanted into animal models, they demonstrate the potential of applying gene-editing technologies for clinical transplantation. Furthermore, in an inverse approach, specific diseases can be engineered in healthy organoids, with the aims of finding mutation-to-disease causalities and modelling specific pathological aspects114–116. For example, the successive induction of single genetic mutations in healthy colorectal organoids allows the simulation of cancer phenotypes. Interestingly, the number of induced mutations correlates with the tumorigenic capacity upon xenotransplantation114; in addition, loss of the adenomatous polyposis coli and p53 is sufficient to induce chromosomal instability and aneuploidy, which are both hallmarks of cancer115. Similar experiments could be performed to establish various relevant disease models in vitro and to highlight or identify causal effects of specific genes on congenital or acquired pathologies. Such experiments would not be physiologically or pathologically relevant in 2D cell cultures, because effects may be dependent on cell types that are missing, and analogous animal studies are associated with technical and ethical challenges1,2.
Genome-editing technologies cannot only be employed to target single genes but also to integrate entire artificial genetic circuits107,108. In particular, synthetic biology enables the engineering of such genetic circuits to generate multicellular logic responses and self-organization through feedback loops. Integration of artificial gene circuits would also benefit the organoid field, albeit such studies have not yet been conducted. Nonetheless, synthetic biology approaches have been used to program cell aggregation in simplified cellular systems117–119. Inspired by the processes in the developing mammalian embryo, simple pattern formation can be engineered using a Turing reaction–diffusion system, composed of the two diffusible ligands Nodal and Lefty. Owing to their mutual activation and repression, characteristic spot-like patterns can be formed in an otherwise homogeneous cell population117. Furthermore, genetically engineered stimuli have the potential to bias cell fate choices; for example, a pulse of the transcription factor GATA6 expression is sufficient to initiate germ layer emergence and symmetry breaking in homogeneous clusters of iPSCs118.
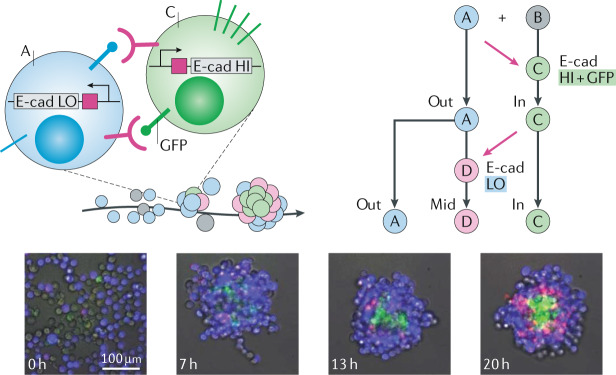
Genetic circuitry can also be combined with cell fate choices, for example, in the engineered Notch-based system synNotch120. In this system, the extracellular domain of the Notch receptor is altered to recognize user-defined ligands, allowing experimental control over the inputs of the normal Notch signal transduction cascade of the cell. Crucially, the intracellular domain of synNotch receptors can also be engineered to activate subcellular signalling factors. Input and output can be controlled, for example, for the contact-dependent transcriptional triggering of fibroblast-to-myoblast transdifferentiation120. The synNotch system has also been incorporated in cadherin-based cell–cell adhesion-controlling circuits in multiple groups of cells121 (Fig. 4). The cells of one group express the CD19 surface ligand and the cells of a second group express a synNotch receptor engineered to respond to CD19 (that is, to the first group), triggering E-cadherin expression. The resulting increase in E-cadherin expression results in cell clustering, owing to increased mutual binding affinity. Interestingly, by introducing a second synNotch receptor in the first cell population, the crosstalk between the two populations leads to self-sorting. The clustered second population activates adjacent cells of the first population to express low concentrations of E-cadherin as well as other genes (that is, a fluorescent reporter). Owing to the different affinities for cell–cell binding, a three-layer self-organized structure with autonomous symmetry breaking and cell type divergence is formed. This system is also able to regenerate patterns upon perturbation, beautifully exemplifying the integration of artificial gene networks in multicellular settings and highlighting that basic morphogenesis can be triggered by a purely synthetic biology approach.
Fig. 4. Engineering cells for guided morphogenesis.
Two populations of genetically engineered cells self-organize into multilayer tissues when mixed together. The microscopy images show cells of type A (blue, BFP+) and cell type B (grey) mixed together and their development into a three-layered structure with cells of type A, C (green, GFP+) and D (pink, mCherry+) by mutual activation and repression of E-cadherin (E-cad) expression. First, in the presence of A-type cells, B-type cells are converted to C-type cells that self-aggregate owing to their high E-cad expression. These cells present GFP at their surface, in turn, leading adjacent A-type cells to express low levels of E-cad and mCherry protein through activation of their synthetic anti-GFP receptor, which will lead to their conversion to D-type cells. BFP, blue fluorescent protein; GFP, green fluorescent protein; HI, high; LO, low. Microscopy images reprinted with permission from ref.121, AAAS.
The engineering of cell-intrinsic properties will certainly gain more traction in the organoid field, not only for fundamental research but also as a versatile toolbox for the precise control of cell fate and, possibly, tailored morphogenesis. Furthermore, the incorporation of advanced techniques, such as optogenetics122, will likely expand the customizability of logic circuits and their application in organoids, by allowing precise spatiotemporal control.
Engineering the niche
Since the establishment of the first ASC-derived organoids9, it has been evident that simulation of the stem cell niche is key for the successful self-organized growth of stem cells in culture. Many features of the stem cell niche, including the ECM, neighbouring cells and their secreted signalling molecules, have been recapitulated; however, such isotropically applied stimuli are not sufficient to grow organoids with high resemblance to their in vivo counterparts for biomedical applications. Indeed, organoid culture conditions suffer from limited cell-responsiveness and batch-to-batch variability. Thus, engineered biomimetic platforms, which provide more faithful proxies of the in vivo niche, may improve control over the growth and differentiation of cells, leading to more physiologically relevant model systems that are amenable for clinical translation.
Designer matrices
The ECM in the stem cell niche is an important parameter for stem cell self-renewal and differentiation, and alterations in ECM composition are a hallmark of many diseases123,124. Therefore, the ECM is the basis of virtually all organoid culture systems. Matrigel (or similar commercially available products such as Geltrex), a laminin-111-rich basement membrane extracted from Engelbreth-Holm-Swarm mouse sarcoma, is the most commonly used matrix for organoids, providing a complex set of ECM signalling inputs and an appropriate mechanical context to organoids in vitro125,126. However, Matrigel is not well defined (which is not surprising, considering that it is composed of nearly 2,000 unique proteins125,126) and offers limited possibility for customization, hindering its incorporation in customized organoid-based assays. In addition, Matrigel is animal-derived, which hampers translation to clinical settings. Therefore, efforts have been made to generate defined hydrogels that overcome these issues, following two general strategies127: matrices generated from naturally occurring materials, such as fibrin128, collagen8,129 or hyaluronic acid130, and synthetic hydrogels126,131,132.
Protein-based or polysaccharide-based biopolymers can be recombinantly produced, reducing ethical concerns and batch-to-batch variability, as compared with animal-derived matrices. Alternatively, hydrogels with synthetic backbones can be decorated with a wide range of bioactive molecules, including ECM molecules or ECM fragments126. For example, poly(ethylene glycol) (PEG)-based hydrogels functionalized with the basement membrane protein laminin enable reproducible intestinal organoid formation131. In these chemically defined hydrogels, intestinal organoid generation relies on the gradual softening of the gel provided by matrix degradation over time. Completely synthetic growth matrices can be generated by functionalization with peptides that match short key amino acid sequences of ECM proteins instead of full-length proteins, for example, peptides corresponding to the fibronectin motif RGD, the collagen motif GFOGER and the laminin motif IKVAV132. Such matrices have shown some success in supporting intestinal organoid formation from single intestinal stem cells, yet, they remain inferior to matrices containing full-length laminin131. However, the viability of PSC-derived intestinal organoids in such synthetic matrices is comparable to viability in Matrigel132.
Synthetic matrices also provide the opportunity to experimentally decouple stiffness, variability and bioactivity of the organoid growth environment, opening up the possibility for screening approaches to investigate the effects of each parameter on stem cell fate131,133. Moreover, synthetic matrices can be designed for specific organoid applications. Indeed, physical properties, in addition to more evident biological features, should be considered for the engineering of new matrices for 3D cell culture. Stiffness is a critical niche parameter influencing stem cell behaviour and appears to be the key determinant of the differentiation of mesenchymal stem cells towards different lineages134. Thus, engineered matrices for organoid cultures should take such physical considerations into account, including stiffness, matrix viscoelasticity and degradability, which have to be optimized for each specific organoid system. Native ECMs are highly dynamic, stress-relaxing matrices — properties that could be engineered in chemically defined hydrogels. In particular, the stress relaxation profile of matrices is directly linked to their viscoelastic and viscoplastic properties, ultimately defining the mechanical confinement of growing cell structures, such as organoids135. To generate biomaterials with tailored relaxation properties, dynamic polymer chemistries can be applied that allow the coexisting of both covalent and weak, reversible crosslinks. Such mechanically sophisticated biomaterials have already shown exciting results for 3D cell culture systems135. For example, the fate of mesenchymal stem cells in tunable 3D alginate hydrogels was shown to be highly dependent on the stress relaxation of matrices, independent of the initial elastic modulus, cell-adhesion-ligand density and degradation136.
Shape-guided morphogenesis
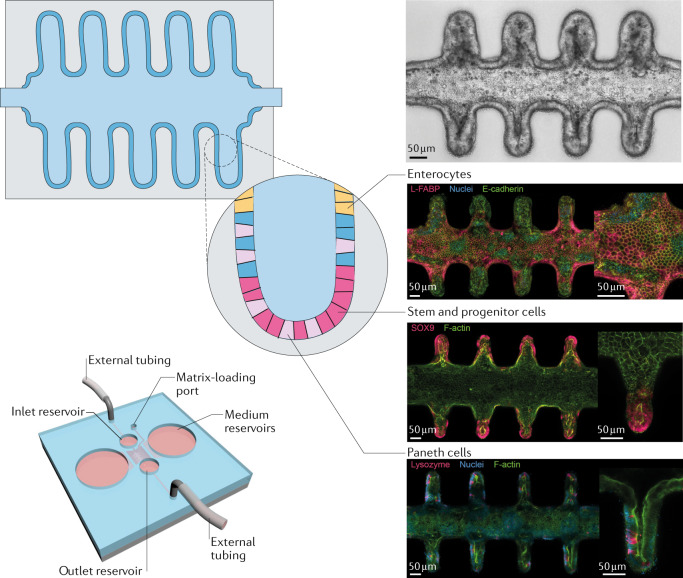
During the development and homeostasis of organs in vivo, physical boundaries play a key role for cellular and tissue organization. To provide such geometrical cues as artificial physical boundaries in vitro, technologies such as micromanufacturing, 3D printing and laser cutting could be used. For example, mammary epithelial cells can be grown in collagen hydrogels with predefined shapes to recapitulate the branching process of mammary epithelial tubules137. The probability of branching and forming new tubules depends on the local geometry induced in the epithelial monolayer. This process is mediated by the diffusion of an autocrine inhibitory morphogen137. In intestinal organoids, the formation of crypt-like structures is a highly stochastic process, which occurs at random orientations and in variable numbers. Microengineered scaffolds allow the robust patterning of intestinal organoids in a predefined manner. For example, a poly(dimethylsiloxane) (PDMS) stamp can be used to pattern a collagen scaffold, onto which organoid-derived cells can then be seeded. In combination with the application of a morphogen gradient akin to gradients observed in vivo, an epithelium can be generated that matches the crypt-villus structure of the template with corresponding proliferative and differentiated regions138. Alternatively, laser-shaped matrices can be applied to grow intestinal organoids into predefined shapes, allowing apical and basal access, crypt-villus-like patterning and, most importantly, long-term homeostasis139 (Fig. 5). In this case, the organoids are shaped into an accessible tubular architecture, enabling the removal of apically shed dead cells as they accumulate over time. The longevity of these ‘mini-intestines’ further allows the development of rare specialized cell types that are absent in short-lived organoids. Intriguingly, the observed cellular patterning reminiscent of the crypt and villus regions is established even in the absence of externally applied biochemical gradients, highlighting the morphogenic guidance potential of scaffold geometry139.
Fig. 5. Engineering the niche.
Intestinal organoids grown in a patterned tubular matrix organize into a crypt-like structure akin to organoids, following the predefined architecture. Stem and progenitor cells (pink, Sox9+), as well as Paneth cells (light pink, Lysozyme+), are found in the crypt structures, whereas enterocytes (yellow, L-FABP+) are present towards the lumen. The integration into a microfluidic device allows the growth for extended periods of time. FABP, fatty-acid-binding protein. Adapted from ref.139, Springer Nature Limited.
Instead of forcing organoids into a final shape, scaffolds can also be used to guide the shape of PSC-derived organoids during their development, that is, at the stage of embryoid bodies140. For example, organoids grown in chemically defined microfilament scaffolds adopt an elongated shape, which leads to the reproducible generation of a neuroectoderm and, ultimately, to improved cortical development compared with standard organoids generated from spherical aggregates140. Confining growing brain organoids to a thin disk shape enables the investigation of the biophysical forces driving the establishment of brain folds141. Interestingly, by employing LIS1-mutated cells, the key features of lissencephaly, a pathological condition showing partial absence of these folds in the brain, could be recapitulated.
Spatiotemporal control
Organogenesis is driven by the intrinsic capacity of stem cells to organize themselves and their progeny. However, organoid studies have shown that the self-organizing ability of stem cells is not sufficient to generate fully functional and mature organs. In vivo, tissue development is subject to external stimuli supplied in a precise spatial and temporal order, which is often not reproduced in traditional 3D culture systems, in which cells are embedded in isotropic matrices and homogeneously flooded with biochemical niche signals. This limitation can be addressed by 3D culture matrices that can release or present biomolecules under spatiotemporal control142–144. The delivery of morphogen gradients by microfluidic devices can further induce controlled symmetry breaking, enabling reproducible organoid formation145–148. For example, a microfabricated device can be used to generate stable opposing gradients of a sonic hedgehog (SHH) agonist and bone morphogenic protein (BMP), mimicking in vivo morphogen gradients important in vertebrae neural tube patterning during spinal cord development. This matrix enables the spatially controlled differentiation of ESCs, akin to development in vivo147. SHH can also be delivered to a forebrain organoid system by genetically engineering human PSC cells to express SHH. These PSCs can then be positioned at one side of the developing brain organoids149 to generate a morphogen gradient, which, ultimately, leads to dorsoventral-like and anteroposterior-like polarization of the forebrain organoids149.
Spatiotemporal control over the presentation of biochemical cues can also be achieved by direct patterning of the growth matrix using engineering methods, which allow greater macroscale control compared with microfluidic-induced gradients127. For example, multiple growth factors promoting cell migration and differentiation could be independently spatiospecifically immobilized within an agarose hydrogel by employing two-photon photochemistry150. Moreover, ECM composition has a drastic impact on the differentiation of stem cells; for example, laminin and fibronectin promote cardiomyocyte and endothelial differentiation of ESCs in vitro, respectively151. Similarly, ECM ligand composition requirements are changing with increasing intestinal organoid maturity131. In the native intestine, the crypt-villus axis is patterned with gradients of ECM proteins, and individual ECM components play specific roles in intestinal stem cell homeostasis and differentiation152–154. Therefore, bioengineering approaches, such as 3D matrix deposition, are likely to further improve crypt-villus regionalization in intestinal organoid cultures or analogous symmetry-breaking events in other organoid systems by guiding patterns in a predefined way.
In addition to the spatial delivery of cues, temporal control may also be necessary to reproducibly generate organoids with a high level of maturation. For example, photochemistry allows the design of matrices that locally release soluble chemical factors (or expose masked ones) in response to light stimulation155,156. Despite the development of a wide variety of photochemistries, the use of such materials for organoid cultures remains underexplored. In intestinal organoids, the photoactivated local release of morphogens could potentially enable spatiotemporal control over the formation of crypt-like buds. Similarly, the mechanical properties of ECMs constantly change during intestinal organoid development131, which could be addressed by introducing light-mediated changes of the mechanical properties of synthetic growth matrices, for example, using photodegradable hydrogels. Likewise, the geometry of the scaffold could be changed over time to guide the shape of emergent organoids, for example, by light-dependent in situ laser ablation157.
In summary, although basic niche mimicry has advanced the generation and maintenance of organoid culture systems, these organoids often exhibit highly heterogeneous shapes and stochastic variability in their level of maturity and functionality. Engineered matrices promise improved control over supplied morphogenic signals in space and time, and, thus, provide the opportunity to mimic the tissue microenvironment not only in its constituent elements but also in its dynamics. Therefore, next-generation engineered materials are expected to further expand the plethora of organoids that can be grown in vitro and increase the developmental robustness of already established cultures.
Engineering the organism-level context
In vivo, organogenesis is regulated by stimuli from the immediate niche, in which the cells are growing. However, organ development is also guided by higher-order inputs, provided by the surrounding tissues and their functions, as well as by system-level parameters, such as fluid flow, mechanical forces or changes in pH or oxygen levels158,159. Organoids are not easily amenable to integration within a whole-organism context; however, such integration would be required to grow physiologically relevant models of homeostatic organs.
Accessibility
Owing to their closed cystic architecture and their continuous expansion, organoids, for example, gastrointestinal organoids, need to be regularly broken up and reseeded to new cultures, which makes them inherently incompatible with long-term studies. Furthermore, functional in situ readouts and experiments involving pathogens require access to the apical side of organoids. Inaccessibility of organoids can be overcome by applying low-throughput microinjections8,17,18,55,160–162 or by using inverted, inside-out versions of organoids163; however, these strategies remain challenging and suffer from low efficiency. Alternatively, polarized intestinal and colonic cell monolayers can be generated on 2D permeable polymer membranes164–166; however, this approach precludes the modelling of 3D tissue biology. The problem of inaccessibility may be better addressed by leveraging engineering approaches to design an organoid system that allows easy experimental access and long-term maintenance of an epithelium. For example, microfluidic devices are promising tools to integrate access channels for waste removal and nutrient supply within organoids, and to enable independent control over experimental conditions95,167.
Integration of mechano-physiological parameters
Mechanical parameters, such as flow, shear stresses, pressure and movements, also have an impact on morphogenesis and tissue homeostasis158,159. For example, blood-flow-induced shear stresses on the endothelium triggers terminal maturation of endothelial cells92. Similarly, diverse physical forces are involved in normal and pathologic intestinal function168. Such tissue-level biomechanical stimuli are lacking in traditional organoid cultures and cannot easily be implemented in current setups. Thus, the incorporation of mechano-physiological parameters calls for a review of the very design principle of organoids, considering combinatorial approaches that involve self-organizing structures and their integration within devices that allow the generation of forces and movements.
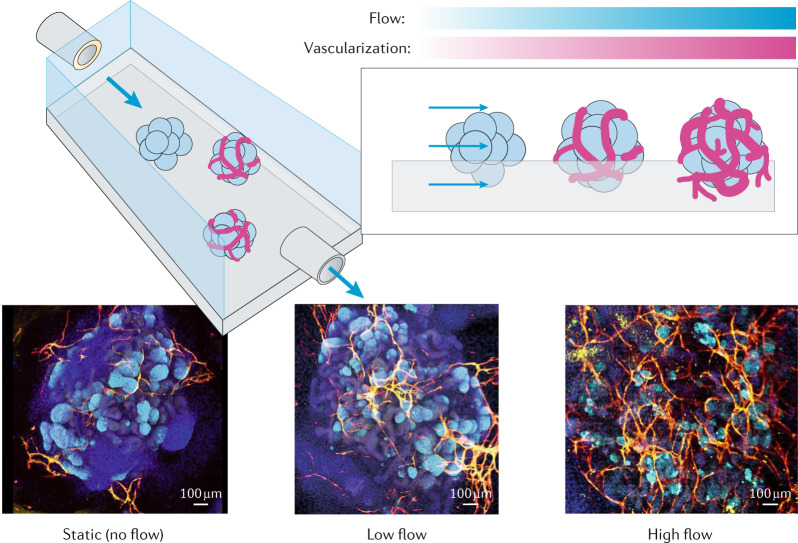
Organ-on-a-chip technology has introduced many techniques for the generation of forces and maintenance of fluid flow; however, the relevance of these systems has often been questioned because the cell lines used did not exhibit cellular diversity95. Taking advantage of organ-on-a-chip setups, single stomach organoids have been cultured on chips, on which the luminal compartment can be accessed with micropipettes to apply luminal flow and pressure cycles to induce peristaltic-like motility169. Although the effect of these external stimuli on cells has not yet been investigated, this system illustrates the possibility of engineering approaches to integrate biophysical factors in organoid cultures. Similarly, kidney organoids have been grown on a chip and exposed to shear stress by application of fluid flow170. The presence of fluid flow not only improves the maturation of the kidney organoids, including tubular and glomerular compartments, but also favours the generation of a vascular network with perfusable lumens (Fig. 6).
Fig. 6. Systemic context engineering for organoid cultures.
Kidney organoids grown in the presence of flow-induced shear stresses show substantially enhanced vascularization and maturation. Confocal microscopy images show organoids under different flow conditions: nuclei (DAPI, blue), vasculature (MCAM, yellow, and PECAM1, red) and podocytes (PODXL, cyan) are stained. DAPI, 4′,6-diamidino-2-phenylindole; MCAM, melanoma cell adhesion molecule; PECAM1, platelet and endothelial cell adhesion molecule 1; PODXL, podocalyxin-like protein 1. Reprinted from ref.170, Springer Nature Limited.
Organ-on-a-chip-like devices also allow the growth of epithelia on 2D membranes with microfluidic channels on both apical and basal sides, providing bilateral accessibility and the ability to apply fluid flow164–166. Intestinal organoid-forming cells, seeded on such a microchip, grow into a functional epithelium with villi-like folds, if also exposed to mechanical stretch cycles mimicking intestinal peristalsis and luminal flow171. Such microfabrication-based devices provide the opportunity to combine biophysical and biochemical stimuli and, thus, increase the physiological relevance of in vitro models and the robustness of their generation protocols.
Multi-organ systems
Many physiological functions and pathological conditions are not attributable to one specific organ but, rather, emerge from the interaction of multiple systems. Current organoids are not capable of modelling biological interactions at higher levels of organization (for example, tissue–tissue or multi-organ interactions). The field of organs-on-a-chip has laid the groundwork for engineering multi-organ systems combined with organoid technology. For example, a microfabricated system combined models of breast cancer with intestine and liver models, simulating the sequential absorption, metabolism and targeted bioactivity of anticancer drugs172. This approach has been extended to the fabrication of an engineered 13-organ complex, called ‘body-on-a-chip’173, showing impressive tissue viability and basic physiological functions (although only five cell lines were used as a proof of concept). Synergistic approaches integrating organ-on-a-chip setups and organoids can approximate the native cellular repertoire and show (patho)physiological relevance. For example, in a microfluidic-based tripartite culture comprising stomach, intestinal and liver organoids, paracrine signalling-dependent regulation of bile acid production could be achieved, recapitulating physiological organ–organ interactions174. Similarly, a heart–lung–liver model was built using bioprinting and microengineering approaches, allowing inter-organ crosstalk175. Interestingly, this setup has provided evidence for lung-dependent cardiotoxicity of an anticancer drug, emphasizing the necessity of multi-organ approaches in pharmacokinetic screenings.
The integration of immune components into organoid systems remains rather unexplored. The immune system does not only play a central role in infectious diseases but also in other pathologies and in normal homeostasis. Thus, the integration of immune components in (disease) model systems substantially improves the degree of their relevance. Viral (for example, Zika virus in brain organoids78,176–178), bacterial (for example, H. pylori in stomach organoids17–19,179,180) or parasitic infections (for example, Cryptosporidium in intestinal organoids160) have been modelled in organoids; however, these studies may lack relevance, owing to the complete or partial absence of important immune components. For example, gastric infections with H. pylori typically persist asymptomatically over decades, which, ultimately, increases the risk of gastric ulcer diseases and gastric cancer181. Such long persistence is enabled by tight host–microorganism interactions; for example, bacteria can evade the immune system by interfering with the differentiation of T cells towards an anti-inflammatory regulatory phenotype182. This immune evasion mechanism could be studied in a triple co-culture of epithelial cells, cytotoxic T cells and bacteria, which led to the identification of H. pylori-induced induction of the checkpoint inhibitor programmed cell death ligand 1 (PDL1)180. In non-infectious diseases, such as IBD, interactions between microbiota, epithelial cells and the immune system play an equally crucial role. Thus, organoids are promising platforms for the elucidation of the mechanisms underlying IBD183. For example, a platform containing a colon-organoid-derived epithelial layer can be complemented with monocyte-derived macrophages as immune compartment using a collagen-containing bioengineered scaffold184. Mimicking Escherichia coli-induced epithelial inflammation results in an increase in macrophage migration and the production of pro-inflammatory cytokines, which are known to be involved in IBD. Protocols for the long-term maintenance of immune cells within organoid systems have yet to be established; however, these pioneering studies underline the potential of organoids in immunological research.
Taken together, approaches towards superior organoids must not only consider aspects of the local niche of a specific organ but also the system-level context. Approximation of systemic parameters promises to yield organoids with longer lifespans, more mature phenotypes, higher cellular diversity and, ultimately, higher clinical predictivity compared with traditional organoid systems. Such systemic parameters include biophysical forces and strains, biochemical signalling with other cell and organoid types, as well as nutrient supply and waste removal.
Improving readouts
Maximizing the benefit of organoid systems is challenged by a lack of appropriate functional readouts. Organoid studies have mostly relied on phenotypic readouts thus far (that is, aspect, shape and number of organoids) (Table 1). However, fully executing the promises of organoids will require the integration of continuous, accurate and versatile functional readouts, which can be automated in a high-throughput manner, to enable applications beyond basic research.
Table 1.
Analytical methods in organoid research
| Method | Advantages | Disadvantages | Physical and technical limitations | Selected refs |
|---|---|---|---|---|
| Image-based analysis | ||||
| Live imaging | Live monitoring; no requirement for specialized equipment; minimally invasive | Little functional insights; dependency on reporter lines for assessment of expression of specific markers | Phototoxicity of fluorescent reporters and probes; motion of samples | 112,186,191,212 |
| Light-sheet imaging | Suitable for live imaging; subcellular resolution; spatial information (3D) | Low throughput; laborious sample preparation; limited to one condition | Simultaneous imaging of many organoids not possible owing to small sample volume | 213–215 |
|
Histochemistry Immunostainings with whole-mount confocal microscopy |
Subcellular resolution; spatial information (3D) | Low throughput; low automatization; single time points | Light penetration for large samples; limited specificity of markers; destructive method | 216 |
|
Histochemistry Routine histology staining and immunostaining on tissue sections |
Well-established protocols; easy visualization of cell-secreted analytes (mucins, ECM proteins) | Low throughput; low automatization; single time points; limited set of standard stains | Destructive method; strict requirement for fixing and slicing of samples | 8,41,69 |
| Electrophysiology | Functional assessment; information about cell-intrinsic properties (Vm, Rm, Cm, ion currents) | Low throughput; laborious sample preparation | Only for electrically active cells (neurons, (photo)receptors) | 75,78,79,88,113 |
| Protein identification and quantification | ||||
| Immunoassays (WB, ELISA) | Functional information (protein abundance, protein–protein interactions, PTMs); high specificity (WB) and high sensitivity (ELISA) | Labour-intensive; low automatization; single time points; dependence on analyte-specific antibodies; no spatial information | Destructive for non-secreted analytes | 29,217 |
| In situ electrochemical sensors | Functional information (abundance of secreted proteins, metabolites); live monitoring | Bulky electrodes; requires calibrations and implementation of electronic circuitry | Low analyte concentrations and volumes; requires one sensor per analyte; suitable only for cell-secreted analytes | 46,195–197 |
| Proteomics (MS) | Proteome-wide functional information (protein abundance, protein–protein interactions, PTMs); quantitative (for example, SILAC) | Costly; low throughput; single time points; expertise needed for processing and analysis; no spatial information | Destructive for non-secreted analytes | 41,218 |
| Flow cytometry | Single-cell level; multiple markers can be analysed simultaneously; cell sorting for downstream analysis (for example, cfu) | No spatial information; high operator dependency | Dissociation into single cells indispensable; limited specificity of markers | 25,47,100 |
| Gene expression | ||||
| PCR | Quantitative gene expression levels; high-sensitivity | No information about abundance of proteins or their PTMs | Destructive method; mRNA levels are only a proxy for the functional state of a cell | 17,37,38 |
| Gene expression arrays and RNA sequencing (bulk and scRNAseq) | Whole transcriptome information; scRNAseq has single-cell-level resolution | Costly; low throughput; single time points; expertise needed for processing and analysis | Destructive method; scRNAseq requires pure single-cell preparation (no doublets, no fragments) | 34,96,219,220 |
cfu, colony-forming units; ECM, extracellular matrix; ELISA, enzyme-linked immunosorbent assay; MS, mass spectrometry; PCR, polymerase chain reaction; PTMs, post-translational modifications; scRNAseq, single-cell RNA sequencing; Vm, Rm and Cm, transmembrane potential, resistance and capacitance, respectively; WB, western blot.
Image-based analysis
Optical observation is probably the oldest technique in biological research and remains one of the most powerful analytical methods. However, typical organoids are generated in 3D matrices and grow at uncontrolled locations, which makes automated live imaging challenging. To enable live cell imaging, organoid growth locations can be engineered by using arrays of microwells185,186 or by capturing cells within microfluidic devices187. For example, intestinal organoids can be trapped post-aggregation in a microfabricated pillar array to monitor morphological changes (for example, swelling)187. Similarly, hepatic organoids can be grown and differentiated directly within a chip to allow downstream analysis188. However, although morphological analysis provides some informative insights, more direct evidence about the molecular and functional state of cells would be desirable. For example, genetically engineered reporter lines, which have traditionally been mainly available for murine organoids owing to technical difficulties in developing such reporters in human cells, have recently also been applied for human organoids. Using genome editing, the efficiency of generating knock-ins (that is, of fluorescent reporter sequences) could be increased in human organoids189,190, opening up opportunities for non-invasive, live monitoring of different cell states or even subcellular protein localization.
Several innovative imaging-based strategies have further been applied to measure the physiological parameters of organoids grown in vitro. For example, continuous phosphorescence lifetime imaging microscopy of an O2-sensitive probe allows tracking of oxygen levels, revealing highly heterogeneous oxygenation levels in intestinal organoids191. Engineered Förster resonance energy transfer-based approaches have also been used to study intracellular protein activity (for example, extracellular signal-regulated kinase (ERK))192.
In situ electrochemical probing
Despite the immense opportunities of imaging-based readouts, fundamental optical principles challenge their faithful reporting. In particular, organoids take complex 3D shapes and are embedded in non-homogeneous matrix environments, which, inevitably, leads to optical artefacts. Radiometric methods have been shown to partially compensate for artefacts and, hence, are better than intensity-based approaches; however, their accuracy remains limited, owing to the fact that light scattering and absorption are wavelength-dependent phenomena. In addition, the automation of downstream data processing and analysis pipelines remains challenging, despite recent breakthroughs186,193, owing to often non-uniform imaging parameters and the recurring problem of segmentation of regions of interest (that is, object separation from the background). By contrast, electrochemical sensors provide 1D datasets, enabling simple, unbiased and automatable downstream analysis. Electrochemical sensors are, indeed, accurate and versatile, but their pervasive use in organoid culture systems is hampered by their bulky nature, and miniaturized versions have only recently been integrated194.
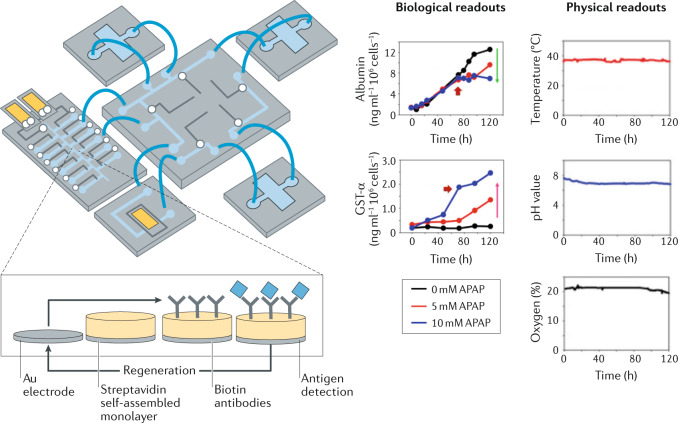
A two-organoid microfluidic device (containing cardiac and hepatic organoids) has been equipped with multiple integrated physical, biochemical and optical sensing capabilities195 (Fig. 7). In addition to sensors for temperature, oxygen and pH levels, the device includes microfabricated immunobiosensors for the continuous monitoring of secreted soluble biomarkers. The detection mechanism is based on a label-free principle, exploiting the change of electron transfer kinetics of a redox probe upon antigen binding to an immobilized antibody. Importantly, a regeneration process makes the sensor reusable for successive measurements over long time periods. Furthermore, all sensors are directly linked to a processing computer, which actuates the sensors and pneumatic valves for fluidic control, demonstrating the possibility of fully automated culture platforms.
Fig. 7. Microengineered platforms for live organoid monitoring.
Automated and continuous in situ monitoring of physical and biological parameters in organoid cultures can be achieved using microfluidic devices. The presented device contains multiple compartments for simultaneous growth of different organoids, as well as sensors to monitor various biological (for example, metabolites and biomarkers) and biophysical (temperature, pH and oxygen levels) parameters. The biosensor was designed to be regenerated, allowing continuous measurements (inset). APAP, acetaminophen; GST, glutathione S-transferase. Adapted with permission from ref.195, National Academy of Sciences.
A main challenge is to measure ultra-low volumes of cell-secreted analytes. Indeed, such data would capture the functional state of cells and would, thus, be particularly interesting for drug toxicity and drug screening assays. Therefore, efforts have been made to incorporate biosensors into small-organ model systems. For example, metabolites, such as lactate196–198, ions199 or protein biomarkers200,201, can be monitored using electrochemical sensors.
Towards high-throughput setups
For drug screening applications, it is particularly important to simultaneously grow and analyse large numbers of organoids to increase statistical power and to screen hundreds to thousands of drug candidates in parallel. By combining imaging methods and microfabrication strategies, a high number of organoids can be analysed at the same time186,188. Microfluidics platforms also allow the preselection of organoids by size187, thus, homogenizing the baseline organoid population and reducing variability. Droplet microfluidics is a promising technique for encapsulating cells but remains rather unexplored in organoid research. Hydrogel capsules can be used for the aggregation, growth and manipulation of organoids202–204. For example, prostate and breast cells embedded in Matrigel beads proliferate and differentiate at temporally and spatially controlled conditions205. Such Matrigel beads containing encapsulated cells were also generated in an automated high-throughput fashion204. This generated a large number of microcapsules with homogeneous size, composition and cell distribution, and opens up the possibility of high-throughput measurements, such as large-particle flow cytometry. These microbeads are easy to handle and, thus, allow the controlled manipulation of single organoids. Similarly, human iPSCs, differentiated into endoderm, pancreatic progenitors and endocrine cells, can be encapsulated in hydrogel capsules202, in which they form functional organoids with unprecedented consistency. These approaches enable the automated generation of a high number of similar organoids, which is an inevitable requirement for automated large-scale screens. Integrated approaches are expected to greatly boost the development of organoid screening platforms, including automated, continuous and functional readout for drug discovery.
Conclusions and perspective
In vitro organoid models can be derived from cells from a variety of organs, often with astonishing resemblances to their in vivo counterparts, both in terms of microscale tissue architecture and recapitulation of cellular diversity and key functions. However, challenges remain to maximize their translational relevance and applicability. Addressing these challenges will require a reconceptualization of the design principles of organoids. One of the biggest hurdles for clinical translation is the fact that growth environments of organoids are ill-defined, which leads to high variability of the resulting organoid phenotypes. Indeed, relying only on cell-intrinsic self-organization leaves little experimental scope for external control of cell fate and morphogenesis. Therefore, environments need to be designed that not only support stem cell maintenance but also allow precise spatiotemporal modulation of bioactive cues to guide organoid growth. For example, hydrogel chemistries and organ-on-a-chip technology can be applied and combined to better mimic the stem cell niche, not only in terms of delivery and presentation of in vivo-based biochemical cues but also by integrating biophysical and topological parameters often absent in traditional culture systems.
At a more fundamental level, genetically engineering the behaviour of cells can increase the functional relevance of organoids, for example, by tailoring responses to specific experimental stimuli or by driving differentiation of rare cell types. Genetically engineered (inducible) promotion of cell differentiation may indeed bypass the lengthy intrinsic differentiation timescales observed in vivo. The integration of organism-level parameters into organoid cultures remains rather underexplored. In vivo, tissues respond to and co-develop in the presence of fluid flow, forces and strains, and, in the context of crosstalk and interplay, with other tissues and organs. Integrating such system-level parameters will help increase the physiological relevance of organoids and enable long-term studies in vitro. In addition, accurate, high-throughput functional readouts are as important as optimization of the organoid culture per se. The precise characterization of organoids is needed at the functional level, otherwise, the multitude of subtle cellular responses, which often do not have an immediate morphological effect in organoids, will be missed, precluding clinical relevance. Therefore, the application of bioengineering strategies to traditional organoid culture protocols will usher in a new era of organoid development and facilitate their translation to real-life applications, fulfilling the many still unmet promises of the organoid field.
The question of the level of complexity remains, that is, how simplistic is complex enough. An appropriate model system needs to be chosen for a given application, keeping in mind that complexity is often increased at the expense of experimental control and throughput. Organoids resemble more physiological details than traditional 2D cell cultures, but are inferior to animal models in terms of system-level processes. Although organoids can be made more robust and more physiologically relevant by using engineering approaches, they will probably never be as robust and reproducible as cell-line-based models, nor will they be able to fully reproduce the complexity of animal models. Therefore, to be able to choose appropriate models, a set of organoids with varying levels of complexity is required. The desired level of complexity should correlate with the scale of the process that needs to be modelled. For example, the study of cellular processes and homogeneous tissue does not require sophisticated organoids; by contrast, studies of stem cell self-organization, brain development or microorganism infections require complex organoids that enable precise experimental control. The cellular complexity of organoids can be increased by incorporating immune or mesenchymal cells to model organs and associated diseases more accurately. The organoid field enjoys tremendous popularity among researchers of various backgrounds and, thus, a variety of protocols are being developed with a tendency towards more complex systems tailored for a specific application. This wealth of novel protocols may, ultimately, lead to superior organoids; however, standardization will be challenging. Therefore, guidelines, quality measures and validation procedures need to be defined for individual organoid systems.
In this Review, we mainly discussed challenges of organoid systems as in vitro models for fundamental research and for applications in drug development. However, organoids are also promising platforms for tissue engineering, which requires the construction of large-scale tissues and organs in vitro for subsequent transplantation to replace damaged or diseased organs. Organoid systems do not yet meet the requirements of engineered tissues, because artificially produced organs must be free of animal components (thus, Matrigel cannot be used), completely safe (no risk of tumorigenicity) and grown on a scale of centimetres to decimetres, with a microscopically and macroscopically controlled architecture. In addition, engineered tissues must exhibit physiologically relevant levels of tissue function. Bottom-up assembly approaches enable the precise control of cell deposition inside a growth-promoting matrix. Indeed, organoids and cellular spheroids have been used as building blocks for large-scale in vitro tissue engineering206,207; however, the precise manipulation of single spheroids and organoids has only recently been achieved by using bioprinting technologies208,209 and microchamber-based approaches210, for example, to grow high-density cardiac microtissues through the fusion of cardiomyocyte-containing and cardiac-fibroblast-containing spheroids208,210, or osteogenic tissue through the fusion of mesenchymal and endothelial spheroids209. To avoid the printing of overly complex 3D structures with preformed patterns, we recently pursued a complementary approach by printing stem cells with organoid-forming capacity to form centimetre-scale multi-organ structures with organoid-like microscale self-organization211. Bioprinting of organoids and organoid-forming cells is still in its infancy; however, it may open up the door for using organoids in tissue engineering, enabling the formation of functional organs and large-scale growth of artificial tissues.
Acknowledgements
The authors thank Stefano Davide Vianello for preparing the illustrations and his much-valued contributions editing the text, and thank Bilge Sen Elci and Antonius Chrisnandy for discussions and comments on the manuscript. M.P.L.’s work in the area of organoid biology and technology is supported by the Swiss National Science Foundation (#310030_179447), École Polytechnique Fédérale de Lausanne (EPFL), the European Union’s Horizon 2020 research and innovation programme (INTENS, #668294), the Personalized Health and Related Technologies initiative from the ETH Board, the National Center of Competence in Research (NCCR) Bio-Inspired Materials, the FreeNovation funding program of the Novartis Research Foundation, the Marie Skłodowska-Curie European Training Network ‘EUROoC’ (H2020-MSCA-ITN-2018) and the Vienna Science and Technology Fund (WWTF). The authors apologize to all researchers and colleagues whose work could not be considered.
Author contributions
M.H. researched data for the article, made substantial contributions to discussions of the content and wrote the article. M.P.L. reviewed and edited the manuscript before submission.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fatehullah A, Tan SH, Barker N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016;18:246–254. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 2.Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 3.Rossi G, Manfrin A, Lutolf MP. Progress and potential in organoid research. Nat. Rev. Genet. 2018;19:671–687. doi: 10.1038/s41576-018-0051-9. [DOI] [PubMed] [Google Scholar]
- 4.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 5.Lancaster MA, Huch M. Disease modelling in human organoids. Dis. Model. Mech. 2019;12:dmm039347. doi: 10.1242/dmm.039347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J, Koo BK, Knoblich JA. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020;21:571–584. doi: 10.1038/s41580-020-0259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCauley HA, Wells JM. Pluripotent stem cell-derived organoids: using principles of developmental biology to grow human tissues in a dish. Development. 2017;144:958–962. doi: 10.1242/dev.140731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ootani A, et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med. 2009;15:701–706. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 10.Sato T, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 11.Jung P, et al. Isolation and in vitro expansion of human colonic stem cells. Nat. Med. 2011;17:1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- 12.Spence JR, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–110. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Workman MJ, et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat. Med. 2017;23:49–59. doi: 10.1038/nm.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai YH, et al. In vitro patterning of pluripotent stem cell-derived intestine recapitulates in vivo human development. Development. 2017;144:1045–1055. doi: 10.1242/dev.138453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barker N, et al. Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 16.Stange DE, et al. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell. 2013;155:357–368. doi: 10.1016/j.cell.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartfeld S, et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148:126–136.e6. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertaux-Skeirik N, et al. CD44 plays a functional role in Helicobacter pylori-induced epithelial cell proliferation. PLoS Pathog. 2015;11:e1004663. doi: 10.1371/journal.ppat.1004663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCracken KW, et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature. 2014;516:400–404. doi: 10.1038/nature13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noguchi TAK, et al. Generation of stomach tissue from mouse embryonic stem cells. Nat. Cell Biol. 2015;17:984–993. doi: 10.1038/ncb3200. [DOI] [PubMed] [Google Scholar]
- 21.McCracken KW, et al. Wnt/β-catenin promotes gastric fundus specification in mice and humans. Nature. 2017;541:182–187. doi: 10.1038/nature21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taguchi A, et al. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell. 2014;14:53–67. doi: 10.1016/j.stem.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 23.Takasato M, et al. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat. Cell Biol. 2014;16:118–126. doi: 10.1038/ncb2894. [DOI] [PubMed] [Google Scholar]
- 24.Takasato M, et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526:564–568. doi: 10.1038/nature15695. [DOI] [PubMed] [Google Scholar]
- 25.Taguchi A, Nishinakamura R. Higher-order kidney organogenesis from pluripotent stem cells. Cell Stem Cell. 2017;21:730–746.e6. doi: 10.1016/j.stem.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 26.Morizane R, et al. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 2015;33:1193–1200. doi: 10.1038/nbt.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freedman BS, et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat. Commun. 2015;6:8715. doi: 10.1038/ncomms9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huch M, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huch M, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160:299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu H, et al. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell. 2018;175:1591–1606.e19. doi: 10.1016/j.cell.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 31.Peng WC, et al. Inflammatory cytokine TNFα promotes the long-term expansion of primary hepatocytes in 3D culture. Cell. 2018;175:1607–1619.e15. doi: 10.1016/j.cell.2018.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sampaziotis F, et al. Reconstruction of the mouse extrahepatic biliary tree using primary human extrahepatic cholangiocyte organoids. Nat. Med. 2017;23:954–963. doi: 10.1038/nm.4360. [DOI] [PubMed] [Google Scholar]
- 33.Takebe T, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 34.Camp JG, et al. Multilineage communication regulates human liver bud development from pluripotency. Nature. 2017;546:533–538. doi: 10.1038/nature22796. [DOI] [PubMed] [Google Scholar]
- 35.Ang LT, et al. A roadmap for human liver differentiation from pluripotent stem cells. Cell Rep. 2018;22:2190–2205. doi: 10.1016/j.celrep.2018.01.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takebe T, et al. Massive and reproducible production of liver buds entirely from human pluripotent stem cells. Cell Rep. 2017;21:2661–2670. doi: 10.1016/j.celrep.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Sampaziotis F, et al. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat. Biotechnol. 2015;33:845–852. doi: 10.1038/nbt.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogawa M, et al. Directed differentiation of cholangiocytes from human pluripotent stem cells. Nat. Biotechnol. 2015;33:853–861. doi: 10.1038/nbt.3294. [DOI] [PubMed] [Google Scholar]
- 39.Huch M, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 2013;32:2708–2721. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greggio C, et al. Artificial three-dimensional niches deconstruct pancreas development in vitro. Development. 2013;140:4452–4462. doi: 10.1242/dev.096628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boj SF, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang L, et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat. Med. 2015;21:1364–1371. doi: 10.1038/nm.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hohwieler M, et al. Human pluripotent stem cell-derived acinar/ductal organoids generate human pancreas upon orthotopic transplantation and allow disease modelling. Gut. 2017;66:473–486. doi: 10.1136/gutjnl-2016-312423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Linnemann JR, et al. Quantification of regenerative potential in primary human mammary epithelial cells. Development. 2015;142:3239–3251. doi: 10.1242/dev.123554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jardé T, et al. Wnt and Neuregulin1/ErbB signalling extends 3D culture of hormone responsive mammary organoids. Nat. Commun. 2016;7:13207. doi: 10.1038/ncomms13207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang L, et al. Establishing estrogen-responsive mouse mammary organoids from single Lgr5+ cells. Cell. Signal. 2017;29:41–51. doi: 10.1016/j.cellsig.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jamieson PR, et al. Derivation of a robust mouse mammary organoid system for studying tissue dynamics. Development. 2017;144:1065–1071. doi: 10.1242/dev.145045. [DOI] [PubMed] [Google Scholar]
- 48.Xin L, Lukacs RU, Lawson DA, Cheng D, Witte ON. Self-renewal and multilineage differentiation in vitro from murine prostate stem cells. Stem Cell. 2007;25:2760–2769. doi: 10.1634/stemcells.2007-0355. [DOI] [PubMed] [Google Scholar]
- 49.Karthaus WR, et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159:163–175. doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Höfner T, et al. Defined conditions for the isolation and expansion of basal prostate progenitor cells of mouse and human origin. Stem Cell Rep. 2015;4:503–518. doi: 10.1016/j.stemcr.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drost J, et al. Organoid culture systems for prostate epithelial and cancer tissue. Nat. Protoc. 2016;11:347–358. doi: 10.1038/nprot.2016.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tadokoro T, et al. IL-6/STAT3 promotes regeneration of airway ciliated cells from basal stem cells. Proc. Natl Acad. Sci. USA. 2014;111:3641–3649. doi: 10.1073/pnas.1409781111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jain R, et al. Plasticity of Hopx+ type I alveolar cells to regenerate type II cells in the lung. Nat. Commun. 2015;6:6727. doi: 10.1038/ncomms7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nikolić MZ, et al. Human embryonic lung epithelial tips are multipotent progenitors that can be expanded in vitro as long-term self-renewing organoids. eLife. 2017;6:e26575. doi: 10.7554/eLife.26575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sachs N, et al. Long- term expanding human airway organoids for disease modeling. EMBO J. 2019;38:e100300. doi: 10.15252/embj.2018100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wong AP, et al. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat. Biotechnol. 2012;30:876–882. doi: 10.1038/nbt.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dye BR, et al. In vitro generation of human pluripotent stem cell derived lung organoids. eLife. 2015;4:e05098. doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dye BR, et al. A bioengineered niche promotes in vivo engraftment and maturation of pluripotent stem cell derived human lung organoids. eLife. 2016;5:e19732. doi: 10.7554/eLife.19732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen Y-W, et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat. Cell Biol. 2017;19:542–549. doi: 10.1038/ncb3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Antonica F, et al. Generation of functional thyroid from embryonic stem cells. Nature. 2012;491:66–71. doi: 10.1038/nature11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kurmann AA, et al. Regeneration of thyroid function by transplantation of differentiated pluripotent stem cells. Cell Stem Cell. 2015;17:527–542. doi: 10.1016/j.stem.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eiraku M, et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472:51–58. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- 63.Nakano T, et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 64.Phillips MJ, et al. Blood-derived human iPS cells generate optic vesicle–like structures with the capacity to form retinal laminae and develop synapses. Investig. Opthalmol. Vis. Sci. 2012;53:2007–2019. doi: 10.1167/iovs.11-9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reichman S, et al. From confluent human iPS cells to self-forming neural retina and retinal pigmented epithelium. Proc. Natl Acad. Sci. USA. 2014;111:8518–8523. doi: 10.1073/pnas.1324212111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhong X, et al. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun. 2014;5:4047. doi: 10.1038/ncomms5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Völkner M, et al. Retinal organoids from pluripotent stem cells efficiently recapitulate retinogenesis. Stem Cell Rep. 2016;6:525–538. doi: 10.1016/j.stemcr.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Capowski EE, et al. Reproducibility and staging of 3D human retinal organoids across multiple pluripotent stem cell lines. Development. 2019;146:dev171686. doi: 10.1242/dev.171686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Camp JG, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl Acad. Sci. USA. 2015;112:15672–15677. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bagley JA, Reumann D, Bian S, Lévi-Strauss J, Knoblich JA. Fused cerebral organoids model interactions between brain regions. Nat. Methods. 2017;14:743–751. doi: 10.1038/nmeth.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Birey F, et al. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545:54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xiang Y, et al. Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell. 2017;21:383–398.e7. doi: 10.1016/j.stem.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kadoshima T, et al. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl Acad. Sci. USA. 2013;110:20284–20289. doi: 10.1073/pnas.1315710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mariani J, et al. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell. 2015;162:375–390. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sakaguchi H, et al. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat. Commun. 2015;6:8896. doi: 10.1038/ncomms9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Paşca AM, et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qian X, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jo J, et al. Midbrain-like organoids from human pluripotent stem cells contain functional dopaminergic and neuromelanin-producing neurons. Cell Stem Cell. 2016;19:248–257. doi: 10.1016/j.stem.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Muguruma K, Nishiyama A, Kawakami H, Hashimoto K, Sasai Y. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 2015;10:537–550. doi: 10.1016/j.celrep.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 81.Qian X, et al. Generation of human brain region–specific organoids using a miniaturized spinning bioreactor. Nat. Protoc. 2018;13:565–580. doi: 10.1038/nprot.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Watson LM, Wong MMK, Vowles J, Cowley SA, Becker EBE. A simplified method for generating purkinje cells from human-induced pluripotent stem cells. Cerebellum. 2018;17:419–427. doi: 10.1007/s12311-017-0913-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eiraku M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 84.Zachos NC, et al. Human enteroids/colonoids and intestinal organoids functionally recapitulate normal intestinal physiology and pathophysiology. J. Biol. Chem. 2016;291:3759–3766. doi: 10.1074/jbc.R114.635995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Di Lullo E, Kriegstein AR. The use of brain organoids to investigate neural development and disease. Nat. Rev. Neurosci. 2017;18:573–584. doi: 10.1038/nrn.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Qian X, Song H, Ming GL. Brain organoids: advances, applications and challenges. Development. 2019;146:dev166074. doi: 10.1242/dev.166074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smits LM, et al. Modeling Parkinson’s disease in midbrain-like organoids. NPJ Parkinsons Dis. 2019;5:5. doi: 10.1038/s41531-019-0078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Monzel AS, et al. Derivation of human midbrain-specific organoids from neuroepithelial stem cells. Stem Cell Rep. 2017;8:1144–1154. doi: 10.1016/j.stemcr.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Assawachananont J, et al. Transplantation of embryonic and induced pluripotent stem cell-derived 3D retinal sheets into retinal degenerative mice. Stem Cell Rep. 2014;2:662–674. doi: 10.1016/j.stemcr.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rashidi H, et al. 3D human liver tissue from pluripotent stem cells displays stable phenotype in vitro and supports compromised liver function in vivo. Arch. Toxicol. 2018;92:3117–3129. doi: 10.1007/s00204-018-2280-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Blackford SJI, et al. Validation of current good manufacturing practice compliant human pluripotent stem cell-derived hepatocytes for cell-based therapy. Stem Cell Transl. Med. 2019;8:124–137. doi: 10.1002/sctm.18-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lu D, Kassab GS. Role of shear stress and stretch in vascular mechanobiology. J. R. Soc. Interface. 2011;8:1379–1385. doi: 10.1098/rsif.2011.0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Abraham G, et al. Growth and differentiation of primary and passaged equine bronchial epithelial cells under conventional and air-liquid-interface culture conditions. BMC Vet. Res. 2011;7:26. doi: 10.1186/1746-6148-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Park JS, et al. The effect of matrix stiffness on the differentiation of mesenchymal stem cells in response to TGF-β. Biomaterials. 2011;32:3921–3930. doi: 10.1016/j.biomaterials.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Park SE, Georgescu A, Huh D. Organoids-on-a-chip. Science. 2019;364:960–965. doi: 10.1126/science.aaw7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Velasco S, et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570:523–527. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim GA, Ginga NJ, Takayama S. Integration of sensors in gastrointestinal organoid culture for biological analysis. Cell. Mol. Gastroenterol. Hepatol. 2018;6:123–131.e1. doi: 10.1016/j.jcmgh.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Collins, S. D. et al. in Hepatocellular Carcinoma Ch. 3 (ed. Tirnitz-Parker, J. E. E.) 47–67 (Codon Publications, 2019). [PubMed]
- 99.Brassard JA, Lutolf MP. Engineering stem cell self-organization to build better organoids. Cell Stem Cell. 2019;24:860–876. doi: 10.1016/j.stem.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 100.Fujimichi Y, Otsuka K, Tomita M, Iwasaki T. An efficient intestinal organoid system of direct sorting to evaluate stem cell competition in vitro. Sci. Rep. 2019;9:20297. doi: 10.1038/s41598-019-55824-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Amaral AJR, Pasparakis G. Cell membrane engineering with synthetic materials: Applications in cell spheroids, cellular glues and microtissue formation. Acta Biomater. 2019;90:21–36. doi: 10.1016/j.actbio.2019.04.013. [DOI] [PubMed] [Google Scholar]
- 102.Rogozhnikov D, O’Brien PJ, Elahipanah S, Yousaf MN. Scaffold free bio-orthogonal assembly of 3-dimensional cardiac tissue via cell surface engineering. Sci. Rep. 2016;6:39806. doi: 10.1038/srep39806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.O’Brien PJ, Luo W, Rogozhnikov D, Chen J, Yousaf MN. Spheroid and tissue assembly via click chemistry in microfluidic flow. Bioconjug. Chem. 2015;26:1939–1949. doi: 10.1021/acs.bioconjchem.5b00376. [DOI] [PubMed] [Google Scholar]
- 104.Luo W, Pulsipher A, Dutta D, Lamb BM, Yousaf MN. Remote control of tissue interactions via engineered photo-switchable cell surfaces. Sci. Rep. 2014;4:6313. doi: 10.1038/srep06313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Akbari E, et al. Engineering cell surface function with DNA origami. Adv. Mater. 2017;29:1703632. doi: 10.1002/adma.201703632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fayol D, et al. Use of magnetic forces to promote stem cell aggregation during differentiation, and cartilage tissue modeling. Adv. Mater. 2013;25:2611–2616. doi: 10.1002/adma.201300342. [DOI] [PubMed] [Google Scholar]
- 107.Xia PF, Ling H, Foo JL, Chang MW. Synthetic genetic circuits for programmable biological functionalities. Biotechnol. Adv. 2019;37:107393. doi: 10.1016/j.biotechadv.2019.04.015. [DOI] [PubMed] [Google Scholar]
- 108.Bashor CJ, Collins JJ. Understanding biological regulation through synthetic biology. Annu. Rev. Biophys. 2018;47:399–423. doi: 10.1146/annurev-biophys-070816-033903. [DOI] [PubMed] [Google Scholar]
- 109.Zhang F. Development of CRISPR-Cas systems for genome editing and beyond. Q. Rev. Biophys. 2019;52:e6. doi: 10.1017/S0033583519000052. [DOI] [Google Scholar]
- 110.Velazquez JJ, et al. Gene regulatory network analysis and engineering directs development and vascularization of multilineage human liver organoids. Cell Syst. 2020;12:41–55. doi: 10.1016/j.cels.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Roper J, Yilmaz ÖH. Breakthrough moments: genome editing and organoids. Cell Stem Cell. 2019;24:841–842. doi: 10.1016/j.stem.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 112.Schwank G, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 113.Deng WL, et al. Gene correction reverses ciliopathy and photoreceptor loss in iPSC-derived retinal organoids from retinitis pigmentosa patients. Stem Cell Rep. 2018;10:1267–1281. doi: 10.1016/j.stemcr.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Matano M, et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 115.Drost J, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 116.Li X, et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat. Med. 2014;20:769–777. doi: 10.1038/nm.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sekine R, Shibata T, Ebisuya M. Synthetic mammalian pattern formation driven by differential diffusivity of Nodal and Lefty. Nat. Commun. 2018;9:5456. doi: 10.1038/s41467-018-07847-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Guye P, et al. Genetically engineering self-organization of human pluripotent stem cells into a liver bud-like tissue using Gata6. Nat. Commun. 2016;7:10243. doi: 10.1038/ncomms10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kunche S, Yan H, Calof AL, Lowengrub JS, Lander AD. Feedback, lineages and self-organizing morphogenesis. PLoS Comput. Biol. 2016;12:e1004814. doi: 10.1371/journal.pcbi.1004814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Morsut L, et al. Engineering customized cell sensing and response behaviors using synthetic notch receptors. Cell. 2016;164:780–791. doi: 10.1016/j.cell.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Toda S, Blauch LR, Tang SKY, Morsut L, Lim WA. Programming self-organizing multicellular structures with synthetic cell-cell signaling. Science. 2018;361:156–162. doi: 10.1126/science.aat0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hartmann J, Krueger D, De Renzis S. Using optogenetics to tackle systems-level questions of multicellular morphogenesis. Curr. Opin. Cell Biol. 2020;66:19–27. doi: 10.1016/j.ceb.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 123.Bateman JF, Boot-Handford RP, Lamandé SR. Genetic diseases of connective tissues: cellular and extracellular effects of ECM mutations. Nat. Rev. Genet. 2009;10:173–183. doi: 10.1038/nrg2520. [DOI] [PubMed] [Google Scholar]
- 124.Cox TR, Erler JT. Remodeling and homeostasis of the extracellular matrix: Implications for fibrotic diseases and cancer. Dis. Model. Mech. 2011;4:165–178. doi: 10.1242/dmm.004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kleinman HK, Martin GR. Matrigel: Basement membrane matrix with biological activity. Semin. Cancer Biol. 2005;15:378–386. doi: 10.1016/j.semcancer.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 126.Aisenbrey EA, Murphy WL. Synthetic alternatives to Matrigel. Nat. Rev. Mater. 2020;5:539–551. doi: 10.1038/s41578-020-0199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kratochvil MJ, et al. Engineered materials for organoid systems. Nat. Rev. Mater. 2019;4:606–622. doi: 10.1038/s41578-019-0129-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Broguiere N, et al. Growth of epithelial organoids in a defined hydrogel. Adv. Mater. 2018;30:1801621. doi: 10.1002/adma.201801621. [DOI] [PubMed] [Google Scholar]
- 129.Jabaji Z, et al. Type I collagen as an extracellular matrix for the in vitro growth of human small intestinal epithelium. PLoS ONE. 2014;9:e107814. doi: 10.1371/journal.pone.0107814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lindborg BA, et al. Rapid induction of cerebral organoids from human induced pluripotent stem cells using a chemically defined hydrogel and defined cell culture medium. Stem Cell Transl. Med. 2016;5:970–979. doi: 10.5966/sctm.2015-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gjorevski N, et al. Designer matrices for intestinal stem cell and organoid culture. Nature. 2016;539:560–564. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- 132.Cruz-Acuña R, et al. Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat. Cell Biol. 2017;19:1326–1335. doi: 10.1038/ncb3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ranga A, et al. 3D niche microarrays for systems-level analyses of cell fate. Nat. Commun. 2014;5:4324. doi: 10.1038/ncomms5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 135.Chaudhuri O, Cooper-White J, Janmey PA, Mooney DJ, Shenoy VB. Effects of extracellular matrix viscoelasticity on cellular behaviour. Nature. 2020;584:535–546. doi: 10.1038/s41586-020-2612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chaudhuri O, et al. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat. Mater. 2016;15:326–334. doi: 10.1038/nmat4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nelson CM, VanDuijn MM, Inman JL, Fletcher DA, Bissell MJ. Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures. Science. 2006;314:298–300. doi: 10.1126/science.1131000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang Y, et al. A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials. 2017;128:44–55. doi: 10.1016/j.biomaterials.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nikolaev M, et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature. 2020;585:574–578. doi: 10.1038/s41586-020-2724-8. [DOI] [PubMed] [Google Scholar]
- 140.Lancaster MA, et al. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 2017;35:659–666. doi: 10.1038/nbt.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Karzbrun E, Kshirsagar A, Cohen SR, Hanna JH, Reiner O. Human brain organoids on a chip reveal the physics of folding. Nat. Phys. 2018;14:515–522. doi: 10.1038/s41567-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mosiewicz KA, et al. In situ cell manipulation through enzymatic hydrogel photopatterning. Nat. Mater. 2013;12:1072–1078. doi: 10.1038/nmat3766. [DOI] [PubMed] [Google Scholar]
- 144.Ruskowitz ER, Deforest CA. Photoresponsive biomaterials for targeted drug delivery and 4D cell culture. Nat. Rev. Mater. 2018;3:17087. doi: 10.1038/natrevmats.2017.87. [DOI] [Google Scholar]
- 145.Attayek PJ, et al. In vitro polarization of colonoids to create an intestinal stem cell compartment. PLoS ONE. 2016;11:e153795. doi: 10.1371/journal.pone.0153795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tabata Y, Lutolf MP. Multiscale microenvironmental perturbation of pluripotent stem cell fate and self-organization. Sci. Rep. 2017;7:44711. doi: 10.1038/srep44711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Demers CJ, et al. Development-on-chip: In vitro neural tube patterning with a microfluidic device. Development. 2016;143:1884–1892. doi: 10.1242/dev.126847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Manfrin A, et al. Engineered signaling centers for the spatially controlled patterning of human pluripotent stem cells. Nat. Methods. 2019;16:640–648. doi: 10.1038/s41592-019-0455-2. [DOI] [PubMed] [Google Scholar]
- 149.Cederquist GY, et al. Specification of positional identity in forebrain organoids. Nat. Biotechnol. 2019;37:436–444. doi: 10.1038/s41587-019-0085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wylie RG, et al. Spatially controlled simultaneous patterning of multiple growth factors in three-dimensional hydrogels. Nat. Mater. 2011;10:799–806. doi: 10.1038/nmat3101. [DOI] [PubMed] [Google Scholar]
- 151.Battista S, et al. The effect of matrix composition of 3D constructs on embryonic stem cell differentiation. Biomaterials. 2005;26:6194–6207. doi: 10.1016/j.biomaterials.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 152.Teller IC, et al. Laminins in the developing and adult human small intestine: Relation with the functional absorptive unit. Dev. Dyn. 2007;236:1980–1990. doi: 10.1002/dvdy.21186. [DOI] [PubMed] [Google Scholar]
- 153.Groulx JF, et al. Collagen VI is a basement membrane component that regulates epithelial cell–fibronectin interactions. Matrix Biol. 2011;30:195–206. doi: 10.1016/j.matbio.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 154.Benoit YD, Groulx J-F, Gagné D, Beaulieu J-F. RGD-dependent epithelial cell-matrix interactions in the human intestinal Crypt. J. Signal Transduct. 2012;2012:248759. doi: 10.1155/2012/248759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Broguiere N, et al. Morphogenesis guided by 3D patterning of growth factors in biological matrices. Adv. Mater. 2020;32:1908299. doi: 10.1002/adma.201908299. [DOI] [PubMed] [Google Scholar]
- 156.Torgersen J, et al. Hydrogels for two-photon polymerization: A toolbox for mimicking the extracellular matrix. Adv. Funct. Mater. 2013;23:4542–4554. doi: 10.1002/adfm.201203880. [DOI] [Google Scholar]
- 157.Pradhan S, Keller KA, Sperduto JL, Slater JH. Fundamentals of laser-based hydrogel degradation and applications in cell and tissue engineering. Adv. Healthc. Mater. 2017;6:1700681. doi: 10.1002/adhm.201700681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mammoto T, Ingber DE. Mechanical control of tissue and organ development. Development. 2010;137:1407–1420. doi: 10.1242/dev.024166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vianello S, Lutolf MP. Understanding the mechanobiology of early mammalian development through bioengineered models. Dev. Cell. 2019;48:751–763. doi: 10.1016/j.devcel.2019.02.024. [DOI] [PubMed] [Google Scholar]
- 160.Heo I, et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol. 2018;3:814–823. doi: 10.1038/s41564-018-0177-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Leslie JL, et al. Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect. Immun. 2015;83:138–145. doi: 10.1128/IAI.02561-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Schumacher MA, et al. The use of murine-derived fundic organoids in studies of gastric physiology. J. Physiol. 2015;593:1809–1827. doi: 10.1113/jphysiol.2014.283028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Co JY, et al. Controlling epithelial polarity: a human enteroid model for host-pathogen interactions. Cell Rep. 2019;26:2509–2520.e4. doi: 10.1016/j.celrep.2019.01.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Huh D, et al. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kim HJ, Huh D, Hamilton G, Ingber DE. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12:2165–2174. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 166.Musah S, et al. Mature induced-pluripotent-stem-cell-derived human podocytes reconstitute kidney glomerular-capillary-wall function on a chip. Nat. Biomed. Eng. 2017;1:0069. doi: 10.1038/s41551-017-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Akhtar AA, Sances S, Barrett R, Breunig JJ. Organoid and organ-on-a-chip systems: new paradigms for modeling neurological and gastrointestinal disease. Curr. Stem Cell Rep. 2017;3:98–111. doi: 10.1007/s40778-017-0080-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gayer CP, Basson MD. The effects of mechanical forces on intestinal physiology and pathology. Cell. Signal. 2009;21:1237–1244. doi: 10.1016/j.cellsig.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lee KK, et al. Human stomach-on-a-chip with luminal flow and peristaltic-like motility. Lab Chip. 2018;18:3079–3085. doi: 10.1039/C8LC00910D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Homan KA, et al. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods. 2019;16:255–262. doi: 10.1038/s41592-019-0325-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kasendra M, et al. Development of a primary human small intestine-on-a-chip using biopsy-derived organoids. Sci. Rep. 2018;8:2871. doi: 10.1038/s41598-018-21201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Imura Y, Sato K, Yoshimura E. Micro total bioassay system for ingested substances: Assessment of intestinal absorption, hepatic metabolism, and bioactivity. Anal. Chem. 2010;82:9983–9988. doi: 10.1021/ac100806x. [DOI] [PubMed] [Google Scholar]
- 173.Miller PG, Shuler ML. Design and demonstration of a pumpless 14 compartment microphysiological system. Biotechnol. Bioeng. 2016;113:2213–2227. doi: 10.1002/bit.25989. [DOI] [PubMed] [Google Scholar]
- 174.Jin Y, et al. Vascularized liver organoids generated using induced hepatic tissue and dynamic liver-specific microenvironment as a drug testing platform. Adv. Funct. Mater. 2018;28:1801954. doi: 10.1002/adfm.201801954. [DOI] [Google Scholar]
- 175.Skardal A, et al. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci. Rep. 2017;7:8837. doi: 10.1038/s41598-017-08879-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Garcez PP, et al. Zika virus: Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352:816–818. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- 177.Dang J, et al. Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell. 2016;19:258–265. doi: 10.1016/j.stem.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Gabriel E, et al. Recent Zika virus isolates induce premature differentiation of neural progenitors in human brain organoids. Cell Stem Cell. 2017;20:397–406.e5. doi: 10.1016/j.stem.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 179.Huang JY, et al. Chemodetection and destruction of host urea allows Helicobacter pylori to locate the epithelium. Cell Host Microbe. 2015;18:147–156. doi: 10.1016/j.chom.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Holokai L, et al. Increased programmed death-ligand 1 is an early epithelial cell response to Helicobacter pylori infection. PLoS Pathog. 2019;15:e1007468. doi: 10.1371/journal.ppat.1007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Burkitt MD, Duckworth CA, Williams JM, Pritchard DM. Helicobacter pylori-induced gastric pathology: Insights from in vivo and ex vivo models. Dis. Model. Mech. 2017;10:89–104. doi: 10.1242/dmm.027649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mejías-Luque R, Gerhard M. Immune evasion strategies and persistence of Helicobacter pylori. Curr. Top. Microbiol. Immunol. 2017;400:53–71. doi: 10.1007/978-3-319-50520-6_3. [DOI] [PubMed] [Google Scholar]
- 183.Ye W, Luo C, Li C, Huang J, Liu F. Organoids to study immune functions, immunological diseases and immunotherapy. Cancer Lett. 2020;477:31–40. doi: 10.1016/j.canlet.2020.02.027. [DOI] [PubMed] [Google Scholar]
- 184.Roh TT, Chen Y, Paul HT, Guo C, Kaplan DL. 3D bioengineered tissue model of the large intestine to study inflammatory bowel disease. Biomaterials. 2019;225:119517. doi: 10.1016/j.biomaterials.2019.119517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Czerniecki SM, et al. High-throughput screening enhances kidney organoid differentiation from human pluripotent stem cells and enables automated multidimensional phenotyping. Cell Stem Cell. 2018;22:929–940.e4. doi: 10.1016/j.stem.2018.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Brandenberg N, et al. High-throughput automated organoid culture via stem-cell aggregation in microcavity arrays. Nat. Biomed. Eng. 2020;4:863–874. doi: 10.1038/s41551-020-0565-2. [DOI] [PubMed] [Google Scholar]
- 187.Jin BJ, et al. Microfluidics platform for measurement of volume changes in immobilized intestinal enteroids. Biomicrofluidics. 2014;8:024106. doi: 10.1063/1.4870400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wang Y, et al. In situ differentiation and generation of functional liver organoids from human iPSCs in a 3D perfusable chip system. Lab Chip. 2018;18:3606–3616. doi: 10.1039/C8LC00869H. [DOI] [PubMed] [Google Scholar]
- 189.Artegiani B, et al. Fast and efficient generation of knock-in human organoids using homology-independent CRISPR–Cas9 precision genome editing. Nat. Cell Biol. 2020;22:321–331. doi: 10.1038/s41556-020-0472-5. [DOI] [PubMed] [Google Scholar]
- 190.Sun, D., Evans, L. D. & Rawlins, E. L. Organoid Easytag: an efficient workflow for gene targeting in human organoids. Preprint at bioRxiv 10.1101/2020.05.04.076067 (2020).
- 191.Okkelman IA, Foley T, Papkovsky DB, Dmitriev RI. Live cell imaging of mouse intestinal organoids reveals heterogeneity in their oxygenation. Biomaterials. 2017;146:86–96. doi: 10.1016/j.biomaterials.2017.08.043. [DOI] [PubMed] [Google Scholar]
- 192.Muta Y, et al. Composite regulation of ERK activity dynamics underlying tumour-specific traits in the intestine. Nat. Commun. 2018;9:2174. doi: 10.1038/s41467-018-04527-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lukonin I, et al. Phenotypic landscape of intestinal organoid regeneration. Nature. 2020;586:275–280. doi: 10.1038/s41586-020-2776-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Adam Kratz SR, Höll G, Schuller P, Ertl P, Rothbauer M. Latest trends in biosensing for microphysiological organs-on-a-chip and body-on-a-chip systems. Biosensors. 2019;9:110. doi: 10.3390/bios9030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zhang YS, et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc. Natl Acad. Sci. USA. 2017;114:E2293–E2302. doi: 10.1073/pnas.1612906114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Bavli D, et al. Real-time monitoring of metabolic function in liver-onchip microdevices tracks the dynamics of mitochondrial dysfunction. Proc. Natl Acad. Sci. USA. 2016;113:E2231–E2240. doi: 10.1073/pnas.1522556113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Misun PM, Rothe J, Schmid YRF, Hierlemann A, Frey O. Multi-analyte biosensor interface for real-time monitoring of 3D microtissue spheroids in hanging-drop networks. Microsyst. Nanoeng. 2016;2:16022. doi: 10.1038/micronano.2016.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Weltin A, et al. Accessing 3D microtissue metabolism: Lactate and oxygen monitoring in hepatocyte spheroids. Biosens. Bioelectron. 2017;87:941–948. doi: 10.1016/j.bios.2016.07.094. [DOI] [PubMed] [Google Scholar]
- 199.Taurino I, et al. Platinum nanopetal-based potassium sensors for acute cell death monitoring. RSC Adv. 2016;6:40517–40526. doi: 10.1039/C6RA01664B. [DOI] [Google Scholar]
- 200.Caluori G, et al. Non-invasive electromechanical cell-based biosensors for improved investigation of 3D cardiac models. Biosens. Bioelectron. 2019;124–125:129–135. doi: 10.1016/j.bios.2018.10.021. [DOI] [PubMed] [Google Scholar]
- 201.Shin SR, et al. Label-free and regenerative electrochemical microfluidic biosensors for continual monitoring of cell secretomes. Adv. Sci. 2017;4:1600522. doi: 10.1002/advs.201600522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Liu H, et al. A droplet microfluidic system to fabricate hybrid capsules enabling stem cell organoid engineering. Adv. Sci. 2020;7:1903739. doi: 10.1002/advs.201903739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wang Y, et al. One-step synthesis of composite hydrogel capsules to support liver organoid generation from hiPSCs. Biomater. Sci. 2020;8:5476–5488. doi: 10.1039/D0BM01085E. [DOI] [PubMed] [Google Scholar]
- 204.Laperrousaz B, et al. Direct transfection of clonal organoids in Matrigel microbeads: A promising approach toward organoid-based genetic screens. Nucleic Acids Res. 2018;46:e70. doi: 10.1093/nar/gky030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Dolega ME, Abeille F, Picollet-D’hahan N, Gidrol X. Controlled 3D culture in Matrigel microbeads to analyze clonal acinar development. Biomaterials. 2015;52:347–357. doi: 10.1016/j.biomaterials.2015.02.042. [DOI] [PubMed] [Google Scholar]
- 206.Sachs N, et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172:373–386.e10. doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 207.Reid JA, Mollica PM, Bruno RD, Sachs PC. Consistent and reproducible cultures of large-scale 3D mammary epithelial structures using an accessible bioprinting platform. Breast Cancer Res. 2018;20:122. doi: 10.1186/s13058-018-1045-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Daly AC, Davidson MD, Burdick JA. 3D bioprinting of high cell-density heterogeneous tissue models through spheroid fusion within self-healing hydrogels. Nat. Commun. 2021;12:753. doi: 10.1038/s41467-021-21029-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ayan B, et al. Aspiration-assisted bioprinting for precise positioning of biologics. Sci. Adv. 2020;6:eaaw5111. doi: 10.1126/sciadv.aaw5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kim TY, et al. Directed fusion of cardiac spheroids into larger heterocellular microtissues enables investigation of cardiac action potential propagation via cardiac fibroblasts. PLoS ONE. 2018;13:e0196714. doi: 10.1371/journal.pone.0196714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Brassard JA, Nikolaev M, Hübscher T, Hofer M, Lutolf MP. Recapitulating macro-scale tissue self-organization through organoid bioprinting. Nat. Mater. 2021;20:22–29. doi: 10.1038/s41563-020-00803-5. [DOI] [PubMed] [Google Scholar]
- 212.Dekkers JF, et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 2013;19:939–945. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- 213.Cora V, et al. A cleared view on retinal organoids. Cells. 2019;8:391. doi: 10.3390/cells8050391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Li Y, et al. Induction of expansion and folding in human cerebral organoids. Cell Stem Cell. 2017;20:385–396.e3. doi: 10.1016/j.stem.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Serra D, et al. Self-organization and symmetry breaking in intestinal organoid development. Nature. 2019;569:66–72. doi: 10.1038/s41586-019-1146-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Dekkers JF, et al. High-resolution 3D imaging of fixed and cleared organoids. Nat. Protoc. 2019;14:1756–1771. doi: 10.1038/s41596-019-0160-8. [DOI] [PubMed] [Google Scholar]
- 217.Choi SH, et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature. 2014;515:274–278. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Qin X, et al. Cell-type-specific signaling networks in heterocellular organoids. Nat. Methods. 2020;17:335–342. doi: 10.1038/s41592-020-0737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Kanton S, et al. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature. 2019;574:418–422. doi: 10.1038/s41586-019-1654-9. [DOI] [PubMed] [Google Scholar]
- 220.Khrameeva E, et al. Single-cell-resolution transcriptome map of human, chimpanzee, bonobo, and macaque brains. Genome Res. 2020;30:776–789. doi: 10.1101/gr.256958.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Rheinwald JG, Green H. Serial cultivation of strains of human epidermal keratinocytes in defined clonal and serum-free culture. J. Invest. Dermatol. 1975;6:331–344. doi: 10.1016/s0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
- 222.Li ML, et al. Influence of a reconstituted basement membrane and its components on casein gene expression and secretion in mouse mammary epithelial cells. Proc. Natl Acad. Sci. USA. 1987;84:136–140. doi: 10.1073/pnas.84.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Clevers H, Watt FM. Defining adult stem cells by function, not by phenotype. Annu. Rev. Biochem. 2018;87:1015–1027. doi: 10.1146/annurev-biochem-062917-012341. [DOI] [PubMed] [Google Scholar]
- 224.Takebe T, Zhang B, Radisic M. Synergistic engineering: organoids meet organs-on-a-chip. Cell Stem Cell. 2017;21:297–300. doi: 10.1016/j.stem.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 225.Artegiani B, et al. Probing the tumor suppressor function of BAP1 in CRISPR-engineered human liver organoids. Cell Stem Cell. 2019;24:927–943.e6. doi: 10.1016/j.stem.2019.04.017. [DOI] [PubMed] [Google Scholar]
- 226.Seino T, et al. Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell. 2018;22:454–467.e6. doi: 10.1016/j.stem.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 227.Dekkers JF, et al. Modeling breast cancer using CRISPR-Cas9-mediated engineering of human breast organoids. J. Natl Cancer Inst. 2020;112:540–544. doi: 10.1093/jnci/djz196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Zhang W, et al. Cerebral organoid and mouse models reveal a RAB39b–PI3K–mTOR pathway-dependent dysregulation of cortical development leading to macrocephaly/autism phenotypes. Genes Dev. 2020;34:580–597. doi: 10.1101/gad.332494.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Park J, et al. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer’s disease. Nat. Neurosci. 2018;21:941–951. doi: 10.1038/s41593-018-0175-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Kim H, et al. Modeling G2019S-LRRK2 sporadic Parkinson’s disease in 3D midbrain organoids. Stem Cell Rep. 2019;12:518–531. doi: 10.1016/j.stemcr.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Suzuki K, Murano T, Shimizu H, Ito G. Single cell analysis of Crohn’ s disease patient-derived small intestinal organoids reveals disease activity-dependent modification of stem cell properties. J. Gastroenterol. 2018;53:1035–1047. doi: 10.1007/s00535-018-1437-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Howell KJ, et al. DNA methylation and transcription patterns in intestinal epithelial cells from pediatric patients with inflammatory bowel diseases differentiate disease subtypes and associate with outcome. Gastroenterology. 2018;154:585–598. doi: 10.1053/j.gastro.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.d’Aldebert E, et al. Characterization of human colon organoids from inflammatory bowel disease patients. Front. Cell Dev. Biol. 2020;8:363. doi: 10.3389/fcell.2020.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Zhang W, et al. Modeling microcephaly with cerebral organoids reveals a WDR62–CEP170–KIF2A pathway promoting cilium disassembly in neural progenitors. Nat. Commun. 2019;10:2612. doi: 10.1038/s41467-019-10497-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Raja WK, et al. Self-organizing 3D human neural tissue derived from induced pluripotent stem cells recapitulate Alzheimer’s disease phenotypes. PLoS ONE. 2016;11:e0161969. doi: 10.1371/journal.pone.0161969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Gonzalez C, et al. Modeling amyloid beta and tau pathology in human cerebral organoids. Mol. Psychiatry. 2018;23:2363–2374. doi: 10.1038/s41380-018-0229-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Chumarina M, et al. Cellular alterations identified in pluripotent stem cell-derived midbrain spheroids generated from a female patient with progressive external ophthalmoplegia and parkinsonism who carries a novel variation (p.Q811R) in the POLG1 gene. Acta Neuropathol. Commun. 2019;7:208. doi: 10.1186/s40478-019-0863-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Buskin A, et al. Disrupted alternative splicing for genes implicated in splicing and ciliogenesis causes PRPF31 retinitis pigmentosa. Nat. Commun. 2018;9:4234. doi: 10.1038/s41467-018-06448-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Parfitt DA, et al. Identification and correction of mechanisms underlying inherited blindness in human iPSC-derived optic cups. Cell Stem Cell. 2016;18:769–781. doi: 10.1016/j.stem.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Li G, et al. Generation and characterization of induced pluripotent stem cells and retinal organoids from a Leber’s congenital amaurosis patient with novel RPE65 mutations. Front. Mol. Neurosci. 2019;12:212. doi: 10.3389/fnmol.2019.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Freedman BS, et al. Reduced ciliary polycystin-2 in induced pluripotent stem cells from polycystic kidney disease patients with PKD1 mutations. J. Am. Soc. Nephrol. 2013;24:1571–1586. doi: 10.1681/ASN.2012111089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Cruz NM, et al. Organoid cystogenesis reveals a critical role of microenvironment in human polycystic kidney disease. Nat. Mater. 2017;16:1112–1119. doi: 10.1038/nmat4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Akbari S, et al. Robust, long-term culture of endoderm-derived hepatic organoids for disease modeling. Stem Cell Rep. 2019;13:627–641. doi: 10.1016/j.stemcr.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Guan Y, et al. Human hepatic organoids for the analysis of human genetic diseases. JCI Insight. 2017;2:e94954. doi: 10.1172/jci.insight.94954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Ouchi R, et al. Modeling steatohepatitis in humans with pluripotent stem cell-derived organoids. Cell Metab. 2019;30:374–384.e6. doi: 10.1016/j.cmet.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Rodansky ES, Johnson LA, Huang S, Spence JR, Higgins PDR. Intestinal organoids: A model of intestinal fibrosis for evaluating anti-fibrotic drugs. Exp. Mol. Pathol. 2015;98:346–351. doi: 10.1016/j.yexmp.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Woo D, et al. Enhancing a Wnt-telomere feedback loop restores intestinal stem cell function in a human organotypic model of dyskeratosis congenita. Cell Stem Cell. 2016;19:397–405. doi: 10.1016/j.stem.2016.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Fan H, Demirci U, Chen P. Emerging organoid models: leaping forward in cancer research. J. Hematol. Oncol. 2019;12:142. doi: 10.1186/s13045-019-0832-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Clevers H, Tuveson DA. Organoid models for cancer research. Annu. Rev. Cancer Biol. 2019;3:223–234. doi: 10.1146/annurev-cancerbio-030518-055702. [DOI] [Google Scholar]
- 250.Fujii M, et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell. 2016;18:827–838. doi: 10.1016/j.stem.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 251.Broutier L, et al. Human primary liver cancer–derived organoid cultures for disease modeling and drug screening. Nat. Med. 2017;23:1424–1435. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Cao W, et al. Modeling liver cancer and therapy responsiveness using organoids derived from primary mouse liver tumors. Carcinogenesis. 2019;40:145–154. doi: 10.1093/carcin/bgy129. [DOI] [PubMed] [Google Scholar]
- 253.Gao D, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176–187. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Kim M, et al. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun. 2019;10:3991. doi: 10.1038/s41467-019-11867-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Vlachogiannis G, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Driehuis E, et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc. Natl Acad. Sci. USA. 2019;116:26580–26590. doi: 10.1073/pnas.1911273116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Yan HHN, et al. A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell. 2018;23:882–897.e11. doi: 10.1016/j.stem.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 258.Bruna A, et al. A biobank of breast cancer explants with preserved intra-tumor heterogeneity to screen anticancer compounds. Cell. 2016;167:260–274.e22. doi: 10.1016/j.cell.2016.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Zhou T, et al. High-content screening in hPSC-neural progenitors identifies drug candidates that inhibit Zika virus infection in fetal-like organoids and adult brain. Cell Stem Cell. 2017;21:274–283.e5. doi: 10.1016/j.stem.2017.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Leite SB, et al. Novel human hepatic organoid model enables testing of drug-induced liver fibrosis in vitro. Biomaterials. 2016;78:1–10. doi: 10.1016/j.biomaterials.2015.11.026. [DOI] [PubMed] [Google Scholar]
- 261.Liu F, Huang J, Liu Z. Vincristine impairs microtubules and causes neurotoxicity in cerebral organoids. Neuroscience. 2019;404:530–540. doi: 10.1016/j.neuroscience.2018.12.047. [DOI] [PubMed] [Google Scholar]
- 262.Qian X, Nguyen HN, Jacob F, Song H, Ming GL. Using brain organoids to understand Zika virus-induced microcephaly. Development. 2017;144:952–957. doi: 10.1242/dev.140707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Dutta D, Heo I, Clevers H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol. Med. 2017;23:393–410. doi: 10.1016/j.molmed.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 264.Zhou J, et al. Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc. Natl Acad. Sci. USA. 2018;115:6822–6827. doi: 10.1073/pnas.1806308115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Hui KPY, et al. Tropism, replication competence, and innate immune responses of influenza virus: an analysis of human airway organoids and ex-vivo bronchus cultures. Lancet Respir. Med. 2018;6:846–854. doi: 10.1016/S2213-2600(18)30236-4. [DOI] [PubMed] [Google Scholar]
- 266.Monteil V, et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Lamers MM, et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Zhou J, et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020;26:1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- 269.Salahudeen AA, et al. Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature. 2020;588:670–675. doi: 10.1038/s41586-020-3014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Han Y, et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589:270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Ettayebi K, et al. Replication of human noroviruses in stem cell–derived human enteroids. Science. 2016;353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Zhang Y-G, Wu S, Xia Y, Sun J. Salmonella-infected crypt-derived intestinal organoid culture system for host–bacterial interactions. Physiol. Rep. 2014;2:e12147. doi: 10.14814/phy2.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Wilson SS, Tocchi A, Holly MK, Parks WC, Smith JG. A small intestinal organoid model of non-invasive enteric pathogen-epithelial cell interactions. Mucosal Immunol. 2015;8:352–361. doi: 10.1038/mi.2014.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Forbester JL, et al. Interaction of Salmonella enterica serovar Typhimurium with intestinal organoids derived from human induced pluripotent stem cells. Infect. Immun. 2015;83:2926–2934. doi: 10.1128/IAI.00161-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Finkbeiner SR, et al. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. mBio. 2012;3:e00159-12. doi: 10.1128/mBio.00159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Porotto M, et al. Authentic modeling of human respiratory virus infection in human pluripotent stem cell-derived lung organoids. mBio. 2019;10:e00723-19. doi: 10.1128/mBio.00723-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Ciancanelli MJ, et al. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science. 2015;348:448–453. doi: 10.1126/science.aaa1578. [DOI] [PMC free article] [PubMed] [Google Scholar]