Abstract
An often-stated ecomorphological assumption that has the status of ‘textbook knowledge’ is that the dimensions of the digestive tract correlate with diet, where herbivores—consuming diets of lower digestibility—have longer intestinal tracts than faunivores—consuming diets of higher digestibility. However, statistical approaches have so far failed to demonstrate this link. Here, we collated data on the length of intestinal sections and body mass of 519 mammal species, and test for various relationships with trophic, climatic and other biological characteristics. All models showed a strong phylogenetic signal. Scaling relationships with body mass showed positive allometry at exponents greater than 0.33, except for the caecum, which is particularly large in smaller species. Body mass was more tightly linked to small intestine than to large intestine length. Adding a diet proxy to the relationships increased model fit for all intestinal sections, except for the small intestine when accounting for phylogeny. Thus, the diet has a main effect on the components of the large intestine, with longer measures in herbivores. Additionally, measures of habitat aridity had a positive relationship with large intestine length. The small intestine was longer in species from colder habitats at higher latitudes, possibly facilitating the processing of peak intake rates during the growing season. This study corroborates intuitive expectations on digestive tract anatomy, while the dependence of significant results on large sample sizes and inclusion of specific taxonomic groups indicates that the relationships cannot be considered fixed biological laws.
Keywords: anatomy, digestion, diet, scaling, ecomorphology, convergence
1. Background
Ecomorphological diversity is considered the main driver of species diversity, and diet is considered as one of the most important components of an animal's niche [1]. Across mammalian taxa, the gastrointestinal tract (GIT) exhibits great variation in length, area, volume and shape. Several hypotheses have been formulated to explain this morphological diversity. The most widely accepted one is that there is a link between the trophic niche and GIT morphology, a concept almost universally accepted as ‘textbook knowledge’ [2–4]. Mammals consuming highly digestible diets, such as faunivores (carnivores, insectivores) do not need complex or long GITs; mammals that feed mainly on vegetable matter, especially on the leaves and stems of grass or browse, require large fermentation chambers to digest plant fibre [5]. Therefore, it has been widely claimed that herbivores' intestines are longer than those of carnivores [2,3,5–8].
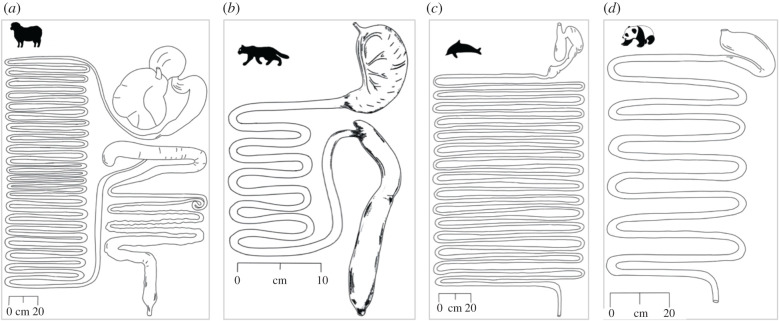
Typically, this claim has been supported by graphical representations of the gut anatomy of strict herbivores, such as a sheep, compared to strict faunivores, such as the domestic cat (figure 1a,b); the difference in length and apparent complexity between the two is striking. However, not all species follow this rule: the giant panda (Ailuropoda melanoleuca), which consumes an exclusive diet of bamboo, has a simple stomach and a short intestine; on the other hand, dolphins feed mainly on fish and squid but have complex stomachs and very long intestines (figure 1c,d).
Figure 1.
Examples of digestive tracts of mammals. (a) The domestic sheep (Ovis aries), a ‘typical’ herbivore, with a long intestine; (b) the domestic cat (Felis catus), a ‘typical’ faunivore, with a short intestine; (c) a dolphin (Larnorhynchus acutus), an ‘atypical’ faunivore with a long intestine; (d) a giant panda, an ‘atypical’ herbivore with a short intestine. Modified from [9] and [10]. Note that phylogeny groups the more similar cat and panda (Carnivora) and sheep and dolphin (Cetartiodactyla) together.
Statistical evaluations of the hypothesis that GIT morphology reflects trophic niche in mammals are seldomly reported. The most often-cited work to back this claim is that of Chivers & Hladik [5]; in citing this study, the large overlap in GIT morphology between trophic groups and several other issues (see electronic supplementary material, S1) are often not mentioned, as well as the fact that these analyses were done without accounting for phylogeny. A large number of original studies that investigated digestive tract anatomy came to supportive conclusions, albeit always necessarily on very small datasets [11–16], and generally also without accounting for phylogeny. By contrast, large-scale studies that accounted for phylogeny did not confirm an association between diet and intestinal length [17–21] or GIT complexity [22]. Other factors than diet thought to influence GIT anatomy include special adaptations to a volant [17,23,24] or a marine [25–27] lifestyle, or the aridity of the habitat [18,28,29].
Given that digestive tract anatomy and function have been instrumental in mechanistically linking mammalian ecology and evolutionary diversification [3], we sought to resolve the contributions of trophic and habitat niche components to GIT variation. Based on previous findings, we expected a significant phylogenetic signal; a scaling at an exponent higher than expected from geometry (i.e. positive allometry); an effect of diet particularly on parts of the large intestine with shorter lengths in more faunivorous species; and longer large intestines in animals from xeric habitats.
2. Methods
Relevant publications were collated using published datasets [17–20,25] as starting points and traced back to the original articles cited in the publications. Additionally, publications were actively searched for using the search engines Google Scholar, PubMed and Web of Science, with taxon names and ‘anatomy’, ‘morphometry’, ‘digestive tract’ ‘intestine’, ‘length’, as search terms. Data were only used if the publication included body mass and provided length measurements of the gut that included the small intestine (SI), the caecum, the colon–rectum complex (colon), the large intestine (LI, colon and caecum) and/or the total intestine (TI). Publications that reported estimated body masses were generally not included; however, some data on bats were included even when the body masses were not from the same animals as the intestine lengths [30,31], as this appeared the only way to include these species. If the literature included data for juveniles and adults, juvenile data were excluded. Additional unpublished data were obtained from post-mortem examinations carried out by M.C. and M.S.E. over the last decade, and more recently by M.D.
The information included the number of sampled animals, body mass and length of the total intestine, small intestine, large intestine, caecum and the colon/rectum. The sum of caecum and colon/rectum was taken as ‘large intestine’, and the sum of small and large intestine as ‘total intestine’. Not all data were available for each species, with total intestine information reaching the largest sample size. Weighted means (correcting for sample size) were calculated of each morphometrical parameter and the corresponding body mass. For example, if more data were available for small intestine than for caecum length of a species, then the body mass used for associations with small intestine length was different from the one used in the same species for associations with caecum length.
Various biological traits were added to the dataset. Data on the diet consumed in the wild was obtained from [32]. If a species was not included in that dataset, the diet of the closest taxonomic relative was used. The dataset gives quantitative information (in %) on the amount of prey animals, fruits, nectar, seeds and other plant parts. In addition to these quantities, we classified species into faunivore, omnivore or herbivore using two different cut-offs. One classification ascribed an extreme category (faunivore or herbivore) if 90% or more of the diet consisted of the corresponding sources, with omnivores being all other species. The second classification used 70% as the respective cut-off. The diet for Laonastes aenigmamus was taken from [33] and [34].
All species were categorized into volant (only those that perform active flying) or non-volant, terrestrial or marine [35], and whether their digestive system includes a non-glandular forestomach or not [36]. Environmental variables for the habitat occupied by each species included mid-latitude (used as absolute latitude), precipitation, temperature and actual evapotranspiration (AET) were obtained from the PanTHERIA database [37]. The fully referenced dataset is provided as electronic supplementary material.
The phylogenetic tree was built following Upham, Esselstyn [38]. A consensus supertree inclusive of 5911 mammalian species with time calibration (MamPhy_fullPosterior_BDvr_Completed_5911sp_topoCons_NDexp_MCC_v2_target.tre) was directly downloaded from (http://vertlife.org/phylosubsets/). The supertree was pruned in R using scripts from the library ‘ape’ [39] and ‘tidyverse’ [40] in order to obtain a final tree inclusive only of the 519 species for which GIT data and body masses were available.
Statistical analyses were done on (i) all available data (i.e. at different sample size for the different intestine sections—generally larger samples for the total intestine than for individual sections), and on two subsets that comprised (ii) those species for which both small and large intestine length was available and (iii) those species for which small intestine, caecum and colon/rectum data were available and (iv) various individual taxonomic groups. Analyses were done for full datasets, and for those species for which climate information was available. The factors volant, marine and forestomach presence were only assessed in the larger datasets comprising total and small intestine.
First, the allometric relationships with body mass were determined, and it was assessed which intestine section showed the best fit with body mass. Scaling exponents were termed ‘more’ or ‘less than geometric’ if they were above or below the expected isometry of 0.33 [41]. Then, the effect of diet was evaluated, using different dietary descriptors as cofactors or covariables with total intestine length, to decide which diet proxy would be used from there onwards (leading to the use of %faunivory, see electronic supplementary material). Next, the effect of being a volant or marine species, and forestomach presence was evaluated, together with the effect of adding the diet proxy to body mass relationships. Finally, in the subset with climate proxies, the additional effect of these was assessed. Allometric regressions were performed as linear regressions on log-transformed data, because we are not aware of another method to which we can apply phylogenetic generalized least squares (see below). Linear regression on log-transformed data has been criticized [42]; therefore, we inspected the fit of the resulting equations to the un-transformed data.
Comparative analyses need to consider the phylogenetic structure of the datasets that are analysed [43–45]. Here, all analyses were performed using generalized least squares (GLS) and phylogenetic generalized least squares (PGLS), recording the 95% confidence interval for parameter estimates, using the R packages ‘caper’ [46] and ‘nlme’ [47]. In all PGLS models, as phylogenetic signal, lambda (λ) was estimated by maximum likelihood. Additionally, we used the R package ‘phytools’ [48] to estimate the phylogenetic signals Blomberg's K [49] and Pagel's λ [50] for the complete datasets and the dataset of those species for which small intestine, caecum and colon/rectum data were available. The significance level was set to 0.05. Different models applied to a certain dataset (separately for GLS and PGLS) were compared using the small sample corrected Akaike's information criterion (AICc) [51], considering models that differed by more than 2 (ΔAICc > 2) as providing a different fit to the data.
3. Results
A final database comprised length data for the total intestinal tract (519 species), the small intestine (397 species), the large intestine (387 species), the caecum (352 species) and the colon/rectum (370 species). Generally, the small intestine represented the longest intestinal section, followed by the colon/rectum, and the caecum (electronic supplementary material, figure S1). These data were regressed against a set of predictor variables including body mass, trophic niche and climate and other characteristics of the natural habitat. Both Pagel's λ (at values of 0.97 to 0.99) and Blomberg's K (0.58 to 0.76) indicated a distinct phylogenetic pattern in the data; the two methods differed only for the caecum length, for which K, but not λ, indicated lower values than for the other intestinal sections (electronic supplementary material, table S1). In all PGLS analyses, there was a strong phylogenetic signal (λ > 0.9), indicating significant phylogenetic structure in all datasets (electronic supplementary material, tables S2–S12). There was no evident spacing in intestinal length between marsupials and placentals (electronic supplementary material, figure S2A). Afrotheria and Xenarthra had comparatively short intestines (electronic supplementary material, figure S2B), and phylogenetic clustering was evident both within the Laurasiatheria and the Euarchontoglires (electronic supplementary material, figure S2C–E).
(a). Allometry
Regardless of the dataset analysed, intestinal lengths scaled more-than-geometrically (positive allometry) throughout, except for the caecum, which scaled less-than-geometrically (negative allometry). Regardless of the phylogenetic signal, the simple scaling relationships were generally similar in generalized least squares (GLS) and PGLS (electronic supplementary material, table S2 and figure S3). A visual inspection of the fit of the regression line on non-transformed data for the total intestine did not indicate a relevant mismatch (electronic supplementary material, figure S4). When using only species for which all respective data were available, the small intestine–body mass relationship achieved lower AICc than the large intestine–body mass relationship (ΔAICc GLS = 357, PGLS = 135), or than the caecum–body mass and colon/rectum–body mass relationships (ΔAICc GLS > 308, PGLS > 178), suggesting that the large intestine is more subjected to additional influence factors (electronic supplementary material, table S2). Body mass was part of all subsequent models.
(b). Trophic level
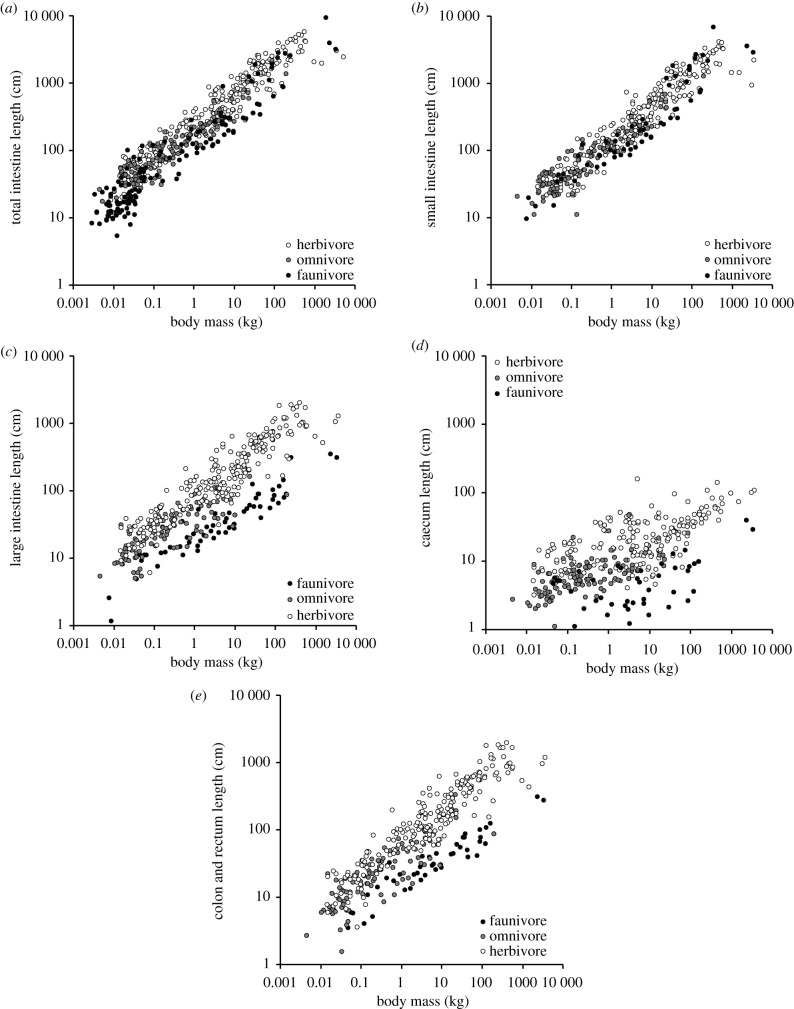
Any trophic proxy increased the data fit for the total intestine, with %faunivory showing the best fit (electronic supplementary material, table S3). The addition of %faunivory yielded a far better data fit (ΔAICc 19–316) for all intestinal sections in GLS; the difference from the model without trophic proxy was least for the small intestine (electronic supplementary material table S4). In PGLS, the same was true for the large intestine (ΔAICc 15–25), but for the small intestine, the model including the trophic proxy was even slightly less supported than the one without it (ΔAICc = 1.7), and the trophic proxy was not significant, suggesting that phylogeny accounted for differences in small intestine length between trophic groups (electronic supplementary material, table S4). For all intestine sections, %faunivory was negatively related to length (figure 2).
Figure 2.
Relationship of body mass and intestinal length for (a) total intestine (n = 519 species), (b) small intestine (n = 397), (c) large intestine (caecum, colon and rectum) (n = 387), (d) caecum (n = 352), (e) colon and rectum (n = 370) by trophic groups. For statistics, see electronic supplementary material, table S4. Note that for statistics, %faunivory was used as a continuous variable, whereas it is depicted for different groups here, using a 10 and 90% threshold to separate herbivores, omnivores and faunivores.
The effect of the trophic level was not consistent across different taxonomic groups. For the total intestine, the large groups of Eutheria, Boroeutheria, Euarchontoglires and Laurasiatheria showed significant effects of diet in GLS but not PGLS, indicating that taxa within these groups differ systematically by total intestine length and diet. By contrast, the large intestine showed a clear diet relationship in all these groups (electronic supplementary material, table S5). Clear diet effects for the total and large intestine were evident in the samples of Marsupialia and Afrotheria, and for the total intestine only (as large intestine data were lacking for this group) in Chiroptera. No effect for the total intestine but an effect on the large intestine was observed in Rodentia. No diet effect at all was evident within Primates, Eulipotyphla, Carnivora and Artiodactyla (electronic supplementary material, table S5)—groups with comparatively uniform diets at the level of diet resolution of the present study.
(c). Volant/marine/forestomach
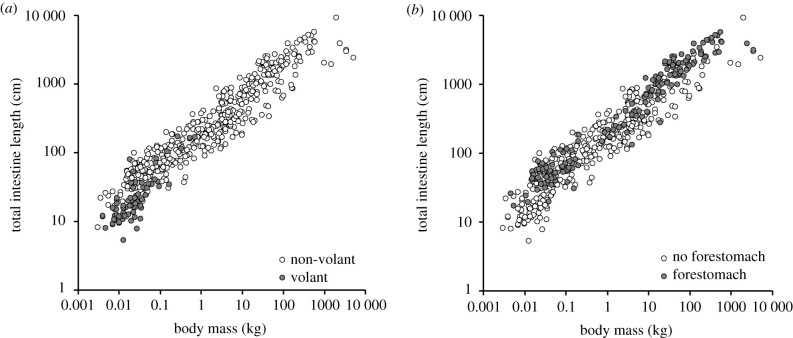
When assessed individually with body mass, being volant had a negative relationship with total intestine length in both GLS and PGLS (figure 3a); being marine was not a significant factor and having a forestomach had a positive relationship with total intestine length in GLS but not in PGLS (figure 3b, electronic supplementary material table S6 and table S6). In GLS, a model that included diet and all three factors (volant, marine, forestomach) was the best-supported (ΔAICc to the next-best model = 9), with all factors being significant (here, being marine had a positive effect on length). In PGLS, this model had similar support as other models that included diet and being volant (ΔAICc < 2); neither being marine nor having a forestomach was significant in these or other PGLS models (electronic supplementary material, table S6).
Figure 3.
Relationships between total intestine length and body mass (a) non-volant versus volant mammals, (b) mammals without and with a forestomach. For statistics, see electronic supplementary material, table S6.
For the small intestine, for which hardly any data for bats existed, models including diet, being marine and having a forestomach were the best-supported in GLS (ΔAICc to next-best model = 8); both factors were positively related to length. In PGLS, the best-supported model only included being marine (ΔAICc to next-best model = 2; electronic supplementary material, table S7). The pattern, however, does not appear convincing at the visual inspection, being based on rather few species (electronic supplementary material, figure S5).
(d). Environment
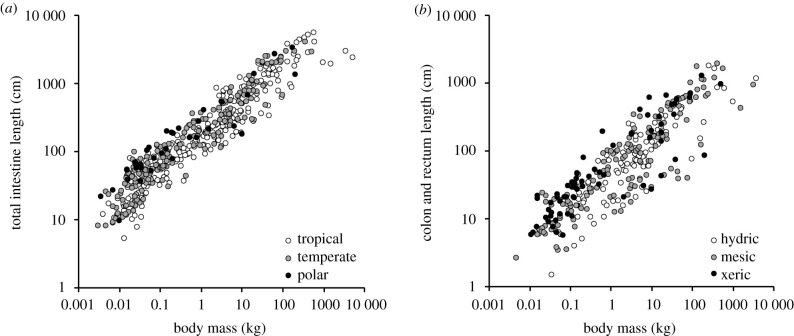
For the total intestine, the best model included diet and absolute latitude in GLS (ΔAICc to next-best model = 4 [temperature] and greater than 17 to other models). In PGLS, the best model included diet and temperature (ΔAICc to next-best model = 7 [latitude] and greater than 18 to other models) (electronic supplementary material, table S8). Latitude was positively related to intestine length (figure 4a) and temperature negatively. For the small intestine, the models with diet and either latitude or temperature were equally supported in GLS (ΔAICc < 2; ΔAICc to other models > 7). In PGLS, the model with temperature had more support than the one with latitude (ΔAICc = 3; ΔAICc to other models > 8) (electronic supplementary material, table S9).
Figure 4.
Relationships between mammalian intestinal length and body mass (a) for the total intestine with species separated by mean latitude of origin, (b) for the colon/rectum and species separated by the dryness of their habitat. For statistics, see electronic supplementary material, tables S8 and S11. Note that for statistics, latitude and actual evapotranspiration were used as continuous variables, whereas they are depicted for different groups here, using 1400 mm and 500 mm thresholds to separate animals from hydric (moist), mesic (intermediate) and xeric (arid) climates.
For both the large intestine and the colon/rectum, the best models included diet and either AET or precipitation in GLS (ΔAICc < 2; ΔAICc to other models > 5). In PGLS, the best models included diet and either AET or temperature for the large intestine (ΔAICc < 2; ΔAICc to precipitation = 2, to other models > 4), but only diet and AET for the colon/rectum (ΔAICc to other models greater than 2) (electronic supplementary material, tables S10 and S11). Higher precipitation or AET (i.e. a habitat with more moisture) were linked to shorter intestinal lengths (figure 4b).
For the caecum, the models including latitude and temperature were equally supported in GLS (ΔAICc < 2; ΔAICc to other models > 10). In PGLS, the model with diet only, with AET and with precipitation were all equally supported (ΔAICc < 2), yet the models with temperature and latitude nearly had equal support (ΔAICc = 3); none of the environmental parameters was significant in these models (electronic supplementary material, table S12).
4. Discussion
The present study provides a comprehensive data collection on mammalian intestinal length, which corroborates previously stated concepts on intestinal allometric scaling, and intuitive concepts about the relationships between digestive tract anatomy, diet and environmental aridity. Additional, existing concepts on the effect of being a volant or marine species are also supported, and some new findings on associations are provided with having a forestomach and with the mid-latitude of the species' current geographic range. Throughout, the results emphasize that it is reasonable to consider different sections of the intestinal tract individually, as they are linked to the different biological factors to varying degrees. The scatter evident from plots recommend that while macroevolutionary trends can be stated, they should not be considered fixed biological laws, and that results may depend to a large extent on the size and composition of the investigated sample.
(a). Limitations of the present study
The typical constraints of large literature compilations apply [52] that are not re-iterated here. Given the state of both the published literature and the information available on dissected specimens, it was not possible to account for the diet actually ingested by the animals either within the last months before measurements, or during their ontogeny. Intestinal anatomy has varying degrees of flexibility in different species; this has particularly been investigated in small mammals [53], whereas there is less evidence in larger mammals [54]. In a recent study on the intra-specific variation of intestinal length measurements linked to material stored frozen or in formalin, no difference between the methods was evident [55]. However, given probable differences between individual studies, it is recommended that the present data compilation is used to investigate broad patterns across many species, but should not form the basis of a comparison of a specific pair of species.
An important limitation in the current dataset is that the functional units of the colon cannot be separated. Just like the caecum, to which it is adjacent, the proximal colon is a site of microbial action (fermentation). The major function of the subsequent parts of the colon, however, is water reabsorption [8]. In most species, it is not possible to distinguish these two colon parts macroscopically, and therefore, the length of the colon/rectum might reflect adaptations to both herbivory and arid environments.
Making absolute statements, even based on comprehensive datasets, is something our results caution against. Based on the complete dataset of 519 species, the PGLS model that related total intestinal length to body mass and diet had better support than the model with body mass alone (ΔAIC = 3, electronic supplementary material, table S3). However, in the reduced datasets of 387 or 351 species, both models were equally supported (ΔAIC = 1.9 and 0, respectively, electronic supplementary material, table S3). Thus, a comparatively large dataset (351 species) did not indicate an effect of trophic level on total intestinal length, whereas a yet distinctively larger dataset (519 species) did. Although the result of the model using the larger sample size corresponds to our expectations, one might question how generalizable a result is that requires such immense sample sizes.
Our results also indicate that it is important to reference the taxonomic level on which a statement is based. The fact that there is a diet effect on the total intestinal tract length across all mammals, or within Chiroptera, contrasts with the absence of such an effect in Primates, Carnivora or Artiodactyla (electronic supplementary material, table S4). Hence, depending on the sample composition, the diet hypothesis would be confirmed or rejected. The problem of defining ‘diet’ in a way that is applicable across taxa is evident. Whereas in the Carnivora, a distinction between large- and small-prey feeders might be appropriate [56], in the Artiodactyla, a separation along the browser–grazer spectrum would make more sense [57].
(b). Phylogeny
Both phylogenetic signals, K and λ, indicated that closely related species share a common intestinal morphology. Although it has been recommended that only results using PGLS or another method to account for phylogeny should be considered [58], a comparison between GLS and PGLS can often be instructive [45,59,60]. A factor that contributes significantly to variation in GLS, but does not do so in PGLS, is likely distributed unequally across the phylogeny; phylogenetic diversification then reflects the diversity in this factor. A relationship that is significant in GLS but not in PGLS shows no convergence across taxa; however, the functional association between the variables should not be discarded based on the PGLS result alone. Whereas in GLS, all diet descriptors were significantly related to total intestine length, this was not the case for several of them in PGLS (electronic supplementary material, table S2), suggesting phylogenetic specialization on either easily digestible or less easily digestible plant parts. Whereas in GLS, having a forestomach was a significant factor for a longer small intestine, this was not significant in PGLS, most likely because the presence of a forestomach is not evenly distributed across taxa but represents a hallmark of specific taxa [36,61]. For the same reason, we expected that a volant lifestyle, which was exclusively represented by bats in the mammalian dataset, should not yield a significant signal in PGLS—similar to other examples where a dichotomous distribution of traits across a phylogeny led to non-significance when accounting for that phylogeny [60,62]. However, it has been stated that methods like PGLS are sometimes susceptible to indicating significant relationships even in such dichotomic cases; in these instances, ‘unreplicated differences colocalized on a single [phylogenetic] branch provide only weak evidence of a causal relationship between traits’ [63]. Yet, that the evolution of flight requires a body plan with light organs, including a short intestinal tract, is physically plausible, and gains support from the convergence with birds [17,23,24].
(c). Allometry
As previously described for different datasets [17–19,25,28] except for a study in rodents [20], intestinal lengths scale at a higher exponent than expected based on simple geometry (i.e. positive allometry at an exponent > 0.33). This applied to the small intestine, the colon/rectum and the summative measures large and total intestine. This has been explained by geometry—that intestinal surface scales geometrically—and the necessity to keep diffusion distances short, so that intestinal diameter should not scale geometrically, but lower. Consequently, the length must scale higher than geometrically to compensate [28]. A comprehensive dataset on intestinal diameter would be required to test this.
The scaling of the caecum differed from that of the other intestinal sections, with a lower exponent than expected (i.e. negative allometry at an exponent < 0.33). Based on these scaling relationships, larger mammals have, on average, a relatively shorter caecum. We hypothesize that the reason for this is not to be sought in a constraint on caecum length at higher body masses. Rather, we suggest that the ‘shallower’ scaling is an effect of particularly long caeca in small species of the Lagomorpha and Rodentia (electronic supplementary material, figure S6). In these species, the digestive strategy of coprophagy is common [64–66], for which a voluminous caecum is one of the prerogatives. This strategy is dependent on a colonic mechanism that separates microbial matter from indigestible components of the digesta [67], which most likely is limited by colonic diameters and hence not feasible above a certain body size.
(d). Reasons for intestine length
Two basic arguments are used to explain the need for a longer intestinal section: (i) a niche that constantly requires more of the intestine's action, like a diet of lower digestibility [2,3,5–8], an arid environment [18,28,29], or a functional link with the strategy of coprophagy outlined above; or a niche that does not allow a long intestine due to other constraints, as in volant animals; (ii) a niche that does not imply the intestine to function consistently, but requires it to adapt to peak bursts of action. This second explanation has so far only been applied to the exceptionally long small intestines of diving marine predators [25–27]. Our results suggest the hypothesis that major constant differences in dietary or humidity niches are mainly reflected in the length of the caecum and colon, whereas differences in the constancy in intake and digestion are reflected in the length of the small intestine.
(i). Consistently different modes of action—diet niches
In broad terms, faunivores as well as herbivores specialized on nectar or seeds have highly digestible diets (80–90%), while herbivores consuming leaves and stem parts of plants have poorly digestible diets (50–70%) [2,3]. The lower digestibility is mainly an effect of the plant cell wall (fibre). Overall digestibility is typically negatively related to the diet's fibre content [4,68]. When considering the intestine, fibre is fermented with the assistance of a microbiome (allo-enzymatically) in the caecum and the proximal colon. Therefore, it is plausible that in animals that consume higher proportions of plant material, these sections are generally longer, as documented in the present study, and more complex [22]. As a side effect, mammalian herbivores require more voluminous body cavities [69] to harbour the longer, more complex digestive tract.
By contrast, the digestive action in the small intestine is not related to fibre fermentation, but to auto-enzymatic digestion of proteins, fats and easily digested carbohydrates. Available data do not suggest a difference in digestive processes for protein and fat between herbivores, omnivores or carnivores [70], and therefore, major differences between the trophic guilds need not to be expected for the small intestine. Possibly, plant fibre here only acts as a dilutant, for which herbivores might compensate with longer small intestines. However, the finding that diet did not significantly explain small intestine length in PGLS indicates that within taxonomic groups, no such relationship was evident.
The GLS finding that animals with a forestomach had longer small intestines was surprising. Forestomachs do not only occur in large mammalian herbivores of various taxa [61], but also in muroid rodents and cetacea [36,71,72]. The presence of a forestomach follows clear phylogenetic boundaries, which explains the absence of a significant signal in PGLS. We can only speculate that the enzymatic digestion of microbes, which grow in the forestomach and ultimately pass through the glandular stomach into the small intestine, requires additional intestinal capacity.
(ii). Consistently different modes of action—xeric environments
The findings that both precipitation and actual evapotranspiration were negatively related with colon/rectum and hence large intestine length support the concept that animals in more xeric environments need more intestinal capacity for water reabsorption [18,28,29], even though this pattern may not be consistent within specific mammal groups [20]. Future work could assess whether renal adaptations to aridity [73] occur in parallel, or in a compensatory manner, with colon length.
(iii). Consistently different modes of action—volant lifestyle
Similar to previous findings, volant mammals had shorter total intestines than terrestrial mammals [17,23,24], most likely to reduce the overall weight. In birds, this does not necessarily apply to the caecum [74]. Yet, active flight is an energetically demanding form of locomotion [75] for which sufficient energy must be absorbed from the shorter intestines. Therefore, flying mammals acquired morpho-physiological adaptations such as an enlarged intestinal absorptive area by increased microvillous amplification and epithelial folding [76]. Additionally, increased paracellular absorption compared to other mammals compensates for shorter intestines [24].
(iv). Irregular modes of action—marine habitats
Similar to previous reports, marine mammals were indicated to have longer small intestines in the present study. This additional capacity is required to compensate for a lack of intestinal function during diving [25–27].
(v). Irregular modes of action—seasonal habitats
A similar logic might apply for animals living in seasonal habitats. Small intestine length increased at increasing (absolute) latitude. In seasonal environments, many mammals have to incur the costs of reproduction in the growing season and also build up body reserves, which are then used in the dormant season [77]. Rather than experiencing a constant food intake, their digestive tract thus has to be able to cope with seasonally high intakes. On the one hand, it has been proposed for small mammals that increased plasticity in the digestive tract is linked to high-latitude habitats, so that more intestinal tissue can be made available when required [53]; on the other hand, our findings might indicate that a long small intestine itself is an adaptive feature for life at high latitudes. Given that seasonal environments have been linked to an increase in the pace of life [78], increased intestinal capacity might thus facilitate intense resource processing during periods of resource limitation.
5. Conclusion
Our investigation demonstrates associations between intestinal anatomy and dietary niches that have been claimed in the biological literature for long. At the same time, they indicate that these associations—or convergences—cannot be considered ubiquitous or ‘fixed laws’. Rather, data scatter suggests that different morpho-physiological solutions exist for the same ecological challenge—a typical finding of ecomorphological studies. The effects of dietary niches are particularly evident in sections of the large intestine, the caecum and the colon, whereas a trophic differentiation of the small intestine follows mammalian phylogeny and yields no significant signal when accounting for phylogeny. Increased habitat aridity is linked to a longer colon, and habitats with the colder temperature at higher latitudes are linked to longer small intestines. Our findings emphasize that different sections of the intestinal tract fulfil different functions during digestion.
Supplementary Material
Acknowledgements
We thank Barbara Schneider and Jacqueline Wick for tireless support in literature acquisition. The contributions to this project by the Disease Investigations and Animal Care staff at San Diego Zoo Global, Charles Paddock Zoo and Pacific Wildlife Care, as well as numerous student assistants, are greatly appreciated. We thank Emilia Clauss for drawing figure 1.
Data accessibility
The data collection including all individually recorded data and the species average values, together with the biological characteristics, the corresponding literature references and the phylogenetic tree used are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.z8w9ghxb8 [79]. The R code used in the statistical procedures has been fully referenced in the method section and is given in the electronic supplementary material.
Authors' contributions
M.J.D.C. participated in data collection and manuscript writing. D.C. performed the statistical analyses and critically revised the manuscript. C.M. compiled the phylogenetic tree and critically revised the manuscript. A.M., C.S. and M.S.E. participated in data collection and critically revised the manuscript. M.C. conceived of the study, designed the study, participated in the data collection and manuscript writing. All authors gave final approval for publication and agree to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by the Science National Foundation as part of the project CRSII5_189970/1.
References
- 1.Sahney S, Benton MJ, Ferry PA. 2010. Links between global taxonomic diversity, ecological diversity and the expansion of vertebrates on land. Biol. Lett. 6, 544–547. ( 10.1098/rsbl.2009.1024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Withers PC, Cooper CE, Maloney SK, Bozinovic F, Cruz-Neto AP. 2016. Ecological and environmental physiology of mammals. Oxford, UK: Oxford University Press. [Google Scholar]
- 3.Karasov WH, Martinez del Rio C, Caviedes-Vidal E. 2011. Ecological physiology of diet and digestive systems. Annu. Rev. Physiol. 73, 69–93. ( 10.1146/annurev-physiol-012110-142152) [DOI] [PubMed] [Google Scholar]
- 4.Karasov WH, Martínez del Rio C. 2007. Physiological ecology: how animals process energy, nutrients, and toxins. Princeton, NJ: Princeton University Press. [Google Scholar]
- 5.Chivers DJ, Hladik CM. 1980. Morphology of the gastrointestinal tract in primates: comparisons with other mammals in relation to diet. J. Morphol. 166, 337–386. ( 10.1002/jmor.1051660306) [DOI] [PubMed] [Google Scholar]
- 6.Eisenberg JF 1981. The mammalian radiations: an analysis of trends in evolution, adaptation, and behavior. Chicago, IL: University of Chicago Press. [Google Scholar]
- 7.Orr RT 1976. Vertebrate biology. Philadelphia, PA: WB Saunders. [Google Scholar]
- 8.Stevens CE, Hume ID. 1995. Comparative physiology of the vertebrate digestive system. New York, NY: Cambridge University Press. [Google Scholar]
- 9.Stevens CE, Hume ID. 1998. Contributions of microbes in vertebrate gastrointestinal tract to production and conservation of nutrients. Physiol. Rev. 78, 393–427. ( 10.1152/physrev.1998.78.2.393) [DOI] [PubMed] [Google Scholar]
- 10.Davis DD 1964. The giant panda: a morphological study of evolutionary mechanisms. Fieldiana (Zoology Memoirs) 3, 1–339. [Google Scholar]
- 11.Korn H 1992. Intestine lengths of Southern African savanna rodents and insectivores: intra- and interspecific comparisons. J. Zool. 228, 455–460. ( 10.1111/j.1469-7998.1992.tb04448.x) [DOI] [Google Scholar]
- 12.Perrin MR, Curtis BA. 1980. Comparative morphology of the digestive system of 19 species of Southern African myomorph rodents in relation to diet and evolution. South African J. Zool. 15, 22–33. ( 10.1080/02541858.1980.11447680) [DOI] [Google Scholar]
- 13.Fooden J 1964. Stomach contents and gastro-intestinal proportions in wild-shot Guianan monkeys. Am. J. Phys. Anthropol. 22, 227–231. ( 10.1002/ajpa.1330220243) [DOI] [PubMed] [Google Scholar]
- 14.Henke Z, Sahd L, Matthee S, Kotzé SH. 2018. Morphometric analysis of the gastrointestinal tract of four African muroid rodent species (Rhabdomys dilectus, Rhabdomys pumilio, Aethomys chrysophilus, and Lemniscomys rosalia). J. Morphol. 279, 1282–1289. ( 10.1002/jmor.20856) [DOI] [PubMed] [Google Scholar]
- 15.Mitsuzuka W, Oshida T. 2018. Feeding adaptation of alimentary tract length in arboreal squirrels. Mamm. Stud. 43, 125–131. ( 10.3106/ms2017-0079) [DOI] [Google Scholar]
- 16.Osawa R WP 1992. A comparative study of macroscopic and microscopic dimensions of the intestines in five macropods. II. Relationship with feeding habits and fibre content of the diet. Aust. J. Zool. 40, 99–113. ( 10.1071/ZO9920099) [DOI] [Google Scholar]
- 17.Lavin SR, Karasov WH, Ives AR, Middleton KM, Garland T. 2008. Morphometrics of the avian small intestine compared with that of nonflying mammals: a phylogenetic approach. Physiol. Biochem. Zool. 81, 526–550. ( 10.1086/590395) [DOI] [PubMed] [Google Scholar]
- 18.McGrosky A, Codron D, Müller D.WH, Navarrete A, Isler K, Hofmann RR, Clauss M. 2019. Gross intestinal morphometry and allometry in ruminants. J. Morphol. 280, 1254–1266. ( 10.1002/jmor.21028) [DOI] [PubMed] [Google Scholar]
- 19.McGrosky A, Meloro C, Navarrete A, Heldstab SA, Kitchener AC, Isler K, Clauss M. 2019. Gross intestinal morphometry and allometry in primates. Am. J. Primatol. 81, e23035 ( 10.1002/ajp.23035) [DOI] [PubMed] [Google Scholar]
- 20.Lovegrove BG 2010. The allometry of rodent intestines. J. Comp. Physiol. B 180, 741–755. ( 10.1007/s00360-009-0437-2) [DOI] [PubMed] [Google Scholar]
- 21.Smith HF, Parker W, Kotzé SH, Laurin M. 2017. Morphological evolution of the mammalian cecum and cecal appendix. C.R. Palevol. 16, 39–57. ( 10.1016/j.crpv.2016.06.001) [DOI] [Google Scholar]
- 22.Langer P, Clauss M. 2018. Morphological adaptation of the eutherian gastrointestinal tract to diet. Vertebr. Zool. 68, 237–252. [Google Scholar]
- 23.Price ER, Brun A, Caviedes-Vidal E, Karasov WH. 2015. Digestive adaptations of aerial lifestyles. Physiology 30, 69–78. ( 10.1152/physiol.00020.2014) [DOI] [PubMed] [Google Scholar]
- 24.Caviedes-Vidal E, McWhorter TJ, Lavin SR, Chediack JG, Tracy CR, Karasov WH. 2007. The digestive adaptation of flying vertebrates: high intestinal paracellular absorption compensates for smaller guts. PNAS 104, 19 132–19 137. ( 10.1073/pnas.0703159104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGrosky A, Navarrete A, Isler K, Langer P, Clauss M. 2016. Gross intestinal morphometry and allometry in Carnivora. Eur. J. Wildl. Res. 62, 395–405. ( 10.1007/s10344-016-1011-3) [DOI] [Google Scholar]
- 26.Mårtensson PE, Nordoy ES, Messelt EB, Blix AS. 1998. Gut length, food transit time and diving habit in phocid seals. Polar Biol. 20, 213–217. ( 10.1007/s003000050298) [DOI] [Google Scholar]
- 27.Krockenberger MB, Bryden MM. 1994. Rate of passage of digesta through the alimentary tract of southern elephant seals (Mirounga leonina). J. Zool. 234, 229–237. ( 10.1111/j.1469-7998.1994.tb06071.x) [DOI] [Google Scholar]
- 28.Woodall PF, Skinner JD. 1993. Dimensions of the intestine, diet and faecal water loss in some African antelope. J. Zool. 229, 457–471. ( 10.1111/j.1469-7998.1993.tb02648.x) [DOI] [Google Scholar]
- 29.Cain JW, Krausman PR, Rosenstock SS, Turner JC. 2006. Mechanisms of thermoregulation and water balance in desert ungulates. Wildl. Soc. Bull. 34, 570–581. ( 10.2193/0091-7648(2006)34[570:MOTAWB]2.0.CO;2) [DOI] [Google Scholar]
- 30.Cruz-Neto AP, Jones KE, Zubaid A, McCracken GF, Kunz TH. 2006. Exploring the evolution of basal rate of metabolism in bats. In Functional and evolutionary ecology of bats (eds Zubaid G, McCracken G, Kunz T), pp. 56–89. Oxford, UK: Oxford University Press. [Google Scholar]
- 31.Jones KE, MacLarnon AM. 2004. Affording larger brains: testing hypotheses of mammalian brain evolution on bats. Am. Nat. 164, E20-E31. ( 10.1086/421334) [DOI] [PubMed] [Google Scholar]
- 32.Wilman H, Belmaker J, Simpson J, de la Rosa C, Rivadeneira MM, Jetz W.. 2014. EltonTraits 1.0: species-level foraging attributes of the world's birds and mammals. Ecology 95, 2027 ( 10.1890/13-1917.1) [DOI] [Google Scholar]
- 33.Jenkins PD, Kilpatrick CW, Robinson MF, Timmins RJ. 2005. Morphological and molecular investigations of a new family, genus and species of rodent from Lao PDR. Syst. Biodivers. 2, 419–454. ( 10.1017/S1477200004001549) [DOI] [Google Scholar]
- 34.Keovichit PK, Nicolas V, Hugot J-P. 2011. Laonastes aenigmamus, rongeur énigmatique, récemment découvert au Laos. Bulletin de l'Académie Vétérinaire de France 164, 143–148. ( 10.4267/2042/48080) [DOI] [Google Scholar]
- 35.Nowak RM 1999. Walkeŕs mammals of the world, 6th edn Baltimore, MD: Johns Hopkins University Press. [Google Scholar]
- 36.Langer P 2017. Comparative anatomy of the gastrointestinal tract in eutheria: taxonomy, biogeography and food. Vol I: afrotheria, xenarthra and euarchontoglires. Vol II: laurasiatheria, general discussion. Berlin, Germany: De Gruyter. [Google Scholar]
- 37.Jones KE, et al. 2009. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology 90, 2648 ( 10.1890/08-1494.1) [DOI] [Google Scholar]
- 38.Upham NS, Esselstyn JA, Jetz W. 2019. Inferring the mammal tree: species-level sets of phylogenies for questions in ecology, evolution, and conservation. PLoS Biol. 17, e3000494 ( 10.1371/journal.pbio.3000494) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. ( 10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- 40.Wickham H, et al. 2019. Welcome to the Tidyverse. J. Open Source Softw. 4, 1686 ( 10.21105/joss.01686) [DOI] [Google Scholar]
- 41.Calder WA 1996. Size, function and life history. Cambridge, MA: Havard University Press. [Google Scholar]
- 42.Packard GC, Birchard GF, Boardman TJ. 2011. Fitting statistical models in bivariate allometry. Biol. Rev. 86, 549–563. ( 10.1111/j.1469-185X.2010.00160.x) [DOI] [PubMed] [Google Scholar]
- 43.Garland T, Bennett AF, Rezende EL. 2005. Phylogenetic approaches in comparative physiology. J. Exp. Biol. 208, 3015–3035. ( 10.1242/jeb.01745) [DOI] [PubMed] [Google Scholar]
- 44.Rezende RP, Diniz-Filho J. 2012. Phylogenetic analyses: comparing species to infer adaptations and physiological mechanisms. Compr. Physiol. 2, 645–674. ( 10.1002/cphy.c100079) [DOI] [PubMed] [Google Scholar]
- 45.Clauss M, Dittmann MT, Müller D.WH, Zerbe P, Codron D. 2014. Low scaling of a life history variable: analysing eutherian gestation periods with and without phylogeny-informed statistics. Mamm. Biol. 79, 9–16. ( 10.1016/j.mambio.2013.01.002) [DOI] [Google Scholar]
- 46.Orme D 2013. The caper package: comparative analysis of phylogenetics and evolution in R. R package version 0.5.2. See http://CRAN.R-project.org/package=caper.
- 47.Pinheiro J, Bates D, DebRoy S, Sarkar D, Core Team R. 2016. nlme: linear and nonlinear mixed effects models. R package version 3.1-128. See http://CRAN.R-project.org/package=nlme.
- 48.Revell LJ 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223. ( 10.1111/j.2041-210X.2011.00169.x) [DOI] [Google Scholar]
- 49.Blomberg SP, Garland T, Ives A. 2003. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57, 717–745. ( 10.1111/j.0014-3820.2003.tb00285.x) [DOI] [PubMed] [Google Scholar]
- 50.Pagel M 1999. Inferring the historical patterns of biological evolution. Nature 401, 877–884. ( 10.1038/44766) [DOI] [PubMed] [Google Scholar]
- 51.Burnham KP, Anderson DR. 2002. Model selection and multimodel inference: a practical information-theoretic approach. New York: NY: Springer. [Google Scholar]
- 52.Ehrlich C, Codron D, Hofmann RR, Hummel J, Clauss M. 2019. Comparative omasum anatomy in ruminants: relationships with natural diet, digestive physiology, and general considerations on allometric investigations. J. Morphol. 280, 259–277. ( 10.1002/jmor.20942) [DOI] [PubMed] [Google Scholar]
- 53.Naya DE, Bozinovic F, Karasov WH. 2008. Latitudinal trends in digestive flexibility: testing the climatic variability hypothesis with data on the intestinal length of rodents. Am. Nat. 172, E122–E134. ( 10.1086/590957) [DOI] [PubMed] [Google Scholar]
- 54.Tahas SA, Martin Jurado O, Hammer S, Arif A, Reese S, Hatt J.-M, Clauss M. 2017. Gross measurements of the digestive tract and visceral organs of addax antelope (Addax nasomaculatus) following a concentrate or forage feeding regime. Anatomia Histologia Embryologia 46, 282–293. ( 10.1111/ahe.12268) [DOI] [PubMed] [Google Scholar]
- 55.Clauss M, Trümpler J, Ackermans NL, Kitchener AC, Hantke G, Stagegaard J, Takano T, Shintaku Y, Matsuda I. 2021. Macroscopic digestive anatomy of ring-tailed lemurs (Lemur catta), including a comparison of frozen and formalin-stored specimens. Primates (online) ( 10.1007/s10329-020-00873-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Cuyper A, Clauss M, Carbone C, Codron D, Cools A, Hesta M, Janssens GPJ. 2019. Predator size and prey size–gut capacity ratios determine kill frequency and carcass production in terrestrial carnivorous mammals. Oikos 128, 13–22. ( 10.1111/oik.05488) [DOI] [Google Scholar]
- 57.Codron D, Hofmann RR, Clauss M. 2019. Morphological and physiological adaptations for browsing and grazing. In The ecology of browsing and grazing II (eds Gordon IJ, Prins HHT), pp. 81–125. Berlin, Germany: Springer. [Google Scholar]
- 58.Freckleton RP 2009. The seven deadly sins of comparative analysis. J. Evol. Biol. 22, 1367–1375. ( 10.1111/j.1420-9101.2009.01757.x) [DOI] [PubMed] [Google Scholar]
- 59.Clauss M, Dittmann MT, Müller D.WH, Meloro C, Codron D. 2013. Bergmann's rule in mammals: a cross-species interspecific pattern. Oikos 122, 1465–1472. ( 10.1111/j.1600-0706.2013.00463.x) [DOI] [Google Scholar]
- 60.Carbone C, Codron D, Scofield C, Clauss M, Bielby J. 2014. Geometric factors influencing the diet of vertebrate predators in marine and terrestrial environments. Ecol. Lett. 17, 1553–1559. ( 10.1111/ele.12375) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langer P 1988. The mammalian herbivore stomach. Stuttgart, Germany: Gustav Fischer Verlag. [Google Scholar]
- 62.Garland T, Dickerman AW, Janis CM, Jones JA. 1993. Phylogenetic analysis of covariance by computer simulation. Syst. Biol. 42, 265–292. ( 10.1093/sysbio/42.3.265) [DOI] [Google Scholar]
- 63.Uyeda JC, Zenil-Ferguson R, Pennell MW. 2018. Rethinking phylogenetic comparative methods. Syst. Biol. 67, 1091–1109. ( 10.1093/sysbio/syy031) [DOI] [PubMed] [Google Scholar]
- 64.Hirakawa H 2001. Coprophagy in leporids and other mammalian herbivores. Mamm. Rev. 31, 61–80. ( 10.1046/j.1365-2907.2001.00079.x) [DOI] [Google Scholar]
- 65.Hirakawa H 2002. Supplement: coprophagy in leporids and other mammalian herbivores. Mamm. Rev. 32, 150–152. ( 10.1046/j.1365-2907.2002.00105.x) [DOI] [Google Scholar]
- 66.Clauss M, Besselmann D, Schwarm A, Ortmann S, Hatt J.-M. 2007. Demonstrating coprophagy with passage markers? The example of the plains viscacha (Lagostomus maximus). Comp. Biochem. Physiol. A 147, 453–459. ( 10.1016/j.cbpa.2007.01.013) [DOI] [PubMed] [Google Scholar]
- 67.Björnhag G, Snipes RL. 1999. Colonic spearation mechanism in lagomorph and rodent species—a comparison. Zoosyst. Evol. 75, 275–281. ( 10.1002/mmnz.19990750208) [DOI] [Google Scholar]
- 68.Hagen KB, Tschudin A, Liesegang A, Hatt J.-M, Clauss M. 2015. Organic matter and macromineral digestibility in domestic rabbits (Oryctolagus cuniculus) as compared to other hindgut fermenters. J. Anim. Physiol. Anim. Nutr. 99, 1197–1209. ( 10.1111/jpn.12323) [DOI] [PubMed] [Google Scholar]
- 69.Clauss M, Nurutdinova I, Meloro C, Gunga H-C, Jiang D, Koller J, Herkner B, Sander PM, Hellwich O. 2017. Reconstruction of body cavity volume in terrestrial tetrapods. J. Anat. 230, 325–336. ( 10.1111/joa.12557) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Richard OK, Codron D, Hagen KB, Südekum K.-H, Clauss M. 2017. Little differences in digestive efficiency for protein and fat in mammals of different trophic guilds and digestive strategies: data constraints or fundamental functional similarity? J. Anim. Physiol. Anim. Nutr. 101(Suppl. 1), 127–141. ( 10.1111/jpn.12657) [DOI] [PubMed] [Google Scholar]
- 71.Vorontsov NN 1967/1969 evolution of the alimentary system in rodents. Novosibirsk: Nauka; (English translation by Indian Scientific Documentation Centre, New Delhi). [Google Scholar]
- 72.Carleton MD 1973. A survey of gross stomach morphology in New World Cricetinae (Rodentia, Muroidea), with comments on functional interpretations. Miscellaneous Publications of the Museum of Zoology, University of Michigan 146, 1–43. [Google Scholar]
- 73.Beuchat CA 1996. Structure and concentrating ability of the mammalian kidney: correlations with habitat. Am. J. Physiol. 271, R157-R179. [DOI] [PubMed] [Google Scholar]
- 74.Hunt A, Al-Nakkash L, Lee AH, Smith HF. 2019. Phylogeny and herbivory are related to avian cecal size. Sci. Rep. 9, 4243 ( 10.1038/s41598-019-40822-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maina JN 2000. What it takes to fly: the structural and functional respiratory refinements in birds and bats. J. Exp. Biol. 203, 3045–3064. [DOI] [PubMed] [Google Scholar]
- 76.Makanya AN, Maina JN, Mayhew TM, Tschanz SA, Burri PH. 1997. A stereological comparison of villous and microvillous surfaces in small intestines of frugivorous and entomophagous bats: species, inter-individual and craniocaudal differences. J. Exp. Biol. 200, 2415–2423. [DOI] [PubMed] [Google Scholar]
- 77.Lindstedt SL, Boyce MS. 1985. Seasonality, fasting endurance, and body size in mammals. Am. Nat. 125, 873–878. ( 10.1086/284385) [DOI] [Google Scholar]
- 78.Clauss M, Zerbe P, Bingaman Lackey L, Codron D, Müller DW.H. 2020. Basic considerations on seasonal breeding in mammals including their testing by comparing natural habitats and zoos. Mamm. Biol. (online) ( 10.1007/s42991-020-00078-y). [DOI] [Google Scholar]
- 79.Duque-Correa MJ, Codron D, Meloro C, McGrosky A, Schiffmann C, Edwards MS, Clauss M. 2021. Data from: Mammalian intestinal allometry, phylogeny, trophic level and climate Dryad Digital Repository. ( 10.5061/dryad.z8w9ghxb8) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Duque-Correa MJ, Codron D, Meloro C, McGrosky A, Schiffmann C, Edwards MS, Clauss M. 2021. Data from: Mammalian intestinal allometry, phylogeny, trophic level and climate Dryad Digital Repository. ( 10.5061/dryad.z8w9ghxb8) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The data collection including all individually recorded data and the species average values, together with the biological characteristics, the corresponding literature references and the phylogenetic tree used are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.z8w9ghxb8 [79]. The R code used in the statistical procedures has been fully referenced in the method section and is given in the electronic supplementary material.