Abstract
Differentiated sex chromosomes are believed to be evolutionarily stable, while poorly differentiated sex chromosomes are considered to be prone to turnovers. With around 1700 currently known species forming ca 15% of reptile species diversity, skinks (family Scincidae) are a very diverse group of squamates known for their large ecological and morphological variability. Skinks generally have poorly differentiated and cytogenetically indistinguishable sex chromosomes, and their sex determination was suggested to be highly variable. Here, we determined X-linked genes in the common sandfish (Scincus scincus) and demonstrate that skinks have shared the same homologous XX/XY sex chromosomes across their wide phylogenetic spectrum for at least 85 million years, approaching the age of the highly differentiated ZZ/ZW sex chromosomes of birds and advanced snakes. Skinks thus demonstrate that even poorly differentiated sex chromosomes can be evolutionarily stable. The conservation of sex chromosomes across skinks allows us to introduce the first molecular sexing method widely applicable in this group.
Keywords: genome, molecular sexing, sex determination, sex chromosomes, qPCR, vertebrates
1. Introduction
Organisms do not share a common mechanism for sex determination. Under genotypic sex determination (GSD), the sex of an individual is set at conception by its sex-specific genotype, i.e. by the sex-specific combination of sex chromosomes. GSD is very common in animals and has evolved in them multiple times. It is estimated that GSD has occurred independently up to 40 times just within amniotes [1]. Sex chromosomes are thus a notable example of convergent evolution. Surprisingly, in spite of over a century of research [2], the adaptive significance and consequences of sex chromosomes and their differentiation, the progressive cessation of recombination and divergence of sequences between chromosomes in a sex chromosome pair, are still rather poorly understood. Sex chromosomes primarily determine the sex of an individual and ensure a stable sex ratio at conception; however, the sex-specific specialization, and particularly the role in the resolution of the conflict between sexes over trait expression, have been considered crucial for differentiation and evolutionary stability of sex chromosomes [1,3]. Nevertheless, there are more ways to resolve sexual conflict, the most important in vertebrates being the control of the expression of sex-specific traits by sex hormones [4,5]. Species without sex chromosomes such as vertebrates with environmental sex determination (ESD) are considerably sexually dimorphic as well [6], and the importance of sex chromosomes for the resolution of sexual conflict is still highly debated [7].
Next to the above-mentioned adaptive advantages, organisms with differentiated sex chromosomes also incur important costs. In animal lineages, a higher mortality and a reduced lifespan in individuals of the heterogametic sex leading to unequal adult sex ratio was explained by the negative consequences of the Y and W degeneration (e.g. by a loss of functional gene copies, altered gene expression, heterochromatinization and accumulation of repetitive elements) [8,9].
Vertebrate lineages differ in the degree of differentiation of sex chromosomes. The causes of the unequal rate of differentiation of sex chromosomes among lineages are still unresolved. The different selection pressures in males compared to females led to the traditional theoretical predictions that sex chromosomes should differentiate faster under male heterogamety (XX/XY) than under female heterogamety (ZZ/ZW). The expectation of the faster differentiation (or degeneration) of Y was based on assumptions of a stronger sexual selection in males, male mutation bias or a smaller effective population size of the Y chromosome with a decreasing male to female ratio in a population [10–12]. However, the very recent model stressing differences in the recombination rates between sexes and among lineages suggested just the opposite pattern: inversions reducing or arresting recombination should be fixed more frequently on Z and W than X and Y chromosomes, and it is therefore expected that the W chromosome degenerates faster than the Y [13]. We clearly need more empirical data on the rate of differentiation of sex chromosomes in different lineages with the opposite heterogamety to solve this controversy.
Among vertebrates, sex chromosomes were traditionally believed to be stable in endotherms, i.e. in mammals and birds, but unstable in fish, amphibians and non-avian reptiles [14]. For reptiles, multiple transitions between ESD and GSD in both directions were expected [15]. This traditional paradigm was shaken only recently by a series of molecular studies documenting that many reptile lineages, specifically advanced snakes, lacertid lizards, anguimorphan reptiles, iguanas and softshell turtles possess in fact highly differentiated and long-term, stable sex chromosomes [16–22]. The minimal age of sex chromosomes in these lineages is approximately between 80 and 180 MY and thus comparable to the minimal age of sex chromosomes of birds (100–120 Ma) and viviparous mammals (165 Ma) [23,24]. For squamates, it was suggested that sex chromosomes evolved a long time ago independently, possibly from the ancestral ESD, in most of their major clades. Only dragon lizards, geckos and possibly skinks were considered exceptional due to extensive variability in sex-determination systems [1,25].
With around 1700 currently known species forming ca 15% of reptile species diversity, skinks (family Scincidae) are a very diverse, nearly cosmopolitan group of squamates known for their large ecological and morphological variability [26]. They include terrestrial, subterranean, arboreal and semiaquatic forms with numerous transitions to viviparity, limblessness and nocturnality and significant variability in body size [27]. In spite of decades of cytogenetic research, sex chromosomes have been determined only in about a dozen skink species across their enormous diversity [28–31]. Variability of sex determination in skinks has been suggested for a long time. Previously Donnellan [32] suggested that sex chromosomes are not homologous across skink lineages based on chromosome morphology. However, chromosome morphology is not the decisive criterion of (non)homology of sex-determination systems. The situation was complicated further by reports of an environmental influence on sex in certain species of skinks, leading to the conclusion that some members of this lineage have ESD and hence no sex chromosomes [33–35]. Nevertheless, many earlier reports of ESD were found to be unreliable based on recent cytogenetic or molecular evidence [36–39]. These erroneous reports of ESD in actually GSD species caused an overestimation of the number of GSD to ESD transitions among amniotes and undermined the long-term stability of GSD.
We decided to shed light on this long-lasting debate. By comparison of the male and female genomes, we identified X-specific genes, i.e. genes present on X but missing on the Y chromosome, in the common sandfish (Scincus scincus), one of the few skink species with cytogenetic evidence for sex chromosomes [40]. After validation of X-specificity by quantitative PCR (qPCR) in this species, we performed a molecular test of the homology of sex chromosomes across skinks and their nearest outgroups.
2. Material and methods
(a). Studied material
Tissue samples (blood from living individuals and muscle or tissue from the tip of the tail from preserved specimens) were collected from one male and one female individual of 13 representative species of skinks and their close outgroups represented by members of the families Cordylidae, Gerrhosauridae and Xantusiidae, who along with skinks form the clade Scincoidea (electronic supplementary material, table S1). Selection of species was done according to their phylogenetic position to cover all major skink lineages [41] and availability of high-quality tissue samples of both sexes for DNA isolation. We were not able to include the African subterranean subfamily Acontinae, where sexed material was not available to us. Genomic DNA was isolated from blood or tail tissue using a Qiagen DNeasy Blood and Tissue kit (Qiagen). All experimental procedures were carried out under the supervision and with the approval of the Ethics Committee of the Faculty of Science, Charles University in Prague, followed by the Committee for Animal Welfare of the Ministry of Agriculture of the Czech Republic (permission no. 29555/2006-30).
(b). Chromosome preparations and cytogenetic analysis of Scincus scincus and Tropidophorus baconi
Metaphase chromosomes were prepared from whole blood cell cultures according to the protocol described in Mazzoleni et al. [42] and Pokorná et al. [43] in widely phylogenetically distant skinks Scincus scincus (Scincinae) and Tropidophorus baconi (Lygosominae). The chromosomes were Giemsa-stained and photographed using a Provis AX70 fluorescence microscope equipped with a digital camera (Olympus DP30BW). Karyograms were prepared using Ikaros software (MetaSystems, Germany). To identify sex-specific differences in sequence composition, we performed comparative genomic hybridization (CGH) using a previously described protocol [36]. Also, we examined the distribution of the rDNA loci as the sex chromosomes of Scincus scincus were identified by differences in numbers of nucleolar organizing regions (NORs) between sexes [40], using fluorescence in situ hybridization (FISH). The probe for the rDNA loci was prepared from a plasmid (pDm r.a51#1) with an 11.5-kb insertion, encoding the 18S and 28S rRNA units of Drosophila melanogaster [44]; this protocol is explained in detail in Rovatsos et al. [36].
(c). Genome coverage analysis in Scincus scincus
Genomic DNA isolated from blood samples of one male and one female of Scincus scincus was sequenced at high coverage by Novogene (Cambridge, UK) in the Illumina HiSeq2500 platform, with 150 base pairs (bp) pair-end option (DNA-seq). The raw Illumina reads are deposited in GenBank under the NCBI BioProject PRJNA660179. Adapters and low-quality bases from raw reads were trimmed by Trimmomatic [45] and reads shorter than 50 bp were removed. Trimmed reads were checked in FASTQC [46] and MULTIQC [47].
The X-specific genes have half the copy numbers in the genomes of XY males in comparison to XX females. The differences in the copy numbers of X-specific genes between sexes are expected to be proportional to the differences in coverage of the reads from DNA sequencing in Illumina HiSeq platforms [48,49]. Therefore, these loci are expected to have half read coverage in males compared to females, while autosomal and pseudoautosomal loci should have equal read coverage in both sexes. We independently mapped the trimmed Illumina reads from the male and the female sandfish using Geneious Prime (https://www.geneious.com) to a reference dataset of 237 298 exons of the common wall lizard (Podarcis muralis), a lizard with a genome assembled to chromosome level and high-quality gene annotation [50]. The average read coverage per gene was calculated in each specimen after filtering all exons with unexpectedly high or low coverage (i.e. fourfold difference from the average coverage). Subsequently, we calculated the ratio of female to male read coverage for each gene, normalized to the total number of assembled reads per specimen (electronic supplementary material, table S2). We identified X-specific genes as genes with a male to female coverage ratio between 0.35 and 0.65.
Also, X-specific, single-copy genes are hemizygous in the heterogametic sex (i.e. in males in S. scincus). Therefore, such loci should not have single nucleotide polymorphisms (SNPs) in our map-to-reference assembly from males. We calculated the presence/absence of SNPs in the assembly of the male sandfish in order to validate the X-specificity of genes uncovered by the comparative coverage analysis (electronic supplementary material, table S2).
(d). Validation of X-specific loci and test of homology of sex chromosomes across skinks by qPCR
The differences in gene copy numbers of the X-specific genes between sexes can be measured by qPCR applied to genomic DNA [16–22]. We applied this technique to further validate X-specificity of a subset of X-specific genes uncovered from bioinformatic analyses. Subsequently, as a test of homology of sex chromosomes across skinks, we tested whether the X-specific genes in S. scincus are also X-specific in other skinks and their close outgroups. For qPCR, we designed specific primers using Primer3 software [51] for the amplification of a 120–200 bp fragment of 10 X-specific genes, i.e. genes with the male to female genome coverage ratio around 0.5 lacking SNPs in the male of S. scincus, (electronic supplementary material, table S3). In addition, we amplified by qPCR three single-copy autosomal genes (abarb2, eef1a1, mecom) [39], for normalization of the quantification values and for autosomal controls.
The qPCR was carried out in a LightCycler II 480 (Roche Diagnostics) with all samples run in triplicate. The detailed qPCR protocol and the description of the calculation of the relative gene dose between sexes are available in our previous reports [17,52]. Briefly, the gene dosage of each target gene was calculated from crossing point values and was subsequently normalized to the dose of the single-copy reference gene eef1a1 or mecom from the same DNA sample. The relative gene dosage ratio (r) between sexes of a given species for each target gene is expected to be close to 0.5 for X-specific genes, 1.0 for (pseudo)autosomal genes and potentially 2.0 for Z-specific genes.
3. Results and discussion
(a). Cytogenetic analysis
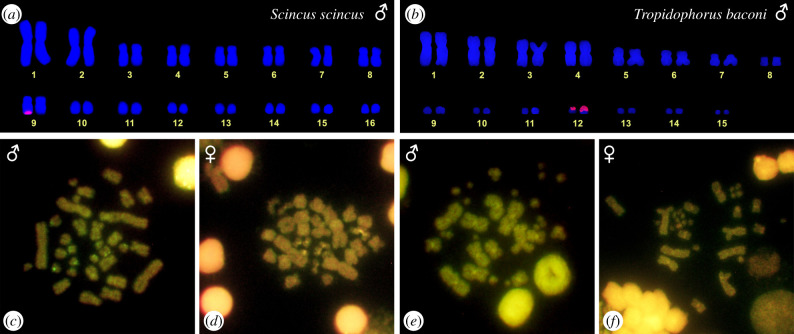
Caputo et al. [40] reported the sex-associated NOR polymorphism in S. scincus with two active NORs in females, while only one in males. We confirmed this observation by FISH with the probe specific for rDNA sequences, demonstrating that the sex chromosomes in this species differ both in the transcription activity (NORs) and the number of rDNA repeats (figure 1a). Nevertheless, the sex-related polymorphism in the presence of rDNA loci is not present in the genera Eumeces and Plestiodon closely related to S. scincus [40], nor in Tropidophorus baconi (figure 1b). The studied male of T. baconi (figure 1b) possessed a small difference in the accumulation of rDNA loci between the two chromosomes of the pair; however, the variability in the number of rDNA copies within a species is common in vertebrates [19], and the presence/absence polymorphism of rDNA loci is clearly not present in this species. Thus, it seems that the polymorphism enabling us to distinguish sex chromosomes cytogenetically is an apomorphy of the sandfish lineage. Surprisingly, sex-specific differences in the genome were not detected by CGH in either S. scincus (figure 1c,d), or in Tropidophorus baconi (figure 1e,f). The lack of sex-specific signal in CGH indicates that the sex chromosomes (if present in Tropidophorus baconi) are homomorphic and poorly differentiated.
Figure 1.
Karyogram reconstruction from metaphase after FISH with the probe for rDNA loci in male individuals of Scincus scincus (a) and Tropidophorus baconi (b). Signal of rDNA loci was detected in a single chromosome from the 9th pair in males of S. scincus, but in both chromosomes of the 12th pair in T. baconi. CGH revealed differences between sexes neither in males, nor in females in both S. scincus (c,d) and T. baconi (e,f). (Online version in colour.)
(b). X-linked genes in Scincus scincus
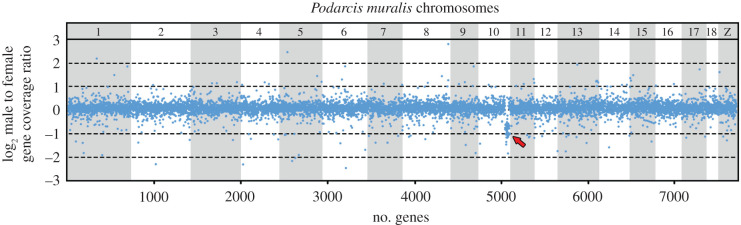
The analysis of exonic genome regions in the common sandfish revealed 560 genes with roughly half coverage in the male in comparison to the female genome (male to female ratio between 0.35 and 0.65), which is expected for X-specific genes missing on the Y chromosome (figure 2; electronic supplementary material, table S2). Notably, 169 out of 560 candidate X-specific genes were lacking polymorphisms in males (determined as the absence of SNPs in at least 80% of exons in a given gene), which is in accordance with the expected hemizygous state. The homologues of these 169 genes are scattered across all P. muralis chromosomes. Notably, 37 of these genes are linked to P. muralis chromosome 10 (PMU10), covering a chromosomal region of approximately 7 million base pairs (electronic supplementary material, table S2). The X-specificity of a subset of 10 genes from PMU10 was validated by comparison of copy number variation between the sexes by quantitative PCR (qPCR) (figure 3; electronic supplementary material, table S3).
Figure 2.
Log2-transformed male to female ratios of DNA-seq read coverage per gene in Scincus scincus. The genes are illustrated according to the position of their orthologues in the chromosomes of Podarcis muralis. Note that the region homologous to PMU10 (indicated by a red arrow) possesses half male to female ratio in read coverage depth, demonstrating that this genome part is X-specific in Scincus scincus. (Online version in colour.)
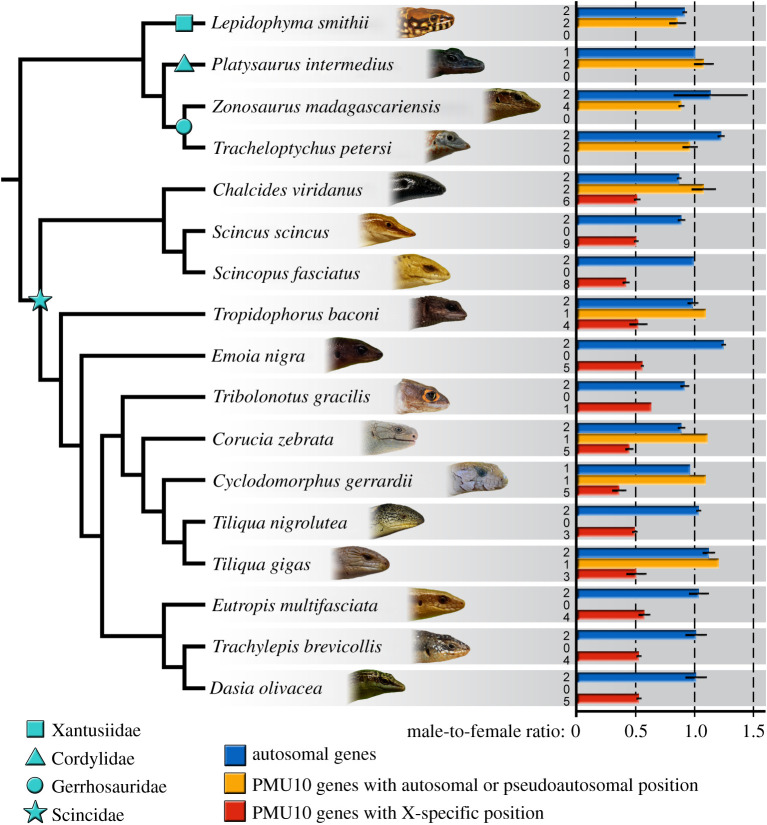
Figure 3.
Average relative gene dose ratios between males and females across 13 species of skinks (Scincidae) and four outgroup species from the families Cordylidae, Gerrhosauridae and Xantusiidae. Blue bars represent autosomal control genes. The genes from the X-specific region of S. scincus have either ratio around 0.5 consistent with the X-specificity (red bars), or around 1.0 consistent with pseudoautosomal or autosomal position (orange bars). Numbers of successfully tested genes in each category are presented. All data are presented in electronic supplementary material, table S3. (Online version in colour.)
The X-linked genes from the region homologous to a part of PMU10 have homologues linked to a region of chicken chromosome 1 (GGA1). As far as is known, the PMU10/GGA1 syntenic block is among amniotes involved in sex chromosomes only in skinks and the phylogenetically distant geckos from the genus Coleonyx [22,53] supporting the hypothesis that the sex chromosomes in skinks evolved independently from other amniote sex chromosomes. Notably, this chromosomal region contains several genes, which are involved in gonad development or in pathological conditions (dmc1, ep300, igf1, kitlg, nup107, pdgfb, sbf1, sox10, stra8, sycp3) and can potentially act as the sex-determining gene(s) in skinks. Among these genes, prominent candidates could be the genes ep300 (E1A binding protein p300), a histone acetyltransferase that regulates transcription via chromatin remodelling, and sox10 (sry-box transcription factor 10), a member of the sox (sry-related hmg-box) family of transcription factors involved in the regulation of embryonic development. Mouse embryos lacking functional copies of ep300 exhibit complete XY gonadal sex reversal [54]. In pathological conditions, the ectopic expression of sox10 in embryonic gonad leads to upregulation of transcriptional targets of the sox9 gene, triggering the male differentiation pathway and resulting in sex-reversed XX males in humans and mice [55,56]. A third potential sex-determining gene is sbf1 (SET-binding factor 1), a member of the protein-tyrosine phosphatase family involved in cell growth and differentiation which is highly expressed in the brain and testes. Alterations in the sbf1 sequence or splicing lead to male infertility, impaired spermatogenesis and azoospermia in humans, mice and rats [57–60]. Notably, sbf1 is nested in the 7 million base pair chromosomal region, enriched with X-specific genes in S. scincus.
(c). Poorly differentiated, but highly conserved sex chromosomes across skinks
The copy number variation in the orthologues of X-specific genes of the common sandfish was consistent with their X-specificity in all 13 tested skinks covering a phylogenetic spectrum of 1665 extant species (figure 3 and electronic supplementary material, table S3). The genes from the X-specific region of S. scincus determined by the coverage analysis and tested by qPCR showed in skinks clearly bimodal pattern in the male to female ratios. Most of them had the ratios around 0.5 consistent with the X-specificity (red bars in figure 3). However, in some species few such genes had ratios around 1.0 consistent with autosomal or pseudoautosomal position (orange bars in figure 3), which can be explained by translocations of genes from the X-specific to autosomal or pseudoautosomal positions, or by rare recombination events between otherwise non-recombining regions of the X and the Y chromosomes. Evidence for such rearrangements can be expected in comparisons of species across old radiations (see similar cases e.g. in lacertid lizards [20,39]).
Our results support that sex chromosomes are highly conserved across the skink radiation (figure 3), in spite of their poor differentiation. We confirmed the poor stage of differentiation in Tropidophorus baconi, where cytogenetic techniques including CGH failed to reveal sex chromosomes, although qPCR confirmed that this species shares XX/XY sex chromosomes with the common sandfish. In support of our results, a recent study in the water skink Eulamprus heatwolei revealed an XX/XY sex-determination system, with sex chromosomes homologous to the same part of GGA1 [61]. Sex chromosomes in skinks are typically homomorphic, and considering the relatively small X-specific region within otherwise quite large sex chromosomes uncovered in S. scincus (this study) and Eulamprus heatwolei [61], they can be indeed assigned as poorly differentiated, although this term is rather subjective and the objective criteria for the degree of differentiation are highly discussed [62]. The small X-specific region of skinks seems to be beyond the detection efficiency of molecular cytogenetic methods such as CGH, which explains why sex chromosomes in most skinks were not detected up to now despite several careful cytogenetic studies [63,64]. The orthologues of X-specific genes of the common sandfish showed a pseudoautosomal or autosomal pattern in the outgroups from the three families (Cordylidae, Gerrhosauridae and Xantusiidae) which along with skinks form the clade Scincoidea. ZZ/ZW sex chromosomes containing genes from other regions were reported in a night lizard from the family Xantusiidae [65], while sex determination is not known up to now in any cordylid or gerrhosaurid lizards. The origin of sex chromosomes in skinks can therefore be dated between the split of the monophylum of the three other scincoidean families and skinks around 150 Ma and the divergence of the subfamilies Lygosominae and Scincinae living ca 85 Ma [66,67].
(d). Towards resolution of controversies on sex determination in skinks and other reptiles
At least two species of skinks (Bassiana duperreyi, Niveoscincus ocellatus) previously reported as ESD, seem to have GSD. The effect of environmental conditions leading to unequal sex ratio is probably due to sex reversals in Bassiana duperreyi [68] and sex-linkage of anonymous molecular markers consistent with XX/XY sex chromosomes were identified in Niveoscincus ocellatus [69]. Of course, we cannot exclude the possibility that some species of skinks do not share the same sex chromosomes. Rare exceptions from the conserved pattern with likely derived sex chromosomes were found in iguanas, anguimorphan lizards and mammals [18,21,70,71]. However, the current results show that the sex chromosomes homologous to the sandfish are conserved for a long time across most of the phylogenetic diversity of skinks. Assuming that all species derived from the common ancestor of Lygosominae and Scincinae share the same chromosomes, we can estimate that roughly at least 15% out of nearly 11 000 currently recognized squamate species share sex chromosomes with the sandfish, and that about 60% of squamate species are members of the five lineages with adequate molecular evidence for conservation of sex chromosomes (figure 4). We assume that GSD will also be frequent in the remaining 40% of squamate species, with sex chromosomes being reported for example in chameleons, agamids and many geckos [22,25,36,37,72]. Against older predictions [73], ESD seems to be rather rare representing approximately 5% of species diversity in non-avian reptiles. Future studies should further verify and increase the precision of these estimations.
Figure 4.
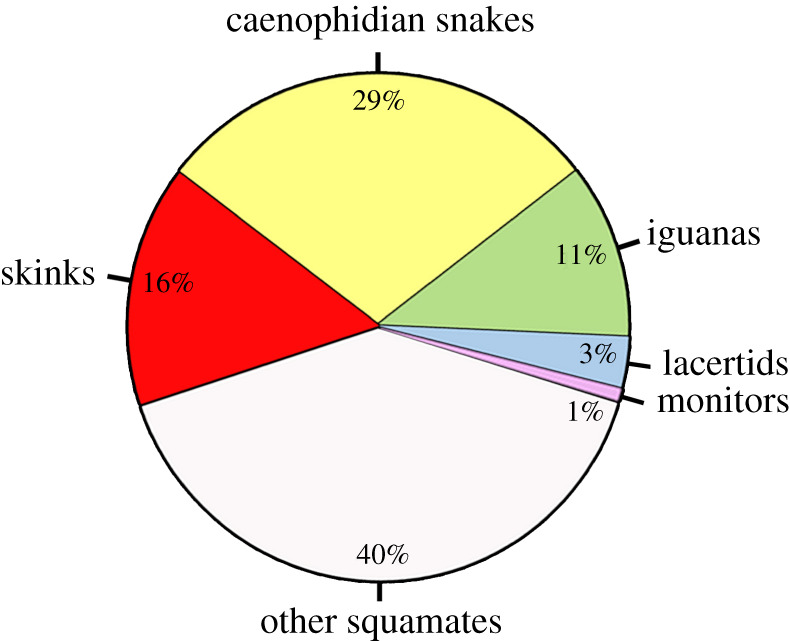
Although sex determination was traditionally assumed to be highly unstable in squamate reptiles, around 60% of squamates are members of five clades with sex chromosomes conserved for dozens of millions of years [current study; 16–18,20,21]. A molecular sexing method based on qPCR was developed for each of these clades, allowing the accurate identification of the sex in approximately 6000 species of squamates. (Online version in colour.)
(e). Molecular sexing of skinks
The technique based on quantitative PCR for testing the homology of sex chromosomes across skinks can be used as a widely applicable method for molecular sexing [17]. Such methods were not available up to now in skinks, although it is very much needed. Molecular sexing is essential for many developmental and ecological studies requiring the determination of the sex of embryos, which were previously hampered in this important group by the lack of a molecular sexing method. Moreover, many skinks are extremely difficult to sex based on external morphology even as adults. Fifty-six skink species are critically endangered and 77 species are endangered worldwide [74]. Our technique for molecular sexing could be crucial for the success of conservation projects. Furthermore, molecular sexing is a valuable tool for studies in developmental biology. This molecular sexing method based on qPCR is now available for potentially 60% of squamate species (figure 4).
4. Conclusion
The observed homology of sex chromosomes in skinks was not expected based on recent models of sex chromosome evolution which postulated that poorly differentiated sex chromosomes of ectotherms are young and unstable [14], i.e. prone to turnovers or even transitions to ESD where sex chromosomes are not present. Skinks can be considered as another reptile group (figure 4) with the stability of sex chromosomes. Skinks provide further support that the phylogenetic border between unstable and stable sex chromosomes does not lay between ectotherms and endotherms. Recent work showing high-evolutionary stability in sturgeons [75] suggests that the dichotomy is neither between amniotes and anamniotes as was previously suggested [20]. The age of the poorly differentiated XX/XY sex chromosomes in skinks is similar to the differentiated ZZ/ZW sex chromosomes in birds and advanced snakes [16,23], which opposes the traditional theoretical predictions that the Y chromosome should degenerate faster than the W chromosome. The traditional predictions are also not supported by the empirical data in other squamate lineages. A higher degree of differentiation in lineages with female heterogamety in comparison to male heterogamety was observed within snakes [76], chameleons [36,37] and between closely related teiid and lacertid lizards [20,77]. On the other hand, XX/XY sex chromosomes are highly differentiated in viviparous mammals and iguanas (with the exception of basilisks [71,78]); however, sex chromosomes in these two groups are among the oldest sex chromosomes uncovered to date in amniotes, and they thus potentially had a longer time to differentiate [79,80]. Moreover, these two lineages are the only known amniote lineages with a chromosome-wide dosage compensation mechanism [81–83], which might influence the rate of differentiation of sex chromosomes. Processes keeping sex chromosomes at a poorly differentiated state for a long time as we observe in skink may include a high recombination rate between sex chromosomes, potentially facilitated by thermally induced sex reversal [84]. In any case, reasons for the differences in the differentiation rate of sex chromosomes among lineages remain unclear and represent a challenging and fruitful research field [14,62,84].
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We would like to express our gratitude to Jana Thomayerová for technical assistance and Amy Wilson for the linguistic improvement of the manuscript. Blood or tissue samples for this study were kindly provided by Daniel Frynta, Jiří Šmíd and Jan Suchánek. Photos of skinks for figure 3 were kindly provided by Anna Bauerová, Arthur Georges, Lukáš Kubička, Tomáš Peš, Jiří Šmíd and Jana Thomayerová.
Data accessibility
The raw Illumina reads from DNA-seq of all studied individuals are deposited into the NCBI BioProject database with ID PRJNA660179.
Authors' contributions
A.K. was involved in cytogenetic and molecular work; L.K. was involved in statistics; M.R. was involved in bioinformatics; M.R. and L.K. conceived the project. All authors contributed to the final form of the manuscript and are responsible for its content.
Competing interests
We declare we have no competing interests.
Funding
The project was supported by the Grant Agency of the Czech Republic (GAČR 19-19672S), the Charles University Primus Research Program (PRIMUS/SCI/46), the Charles University Research Centre programme (204069) and the Charles University Grant Agency (1518119).
References
- 1.Johnson Pokorná M, Kratochvíl L. 2014. What was the ancestral sex-determining mechanism in amniote vertebrates? Biol. Rev. 91, 1–12. ( 10.1111/brv.12156) [DOI] [PubMed] [Google Scholar]
- 2.Abbott JK, Nordén AK, Hansson B.. 2017. Sex chromosome evolution: historical insights and future perspectives. Proc. R. Soc. B 284, 20162806 ( 10.1098/rspb.2016.2806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bull JJ, Charnov EL. 1985. On irreversible evolution. Evolution 39, 1149–1155. ( 10.2307/2408742) [DOI] [PubMed] [Google Scholar]
- 4.Rinn JL, Snyder M. 2005. Sexual dimorphism in mammalian gene expression. Trends Genet. 21, 298–305. ( 10.1016/j.tig.2005.03.005) [DOI] [PubMed] [Google Scholar]
- 5.Norris DO, Lopez KH. 2011. Hormones and reproduction of vertebrates San Diego, CA: Elsevier Academic Press Inc. [Google Scholar]
- 6.Viets BE, Ewert MA, Talent LG, Nelson CE. 1994. Sex-determining mechanisms in squamate reptiles. J. Exp. Zool. 270, 45–56. ( 10.1002/jez.1402700106) [DOI] [Google Scholar]
- 7.Bergero R, Gardner J, Bader B, Yong L, Charlesworth D. 2019. Exaggerated heterochiasmy in a fish with sex-linked male coloration polymorphisms. Proc. Natl Acad. Sci. USA 116, 6924–6931. ( 10.1073/pnas.1818486116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pipoly I, Bókony V, Kirkpatrick M, Donald PF, Székely T, Liker A. 2015. The genetic sex-determination system predicts adult sex ratios in tetrapods. Nature 527, 91–94. ( 10.1038/nature15380) [DOI] [PubMed] [Google Scholar]
- 9.Xirocostas ZA, Everingham SE, Moles AT. 2020. The sex with the reduced sex chromosome dies earlier: a comparison across the tree of life. Biol. Lett. 16, 20190867 ( 10.1098/rsbl.2019.0867) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachtrog D, Kirkpatrick M, Mank JE, McDaniel SF, Pires JC, Rice W, Valenzuela N. 2011. Are all sex chromosomes created equal? Trends Genet. 27, 350–357. ( 10.1016/j.tig.2011.05.005) [DOI] [PubMed] [Google Scholar]
- 11.Mank JE 2012. Small but mighty: the evolutionary dynamics of W and Y sex chromosomes. Chromosome Res. 20, 21–33. ( 10.1007/s10577-011-9251-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson Sayres MA 2018. Genetic diversity on the sex chromosomes. Genome Biol. Evol. 10, 1064–1078. ( 10.1093/gbe/evy039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sardell JM, Kirkpatrick M. 2020. Sex differences in the recombination landscape. Am. Nat. 195, 361–379. ( 10.1086/704943) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grossen C, Neuenschwander S, Perrin N. 2011. Temperature-dependent turnovers in sex-determination mechanisms: a quantitative model. Evolution 65, 64–78. ( 10.1111/j.1558-5646.2010.01098.x) [DOI] [PubMed] [Google Scholar]
- 15.Pennell MW, Mank JE, Peichel CL. 2018. Transitions in sex determination and sex chromosomes across vertebrate species. Mol. Ecol. 27, 3950–3963. ( 10.1111/mec.14540) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rovatsos M, Vukić J, Lymberakis P, Kratochvíl L.. 2015. Evolutionary stability of sex chromosomes in snakes. Proc. R. Soc. B 282, 20151992 ( 10.1098/rspb.2015.1992) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rovatsos M, Kratochvíl L. 2017. Molecular sexing applicable in 4000 species of lizards and snakes? From dream to real possibility. Methods Ecol. Evol. 8, 902–906. ( 10.1111/2041-210X.12714) [DOI] [Google Scholar]
- 18.Rovatsos M, Rehák I, Velenský P, Kratochvíl L. 2019. Shared ancient sex chromosomes in varanids, beaded lizards, and alligator lizards. Mol. Biol. Evol. 36, 1113–1120. ( 10.1093/molbev/msz024) [DOI] [PubMed] [Google Scholar]
- 19.Rovatsos M, Praschag P, Fritz U, Kratochvíl L. 2017. Stable Cretaceous sex chromosomes enable molecular sexing in softshell turtles (Testudines: Trionychidae). Sci. Rep. 7, 42150 ( 10.1038/srep42150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rovatsos M, Vukić J, Altmanová M, Johnson Pokorná M, Moravec J, Kratochvíl L. 2016. Conservation of sex chromosomes in lacertid lizards. Mol. Ecol. 25, 3120–3126. ( 10.1111/mec.13635) [DOI] [PubMed] [Google Scholar]
- 21.Rovatsos M, Pokorná M, Altmanová M, Kratochvíl L. 2014. Cretaceous park of sex determination: sex chromosomes are conserved across iguanas. Biol. Lett. 10, 20131093 ( 10.1098/rsbl.2013.1093) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rovatsos M, Farkačová K, Altmanová M, Johnson Pokorná M, Kratochvíl L. 2019. The rise and fall of differentiated sex chromosomes in geckos. Mol. Ecol. 28, 3042–3052. ( 10.1111/mec.15126) [DOI] [PubMed] [Google Scholar]
- 23.Zhou Q, Zhang J, Bachtrog D, An N, Huang Q, Jarvis ED, Gilbert MTP, Zhang G. 2014. Complex evolutionary trajectories of sex chromosomes across bird taxa. Science 346, 1246338 ( 10.1126/science.1246338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marshall Graves JA. 2016. Did sex chromosome turnover promote divergence of the major mammal groups? BioEssays 38, 734–743. ( 10.1002/bies.201600019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gamble T, Coryell J, Ezaz T, Lynch J, Scantlebury DP, Zarkower D. 2015. Restriction site-associated DNA sequencing (RAD-seq) reveals an extraordinary number of transitions among gecko sex-determining systems. Mol. Biol. Evol. 32, 1296–1309. ( 10.1093/molbev/msv023) [DOI] [PubMed] [Google Scholar]
- 26.Uetz P, Hošek J (eds). 2014. The reptile database. See http://www.reptile-database.org (accessed 21 March 2020).
- 27.Vitt LJ, Caldwell JP. 2014. Herpetology: an introductory biology of amphibians and reptiles. Amsterdam, The Netherlands: Academic Press. [Google Scholar]
- 28.Olmo E, Signorino G. 2013. Chromorep: a reptiles chromosomes database. http://chromorep.univpm.it. (accessed 21 March 2020).
- 29.Hutchinson MN, Donnellan SC. 1992. Taxonomy and genetic variation in the Australian lizards of the genus Pseudemoia (Scincidae: Lygosominae). J. Nat. Hist. 26, 215–264. ( 10.1080/00222939200770091) [DOI] [Google Scholar]
- 30.Hardy GS 1979. The karyotypes of two scincid lizards, and their bearing on relationships in genus Leiolopisma and its relatives (Scincidae: Lygosominae). N. Z. J. Zool. 6, 609–612. ( 10.1080/03014223.1979.10428403) [DOI] [Google Scholar]
- 31.Wright JW 1973. Evolution of the X1X2Y sex chromosome mechanism in the scincid lizard Scincella laterale (Say). Chromosoma 43, 101–108. [PubMed] [Google Scholar]
- 32.Donnellan SC 1985. The evolution of sex chromosomes in scincid lizards. PhD thesis, Macquarie University, Sydney, Australia. [Google Scholar]
- 33.Shine R, Elphick MJ, Donnellan S. 2002. Co-occurrence of multiple, supposedly incompatible modes of sex determination in a lizard population. Ecol. Lett. 5, 486–489. ( 10.1046/j.1461-0248.2002.00351.x) [DOI] [Google Scholar]
- 34.Wapstra E, Olsson M, Shine R, Edwards A, Swain R, Joss JMP. 2004. Maternal basking behaviour determines offspring sex in a viviparous reptile. Proc. R. Soc. B 271, S230–S232. ( 10.1098/rsbl.2003.0152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robert KA, Thompson MB. 2001. Viviparous lizard selects sex of embryos. Nature 412, 698–699. ( 10.1038/35089135) [DOI] [PubMed] [Google Scholar]
- 36.Rovatsos M, Johnson Pokorná M, Altmanová M, Kratochvíl L. 2015. Female heterogamety in Madagascar chameleons (Squamata: Chamaeleonidae: Furcifer): differentiation of sex and neo-sex chromosomes. Sci. Rep. 5, 13196 ( 10.1038/srep13196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen SV, Banks JL, Diaz RE Jr, Trainor PA, Gamble T. 2018. Dynamic sex chromosomes in Old World chameleons (Squamata: Chamaeleonidae). J. Evol. Biol. 31, 484–490. ( 10.1111/jeb.13242) [DOI] [PubMed] [Google Scholar]
- 38.Iannucci A, et al. 2019. Conserved sex chromosomes and karyotype evolution in monitor lizards (Varanidae). Heredity 123, 215–227. ( 10.1038/s41437-018-0179-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rovatsos M, Vukić J, Mrugała A, Suwala G, Lymberakis P, Kratochvíl L. 2019. Little evidence for switches to environmental sex determination and turnover of sex chromosomes in lacertid lizards. Sci. Rep. 9, 7832 ( 10.1038/s41598-019-44192-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caputo V, Odierna G, Aprea G. 1994. A chromosomal study of Eumeces and Scincus, primitive members of the Scincidae (Reptilia, Squamata). Boll. Zool. 61, 155–162. ( 10.1080/11250009409355876) [DOI] [Google Scholar]
- 41.Pyron R, Burbrink FT, Wiens JJ. 2013. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol. Biol. 13, 93 ( 10.1186/1471-2148-13-93) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazzoleni S, et al. 2019. Turtles of the genera Geoemyda and Pangshura (Testudines: Geoemydidae) lack differentiated sex chromosomes: the end of a 40-year error cascade for Pangshura. PeerJ 7, e6241 ( 10.7717/peerj.6241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pokorná M, Rens W, Rovatsos M, Kratochvíl L. 2014. A ZZ/ZW sex chromosome system in the thick-tailed gecko (Underwoodisaurus milii; Squamata: Gekkota: Carphodactylidae), a member of the ancient gecko lineage. Cytogenet. Genome Res. 142, 190–196. ( 10.1159/000358847) [DOI] [PubMed] [Google Scholar]
- 44.Endow SA 1982. Polytenization of the ribosomal genes on the X and Y chromosomes of Drosophila melanogaster. Genetics 100, 375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. ( 10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andrews S 2010. FASTQC. A quality control tool for high throughput sequence data See http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 47.Ewels P, Magnusson M, Lundin S, Käller M. 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. ( 10.1093/bioinformatics/btw354) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vicoso B, Emerson JJ, Zektser Y, Mahajan S, Bachtrog D. 2013. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 11, e1001643 ( 10.1371/journal.pbio.1001643) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Picard MAL, Cosseau C, Ferré S, Quack T, Grevelding CG, Couté Y, Vicoso B. 2018. Evolution of gene dosage on the Z-chromosome of schistosome parasites. eLife 7, e35684 ( 10.7554/elife.35684) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andrade P, et al. 2019. Regulatory changes in pterin and carotenoid genes underlie balanced color polymorphisms in the wall lizard. Proc. Natl Acad. Sci. USA 116, 5633–5642. ( 10.1073/pnas.1820320116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden T. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13, 134 ( 10.1186/1471-2105-13-134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rovatsos M, Altmanová M, Johnson Pokorná M, Kratochvíl L. 2014. Novel X-linked genes revealed by quantitative polymerase chain reaction in the green anole, Anolis carolinensis . G3 4, 2107–2113. ( 10.1534/g3.114.014084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pensabene E, Kratochvíl L, Rovatsos M. 2020. Independent evolution of sex chromosomes in eublepharid geckos, a lineage with environmental and genotypic sex determination. Life 10, 342 ( 10.3390/life10120342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carré G-A, Siggers P, Xipolita M, Brindle P, Lutz B, Wells S, Greenfield A. 2017. Loss of p300 and CBP disrupts histone acetylation at the mouse Sry promoter and causes XY gonadal sex reversal. Hum. Mol. Genet. 27, 190–198. ( 10.1093/hmg/ddx398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Falah N, Posey JE, Thorson W, Benke P, Tekin M, Tarshish B, Lupski JR, Harel T. 2017. 22q11.2q13 duplication including SOX10 causes sex-reversal and peripheral demyelinating neuropathy, central dysmyelinating leukodystrophy, Waardenburg syndrome, and Hirschsprung disease. Am. J. Med. Genet. 173, 1066–1070. ( 10.1002/ajmg.a.38109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Polanco JC, Wilhelm D, Davidson T-L, Knight D, Koopman P.. 2009. Sox10 gain-of-function causes XX sex reversal in mice: implications for human 22q-linked disorders of sex development. Hum. Mol. Genet. 19, 506–516. ( 10.1093/hmg/ddp520) [DOI] [PubMed] [Google Scholar]
- 57.Kuzmin A, Jarvi K, Lo K, Spencer L, Chow GYC, Macleod G, Wang Q, Varmuza S. 2009. Identification of potentially damaging amino acid substitutions leading to human male infertility. Biol. Reprod. 81, 319–326. ( 10.1095/biolreprod.109.076000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liška F, Chylíková B, Janků M, Šeda O, Vernerová Z, Pravenec M, Křen V. 2016. Splicing mutation in Sbf1 causes nonsyndromic male infertility in the rat. Reproduction 152, 215–223. ( 10.1530/rep-16-0042) [DOI] [PubMed] [Google Scholar]
- 59.Balla T 2013. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93, 1019–1137. ( 10.1152/physrev.00028.2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Firestein R, Nagy PL, Daly M, Huie P, Conti M, Cleary ML. 2002. Male infertility, impaired spermatogenesis, and azoospermia in mice deficient for the pseudophosphatase Sbf1 . J. Clin. Invest. 109, 1165–1172. ( 10.1172/jci0212589) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cornejo-Páramo P, et al. 2020. Viviparous reptile regarded to have temperature-dependent sex determination has old XY chromosomes. Genome Biol. Evol. 12, 924–930. ( 10.1093/gbe/evaa104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Charlesworth D 2021. The timing of genetic degeneration of sex chromosomes. Phil. Trans. R. Soc. Lond. B, in press ( 10.1098/rstb.2020.0093) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Giovannotti M, Caputo V, O'Brien PCM, Lovell FL, Trifonov V, Nisi Cerioni P, Olmo E, Ferguson-Smith MA, Rens W. 2009. Skinks (Reptilia: Scincidae) have highly conserved karyotypes as revealed by chromosome painting. Cytogenet. Genome Res. 127, 224–231. ( 10.1159/000295002) [DOI] [PubMed] [Google Scholar]
- 64.Donnellan SC 1991. Chromosomes of Australian lygosomine skinks (Lacertilia: Scincidae). Genetica 83, 207–222. ( 10.1007/bf00126227) [DOI] [Google Scholar]
- 65.Nielsen SV, Pinto BJ, Guzmán-Méndez IA, Gamble T. 2020. First report of sex chromosomes in night lizards (Scincoidea: Xantusiidae). J. Hered. 111, 307–313. ( 10.1093/jhered/esaa007) [DOI] [PubMed] [Google Scholar]
- 66.Zheng Y, Wiens JJ. 2016. Combining phylogenomic and supermatrix approaches, and a time-calibrated phylogeny for squamate reptiles (lizards and snakes) based on 52 genes and 4162 species. Mol. Phylogenet. Evol. 94, 537–547. ( 10.1016/j.ympev.2015.10.009) [DOI] [PubMed] [Google Scholar]
- 67.Kumar S, Stecher G, Suleski M, Hedges SB. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819. ( 10.1093/molbev/msx116) [DOI] [PubMed] [Google Scholar]
- 68.Radder RS, Quinn AE, Georges A, Sarre SD, Shine R. 2007. Genetic evidence for co-occurrence of chromosomal and thermal sex-determining systems in a lizard. Biol. Lett. 4, 176–178. ( 10.1098/rsbl.2007.0583) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hill PL, Burridge CP, Ezaz T, Wapstra E. 2018. Conservation of sex-linked markers among conspecific populations of a viviparous skink, Niveoscincus ocellatus, exhibiting genetic and temperature-dependent sex determination. Genome Biol. Evol. 10, 1079–1087. ( 10.1093/gbe/evy042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sutou S, Mitsui Y, Tsuchiya K. 2001. Sex determination without the Y chromosome in two Japanese rodents Tokudaia osimensis osimensis and Tokudaia osimensis spp. Mamm. Genome 12, 17–21. ( 10.1007/s003350010228) [DOI] [PubMed] [Google Scholar]
- 71.Acosta A, Suárez-Varón G, Rodríguez-Miranda LA, Lira-Noriega A, Aguilar-Gómez D, Gutiérrez-Mariscal M, Hernández-Gallegos O, Méndez-de-la-Cruz F, Cortez D. 2019. Corytophanids replaced the pleurodont XY system with a new pair of XY chromosomes. Genome Biol. Evol. 11, 2666–2677. ( 10.1093/gbe/evz196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rovatsos M, Johnson Pokorná M, Altmanová M, Kratochvíl L. 2016. Mixed-up sex chromosomes: identification of sex chromosomes in the X1X1X2X2/X1X2Y system of the legless lizards of the genus Lialis (Squamata: Gekkota: Pygopodidae). Cytogenet. Genome Res. 149, 282–289. ( 10.1159/000450734) [DOI] [PubMed] [Google Scholar]
- 73.Bókony V, Milne G, Pipoly I, Székely T, Liker A. 2019. Sex ratios and bimaturism differ between temperature-dependent and genetic sex-determination systems in reptiles. BMC Evol. Biol. 19, 57 ( 10.1186/s12862-019-1386-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.IUCN. 2020. The IUCN Red List of Threatened Species. Version 2020-1 See https://www.iucnredlist.org (accessed on 19 March 2020).
- 75.Kuhl H, et al. 2021. A 180 My-old female-specific genome region in sturgeon reveals the oldest known vertebrate sex determining system with undifferentiated sex chromosomes. Phil. Trans. R. Soc. Lond. B, in press ( 10.1098/rstb.2020.0089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Augstenová B, Johnson Pokorná M, Altmanová M, Frynta D, Rovatsos M, Kratochvíl L. 2018. ZW, XY, and yet ZW: sex chromosome evolution in snakes even more complicated. Evolution 72, 1701–1707. [DOI] [PubMed] [Google Scholar]
- 77.Carvalho NDM, Arias FJ, da Silva FA, Schneider CH, Gross MC. 2015. Cytogenetic analyses of five amazon lizard species of the subfamilies Teiinae and Tupinambinae and review of karyotyped diversity the family Teiidae. Comp. Cytogenet. 9, 625–644. ( 10.3897/compcytogen.v9i4.5371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nielsen SV, Guzmán-Méndez IA, Gamble T, Blumer M, Pinto BJ, Kratochvíl L, Rovatsos M. 2019. Escaping the evolutionary trap? Sex chromosome turnover in basilisks and related lizards (Corytophanidae: Squamata). Biol. Lett. 15, 20190498 ( 10.1098/rsbl.2019.0498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, Grützner F, Kaessmann H. 2014. Origins and functional evolution of Y chromosomes across mammals. Nature 508, 488–493. ( 10.1038/nature13151) [DOI] [PubMed] [Google Scholar]
- 80.Altmanová M, Rovatsos M, Johnson Pokorná M, Veselý M, Wagner F, Kratochvíl L. 2018. All iguana families with the exception of basilisks share sex chromosomes. Zoology 126, 98–102. ( 10.1016/j.zool.2017.11.007) [DOI] [PubMed] [Google Scholar]
- 81.Rupp SM, Webster TH, Olney KC, Hutchins ED, Kusumi K, Wilson Sayres MA. 2016. Evolution of dosage compensation in Anolis carolinensis, a reptile with XX/XY chromosomal sex determination. Genome Biol. Evol. 9, 231–240. ( 10.1093/gbe/evw263) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Marin R, et al. 2017. Convergent origination of a Drosophila-like dosage compensation mechanism in a reptile lineage. Genome Res. 27, 1974–1987. ( 10.1101/gr.223727.117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marshall Graves JA 2015. Evolution of vertebrate sex chromosomes and dosage compensation. Nat. Rev. Genet. 17, 33–46. ( 10.1038/nrg.2015.2) [DOI] [PubMed] [Google Scholar]
- 84.Perrin N 2009. Sex reversal: a fountain of youth for sex chromosomes? Evolution 63, 3043–3049. ( 10.1111/j.1558-5646.2009.00837.x) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw Illumina reads from DNA-seq of all studied individuals are deposited into the NCBI BioProject database with ID PRJNA660179.