Abstract
In 2020 SARS-CoV-2 reached pandemic status, reaching Brazil in mid-February. As of now, no specific drugs for treating the disease are available. In this work, the possibility of interaction between SARS-CoV-2 viral proteins (open and closed spike protein, isolate spike protein RBD, NSP 10, NSP 16, main protease, and RdRp polymerase) and multiple molecules is addressed through the repositioning of drugs available for the treatment of other diseases that are approved by the FDA and covered by SUS, the Brazilian Public Health System. Three different docking software were used, followed by a unification of the results by independent evaluation. Afterwards, the chemical interactions of the compounds with the targets were inspected via molecular dynamics and analyzed. The results point to a potential effectiveness of Penciclovir, Ribavirin, and Zanamivir, from a set of 48 potential candidates. They may also be multi-target drugs, showing high affinity with more than one viral protein. Further in vitro and in vivo validation is required to assess the suitability of repositioning the proposed drugs for COVID-19.
Keywords: Drug repositioning, SUS, SARS-CoV-2, Molecular docking, COVID19
Graphical abstract
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic was declared by the WHO in 2020. It has spread since late 2019 from the Chinese province of Hubei to the rest of the world at an increasing and alarming rate, causing respiratory complications and a large death toll [1,2].
Therapies against SARS-CoV-2 target the viral structure and the inflammatory storm that is secondary to viral infection [3]. Surface proteins in SARS-CoV-2, called S or Spike proteins, bind to target cells by the angiotensin-converting enzyme 2 (ACE2), which acts as a viral receptor [4]. S proteins have two subunits, S1 and S2, the former harboring the receptor binding domain (RBD) [5]. Host infection maturation involves an extensive cascade of events with protease and RdRp polymerase activity to control viral gene expression and replication. Most cleavage events for maturation of the precursor polyprotein are mediated by the SARS-CoV-2 Main protease (M protease), a three-domain cysteine protease. Two M protease molecules form an active homodimer that cleave Cys-His sites, while N-terminal residues are important for proteolytic protection [6]. In its turn, RdRp polymerase is an oligomer of nonstructural proteins (nsp 12, nsp 7, nsp 8) that have little to no activity when isolated [5]. The massive expression of ACE2 in the lungs justifies the severe respiratory manifestations presented in patients affected by respiratory disease (named COVID-19, “coronavirus disease 2019”). Subjects show an increase in cytokines (such as interleukins 2, 6 and 7), granulocyte colony stimulating factor, gamma interferon inducing protein, monocyte chemotactic protein, inflammatory protein from macrophages 1 and tumor necrosis factor, which causes edema, acute breathing difficulties, secondary infections, acute cardiac damage and, in more extreme cases, death [7,8]. Despite coding for twenty different proteins (of which four are structural), the main therapeutic targets in the SARS-CoV-2 genome are the S protein, the Main protease and the RdRp polymerase [5].
The repositioning (or repurposing) of drugs guided by molecular docking is a structure-based computational strategy based on the complementarity of the target binding site (receptor) and the ligand (drug) [9], the latter of which has already been approved by regulatory agencies. This strategy reduces costs, considering that these drugs have already been characterized and approved during clinical development [10]. In addition, the risks of candidate drugs for repositioning are drastically reduced, as they already have well-established pharmacokinetic, pharmacodynamic, and toxicology profiles [11]. Thus, the repositioning of drugs guided by molecular docking can offer a better risk-benefit trade-off than other methods [11].
Given the urgency of the SARS-CoV-2 outbreak, due to its high rate of virulence and potential to collapse healthcare systems, it is extremely important to privilege the screening of existing molecules that may have antiviral characteristics, and that are already approved and available to the population [12]. In order to optimize the candidate drugs and facilitate the population's access to the compounds, the drugs already authorized by the Food and Drug Administration (FDA) of the United States of America and the drugs available through the Brazilian Public Health System (Sistema Único de Saúde, SUS) were filtered based on current therapeutic experience and literature for the treatment of viral pathologies, such as HIV/AIDS, hepatitis B, hepatitis C, influenza and severe acute respiratory syndrome.
Based on pre-clinical, pharmacokinetic, and pharmacodynamic profiles already known from repositionable drugs, it may be possible to obtain a drug that, at the present time, is more suitable for use in diseases that do not have their clinical needs met, such as SARS-CoV-2 [13]. Therefore, in this work we carried out an analysis of the drugs approved by the FDA and available via SUS that can meet the current emergency demand that the moment and society request, with a special view towards Brazilian population (and populations from other countries with similar socioeconomical profile). The results point to a potential effectiveness of Penciclovir, Ribavirin, and Zanamivir, from a set of 48 potential candidates.
2. Materials and methods
2.1. Screening for viral proteins and drugs
For screening the SARS-CoV-2 viral proteins, the RCSB Protein Databank was used [14]. The Spike protein (protein S) was obtained in different conformation states: open (PDB id: 6VSB), closed (PDB id: 6VXX), and its isolated receptor binding domain (RBD) (PDB id: 6M0J). These variant conformations were used to inspect possible differences in ligand binding modes (e.g. putative conformational stabilizers able to hamper the protein opening). Other proteins were also selected, such as the main protease (M protease) (PDB id: 6LU7), the non-structural proteins 10 (NSP 10) (PDB id: 6W75), and 16 (NSP 16) (PDB id: 6W4H) and the viral RdRp polymerase (PDB id: 7BW4). For the selection of drugs, clinical trials and in silico repositioning studies available in the literature up to the present moment, as well as the FDA and SUS databanks, were considered. Thus, 48 drugs described as potential antivirals effect, which were registered simultaneously in the FDA and in the SUS database, were selected.
The preparation of the viral proteins was performed using the PyMOL software where all non-protein records were removed. Drug structures were obtained from PubChem [15] and were energetically minimized with Avogadro under standard protocols [16]. Bidimensional representations were created with PoseView [17].
2.2. Molecular docking
To increase the degree of reliability of the results, three molecular docking software were used: DockThor [18], Autodock Vina [19], and PatchDock [20]. This strategy was based on combined docking protocols [21], which have been shown to significantly enhance the success rate in virtual screening for drugs and protein complexes [22,23]. In Dockthor and Autodock Vina, grid boxes were inserted at predetermined points (Supplementary Material 1, Table 1), i.e., site of interaction between proteins and a cellular receptor (in the case of spike protein) or active sites of viral proteins (all other proteins). Binding site references for all proteins were taken from current literature, as follows: Spike protein and RBD [24], RdRp Polymerase [25], NSP 16 [26], NSP 10 and Main protease [27]. In the PatchDock server the ligand was free to explore the entire surface area of the target protein (blind docking). At the end of the first analysis phase, the three best candidates for treatment of COVID-19 were docked against all the analyzed proteins. In order to validate the dockings proposed in this work, we also docked zanamivir with its original target protein, neuramidase (1NN2), as a control.
Table 1.
Chemical analysis data of the compounds selected by the cutoff point of match1.
| Drugs | Software | Protein | Binding Strength * | Stability ** | Steric Hindrance*** |
|---|---|---|---|---|---|
| Zanamivir | Patchdock | RBD | 40 | 1 | C |
| Zanamivir | Autodock Vina | RBD | 39 | 2 | B |
| Penciclovir | Autodock Vina | RBD | 39 | 3 | A |
| Galidesivir | Patchdock | Polymerase | 35 | 4 | B |
| Ribavirin | Patchdock | Polymerase | 33 | 5 | B |
| Ribavirin | Autodock Vina | Polymerase | 33 | 6 | B |
| Ganciclovir | Patchdock | RBD | 33 | 7 | A |
| Tenofovir | Patchdock | Polymerase | 29 | 8 | C |
| Ribavirin | Dockthor | Polymerase | 27 | 9 | A |
| Tenofovir | Autodock Vina | Polymerase | 27 | 10 | B |
| Zanamivir | Patchdock | Protein S - closed | 24 | 11 | B |
| Adefovir | Autodock Vina | Polymerase | 23 | 12 | A |
| Zanamivir | Autodock Vina | Protein S - open | 22 | 13 | B |
| Penciclovir | Autodock Vina | Protein S - open | 20 | 14 | A |
| Zanamivir | Patchdock | Protein S - open | 18 | 15 | A |
| Niclosamine | Autodock Vina | Protein S - open | 18 | 16 | A |
| Penciclovir | Dockthor | Protein S - open | 17 | 17 | A |
| Ganciclovir | Patchdock | Protein S - open | 4 | 18 | A |
| Penciclovir | Patchdock | Protein S - closed | 2 | 19 | A |
* Calculated as the sum of: strong H bonds: 10; moderate H bonds: 5; weak H bonds: 2; hydrophobic interactions: 2; Saline bridges: 4, π interactions: 4. ** Ordered from more (1) to less (19) stable complexes. *** A: small hindrance; B: medium hindrance; C: considerable hindrance; D: absolute hindrance.
2.3. Analysis of interaction: forces, stability, and dynamics
Due to the diversity of data generated by each software, the Platinum server [28] was used to reorder the results based on a unified metric. For this purpose we used match1, that is the fraction of lipophilic and hydrophilic match (SLL + SHH/SLL + SLH + SHL + SHH + SLH’ + SHH’ [Å2]); where (SLL [Å2], Lipophilic ligand match Lipophilic receptor Surface; SHH [Å2], Hydrophilic ligand match Hydrophilic receptor surface; SLH, Lipophilic match Hydrophilic receptor surface; SHL, Hydrophilic ligand match Lipophilic receptor Surface; SLH’, Lipophilic ligand match Hydrophilic solvent (water) Surface; SHH’, Hydrophilic ligand match Hydrophilic solvent (water) Surface [Å2]) [28]. Scores were used to standardize the results of all dockings on the different docking strategies. The 0.600 value of match1 was used as a cutoff point to select candidates for the chemical stability analysis. The Platinum match1 cutoff value is adjustable in a case-dependent fashion [28], hence the 0.600 value being based on previous works employing similar methods [[29], [30], [31]]. A total of 18 protein-ligand complexes were selected, which were subsequently evaluated for their propensity to form stable complexes. The analysis was performed using TU Dresden BIOTEC's Protein-Ligand Interaction Profiler (PLIP) [32].
Protein-ligand complexes were tested and ordered for stability (based on the number and nature of interactions) and the ease of formation of the complex was classified based on the structural accessibility of the target to the ligand. At the end of the first round of analyses, the three best candidate drugs for treatment of COVID-19 were evaluated for their chemical interaction with the remaining viral proteins.
To confirm the binding stability of docking results in physiological conditions, molecular dynamics (MD) simulations were carried out for the selected complexes. PDB structural files from the docking step were split into two individual files, one containing the protein structure and the other the ligand structure. Next, each ligand structure was submitted to the LigParGen server [[33], [34], [35]] where OPLS-AA parameters were generated. All molecular dynamics simulations were performed employing GROMACS package version 2020.2 [36,37]. OPLS-AA force field [38] was selected along with the water model SPC [39]. The box geometry was defined as a dodecahedron and the distance between the protein and the box was set as 1.2 nm, under periodic boundary conditions. Ions were added to the system proportionally for both purposes of neutralizing the global net charge and simulating the physiological condition of 0.15 M. For the cationic contribution Na+ ions were selected, whereas Cl− ions were selected for the anionic effect. An energy minimization step was performed employing the Steepest Descent algorithm. Next, equilibration was performed in two phases. Both isothermal-isochoric (NVT) and isothermal-isobaric (NPT) phases were conduct for 100 ps Covalent bonds were constrained using the LINCS algorithm [40], and an integration step of 2 fs was applied. The Particle Mesh Ewald method [41] was employed for the calculation of electrostatic interactions, along with the Parrinello-Rahman barostat [42,43] set with a 2 ps coupling constant. The V-rescale [44] was employed with a coupling constant of τ = 0.1. The production step was performed for 20 ns There were twenty-one independent simulated systems, the combination of seven different protein structures and three different ligands, representing the best overall scoring complex for each case.
3. Results
3.1. Virtual screening, molecular docking and chemical analysis
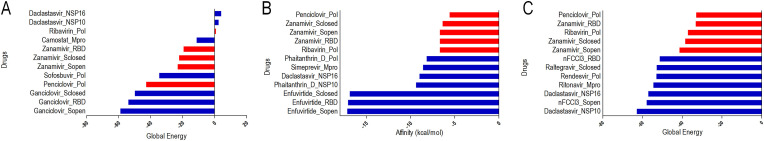
The literature analysis, clinicals trials, and databases (SUS and FDA) pointed to 48 possible drugs that could satisfy the criteria of interest in this investigation (Supplementary Material 1, Table 2). The chemical compounds were docked with targets similar to their original ones. Only Phaitanthrin D was docked with all the viral proteins, due to the unknown nature of its binding target(s) [25]. In addition, all the drugs chosen were analyzed by different software (Dockthor, Autodock Vina, and PatchDock). For Dockthor and Autodock Vina, grid boxes were used to direct the search to regions of interest, while for PatchDock docking was carried out blindly. The docking data shows similar interactions between different software (at least two docking strategies pointed to a drug docked in nearby regions). As each software had a different scoring function, leading to different output results, the initial comparison between all results was hampered (Supplementary Material 1, Table 3). Considering the individual analysis of global energy or estimate of free binding energy (kcal/mol), such as the one provided by Autodock Vina, revealed that the software had variations of the best proposed candidates (Fig. 1 ).
Table 2.
Chemical analysis data of the 3 final compounds, three selected and docked with all the proteins.
| Drug | Software | Protein | Bonding Force * | Stability** | Steric Hindrance*** |
|---|---|---|---|---|---|
| Penciclovir | PatchDock | NSP 16 | 40 | 2 | C |
| Ribavirin | Autodock Vina | Protein S - closed | 39 | 5 | A |
| Zanamivir | DockThor | NSP 16 | 39 | 6 | B |
| Penciclovir | DockThor | Main protease | 35 | 8 | A |
| Penciclovir | DockThor | Protein S - closed | 34 | 9 | B |
| Zanamivir | Autodock Vina | Main protease | 32 | 13 | B |
| Penciclovir | PatchDock | Main protease | 31 | 14 | A |
| Penciclovir | DockThor | NSP 16 | 31 | 15 | B |
| Zanamivir | PatchDock | NSP 16 | 31 | 16 | B |
| Penciclovir | DockThor | Polymerase | 30 | 19 | C |
| Penciclovir | PatchDock | Polymerase | 30 | 20 | C |
| Zanamivir | PatchDock | Main protease | 30 | 17 | B |
| Zanamivir | Autodock Vina | NSP 16 | 30 | 18 | C |
| Zanamivir | DockThor | Polymerase | 28 | 22 | B |
| Penciclovir | Autodock Vina | Polymerase | 26 | 26 | B |
| Zanamivir | DockThor | Protein S - closed | 26 | 25 | A |
| Ribavirin | DockThor | Main protease | 25 | 27 | A |
| Ribavirin | DockThor | NSP 16 | 25 | 28 | B |
| Ribavirin | DockThor | Protein S - closed | 24 | 30 | A |
| Ribavirin | PatchDock | Protein S - closed | 24 | 31 | A |
| Ribavirin | Autodock Vina | NSP 10 | 24 | 32 | B |
| Ribavirin | DockThor | NSP 10 | 24 | 33 | B |
| Ribavirin | Autodock Vina | NSP 16 | 24 | 34 | C |
| Zanamivir | Autodock Vina | Polymerase | 23 | 36 | B |
| Ribavirin | PatchDock | Main protease | 22 | 38 | A |
| Ribavirin | DockThor | Protein S - open | 22 | 39 | A |
| Zanamivir | DockThor | Main protease | 21 | 40 | A |
| Penciclovir | Autodock Vina | NSP 10 | 20 | 42 | A |
| Penciclovir | Autodock Vina | Protein S - closed | 19 | 43 | A |
| Penciclovir | PatchDock | NSP 10 | 19 | 44 | A |
| Zanamivir | DockThor | NSP 10 | 19 | 45 | B |
| Zanamivir | PatchDock | NSP 10 | 19 | 46 | C |
| Zanamivir | DockThor | Protein S - open | 19 | 47 | B |
| Penciclovir | Autodock Vina | Main protease | 18 | 50 | A |
| Penciclovir | Autodock Vina | NSP 16 | 18 | 51 | A |
| Ribavirin | PatchDock | NSP 16 | 18 | 52 | B |
| Penciclovir | DockThor | NSP 10 | 16 | 55 | A |
| Ribavirin | Autodock Vina | Main protease | 16 | 54 | A |
| Ribavirin | PatchDock | NSP 10 | 14 | 56 | B |
| Ribavirin | Autodock Vina | Protein S - open | 13 | 57 | B |
| Zanamivir | PatchDock | Polymerase | 13 | 58 | A |
| Penciclovir | PatchDock | Protein S - open | 8 | 59 | A |
| Ribavirin | PatchDock | Protein S - open | 4 | 61 | A |
*Calculated as the sum of: strong H bonds: 10; moderate H bonds: 5; weak H bonds: 2; hydrophobic interactions: 2; Saline bridges: 4, π interactions: 4. ** Ordered from more (1) to less (19) stable complexes. *** A: small hindrance; B: medium hindrance; C: considerable hindrance; D: absolute hindrance.
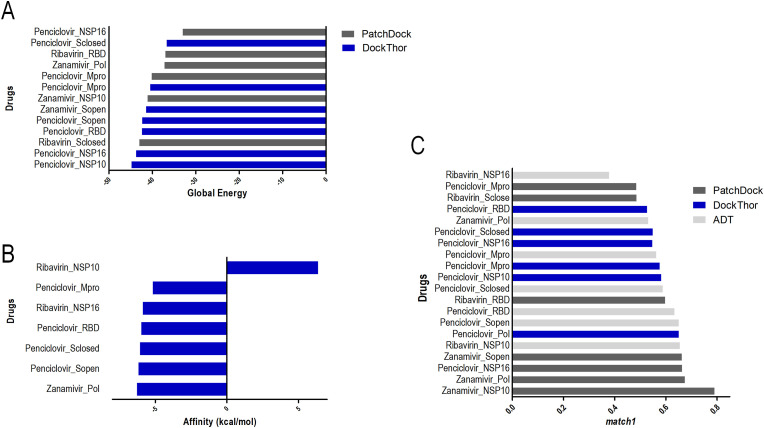
Fig. 1.
Global energy/Affinity data obtained from each docking software. Each bar represents a drug assigned to its putative target protein. In red are highlighted the docking results for complexes involving Penciclovir, Ribavirin, and Zanamivir. Results for Dockthor (A), Autodock Vina (B), and Patchdock (C). Full results are shown in Suppl. Fig. 1. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In addition, the variation in results also indicated the divergence in the classification of the drugs. After the Dockthor analysis, the best candidates listed in decreasing order of affinity were Ganciclovir, Penciclovir, and Sofobuvir (considering that Ganciclovir and Penciclovir have similar action mechanism). For Autodock Vina, the ideal candidates were represented by Enfuvirtide, Dolutegravir, and Phaitanthrin D, while the results of PatchDock point to Daclastasvir, (E) -1- [4 - [(2E) −3,7-Dimethylocta-2,6-dienoxy] phenyl] -3- (4-methoxyphenyl) prop-2-en-1-one (nFCCl3), and Ritonavir as the best candidates.
The absence of agreement among the candidates demonstrates differences in the docking algorithms. To circumvent the biases of each algorithm, the Platinum server was used to rank all results. Following the unification of the results using the match1 data that was generated by Platinum, a cutoff point (0.600) was assigned to select the best candidate drugs. Hence, 18 possible models of ideal protein-ligand complexes were selected, which were subsequently submitted to visual chemical inspection.
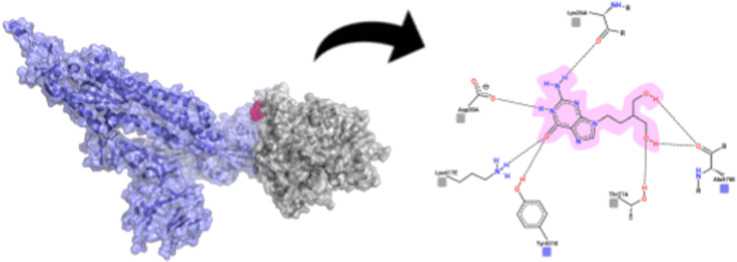
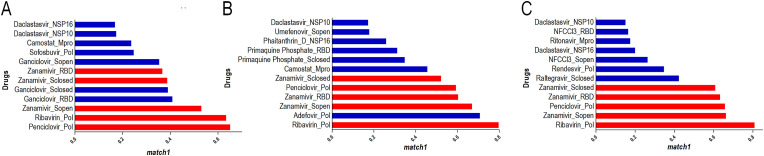
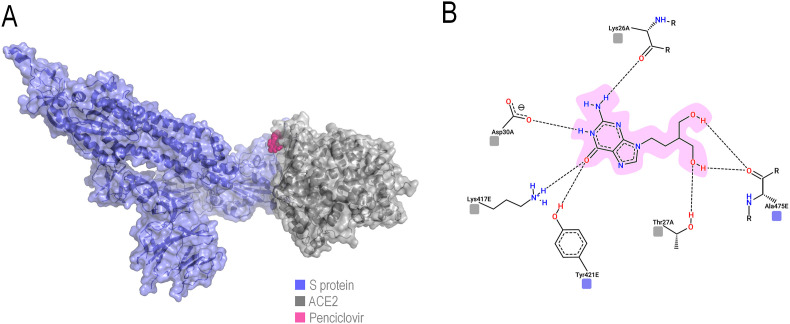
The molecular docking results indicated that, among all the proteins considered as therapeutic targets, both RdRp polymerase and S protein are excellent target candidates, considering that the comparison of match1 values above 0.600 consists essentially of these proteins (Fig. 2 ). Through the analyses, it was shown that the best candidate for S protein inhibition interacts on the ACE2 binding site, indicating a possible physical blockage of the receptor binding (Fig. 3 ), a crucial step in the viral cycle.
Fig. 2.
Match1 ranking for each docking software obtained with Platinum for each software: Dockthor (A), Autodock Vina (B), and Patchdock (C). The threshold for selecting the best complexes was defined as 0.600. In red are highlighted the docking results for complexes involving Penciclovir, Ribavirin, and Zanamivir. Full results are shown in Suppl. Fig. 2. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 3.
Representation of Penciclovir in its best pose for S protein-ACE2 interface blocking. The protein complex is shown in transparent surface with underlying cartoon (A), while the drug-residue interactions are shown in 2D (B) following the same color-coding of (A). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Chemical evaluation of the 18 candidates obtained from the Platinum threshold were performed. The best candidates were chosen based on their stability, number and strength of the non-covalent interactions between target protein and ligand, as well as the accessibility of the binding site based on the surrounding protein structure. For each protein-ligand complex, a total interaction strength score was assigned based on the sum of individual interaction scores, i.e., strong H bonds (D-A distance <2.5 Å): 10; moderate H bonds (D-A distance 2.5–3.2 Å): 5; weak H bonds (D-A distance >3.2) Å: 2; hydrophobic interactions: 2; Saline bridges: 4, π interactions: 4. This generated a ranking of presumed stability among the evaluated complexes. Such analysis was used to make docking results more reliable, and not dependent solely on each software affinity score. The steric availability of the binding sites for each complex was also evaluated separately from the interaction score (Table 1). The detailed evaluation of each putative complex is available in the Supplementary Material 3. The selection criteria for the best candidates were based primarily on their stability and consequent ranking in Table 1 . However, binding site availability and steric hindrances were also taken into consideration.
After the first stage of comparative analysis, the ranking results for binding strength (by match1) and the structural stability were considered. Three adequate ligand candidates were observed: Penciclovir, Ribavirin, and Zanamivir. These drugs are already in use for treatment of herpes virus, hepatitis, and influenza, respectively.
After defining these candidates, their interactions with other viral proteins were also evaluated. We observed that Penciclovir and Ribavirin also exhibited potential for interaction with the other viral proteins being evaluated. The candidate drugs show high values of match1 and good positioning within the chemical analysis ranking (Table 2 , Fig. 4 ).
Fig. 4.
Analysis of the interaction of the best compounds in each docking software. Individual global energy obtained for the best compounds against all viral proteins in each docking software, (DockThor and PatchDock (A), Autodock Vina (B), followed by analysis of Platinum result unification (C). Full results are shown in Suppl. Fig. 3.
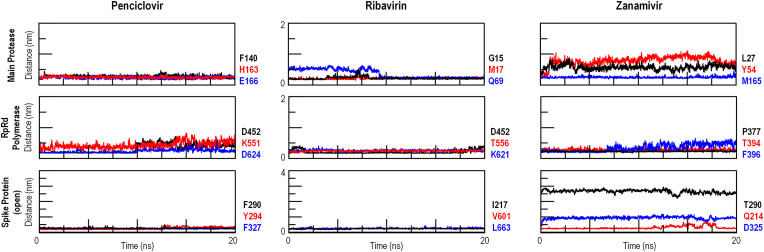
The obtained complexes were subjected to molecular dynamics simulations to inspect the ligand binding stability, considering that docking result are static and do not represent natural binding conditions. The distances between ligands and amino acid residues previously identified as contacting moieties via PLIP were measured during the simulations to assess possible ligand detachment or reorientation. From these measurements (Fig. 5 ) it can be confirmed that all evaluated complexes are stable, having at least one constant interaction throughout the simulation. The one observed exception is the Ribavirin-RBD simulation, in which it was not possible to assess if the ligand was drifting away from the binding site (i.e., unbound) or adopting a new binding pose.
Fig. 5.
Analysis of the interaction between selected drugs and SARS-CoV-2 proteins via molecular dynamics. Distances between ligand (drug molecules) and receptor (contacting amino acids of the target protein) were measured to assess binding stability in physiological, time-dependent conditions. Full results are shown in Suppl. Fig. 4.
4. Discussion
In this work, from the universe of generally available drugs, we were able to select 48 antivirals with potential to inhibit some part of the SARS-CoV-2 viral cycle, based on an availability criterium. These 48 drugs were further reduced to 3 ideal candidates, namely Zanamivir, Ribavirin, and Penciclovir. To account for conformational changes and near-physiological binding conditions, all best-ranking drug-receptor complexes were simulated by molecular dynamics with all-atom resolution. The simulation trajectories confirmed the suitability of the proposed drugs as candidate inhibitors for the virus.
Through the analyses, we corroborated the observation that the Spike protein and the RdRp polymerase are targets of great relevance for SARS-CoV-2 inhibition [5,45,46]. For the Spike glycoprotein to play its role of recognition and fusion between virus and host cell, it undergoes a conformational change (starting from a closed homotrimer state to an open homotrimer state, in which the RBD is exposed), favoring the interaction. Each subunit of the protein has an N-terminal domain named S1 that has receptor binding properties (analogous to RBD) and a C-terminal or S2 domain that is fundamental in virus-cell fusion [47,48].
The main receptor for coronavirus is ACE2, which has the physiological function of hydrolyzing angiotensin 2. This enzyme is abundantly expressed in lung tissue, which explains the tropism of the virus for cells in the respiratory system [49]. Another target of great importance in our work was RNA polymerase, an enzyme essential for viral replication. This dependence has made polymerase a target for drugs in several studies [[50], [51], [52]]. The active site of this enzyme is conserved among several organisms, with two successive aspartate residues that become accessible through a projection in a β-hairpin structure [53].
In view of the importance of the functional impairment of these proteins, the best candidates found here can be taken as pharmacologically relevant. These repositioning candidates are novel in the molecular docking scenario and in the scope of treatment for SARS-CoV-2.
Penciclovir demonstrated high values of match1, as well as suitable chemical interactions with viral proteins. Penciclovir is an antiviral drug used to treat several types of herpes virus [54]. The compound reaches a therapeutic effect by blocking viral replication through the competitive inhibition of the virus polymerase [55]. For the adequate effect of Penciclovir, it must be administered intradermally (favorable in the treatment of herpes virus) or be administered orally via the Famciclovir form (due to the poor absorption of Penciclovir by the oral route) which will then be converted by deacetylation to Penciclovir [[56], [57], [58], [59]]. Enhancing its pharmacological potential, Penciclovir demonstrated only few adverse effects, such as headache, local anesthesia, changes in taste, pruritus, and site-specific allergic reaction [60].
Ribavirin is a guanosine analogue that has properties to cease viral RNA synthesis, thus being a nucleoside inhibitor [61,62]. Nevertheless, at least five other mechanisms of direct and indirect action have been proposed, highlighting the potential of these poorly understood activities [63]. Ribavirin, after the initial screening, was shown to be a drug with multiple activities against the SARS-CoV-2 viral proteins. The higher affinity interaction of this prodrug was with the viral RdRp polymerase, contemplating a Platinum match1 result of 0.6352. This reinforces the mechanism of direct action of Ribavirin on the polymerase, being an agent with the possibility of demonstrating antiviral action also against SARS-CoV-2. In addition, Ribavirin interacted with multiple targets with greater affinity: non-structural protein 16 (NSP16) (match1: 0.6002), non-structural protein 10 (NSP 10) (match1: 0.6562), and the viral protease M protease (match1: 0.6873).
Zanamivir is an approved drug that is consolidated for the treatment of influenza A and B [64,65]. This drug is part of the class of compounds that target neuraminidase, an enzyme expressed on the viral surface [66]. Despite not having neuraminidase activity [5], SARS-CoV-2 presented suitable targets for Zanamivir binding, such as NSP 10 (match1: 0.7914), NSP 16 (match1: 0.6876), and RdRp polymerase (match1 0.6757). Currently there are few published works suggesting Zanamivir as a treatment option for COVID-19 [67,68].
Taken together, the data presented here reinforces the possibility of multi-target drugs in the treatment of COVID-19, while highlighting some compounds (Penciclovir, Ribavirin, Zanamivir) as likely to succeed in further in vitro and in vivo essays. Penciclovir was shown as a putative Spike protein-ACE2 interaction inhibitor besides being a known polymerase inhibitor, highlighting its relevance in ongoing drug repositioning strategies.
5. Conclusion
In this study of drug repositioning guided by molecular docking and molecular dynamics, we identified three putative candidates for COVID-19 therapy, namely Penciclovir, Ribavirin and Zanamivir. Despite Penciclovir being the best candidate found for interfering in the Spike protein-ACE2 interaction, all of them are putatively able to bind to more than one viral protein. These drugs are available in the Unified Health System (SUS) of Brazil, pointing to a possible readily available therapeutic alternative. To confirm the hypotheses raised with this work, further in vitro and in vivo studies are required to verify their potential to inhibit viral replication.
Author contributions
MVCG designed the protocols, conducted most experiments, and wrote the manuscript; AMA performed molecular docking calculations; CFM performed chemical analysis; APAP participated in the standardization of data; IMG and FVFR aided in building a library of viral drugs and proteins, and wrote the manuscript; ESMP, BCF and MD performed molecular dynamics simulations and interpreted data; JCC, ONS, and RLB coordinated the research project, proposed experiments, interpreted data, and wrote the manuscript. All authors carefully reviewed the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) – Finance Code 001 and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). MVCG, APAP, CM, and ESMP received PhD fellowships from CAPES. AMA and IMG received MSc fellowships from CNPq. FVFR received a PhD fellowship from CNPq. BCF received a PostDoc fellowship from CAPES. The authors thank Paula Caruso for her support in the initial stages of this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imu.2021.100539.
Abbreviations
- FDA
Food and Drug Administration (US);
- SUS,
Sistema Único de Saúde (BR)
- CoV
coronavirus
- SARS-CoV-2
Severe Acute Respiratory Coronavirus Syndrome
- ACE2
angiotensin-converting enzyme 2
- M protease
main protease
- COVID-19
coronavirus disease 2019
- protein S
spike protein
- RBD
receptor binding domain
- PLIP
Protein-Ligand Interaction Profiler
- nFCCl3
(E) -1- [4 - [(2E) −3,7-Dimethylocta-2,6-dienoxy] phenyl] -3- (4-methoxyphenyl) prop-2-en-1-one
- HIV
Human Immunodeficiency Virus
- NSP
Non-structural protein
- EC50
semi-maximum effective concentration
- CC50
semi-cytotoxic concentration
- SI
selectivity index
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Velavan T.P., Meyer C.G. The COVID-19 epidemic. Trop Med Int Health: TM & IH. 2020;25(3):278–280. doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Croda J.H.R., Garcia L.P. Resposta imediata da Vigilância em Saúde à epidemia da COVID-19. Epidemiologia e Serviços de Saúde. 2020;29 doi: 10.5123/S1679-49742020000100021. [DOI] [PubMed] [Google Scholar]
- 3.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet (London, England) 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woo P.C., Huang Y., Lau S.K., Yuen K.Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2(8):1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang M.Y., Zhao R., Gao L.J., Gao X.F., Wang D.P., Cao J.M. SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front Cell Infect Microbiol. 2020;10:587269. doi: 10.3389/fcimb.2020.587269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xue X., Yu H., Yang H., Xue F., Wu Z., Shen W., et al. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J Virol. 2008;82(5):2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leng Z., Zhu R., Hou W., Feng Y., Yang Y., Han Q., et al. Transplantation of ACE2(-) mesenchymal stem cells improves the outcome of patients with COVID-19 pneumonia. Aging and disease. 2020;11(2):216–228. doi: 10.14336/AD.2020.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shetty A.K. Mesenchymal stem cell infusion shows promise for combating coronavirus (COVID-19)- induced pneumonia. Aging and disease. 2020;11(2):462–464. doi: 10.14336/AD.2020.0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A., et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18(1):41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 10.Nabirotchkin S., Peluffo A.E., Rinaudo P., Yu J., Hajj R., Cohen D. Next-generation drug repurposing using human genetics and network biology. Curr Opin Pharmacol. 2020;51:78–92. doi: 10.1016/j.coph.2019.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Ashburn T.T., Thor K.B. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3(8):673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 12.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat Rev Drug Discov. 2020;19(3):149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 13.Xu Li JY., Zhang Zhiming, Ren Jing, Peluffo Alex E., Zhang Wen, Zhao Yujie, Yan Kaijing, Cohen Daniel, Wang Wenjia. 2020. Network bioinformatics analysis provides insight into drug repurposing for COVID-2019. Preprints 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., et al. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S., Chen J., Cheng T., et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47(D1):D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeersch T., Zurek E., Hutchison G.R. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Cheminf. 2012 Aug 13;4(1):17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stierand K., Maass P.C., Rarey M. Molecular complexes at a glance: automated generation of two-dimensional complex diagrams. Bioinformatics. 2006;22(14):1710–1716. doi: 10.1093/bioinformatics/btl150. [DOI] [PubMed] [Google Scholar]
- 18.Santos K.B., Guedes I.A., Karl A.L.M., Dardenne L.E. Highly flexible ligand docking: benchmarking of the DockThor program on the LEADS-PEP protein–peptide data set. J Chem Inf Model. 2020;60(2):667–683. doi: 10.1021/acs.jcim.9b00905. [DOI] [PubMed] [Google Scholar]
- 19.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duhovny D., Nussinov R., Wolfson H.J., editors. Efficient unbound docking of rigid molecules. Springer Berlin Heidelberg; Berlin, Heidelberg: 2002. [Google Scholar]
- 21.Poli G., Tuccinardi T. Consensus docking in drug discovery. Curr Bioact Compd. 2020;16:182–190. [Google Scholar]
- 22.Houston D.R., Walkinshaw M.D. Consensus docking: improving the reliability of docking in a virtual screening context. J Chem Inf Model. 2013;53:384–390. doi: 10.1021/ci300399w. [DOI] [PubMed] [Google Scholar]
- 23.Ligabue-Braun R., Real-Guerra R., Carlini C.R., Verli H. Evidence-based docking of the urease activation complex. J Biomol Struct Dyn. 2013;31(8):854–861. doi: 10.1080/07391102.2012.713782. [DOI] [PubMed] [Google Scholar]
- 24.Kirchdoerfer R.N., Wang N., Pallesen J., Wrapp D., Turner H.L., Cottrell C.A., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci Rep. 2018;8(1):15701. doi: 10.1038/s41598-018-34171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tazikeh-Lemeski E., Moradi S., Raoufi R., Shahlaei M., Janlou M.A.M., Zolghadri S. Targeting SARS-COV-2 non-structural protein 16: a virtual drug repurposing study. J Biomol Struct Dyn. 2020;(Jun 23):1–14. doi: 10.1080/07391102.2020.1779133. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pal S., Talukdar A. ChemRxiv; 2020. Compilation of potential protein targets for SARS-CoV-2: preparation of homology model and active site determination for future rational antiviral design. Preprint. [DOI] [Google Scholar]
- 28.Kitchen D.B., Decornez H., Furr J.R., Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov. 2004;3(11):935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 29.Nicolas A., Raguénès-Nicol C., Ben Yaou R., Ameziane-Le Hir S., Chéron A., Vié V., Claustres M., Leturcq F., Delalande O., Hubert J.F., Tuffery-Giraud S., Giudice E., Le Rumeur E. Becker muscular dystrophy severity is linked to the structure of dystrophin. Hum Mol Genet. 2015 Mar 1;24(5):1267–1279. doi: 10.1093/hmg/ddu537. French Network of Clinical Reference Centres for Neuromuscular Diseases (CORNEMUS) [DOI] [PubMed] [Google Scholar]
- 30.Vargiu A.V., Nikaido H. Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc Natl Acad Sci U S A. 2012 Dec 11;109(50):20637–20642. doi: 10.1073/pnas.1218348109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruggerone P., Murakami S., Pos K.M., Vargiu A.V. RND efflux pumps: structural information translated into function and inhibition mechanisms. Curr Top Med Chem. 2013;13(24):3079–3100. doi: 10.2174/15680266113136660220. [DOI] [PubMed] [Google Scholar]
- 32.Salentin S., Schreiber S., Haupt V.J., Adasme M.F., Schroeder M. PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015;43(W1):443–447. doi: 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jorgensen W., et al. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–935. [Google Scholar]
- 34.Dodda L.S., et al. 1.14* CM1A-LBCC: localized bond-charge corrected CM1A charges for condensed-phase simulations. J Phys Chem B. 2017;121(15):3864–3870. doi: 10.1021/acs.jpcb.7b00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dodda L.S., et al. LigParGen web server: an automatic OPLS-AA parameter generator for organic ligands. Nucleic Acids Res. 2017;45:W331–W336. doi: 10.1093/nar/gkx312. W1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abraham M.J., et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. Software. 2015;1:19–25. [Google Scholar]
- 37.Lindahl, Abraham, Hess, van der Spoel GROMACS 2020.2 manual (version 2020.2) 2020, April 30. Zenodo. [DOI]
- 38.Jorgensen W.L., Tirado-Rives J. Potential energy functions for atomic-level simulations of water and organic and biomolecular systems. Proc Natl Acad Sci Unit States Am. 2005;102(19):6665–6670. doi: 10.1073/pnas.0408037102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berweger C.D., van Gunsteren W.F., Müller-Plathe F. Force field parametrization by weak coupling. Re-engineering SPC water. Chem Phys Lett. 1995;232(5–6):429–436. [Google Scholar]
- 40.Hess Berk, et al. LINCS: a linear constraint solver for molecular simulations. J Comput Chem. 1997;18(12):1463–1472. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 41.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅ log (N) method for Ewald sums in large systems. J Chem Phys. 1993;98(12):10089–10092. [Google Scholar]
- 42.Nosé S., Klein M.L. Constant pressure molecular dynamics for molecular systems. Mol Phys. 1983;50(5):1055–1076. [Google Scholar]
- 43.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys. 1981;52(12):7182–7190. [Google Scholar]
- 44.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126(1) doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 45.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science (New York, NY) 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rane J., Chatterjee A., Ray S. 2020. Targeting SARS-CoV-2 spike protein of COVID-19 with naturally occurring phytochemicals: an in silco study for drug development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ibrahim I.M., Abdelmalek D.H., Elshahat M.E., Elfiky A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J Infect. 2020;80(5):554–562. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang J., Song W., Huang H., Sun Q. Pharmacological therapeutics targeting RNA-dependent RNA polymerase, proteinase and spike protein: from mechanistic studies to clinical trials for COVID-19. J Clin Med. 2020;9(4) doi: 10.3390/jcm9041131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elfiky A.A. Zika viral polymerase inhibition using anti-HCV drugs both in market and under clinical trials. J Med Virol. 2016;88(12):2044–2051. doi: 10.1002/jmv.24678. [DOI] [PubMed] [Google Scholar]
- 51.Elfiky A.A., Ismail A. Molecular dynamics and docking reveal the potency of novel GTP derivatives against RNA dependent RNA polymerase of genotype 4a HCV. Life Sci. 2019;238:116958. doi: 10.1016/j.lfs.2019.116958. [DOI] [PubMed] [Google Scholar]
- 52.Elfiky A.A. SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: an in silico perspective. J Biomol Struct Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1761882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doublie S., Ellenberger T. The mechanism of action of T7 DNA polymerase. Curr Opin Struct Biol. 1998;8(6):704–712. doi: 10.1016/s0959-440x(98)80089-4. [DOI] [PubMed] [Google Scholar]
- 54.Meira R.Z.C., Biscaia I.F.B., Nogueira C., Murakami F.S., Bernardi L.S., Oliveira P.R. Solid-state characterization and compatibility studies of penciclovir, lysine hydrochloride, and pharmaceutical excipients. Materials. 2019;12(19):3154. doi: 10.3390/ma12193154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hodge R.A.V., Cheng Y.C. The mode of action of penciclovir. Antiviral Chem Chemother. 1993;4(6_suppl):13–24. [Google Scholar]
- 56.Boyd M.R., Bacon T.H., Sutton D., Cole M. Antiherpesvirus activity of 9-(4-hydroxy-3-hydroxy-methylbut-1-yl)guanine (BRL 39123) in cell culture. Antimicrob Agents Chemother. 1987;31(8):1238–1242. doi: 10.1128/aac.31.8.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Degreef H. Famciclovir, a new oral antiherpes drug: results of the first controlled clinical study demonstrating its efficacy and safety in the treatment of uncomplicated herpes zoster in immunocompetent patients. Int J Antimicrob Agents. 1994;4(4):241–246. doi: 10.1016/0924-8579(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 58.Hodge R.A., Perkins R.M. Mode of action of 9-(4-hydroxy-3-hydroxymethylbut-1-yl)guanine (BRL 39123) against herpes simplex virus in MRC-5 cells. Antimicrob Agents Chemother. 1989;33(2):223–229. doi: 10.1128/aac.33.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mondal D. Elsevier; 2016. Famciclovir. Reference module in biomedical sciences. [Google Scholar]
- 60.Yu A., Guo C., Zhou Y., Cao F., Zhu W., Sun M., et al. Skin irritation and the inhibition effect on HSV-1 in vivo of penciclovir-loaded microemulsion. Int Immunopharm. 2010;10(10):1305–1309. doi: 10.1016/j.intimp.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 61.Sidwell R.W., Huffman J.H., Khare G.P., Allen L.B., Witkowski J.T., Robins R.K. Broad-spectrum antiviral activity of Virazole: 1-beta-D-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science (New York, NY) 1972;177(4050):705–706. doi: 10.1126/science.177.4050.705. [DOI] [PubMed] [Google Scholar]
- 62.Muller W.E., Maidhof A., Taschner H., Zahn R.K. Virazole (1-beta-D-ribofuranosyl-1,2,4-triazole-3-carboxamide; a cytostatic agent. Biochem Pharmacol. 1977;26(11):1071–1075. doi: 10.1016/0006-2952(77)90246-5. [DOI] [PubMed] [Google Scholar]
- 63.Graci J.D., Cameron C.E. Mechanisms of action of ribavirin against distinct viruses. Rev Med Virol. 2006;16(1):37–48. doi: 10.1002/rmv.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bassetti M., Castaldo N., Carnelutti A. Neuraminidase inhibitors as a strategy for influenza treatment: pros, cons and future perspectives. Expet Opin Pharmacother. 2019;20(14):1711–1718. doi: 10.1080/14656566.2019.1626824. [DOI] [PubMed] [Google Scholar]
- 65.Eiland L.S., Eiland E.H. Zanamivir for the prevention of influenza in adults and children age 5 years and older. Therapeut Clin Risk Manag. 2007;3(3):461–465. [PMC free article] [PubMed] [Google Scholar]
- 66.Colman P.M. Influenza virus neuraminidase: structure, antibodies, and inhibitors. Protein Sci. 1994;3(10):1687–1696. doi: 10.1002/pro.5560031007. a publication of the Protein Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hall D.C., Jr., Ji H.F. A search for medications to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2 spike glycoprotein and 3CL protease. Trav Med Infect Dis. 2020:101646. doi: 10.1016/j.tmaid.2020.101646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shah B., Modi P., Sagar S.R. In silico studies on therapeutic agents for COVID-19: drug repurposing approach. Life Sci. 2020;252:117652. doi: 10.1016/j.lfs.2020.117652. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.