Figure 1.
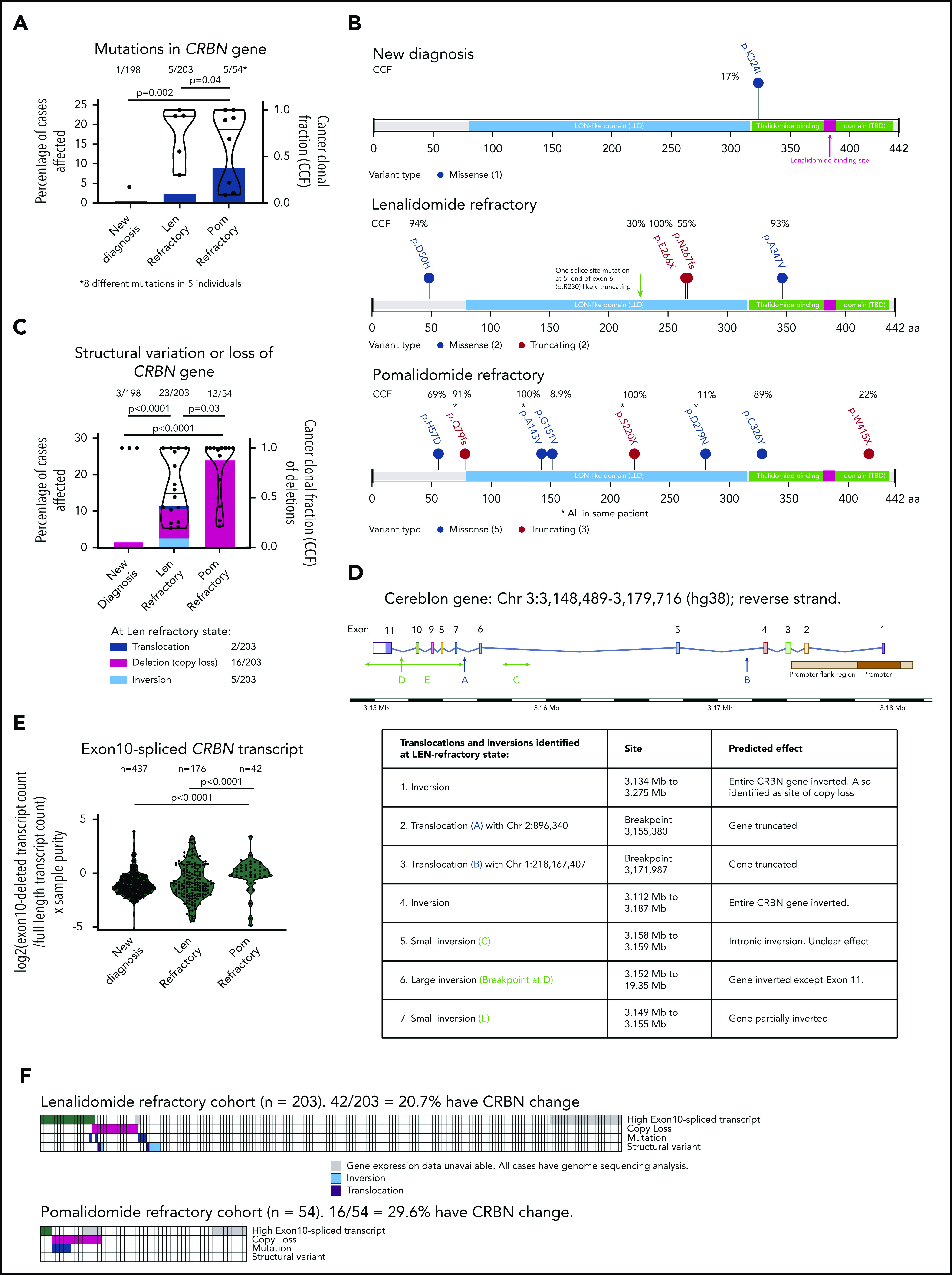
Incidence of mutation, copy loss, structural variation, or high exon 10–spliced transcript of CRBN increases with sequential IMiD refractoriness. (A) Incidence of CRBN mutations (left-handed system [LH] y-axis) and their cancer clonal fractions (CCFs; right-handed system [RH] y-axis) at ND, LEN-, and POM-refractory states. Significance detected by Fisher’s exact test to detect equality of proportions (nonparametric). (B) Amino acid changes resulting from the mutations in CRBN gene found at each state. CCF of each mutation shown. (C) Incidence of copy loss or structural variation (LH y-axis) and their CCFs (RH y-axis; copy loss only) at ND, LEN-, and POM-refractory states. Significance detected by Fisher’s exact test to detect equality of proportions (nonparametric). (D) Diagrammatic representation of site of structural variant breakpoints identified at LEN refractoriness. (E) Change in ratio of exon 10–spliced/full-length transcripts between ND, LEN-, and POM-refractory states; ratio expressed as log2(exon 10–spliced/full-length transcript count) × tumor sample purity. Significant difference detected by Kruskal-Wallis 1-way analysis of variance (nonparametric). (F) Landscape of total and coincidence of 4 types of CRBN aberrations in LEN- and POM-refractory cases analyzed. *Mutations from same patient.
