Abstract.
Purpose: Deep learning is a promising technique for spleen segmentation. Our study aims to validate the reproducibility of deep learning-based spleen volume estimation by performing spleen segmentation on clinically acquired computed tomography (CT) scans from patients with myeloproliferative neoplasms.
Approach: As approved by the institutional review board, we obtained 138 de-identified abdominal CT scans. A sum of voxel volume on an expert annotator’s segmentations establishes the ground truth (estimation 1). We used our deep convolutional neural network (estimation 2) alongside traditional linear estimations (estimation 3 and 4) to estimate spleen volumes independently. Dice coefficient, Hausdorff distance, coefficient, Pearson coefficient, the absolute difference in volume, and the relative difference in volume were calculated for 2 to 4 against the ground truth to compare and assess methods’ performances. We re-labeled on scan–rescan on a subset of 40 studies to evaluate method reproducibility.
Results: Calculated against the ground truth, the coefficients for our method (estimation 2) and linear method (estimation 3 and 4) are 0.998, 0.954, and 0.973, respectively. The Pearson coefficients for the estimations against the ground truth are 0.999, 0.963, and 0.978, respectively (paired -tests produced between 2 and 3, and 2 and 4).
Conclusion: The deep convolutional neural network algorithm shows excellent potential in rendering more precise spleen volume estimations. Our computer-aided segmentation exhibits reasonable improvements in splenic volume estimation accuracy.
Keywords: deep learning, image segmentation, spleen, computed tomography, splenomegaly
1. Introduction
Organ size measurements provide clinical utility in diagnosis and assessing treatment response with different cancers. For instance, reduction in spleen volume is a crucial response element for myeloproliferative neoplasms (MPN),1 and recent studies showed that significant (greater than or equal to 35%) splenic volume reduction (SVR) in myelofibrosis patients is associated with improved overall survival.2 Splenic volume is measured in MPNs as an indicator of extramedullary hematopoiesis, and SVR is a surrogate endpoint used to measure success of a intervention in MPNs.3,4 Quantitative estimates of splenic biomarkers have been of clinical interest in molecular, histologic, radiographic, and physiologic characterization of spleens.5–7
Unique challenges emerge when validating machine learning-generated imaging biomarkers. Changes in imaging hardware, acquisition parameters, and patient positioning during imaging cause significant variations among images. Several groups, such as the NCI-sponsored Quantitative Imaging Network8 and the Radiological Society of North America’s Quantitative Imaging Biomarker Alliance,9 have pursued multicenter imaging trials to address these variabilities. Using machine learning algorithms to compute biomarkers presents additional challenges. Performance from machine learning algorithms is dependent not only on the training dataset but also dataset upon which the algorithms are deployed. Park and Han10 addressed this challenge in their recent evaluation of diagnostic and predictive artificial intelligence technologies. They postulate that validating such algorithms’ performance requires testing in a clinical cohort that adequately represents the target patient population.10
Given an image from ultrasound, magnetic resonance imaging, or computed tomography (CT), manual annotation of spleen segmentation is still the gold standard,11 yet the process is time-intensive and requires domain expertise. Here, computer-assisted spleen labeling could not only reduce resource consumption, but also support investigation into spleen size’s clinical utility as a biomarker.12–14 Several challenges remain, including significant inter-subject variability in spleen size, shape, and orientation. The multi-atlas approaches produced encouraging results,15,16,17 and we recently developed a deep convolutional neural network algorithm that performs significantly better than previous methods.18,19–21 The non-manual measurements are “fit for purpose,” i.e., the rigor and methods utilized should be in accordance with the intended purpose of the biomarker study.22,23 Given this fit for purpose approach, it is difficult to establish absolute benchmark criteria for validation studies. Accordingly, the reporting of validation studies becomes increasingly important to allow investigators to evaluate whether the assay is appropriate for a proposed study.
We hypothesize that automated measures of spleen size can serve as a biomarker for clinicians to better predict MPN disease progression and response to therapy. Investigation into and potential utility of spleen volume as a biomarker is limited due to the time-intensive process in obtaining those measurements. Indeed, Sargent et al.24 defined a set of prerequisite criteria for an imaging biomarker before clinical validation studies can be performed. Notably, the authors highlight the necessity that the technology to assess the biomarker of interest be stable and widely available. With automated spleen volume estimation, it is essential to validate the methods following such criteria for clinical validation. The goal of this study is to evaluate the performance of the proposed state-of-the-art spleen segmentation algorithm in measuring spleen volumes on clinically acquired CT scans from patients with MPNs.
Our validation study includes a complete assessment of the technical performance of the biomarker assay using the state-of-the-art deep neural networks. Such assessment includes measures of assay accuracy, repeatability, reproducibility, technical bias, sensitivity, and specificity. We investigate four pipeline estimates for using the deep learning algorithms on all scans: (1) manual segmentation by expert readers; (2) automatic segmentation using deep learning algorithms; (3) unidimensional measurements, and (4) 3D splenic index measurement. Further, the validation study defines the limits of the detection and quantification for a given assay.7 In the context of imaging biomarkers, the assay includes both the image generation process (the specific imaging protocol) and the subsequent post-processing procedures to yield the biomarker measurement accuracy. Ultimately, through the validation process, we show the agreement and bias proportion to the intended purpose of the biomarker study.
In the cross-validation experiments, our computer-assisted method produced segmentation masks with an averaged dice coefficient of 0.95148 when evaluating against hand labeled masks. Most importantly, our proposed method’s volume estimation achieved coefficient of 0.99800 and Pearson coefficient of 0.99905, indicating a significant improvement over traditional linear estimations.25 This demonstrates the potential of obtaining more accurate spleen volume estimates from state-of-the-art deep learning algorithm.
2. Materials and Methods
2.1. Data Acquisition
Under institutional review board (IRB) approval, we obtained 138 de-identified abdominal CT from patients enrolled in NCT02493530. This is a phase 1 multi-center study of TGR-1202 administered together with ruxolitinib in patients with MPNs. To minimize spectrum bias, we utilized a consecutive series of patients from four study locations (Mayo, Wisconsin, Colorado, and Vanderbilt). Including multi-center data provides evidence to evaluate how this algorithm adapts to the variability inherent in multi-center trials. The spleen was segmented by expert readers from each scan in this dataset to establish the ground truth.
2.2. Manual Segmentation (Estimation 1)
Manual spleen segmentation on all 138 scans establishes baseline splenic volumes. We used open-sourced tool MIPAV software from the National Institutes of Health (NIH) to trace the spleen anatomies. In our study, CT scans from patients with splenomegaly were retrieved. We delineated the outlines on every axial slice and filled the regions enclosed by the tool. A radiologist, certified by the abdominal imaging board, verified all splenic contours on the volumetric investigations. We calculated ground truth spleen volume for each image by directly multiplying unit volume (cc/voxel) with number of voxels inside segmentation region.
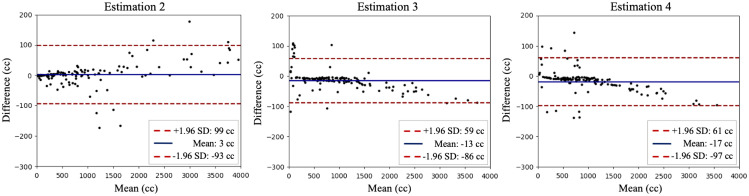
To evaluate the repeatability and reproducibility, we retrieved a subset of 40 patients labeled by a second similarly qualified imaging analyst under the supervision of a radiologist to assess the inter-rater reliability of manual segmentations. Both readers adhered to the same tracing protocol, and the agreement evaluation is shown in the result by the Bland–Atman plot (Fig. 5).
Fig. 5.
The repeatability and reproducibility between different imaging analyst readers on 40 respective studies. Bland–Altman plot between estimations (2 to 4). The mean in difference and a confidence interval of 1.96 standard deviation are shown.
2.3. Deep Convolutional Neural Network Algorithm (Estimation 2)
2.3.1. Stage 1: low-resolution segmentation
Given CT scans with a fine resolution of [], we first downsampled the images and trained a 3D U-Net for segmentation with lower resolution. Each scan slice is downsampled from [] to [] and images were normalized to a consistent voxel resolution of []. We used Dice loss to compare network outputs and ground truth labels. We ignored the background loss in order to increase weights for anatomies. The crude segmentation masks are then upsampled to original resolution with nearest interpolation for later stages. This approach is trained end-to-end, and Figs. 3–5 assess the approach’s segmentation quality. The downsampled volume in low-resolution framework, while lacking detailed structures of anatomies, still preserves complete spatial context in CT scan.
Fig. 3.
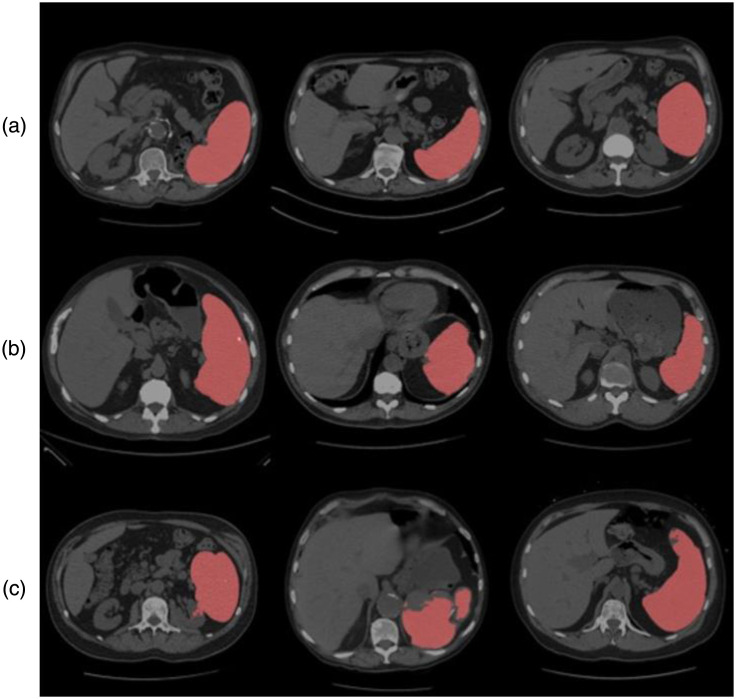
Quality assurance of the deep learning method in estimation 2 with CT. (a) Three representative subjects’ slice above state-of-the-art. (b) Three representative cases with successful segmentation. (c) Failure cases where manual correction was required.
Fig. 4.
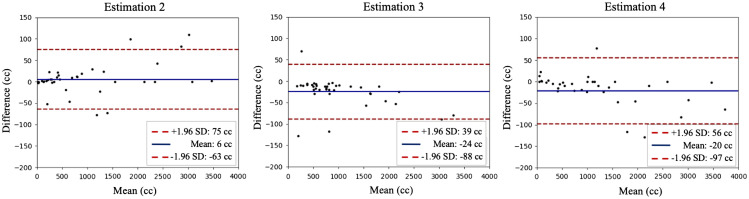
Bland–Altman plot for computer-assisted method (estimation 2), linear estimate with length and splenic index (estimation 3 and 4). On each plot, the -axis indicates the mean volume between the ground truth and the estimation from computer-aided method. The -axis shows the difference in volume. A 1.96 standard deviation is shown as the confidence interval.
2.3.2. Stage 2: random patch selection
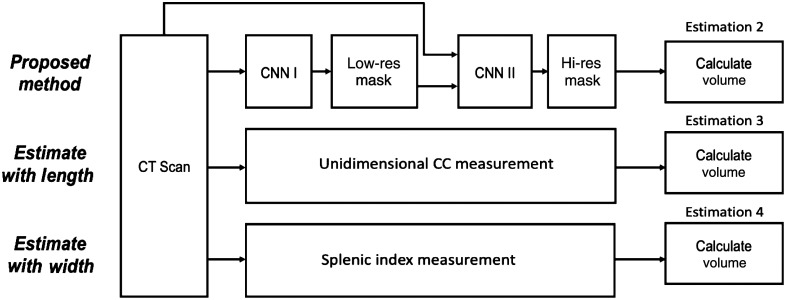
For each CT scan, we randomly selected voxels in the predicted coarse segmentation mask. Fixing the selected voxels as centers, we placed bounding cubes with slight random shifts along all axes. A Gaussian random variable determines the shifting distance. High-resolution patches from original images were cropped according to bounding cubes, and they formed second-stage model inputs (middle panel of Fig. 1). This strategy builds the hierarchy of non-linear features from random patches regardless of 3D contexts, and it employs detailed context at original resolution and incorporates advantages of data augmentation with shifting. We fixed the patch size at in our experiments.
Fig. 1.
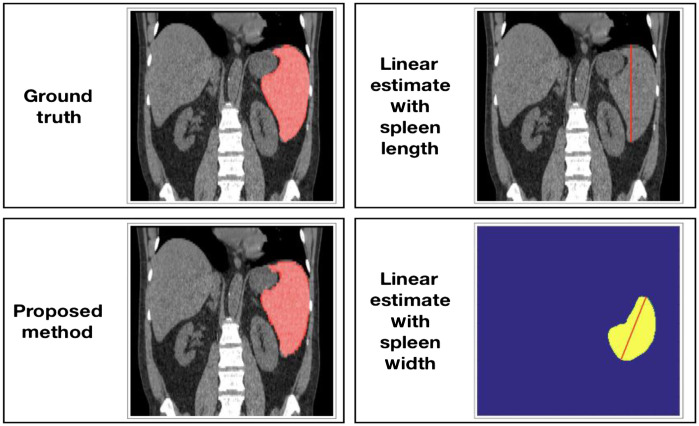
Pipeline for the proposed method and unidimensional linear regression estimation methods. The computer-assisted method in estimation 2 includes two CNN models with a coarse-to-fine framework. Estimation 3 and 4 use measurements of length and width (splenic index) from the ground truth for cc (cubic centimeter) volume estimation.
2.3.3. Stage 3: high-resolution segmentation and label fusion
Using randomly selected high-resolution patches from the prior stage, we trained a second 3D U-Net. Integrating all patches on field of views, we estimated the full field of view. Majority vote is used to merge estimates into a final segmentation yielding the spleen voxels. Specifically, after separating full spatial context to randomly selected subspaces, the overlapped regions provide more than one segmentation label for a voxel. We summarize a single label given a vector of class labels from candidates. We ignore voters outside the image space and related values are excluded in the label fusion.
Architecture
The 3D U-Net is adopted as the segmentation model backbone; the architecture of the network contains an encoder and a decoder with four scales. It employs deconvolution to upsample the lower scale feature maps to the higher scale of dimension. This process enables the efficient denser pixel-to-pixel mappings. Each scale in the encoder has two convolutional layers, followed by rectified linear units and a max pooling of and strides of 2. The decoder has the transpose convolutions of and strides of 2. The last layer is composed by convolution that set the number of output channels to the number of class labels. The Dice loss is used for the spleen segmentation. The baseline low-resolution segmentation uses the largest volume size of to fit maximum memory of a normal 12-GB GPU. The volume size is also employed in baseline hierarchical method for training the first-level model. We used batch size of 1 for all experiments. The instance normalization is employed, which is agnostic to batch size. We adapted the ADAM algorithm with stochastic gradient decent, momentum = 0.9. The initial learning rate was set to 0.001, and it was reduced by a factor of 10 every 10 epochs after 50 epochs. Implementations were performed using NVIDIA Titan X GPU 12-G memory and CUDA 9.0.
2.4. Linear Estimation of Spleen Volume with Spleen Length (Estimation 3 and 4)
Recently, Bezerra et al.25 introduced a helpful approach that estimates spleen volume through unidimensional spleen measurements and 3D splenic index. We obtained maximum spleen length from coronal/frontal plane (L) and maximum spleen width from oblique sagittal/axial plane (W). We calculated spleen volume estimates with linear regression equations specified by Bezerra et al.25 The equations they proposed for maximum spleen length and maximum spleen widths are and . Figure 2 depicts maximum spleen length and maximum spleen width alongside other estimation methods in this study.
Fig. 2.
Demonstration of the measurements from pipelines for estimating spleen volumes. The manual and computer-assisted methods evaluate the spleen volume (estimation 1 and 2). The linear estimates (3 and 4) manually extract splenic diameters along different axes (length and width) from an unlabeled CT scan.
2.5. Baseline Comparisons
2.5.1. Low-resolution architecture
We compared our method with the finest resolution to house the maximum GPU 12-G memory. The baseline method is implemented with a single-step resampling process. Each scan is downsampled to . The entire volume is inputted to the 3D U-Net model.
2.5.2. High-resolution architecture
The high-resolution baseline method is implemented with several connected tiles as input to the model. We kept the original image resolution under dimension of , then cropped the image into adjacent patches to fit the GPU memory. We use the patch of voxels. Patches were extracted without overlap, each tile was padded to fixed size once it exceeded the volume dimension. The final segmentation was acquired by tiling ordered patches.
2.5.3. Multi-atlas segmentation
We compared our current method with the previous multi-atlas approach. The adaptive Gaussian mixture model was used as the atlas selection step. Then, the joint label fusion is implemented to obtain the spleen segmentation.
3. Analysis
3.1. Accuracy
Several metrics have been proposed to evaluate imaging segmentation accuracy.26 We computed the Dice coefficient and average Hausdorff distance to judge segmentations from different methods against the ground truth (Table 1). We compared the different methods’ volume predictions against the ground truth with , Pearson correlation, and absolute and percent deviations.
Table 1.
Summarized statistics for different estimations compared to ground truth.
| Proposed method (estimation 2) | Linear estimation with length (estimation 3) | Linear estimation with width (estimation 4) | |
|---|---|---|---|
| Dice similarity coefficient | N/A | N/A | |
| Hausdorff distance | N/A | N/A | |
| 0.998 | 0.954 | 0.973 | |
| Pearson | 0.999 | 0.963 | 0.978 |
| Absolute deviation of volume () | |||
| Percent difference (%) |
3.2. Bias
Bland–Altman plots (Fig. 4) serve to compare different algorithm’s estimates’ agreement with the ground truth (Fig. 4).
3.3. Reproducibility
Once trained, the automated segmentation algorithm yields same result for a given image. To ascertain the method as a sound replacement for manual segmentation, we manually examined the approach’s reproducibility on a subset of 40 patients. This subset of patient images was labeled simultaneously by a second research associate in order to assess the inter-rater reliability. Assuming the label from expert 2 is the ground truth, we present the reproducibility comparisons in Fig. 5.
4. Results
The mean Dice score between interpreter 1 and interpreter 2 is , the average symmetric Hausdorff distance is . Percent difference between two observers is . Two observers assessed subjects independently without communication.
In Fig. 3, we present examples of predicted segmentation masks on their respective CT scans. The top row shows three examples with exceptional alignment. The second row’s predictions are satisfactory but slightly flawed at the edges, and the bottom row presents some of our failure cases.
As shown in Table 1, our proposed method’s population Dice coefficient statistic is , indicating a high degree of alignment between prediction masks and ground truth masks. The averaged symmetric Hausdorff distance is . The superior volume estimation performance of the proposed method becomes more apparent when we compare estimation 2’s values (0.99800), Pearson coefficient (0.99905), absolute difference in volume estimates (), and percent difference in volume estimates () ( with paired -test between estimation 2 and 3, 2 and 4, respectively) against those of the unidimensional linear regression estimation methods. The Bland–Altman plots in Fig. 4 also illustrates that the deep convolutional neural network algorithm produced superior results.
As shown in Table 2, our method archives consistent improved performance comparing to single step segmentation baselines. The random patch method has the Dice score of 0.951 compared to 0.897 for low-resolution baseline and 0.921 for high-resolution baseline. We also observed the improved Hausdorff distance at 9.385 against 11.847 and 10.458 for low- and high-resolution methods. Comparing to the multi-atlas approach, the deep learning-based methods achieved consistently higher performance, the low-resolution model observed about 5% improvement.
Table 2.
Segmentation metrics comparing to state-of-the-art methods.
| Dice | HD | ASD | |
|---|---|---|---|
| Low resolution (single step) | |||
| High resolution (single step) | |||
| Multi-atlas1 | |||
| Random patches (ours) |
For evaluating the proposed automatic method, we conducted external testing on a splenomegaly dataset.
4.1. ImageVU Splenomegaly
A total of 40 subjects were selected and retrieved from Vanderbilt University Medical Center. The dataset is designed by cases of splenomegaly patients. All CT volumes are subjected to splenomegaly ICD-10 criteria. The abnormal spleens were manually traced following the same framework of annotation.
4.2. Performance
As shown in Table 3, the mean Dice score of the external testing set is 0.949, the clinically acquired scans show the stability of the proposed method. We also observed similar performance in terms of averaged symmetric Hausdorff distance and averaged surface distance.
Table 3.
Segmentation performance of models tested on ImageVU Splenomegaly dataset in mean DSC and variance.
| Dice | HD | ASD | |
|---|---|---|---|
| Low resolution (single step) | |||
| High resolution (single step) | |||
| Multi-atlas1 | |||
| Random patches (ours) |
Note: HD, Hausdorff distance; ASD, averaged surface distance.
5. Discussion
5.1. Main Contributions
The study demonstrates a deep learning algorithm’s ability to produce precise spleen masks and spleen volume estimates from abdominal CT scans. Shown in Bland–Altman plots, estimates 2 to 4 performing on normal patient CT scans and failed to inference accurately where splenomegaly was present. Our proposed method yielded superior results in both cases and was also able to produce empty mask for CT scans after splenectomy. As shown in Table 1, our method achieves a Pearson coefficient of 0.99905 and an average non-significant absolute deviation of 20.604 cc with respect to the ground truth. This approach performs consistent comparable result with resource-intensive manual segmentation with unidimensional measurements. These results show that a deep learning algorithm supervised by manual segmentation can enable generation of higher accuracy in estimation of spleen volumes.
Based on the performance between visual ratings and the automatic segmentations, our method could reliably label clinically acquired CT scans. Figure 3 shows that the abnormal anatomy (e.g., splenomegaly) can lead to a less accurate segmentation performance for baseline estimations. The limitations, for instance, patients with severe red cell inflation, may obtain irregular segmentation. In addition, when the patient is under treatment with drugs or surgery, previous algorithm may segment a small false-positive region when irregular shape appears. However, our method showed the consistent performance evaluating the splenomegaly patients. Above erroneous labeling can be prevented by discarding candidates with the selected spleen volume segmented. Another direct application using our algorithms presented in the observing period at patient treatment. In our trials, each patient can have up to four longitudinal CT scans at different drug period. The automatic segmentation can lead to fast and accurate outcomes for measuring spleen volume, this process can potentially help physicians delivering further step of treatment suggestions. Finally, the model is trained and evaluated using only CT scans with enlarged spleens, we intended to deploy and focus on splenomegaly patients. We presented the reliable and robust method from other domains to delivery quantitative measurements for splenomegaly without extra manual efforts.
In reviewing the performance of inter-rater reproducibility, we found that agreement between experts were highly accurate (mean 6 cc in Bland–Atman plot in Fig. 5). Using the labeling by the second expert as the ground truth, the computer-assisted segmentation achieves slightly higher agreement (mean 3 cc in Fig. 4). Typically, the automatic method observes outliers that include spleens with severe splenomegaly and those under surgery, which these outliers are required by rudimentary visual quality refinement.
5.2. Clinical Improvement
Organ size measurements remain attractive biomarkers for the assessment of disease. However, diagnosis time is always a concern, and the significant time and resource cost associated with the extraction of organ size limit its use. So far, such methods demonstrated limited clinical utility to justify its adoption in clinical workflows, and it is our vision that automated measures of organ size will reduce the cost of obtaining such measures, allowing for the prospective evaluation of organ sizes in the study of disease prognosis and treatment response.
6. Summary
In summary, we proposed a deep convolutional neural network algorithm that produced more accurate spleen volume estimates for abdominal CT scans. Given the importance of spleen volume as a biomarker and considering the superior effectiveness of the algorithm on patients with splenomegaly, we conclude the algorithm can provide sufficiently accurate spleen volume measurements. Given the current inaccurate and computationally expensive algorithms or accurate but laborious manual labeling, this proposed method should exempt expert radiologists from arduous manual labeling of spleens while allowing more precise, and expedient clinical diagnosis and treatment suggestions with better spleen volume estimates.
Acknowledgments
This study was approved by the Vanderbilt University IRB (150459) and registered with the FDA (NCT02493530). This research was supported by the National Science Foundation, NSF CAREER 1452485, and the National Institutes of Health, NIH 3U54CK120058. This study was in part using the resources of the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University, Nashville, Tennessee. We gratefully acknowledge support of NVIDIA Corporation with the donation of the Titan X Pascal GPU used for this research. The imaging dataset(s) used for the analysis described were obtained from ImageVU, a research resource supported by the VICTR CTSA award (ULTR000445 from NCATS/NIH). TG Therapeutics provided funding for the study of TGR1202 in patients with MPNs (NCT02493530). M.R.S. is a clinical scholar of the Leukemia and Lymphoma Society and EP Evans fellow.
Biographies
Yiyuan Yang is an undergraduate student in computer science and math at Vanderbilt University.
Yucheng Tang is a graduate student at Vanderbilt University. He received his BS degree from Tianjin University in 2015, his MS degree in computer science from New York University. He is a member of SPIE.
Biographies of the other authors are not available.
Disclosures
M.S. receives research funding from Astex, Incyte, TG Therapeutics, Takeda; has equity in Karyopharm; consults for Karyopharm and Ryvu; and serves on the advisory board for AbbVie, BMS, Celgene, Sierra Oncology, Takeda, TG Therapeutics. The authors of the paper are directly employed by the institutes or companies provided in this paper. Yiyuan Yang, Yucheng Tang, Riqiang Gao, Yuankai Huo, Shunxing Bao, Richard G. Abramson, and Bennett A. Landman declare no conflicts of interest in the submission of this paper.
Contributor Information
Yiyuan Yang, Email: yiyuan.yang@vanderbilt.edu.
Yucheng Tang, Email: yucheng.tang@vanderbilt.edu.
Riqiang Gao, Email: riqiang.gao@vanderbilt.edu.
Shunxing Bao, Email: shunxing.bao@vanderbilt.edu.
Yuankai Huo, Email: yuankai.huo@vanderbilt.edu.
Matthew T. McKenna, Email: matthew.t.mckenna.1@vumc.org.
Michael R. Savona, Email: michael.savona@vumc.org.
Richard G. Abramson, Email: rgabramson@gmail.com.
Bennett A. Landman, Email: bennett.landman@vanderbilt.edu.
References
- 1.Tefferi A., et al. , “Revised response criteria for myelofibrosis: International Working Group-Myeloproliferative Neoplasms Research and (IWG-MRT) and European LeukemiaNet (ELN) consensus report,” Blood 122, 1395–1398 (2013). 10.1182/blood-2013-03-488098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verstovsek S., et al. , “A double-blind, placebo-controlled trial of ruxolitinib for myelofibrosis,” Engl. J. Med. 366, 799–807 (2012). 10.1056/NEJMoa1110557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang Y., et al. , “Improving splenomegaly segmentation by learning from heterogeneous multi-source labels,” Proc. SPIE 10949, 1094908 (2019). 10.1117/12.2512842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huo Y., et al. , “Splenomegaly segmentation using global convolutional kernels and conditional generative adversarial networks,” Proc. SPIE 10574, 1057409 (2018). 10.1117/12.2293406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abramson R. G., et al. , “Methods and challenges in quantitative imaging biomarker development,” Acad. Radiol. 22, 25–32 (2015). 10.1016/j.acra.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abramson R. G., Yankeelov T. E., “Imaging biomarkers and surrogate endpoints in oncology clinical trials,” in Functional Imaging in Oncology, Luna A., et al., Eds., pp. 29–42, Springer, Berlin, Heidelberg: (2014). [Google Scholar]
- 7.Rosenkrantz A. B., et al. , “Clinical utility of quantitative imaging,” Acad. Radiol. 22, 33–49 (2015). 10.1016/j.acra.2014.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke L. P., et al. , “The quantitative imaging network: NCI’s historical perspective and planned goals,” Transl. Oncol. 7, 1–4 (2014). 10.1593/tlo.13832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buckler A. J., et al. , “A collaborative enterprise for multi-stakeholder participation in the advancement of quantitative imaging,” Radiology 258, 906–914 (2011). 10.1148/radiol.10100799 [DOI] [PubMed] [Google Scholar]
- 10.Park S. H., Han K., “Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction,” Radiology 286, 800–809 (2018). 10.1148/radiol.2017171920 [DOI] [PubMed] [Google Scholar]
- 11.Heymsfield S. B., et al. , “Accurate measurement of liver, kidney, and spleen volume and mass by computerized axial tomography,” Ann. Intern. Med. 90, 185 (1979). 10.7326/0003-4819-90-2-185 [DOI] [PubMed] [Google Scholar]
- 12.Xu Z., et al. , “Efficient abdominal segmentation on clinically acquired CT with SIMPLE context learning,” Proc. SPIE 9413, 94130L (2015). 10.1117/12.2081012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huo Y., et al. , “Robust multicontrast MRI spleen segmentation for splenomegaly using multi-atlas segmentation,” IEEE Trans. Biomed. Eng. 65, 336–343 (2018). 10.1109/TBME.2017.2764752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang O., et al. , “Validation and optimization of multi-organ segmentation on clinical imaging archives,” Proc. SPIE 11313, 113132O (2020). 10.1117/12.2549035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Z., et al. , “Improving spleen volume estimation via computer-assisted segmentation on clinically acquired CT scans,” Acad. Radiol. 23, 1214–1220 (2016). 10.1016/j.acra.2016.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linguraru M. G., et al. , “Automated segmentation and quantification of liver and spleen from CT images using normalized probabilistic atlases and enhancement estimation,” Med. Phys. 37, 771–783 (2010). 10.1118/1.3284530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McAuliffe M. J., et al. , “Medical image processing, analysis and visualization in clinical research,” Proc. 14th IEEE Symp. Comput.-Based Med. Syst., pp. 381–386, IEEE; (2001). 10.1109/CBMS.2001.941749 [DOI] [Google Scholar]
- 18.Huo Y., et al. , “Splenomegaly segmentation on multi-modal MRI using deep convolutional networks,” IEEE Trans. Med. Imaging 38, 1185–1196 (2019). 10.1109/TMI.2018.2881110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huo Y., et al. , “Adversarial synthesis learning enables segmentation without target modality ground truth,” in IEEE 15th Int. Symp. Biomed. Imaging (2017). 10.1109/ISBI.2018.8363790 [DOI] [Google Scholar]
- 20.Huo Y., et al. , “SynSeg-Net: synthetic segmentation without target modality ground truth,” IEEE Trans. Med. Imaging 38(4), 1016–1025 (2019). 10.1109/TMI.2018.2876633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moon H., et al. , “Acceleration of spleen segmentation with end-to-end deep learning method and automated pipeline,” Comput. Biol. Med. 107, 109–117 (2019). 10.1016/j.compbiomed.2019.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J. W., et al. , “Fit-for-purpose method development and validation for successful biomarker measurement,” Pharm. Res. 23, 312–328 (2006). 10.1007/s11095-005-9045-3 [DOI] [PubMed] [Google Scholar]
- 23.Vannucchi A. M., et al. , “A pooled analysis of overall survival in COMFORT-I and COMFORT-II, 2 randomized phase III trials of ruxolitinib for the treatment of myelofibrosis,” Hematological 100(9), 1139–1145 (2015). 10.3324/haematol.2014.119545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sargent D. J., et al. , “Validation of novel imaging methodologies for use as cancer clinical trial end-points,” Eur. J. Cancer 45, 290–299 (2009). 10.1016/j.ejca.2008.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bezerra A. S., et al. , “Determination of splenomegaly by CT: is there a place for a single measurement?” AJR Am. J. Roentgenol. 184, 1510–1513 (2005). 10.2214/ajr.184.5.01841510 [DOI] [PubMed] [Google Scholar]
- 26.Taha A. A., Hanbury A., “Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool,” BMC Med. Imaging 15, 29 (2015). 10.1186/s12880-015-0068-x [DOI] [PMC free article] [PubMed] [Google Scholar]